Abstract
The choline binding proteins (CBPs) are a family of surface proteins noncovalently bound to the phosphorylcholine moiety of the cell wall of Streptococcus pneumoniae by a conserved choline binding domain. Six new members of this family were identified, and these six plus two recently described cell wall hydrolases, LytB and LytC, were characterized for their roles in virulence. CBP-deficient mutants were constructed and tested for adherence to eukaryotic cells, colonization of the rat nasopharynx, and ability to cause sepsis. Five CBP mutants, CbpD, CbpE, CbpG, LytB, and LytC, showed significantly reduced colonization of the nasopharynx. For CbpE and -G this was attributable to a decreased ability to adhere to human cells. CbpG, a putative serine protease, also played a role in sepsis, the first observation of a pneumococcal virulence determinant strongly operative both on the mucosal surface and in the bloodstream.
Streptococcus pneumoniae is currently the major invasive pathogen of children. Pneumococci attach to nasopharynx, lung, and vascular endothelial cells and invade causing pneumonia, bacteremia, and meningitis (23, 28). The pneumococcus has no fimbriae, like gram-negative organisms, and no fibrils, like other streptococci (28). The mechanism by which such a bald surface interacts with human cells is likely to be novel. The surface of S. pneumoniae is decorated with proteins that are covalently and noncovalently attached to the cell wall. These proteins fall into three classes. One well-characterized family of surface proteins, found in both streptococci and staphylococci, employs an LPXTGE motif that serves both as a cleavage site and an anchor for covalent attachment of a secreted protein to the cell wall (22). There are relatively few proteins with this motif in the pneumococcal genome. A second family consists of surface lipoproteins containing an LXXC motif in the N terminus that is cleaved and covalently attached to palmitic acid in the membrane (14, 17). Several members of this family of proteins have been implicated in pneumococcal pathogenesis (17, 18, 31). The most unique group of cell wall-associated proteins in pneumococci are the choline binding proteins (CBPs).
The pneumococcus contains phosphorylcholine on both the cell wall teichoic acid and the membrane-associated lipoteichoic acid (26). The presence of choline in the cell wall was thought to be unique to S. pneumoniae. However, recent data have indicated the presence of choline on the surface of a number of other respiratory tract pathogens, i.e., S. oralis, S. mitis, S. constellatus, Clostridium strain NI-4, C. beijerinckii, Neisseria meningitidis, Pseudomonas aeruginosa, and Haemophilus influenzae (9, 32, 33). In pneumococci, the CBPs bind to the phosphorylcholine of the cell wall noncovalently through a choline binding domain consisting of 2 to 10 repeats of a 20-amino-acid sequence (8, 12, 34). This choline binding motif, first described for pneumococci, has now been identified in other exported proteins, including toxins A and B of C. difficile (2, 6, 30), CspA of C. beijerinckii (21), glucan binding protein of S. mutans, and glycosyltransferases of both S. mutans and S. downei (1, 7, 13, 24, 29). It has been proposed that this domain forms a small ligand binding domain (34).
In pneumococci, six surface proteins that bind the phosphoryl-choline moiety of the cell wall through their choline binding domain have been identified. The major autolysin of pneumococcus, LytA, was the first such protein characterized (9). LytA is required for daughter cell separation and pneumococcal lysis in stationary phase as well as in the presence of penicillin. It has been variably implicated to affect virulence by enabling release of the intracellular toxin pneumolysin (3, 5). Two other cell wall hydrolases, LytB and LytC, have recently been described, but their roles in virulence have not been assessed. LytB plays a role in pneumococcal daughter cell separation (10). LytC is reported to have lysozyme-like activity at 30°C (11). PcpA was cloned recently and is thought to be involved in protein-protein and protein-lipid interactions (20). PspA is a 65-kDa protein that decreases complement deposition on the bacterial surface during sepsis (25, 27). Finally, CbpA (SpsA), the largest and most abundant of the CBPs, functions as a cell surface adhesin and plays a major role in colonization of the nasopharynx in the infant rat model (19). CbpA has also been shown to bind the secretory component of immunoglobulin A and the complement protein C3 (15; B. L. Smith, Q. Cheng, and M. K. Hostetter, Abstr. 98th Gen. Meet. Am. Soc. Microbiol., abstr. D122, 1998). CbpA, LytA, and PspA are subject to phase variation, CbpA and LytA being expressed strongly on mucosal surfaces whereas PspA is upregulated in the bloodstream. A surface-exposed virulence determinant operative in both sites would be a favored potential pneumococcal vaccine candidate.
Previous studies have indicated that many proteins can be eluted from the pneumococcal surface with soluble choline (4, 8, 19, 35). This is confirmed by a search of the pneumococcal genome with the sequence of the conserved choline binding domain. We sought to characterize the family of CBPs with respect to participation in colonization of the nasopharynx and in the pathogenesis of sepsis. In this report, we define a newly recognized role in colonization for two cell wall hydrolases and describe one new CBP, CbpG, active both on the mucosal surface and in the bloodstream.
Nucleotide sequence accession numbers.
Accession numbers for the CBP sequences are AF278686 (CbpD), AF278687 (CbpE), AF278688 (CbpF/G), AF278689 (CbpI), and AF278690 (CbpJ).
MATERIALS AND METHODS
Bacterial strains and plasmids.
S. pneumoniae Norway type 4 is a clinical isolate obtained from MedImmune Inc., Gaithersburg, Md. An unencapsulated derivative, 4R, was obtained from Rodger Novak, St. Jude Children's Hospital. Strain R6 was obtained from the Rockefeller University collection. Cultures were grown without aeration at 37°C in 5% CO2 in a defined semisynthetic medium (C+Y medium) (15a) or plated on tryptic soy agar supplemented with 3% (vol/vol) sheep blood. Pneumococci with integrated plasmids were grown in the presence of appropriate antibiotics (erythromycin [1 μg/ml] and/or chloramphenicol [5 μg/ml]).
Recombinant protein expression, purification, and antibody production.
DNA techniques including PCR, plasmid isolation, chromosomal DNA purification, restriction endonuclease digestion, ligation, and transformation were done according to standard protocols (16, 18). For all CBPs, primers were designed to the N terminus and the C terminus of each cbp gene and used to amplify DNA fragment by PCR (Table 1). PCRs reactions were performed according to the Qiagen Taq DNA polymerase protocol as follows: 94°C for 3 min, then 25 cycles at 94°C for 1 min, 52°C for 1 min, and 72°C for 1.5 min, and a final extension at 72°C for 10 min. The PCR products were digested with BamHI or BglII and SmaI, inserted into pQE30 digested with BamHI and SmaI, and transformed into the E. coli host strain M15[pREP4] (Qiagen). Clones were verified by nucleotide sequencing in St. Jude Children's Hospital Biotechnology Center.
TABLE 1.
Primers used in PCR-based cloning into the E. coli expression system and in insertion duplication mutagenesis
| CBP gene | Primera
|
|
|---|---|---|
| 5′ | 3′ | |
| lytB | gactggatccTAGTGATGGTACTTGGCAAGGAAAACAG | actgctgcagATCTTTGCCACCTAGCTTCTCATTG |
| cggaattccgAGCTCTGCTATTTTTCTTAG | cgggatcccgCATCTTTCCATCTTGGTCA | |
| lytC | gactggatccTGTCGCTGCAAATGAAACTGAAGTAGC | gactaagcttATACCAAACGCTGACATCTACGCG |
| ggaattccATTAGCAAGTATCTGTTTAC | cgggatcccgAGCCTTATAGTAGTTGTCA | |
| cbpD | cgaagatcttcgAAAATTTTACCGTTTATAGCA | tcccccgggggaTGTCAAGGAAACTGCTTACA |
| ggaattcgatcTTTCTTCAACAGGTGGAACT | ggaattcgatcAGCTAGAACCGTCTTTCAG | |
| cbpE | gactggatccGAATGTTCAGGCTCAAGAAAGTTCAGG | gactaagcttTTCCCCTGATGGAGCAAAGTAATACC |
| GTATGGGAATtcTCAGGCTCAAGAAAG | CAACTGGATccGTAGACAGTAATTCAT | |
| cbpF | gactggatccATTTGCAGATGATTCTGAAGGATGG | tcagctgcagCTTAACCCATTCACCATTCTAGTTTAAG |
| cggaattccgTCTTCGGTTTGTTAGCG | cggatcccgCCAATCTGGTCTAAGAG | |
| cbpG | cgcggatccgcgTATACAGATAAGAAACAAG | tcccccgggggaACATTAAATCCACTCA |
| TTCTTGaATTcCCAAGTTGATACTTT | ATAATGGatCCAACCTACCATTTATTTT | |
| cbpI | ctgaggatccGGGGATGGCAGCTTTTAAAAATC | cagtaagcttGTTTACCCATTCACCATTACC |
| ggaattcgatcAATCCTAACAATCAATACAAG | ggaattcgatcCAGTCTGCATGACACCTAA | |
| cbpJ | tcgaggatccGGTTGTCGGCTGGCAATATATCCCGT | CagtaagcttCCGAACCCATTCGCCATTATAGTTGAC |
| ggaattcgatcCTGGCAATATATCCCGTTTC | ggaattcgatcTAGATATTGCCAACCTGTTT | |
Lowercase letters indicate added linkers used for cloning. cbpE and cbpG primers were designed such that restriction sites were incorporated into the coding sequence by changing two of the bases (lowercase letters). Sequences used for insertion duplication mutagenesis are italicized.
Recombinant proteins were purified according to the protocol provided by Qiagen. Escherichia coli strains with recombinant plasmids were grown in 100 ml of Luria broth with ampicillin (100 μg/ml) and kanamycin (25 μg/ml) to an optical density at 600 nm (OD600) of 0.8 to 0.9 at 37°C with vigorous shaking. Cultures were induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 3 h. Cells were harvested by centrifugation at 3,800 × g for 15 min. Pellets were resuspended in lysis buffer (6 M guanidine, 0.1 M NaH2PO4, 0.01 M Tris, 10 mM imidazole [pH 8.0]) and lysed overnight at 4°C. The lysate was centrifuged at 10,000 × g for 30 min at room temperature; 1.5 ml of Ni-nitrilotriacetic acid resin was added to the supernatant and mixed gently by shaking for 1 h. The lysate-resin mixture was loaded onto a 1-ml column and washed with 10 to 15 column volumes of wash buffer (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris, 20 mM imidazole [pH 8.0]). Recombinant protein was eluted four times with 0.5 ml of elution buffer (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris, 150 mM imidazole [pH 8.0]). Protein was quantitated using the Bio-Rad Bradford protein assay, with bovine serum albumin as a standard. Purified recombinant proteins were run on sodium dodecyl sulfate (SDS) 10 to 12% polyacrylamide gels, and bands corresponding to the proteins were cut out and used for production of antisera. Antisera were generated in rabbits by Covance Inc. (Denver, Pa.).
Western blot analysis.
Native CBPs were purified as previously described (19, 35). Pneumococcal cultures (100 to 400 ml) were grown to an OD of 0.4 to 0.6 and centrifuged at 3,800 × g for 10 min. Cells were washed once in phosphate-buffered saline (PBS), and CBPs were eluted by mixing the cells with 5 to 10 ml of PBS containing 2% choline and gently shaking at room temperature for 20 min. The eluate was dialyzed overnight against PBS at 4°C and concentrated on a Centriplus 10 concentrator (Amicon). The Bio-Rad protein assay was used to determine protein concentration. Ten microliters of the eluate (one-fourth of eluate from 108 cells) was added to 2 μl of 5× loading dye; the sample was boiled at 100°C for 5 min and then loaded on a precast SDS–4 to 15% polyacrylamide gel (Bio-Rad). Following separation by gel electrophoresis, samples were transferred to Immobilon-P (Millipore Corp., Bedford, Mass.) and probed with antisera raised against individual CBPs at a dilution of 1:5,000 to 1:10,000. Bands were visualized following incubation with peroxidase-conjugated goat anti-rabbit sera (diluted 1:8,000; Bio-Rad), using a chemiluminescence kit from Amersham.
Insertion duplication mutagenesis.
CBP-deficient mutants were constructed by insertion duplication mutagenesis as previously described (18). PCR was used to amplify 200- to 457-bp fragments from the N-terminal domains of these genes (Table 1). EcoRI and BamHI sites were introduced at the ends of the primers and used to clone the PCR fragments into the pJDC9 vector. For cbpJ, cbpI, and cbpD, fragments spanned amino acids 74 to 217, 7 to 114, and 262 to 379, respectively. For cbpB, cbpC, cbpE, cbpF, and cbpG, the amplified DNA fragments corresponded to amino acid residues 7 to 89, 29 to 122, 20 to 128, 10 to 95, and 15 to 87, respectively. PCR fragments were digested with either EcoRI or EcoRI and BamHI, ligated into pJDC9 digested with either EcoRI or EcoRI and BamHI, and transformed into E. coli. Single transformants containing the insert were identified and plasmid DNA from these clones was transformed into pneumococcal strains serotype 4 and type 4R. Chromosomal integration of the vector at the right locus was verified by PCR, using primers homologous to plasmid sequences (M13 forward −21 and reverse primers) and to sequences upstream of the point of insertion of the plasmid.
In vitro adhesion assays.
Adherence to Detroit nasopharyngeal cells was assessed as previously described (19). Pneumococci were grown to an OD620 of 0.45, centrifuged, resuspended in 0.5 ml of carbonate buffer (0.05 M sodium carbonate, 0.1 M sodium chloride), and labeled with fluorescein isothiocyanate (1 mg/ml; Sigma) for 30 min at room temperature. Labeled bacteria were washed three times in PBS and diluted to 107 CFU/ml in PBS. Cell monolayers established in Terasaki plates were incubated with fluorescein isothiocyanate-labeled bacteria (105) for 30 min at 37°C. Plates were washed four times with PBS and fixed with 2.5% glutaraldehyde; then adherent bacteria were counted. Each strain was tested in six wells per experiment, and experiments were repeated four independent times.
Animal models.
Nasopharyngeal colonization of 1- to 5-day-old infant rats by the Norway T4 parental strain and CBP-deficient mutants was performed as follows. For each experiment, 8 to 10 rat pups were inoculated intranasally with 2.5 × 103 to 8 × 103 CFU of the CBP-deficient mutants or the isogenic parent in PBS. Colonization was assessed at 48 and 96 h postinoculation. The fluid from the nasal washes was diluted and plated, and colony counts were determined. For each mutant, the experiment was repeated at least three independent times. To assess virulence in a model of sepsis, 2- to 5-day-old infant rats were injected intraperitoneally with 2 × 105 to 4 × 105 CFU of the parental strain or a CBP-deficient mutant. Survival was assessed at 24 and 48 h postinjection.
RESULTS
Identification and cloning of CBP DNA fragments.
A 180-amino-acid sequence corresponding to the C-terminal choline binding region of CbpA (amino acids 514 to 694 of S. pneumoniae type 4 strain) was used to search the partially completed genome of a Norway type 4 strain of S. pneumoniae, utilizing the National Center for Biotechnology Information BLAST search engine (www.ncbi.nlm.nih.gov). The search sequence contained all of the choline binding repeats of CbpA but did not include the proline-rich region or any N-terminal sequence. This search identified six discrete contigs containing previously identified cbpA, lytA, lytB, lytC, pspA, or pcpA. Additionally, six other contigs containing seven open reading frames with sequence homology to the choline binding domain of CbpA were identified (Table 2). The cbp genes ranged in size from 426 to 2,034 bp and were predicted to encode for proteins of ∼20 to 80 kDa. The new CBPs did not appear to have a proline-rich linker region or a common CBP promoter. The genes were diversely located throughout the chromosome except for cbpF and cbpG. cbpG was located 13 bp upstream of cbpF on the same contig. cbpH had a large number of stop codons in all frames, suggesting that it did not encode a protein, and thus this locus was not studied further. The predicted proteins had between 2 and 10 choline binding repeats. These repeats were located in the C-terminal domain of all CBPs except for LytB and LytC (at amino acids 206 to 396 and 68 to 175, respectively). The degree of similarity between the choline binding domains of the CBPs and that of CbpA (the search sequence) ranged from 30 to 60%. The N-terminal domain of each protein was distinct, indicating that individual family members likely serve different functions. The diverse N-terminal functional domain of each of these predicted proteins was used individually to search the current protein databases. Significant homology was found only for CbpG, which showed ∼56% similarity over 70% of the N-terminal sequence to a serine proteinase homolog of Enterococcus faecalis.
TABLE 2.
Recombinant and native CBPs and their roles in virulence
| CBP | Contig no.a | Length of open reading frame (bp) | No. of CBD repeats | Predicted no. of amino acids | Protein sizeb (kDa)
|
Virulence of mutants defective in production of CBPc
|
||
|---|---|---|---|---|---|---|---|---|
| Recombinant | Native | Nasopharynx | Sepsis | |||||
| LytB | c4270 | 1,473 | 8 | 678 | 76 | 70 | ↓ | N |
| LytC | c4117 | 2,034 | 4 | 491 | 62 | 60–65 | ↓ | N |
| CbpD | c4374 | 1,347 | 4 | 449 | T | ↓ | N | |
| CbpE | c4139 | 1,884 | 10 | 628 | 70 | 70–75 | ↓ | N |
| CbpF | c4103 | 1,023 | 8 | 341 | 45 | 41 | N | N |
| CbpG | c4103 | 426 | 2 | 154 | 22 | 30–35 | ↓ | ↓ |
| CbpI | c4215 | 636 | 5 | 212 | 29 | — | N | N |
| CbpJ | c4256 | 999 | 7 | 333 | 33 | 38 | N | N |
Based on the February 1998 National Center for Biotechnology Information database.
Approximated based on migration on an SDS-polyacrylamide gradient gel. T, truncated form was expressed; —, the antibody against rCBP does not recognize the native protein.
N, normal activity; arrow, decreased activity.
Expression of CBPs.
A His tag expression system was used to overexpress and purify eight CBPs from E. coli: LytB, LytC, and the six new CBPs. CbpD (which proved lethal to E. coli in full length) and CbpG and were purified as N-terminal truncated forms missing the choline binding domains. Polyclonal antisera were raised against each recombinant protein. Although all of the CBPs harbor a choline binding domain, the antibodies did not cross-react among the CBPs, with one exception. Antibodies raised against recombinant CbpJ (rCbpJ) showed some cross-reactivity with rCbpE (Fig. 1), but this cross-reactivity was not observed with the native proteins. The isolated choline binding domain was exceptionally poorly immunogenic, and its antiserum failed to react with any native CBPs (data not shown), perhaps explaining the lack of cross-reactivity between CBP family members.
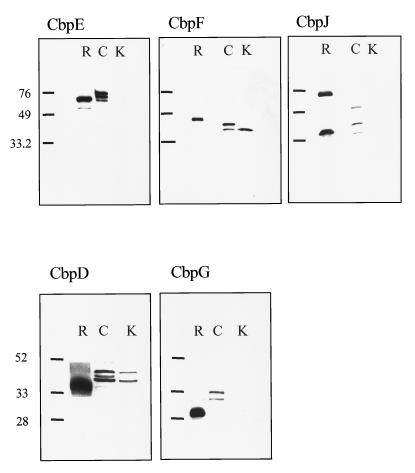
FIG. 1.
Recognition of recombinant and native CBPs by rCBP-specific antisera. The membrane was probed with antisera reactive with the indicated recombinant CBP. R, purified rCBP; C, CBP eluted from strain T4 by choline; K, CBP eluted from isogenic CBP-deficient strains. Sizes are indicated in kilodaltons.
Antibodies raised against individual rCbpE, rCbpF, and rCbpJ and truncated rCbpD and rCbpG recognized native proteins in choline eluates of the wild-type Norway strain (Fig. 1). This indicated that these proteins were expressed and that they could be eluted from the bacterial surface with choline, consistent with membership in the CBP family. Conversely, antiserum to a mixture of native CBPs (19) recognized rLytC, rCbpG, rCbpI, and rCbpJ (data not shown). However, native and recombinant proteins consistently migrated differently, suggesting possible structural or conformational differences. Antibodies against CbpE and CbpG recognized multiple bands which were specific, since mutants deficient in these CBPs lacked all bands. Antibodies against CbpD and CbpF recognized multiple bands, only one of which was specific, as evidenced by its absence in the isogenic knockout strain. No differences were found in the expression of any of these CBPs at 30 and 37°C or during competence (data not shown). Analysis of the expression of the CBPs over a growth cycle indicated that the expression of CbpE increased during logarithmic growth; the levels of other CBPs did not vary with growth phase (data not shown).
Analysis of CBP-defective mutants.
To assess the in vivo roles of LytB, LytC, and the six new CBPs, mutants defective in expression of each CBP were constructed by insertion duplication mutagenesis. CBP-deficient mutants were constructed in both the Norway T4 strain and an isogenic, nonencapsulated derivative, T4R. All genes were distant from surrounding open reading frames, eliminating the possibility of polar effects of significance except for cbpF and cbpG. cbpG is located directly upstream of cbpF, and it is possible that mutations in cbpG are polar onto cbpF. However, since a mutant deficient in only CbpF had no observable phenotype, the phenotype observed for a CbpG-defective strain appears to be attributable to CbpG function alone. Furthermore, Western analysis of the CbpF− and CbpG− mutants indicated normal products expressed for CbpG and CbpF, respectively (data not shown).
There were no differences between the parent strain and any of the CBP-deficient mutants in efficiency of genetic transformation, lysis in stationary phase, or lysis in response to penicillin (data not shown). In particular, confirming results of Garcia et al. (10, 11), no lytic defect was found for the LytB and LytC mutants at 37°C.
An infant rat model was used to determine the role of the CBPs in colonization of the nasopharynx. Compared to the isogenic wild-type strain, loss of function of five of the eight tested CBPs showed a statistically significant reduction in the colonization of the nasopharynx at 48 h (Table 3). Mutants defective in LytB, LytC, CbpD, CbpE, and CbpG showed a 2- to 20-fold reduction in colonization after 48 h. This pattern persisted at 96 h.
TABLE 3.
Effect of loss of CBP function on colonization of the infant rat nasopharynx
| CBP | % of WT CFU at 48 ha (mean ± SD) |
|---|---|
| LytB | 18 ± 8* |
| LytC | 6 ± 5* |
| CbpD | 43 ± 18* |
| CbpE | 25 ± 12* |
| CbpF | 67 ± 22 |
| CbpG | 25 ± 12* |
| CbpI | 60 ± 39 |
| CbpJ | 68 ± 35 |
100% adherence = 243 ± 51 bacteria/10 μl of nasal wash. ∗, P < 0.001 versus wild-type (WT) Norway strain.
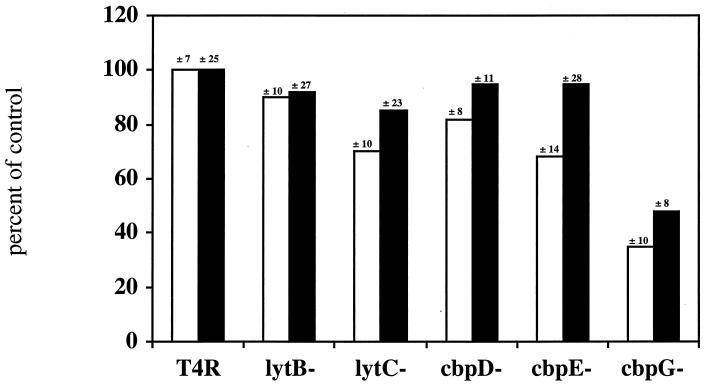
The adherence properties of these mutants were tested against Detroit nasopharyngeal cells at 37 and 30°C, the lower temperature reflective of the ambient value in vivo in the nasopharynx. Loss of function of LytC, CbpE, and CbpG reduced adherence to Detroit cells at 30°C to 70, 68, and 35%, respectively (Fig. 2). At 37°C, only adherence of CbpG was compromised (45%).
FIG. 2.
Adherence of CBP mutants to Detroit nasopharyngeal cells. Mutants were incubated with Detroit cell monolayers at either 30°C (white bars) or 37°C (black bars). Adherent bacteria were quantitated visually. Values are means ± standard deviations for six wells. Data are representative of four experiments. For T4R, the parent strain, 100% = 59 ± 11 bacteria/cell.
The CBP-deficient mutants were also tested in a model for pneumococcus-induced sepsis. Most infant rats injected with the parental strain died within 24 h, and all were dead by 48 h (Table 4). Loss of function of CbpG resulted in reduced virulence at 24 and 48 h (30 to 45% survival). Inoculation with up to 107 CFU of the CbpG− mutant per ml failed to kill any mice, indicating loss of virulence of at least 3 logs. The remaining CBP-deficient mutants did not differ in virulence from the wild type (data not shown).
TABLE 4.
Decreased virulence of a CbpG-deficient strain in an infant rat sepsis model
| Strain | No. of surviving animals/no. of animals injected
|
|||
|---|---|---|---|---|
| Expt 1
|
Expt 2
|
|||
| 24 h | 48 h | 24 h | 48 h | |
| Norway | 3/11 | 0/11 | 6/14 | 0/14 |
| CbpG− mutant | 11/11 | 11/11 | 14/14 | 14/14 |
DISCUSSION
The availability of the pneumococcal genome sequence provided a tool with which to identify additional members of the CBP family. The entire family consisted of 13 loci, 12 of which constitute open reading frames. Like most other CBPs, none exhibited a signal sequence, leaving open the mechanism of secretion of this class of protein. All six new CBP family members reported here were surface expressed and eluted with choline, consistent with the behavior of known CBPs. Antibodies to recombinant CBPs recognized native CBPs eluted from pneumococci by choline. However, the native species uniformly migrated differently than the recombinant species, indicating possible differences in the folding or structure of the native versus recombinant proteins of this entire family.
Analysis of the CBP-deficient mutants indicated that a number of CBPs play a role in adhesion and colonization of the nasopharynx. Previous results have suggested that CbpA accounts for 40 to 50% of the adherence of wild-type bacteria to nasopharyngeal cells (19). A similarly strong phenotype was found for the putative serine protease, CbpG. This correlated with a strong defect in colonization of the nasopharynx in vivo. Mutants in CbpD and CbpE showed significant loss of nasopharyngeal colonization with only modestly decreased adherence in vitro at 30°C. Their primary function in colonization remains to be determined.
The role of cell wall hydrolases in virulence has long been sought because of the general belief that these suicidal enzymes must confer an in vivo advantage in order to be retained in the face of strong negative selection by penicillin. LytA is a well-characterized amidase triggered by treatment with penicillin. LytB has been designated a muramidase (10). LytC has been identified as the first streptococcal lysozyme (11). Deletion of any of the three hydrolases does not alter viability or transformability in vitro (10, 11, 26). Loss of function of LytA modestly affects the course of pneumonia but not sepsis (3, 5). This phenotype is thought to be due to the role of autolysis in the release of pneumolysin and neuraminidase, two well-described virulence determinants. The significant loss of ability to colonize the nasopharynx in both the LytB and LytC mutants with only modest changes in adherence in vitro raises the possibility that the two new hydrolases could also play this same role in toxin release. The suggestion that LytC functions optimally as a hydrolase at 30°C (11) is consistent with a potential role in the nasopharynx, where temperatures are cooler.
PspA is the only previously known CBP with a dominant role in sepsis. The present analysis adds Cbp G to this group. CbpG appears to bear sequence similarity to a serine protease, the substrate of which may relate to its role in adherence to human cells. It is possible that CbpG modifies proteins on the surface of the pneumococcus, enabling them to bind to receptors on the eukaryotic cells. Conversely, CbpG may modify the eukaryotic cell surface, promoting ligand-receptor interactions.
In summary, CBPs appear to be a functionally significant group of pneumococcal surface proteins that are noncovalently and reversibly attached to the choline moiety of the cell wall through their C-terminal choline binding domain. These proteins have unique N-terminal domains that indicate varied functions for these proteins. Clearly a main function of the CBP family is in promoting colonization of the nasopharynx, as evidenced by the clear phenotype of CbpA, CbpD, CbpE, CbpG, LytB, and LytC mutants in the model of nasopharyngeal colonization. For CbpA and CbpG, the role in vivo is strongly implicated to be host cell recognition, and this may also apply to a much lesser extent for CbpB, CbpC, CbpD, and CbpE. Only PspA and CbpG are currently directly implicated in pneumococcal virulence in sepsis. The dual role of CbpG in mucosal and bloodstream compartments suggest that this protein may be excellent candidate for evaluation in pneumococcal vaccines.
ACKNOWLEDGMENTS
This work was supported by NIH grants AI27913 and AI 39482, AI36445 and in part by Cancer Center Support CORE grant P30 CA21765 and the American Lebanese Syrian Associated Charities (ALSAC).
We are grateful to T. Wizemann, S. Johnson, and S. Koenig, MedImmune Inc., for helpful discussion. We thank Micha Ring and John Killmar for technical assistance.
REFERENCES
- 1.Banas J A, Russell R R, Ferretti J J. Sequence analysis of the gene for the glucan-binding protein of Streptococcus mutans Ingbritt. Infect Immun. 1990;58:667–673. doi: 10.1128/iai.58.3.667-673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barroso L A, Wang S Z, Phelps C J, Johnson J L, Wilkins T D. Nucleotide sequence of Clostridium difficile toxin B gene. Nucleic Acids Res. 1990;18:4004. doi: 10.1093/nar/18.13.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry A M, Paton J C, Hansman D. Effect of insertional inactivation of the genes encoding pneumolysin and autolysin on the virulence of Streptococcus pneumoniae type 3. Microb Pathog. 1992;12:87–93. doi: 10.1016/0882-4010(92)90111-z. [DOI] [PubMed] [Google Scholar]
- 4.Briese T, Hakenbeck R. Interaction of the pneumococcal amidase with lipoteichoic acid and choline. Eur J Biochem. 1985;146:417–427. doi: 10.1111/j.1432-1033.1985.tb08668.x. [DOI] [PubMed] [Google Scholar]
- 5.Canvin J, Marvin A, Sivakumaran M, Paton J, Boulnois G, Andrew P, Mitchell T. The role of pneumolysin and autolysin in the pathology of pneumonia and septicemia in mice infected with a type 2 pneumococcus. J Infect Dis. 1995;172:119–123. doi: 10.1093/infdis/172.1.119. [DOI] [PubMed] [Google Scholar]
- 6.Dove C H, Wang S Z, Price S B, Phelps C J, Lyerly D M, Wilkins T D, Johnson J L. Molecular characterization of the Clostridium difficile toxin A gene. Infect Immun. 1990;58:480–488. doi: 10.1128/iai.58.2.480-488.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferretti J J, Huang T T, Russell R R. Sequence analysis of the glucosyltransferase A gene (gtfA) from Streptococcus mutans Ingbritt. Infect Immun. 1988;56:1585–1588. doi: 10.1128/iai.56.6.1585-1588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia J L, Sanchez-Beato A R, Medrano F J, Lopez R. Versatility of choline-binding domain. Microb Drug Resist. 1998;4:25–36. doi: 10.1089/mdr.1998.4.25. [DOI] [PubMed] [Google Scholar]
- 9.Garcia P, Garcia J L, Garcia E, Lopez R. Nucleotide sequence and expression of the pneumococcal autolysin gene from its own promoter in Escherichia coli. Gene. 1986;43:265–272. doi: 10.1016/0378-1119(86)90215-5. [DOI] [PubMed] [Google Scholar]
- 10.Garcia P, Gonzalez M P, Garcia E, Lopez R, Garcia J L. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol Microbiol. 1999;31:1275–1281. doi: 10.1046/j.1365-2958.1999.01238.x. [DOI] [PubMed] [Google Scholar]
- 11.Garcia P, Gonzalez M P, Garcia E, Garcia J L, Lopez R. The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionary mobile domain. Mol Microbiol. 1999;33:128–138. doi: 10.1046/j.1365-2958.1999.01455.x. [DOI] [PubMed] [Google Scholar]
- 12.Giffard P M, Jacques N A. Definition of a fundamental repeating unit in streptococcal glucosyltransferase glucan-binding regions and related sequences. J Dental Res. 1994;73:1133–1141. doi: 10.1177/00220345940730060201. [DOI] [PubMed] [Google Scholar]
- 13.Gilmore K S, Russell R R, Ferretti J J. Analysis of the Streptococcus downei gtfS gene, which specifies a glucosyltransferase that synthesizes soluble glucans. Infect Immun. 1990;58:2452–2458. doi: 10.1128/iai.58.8.2452-2458.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilson E, Alloing G, Schmidt T, Claverys J-P, Dudler R, Hofnung M. Evidence for high affinity binding-protein dependent transport systems in Gram-positive bacteria and in Mycoplasma. EMBO J. 1988;7:3971–3974. doi: 10.1002/j.1460-2075.1988.tb03284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammerschmidt S, Talay S, Brandtzaeg P, Chhatwal G. SpsA, a novel pneumococcal surface protein with specific binding to secretory immunoglobulin A and secretory component. Mol Microbiol. 1997;21:965–971. doi: 10.1046/j.1365-2958.1997.5391899.x. [DOI] [PubMed] [Google Scholar]
- 15a.Lacks S, Hotchkiss R D. A study of the genetic material determining an enzyme activity in pneumococcus. Biochim Biophys Acta. 1960;39:508–517. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 16.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 17.Pearce B J, Naughton A M, Masure H R. Peptide permeases modulate transformation in Streptococcus pneumoniae. Mol Microbiol. 1994;12:881–892. doi: 10.1111/j.1365-2958.1994.tb01076.x. [DOI] [PubMed] [Google Scholar]
- 18.Pearce B J, Yin Y B, Masure H R. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol Microbiol. 1993;9:1037–1050. doi: 10.1111/j.1365-2958.1993.tb01233.x. [DOI] [PubMed] [Google Scholar]
- 19.Rosenow C, Ryan P, Weiser J N, Johnson S, Fontan P, Ortquist A, Masure H R. Contribution of novel choline-binding proteins to adherence, colonization, and immunogenicity of Streptococcus pneumoniae. Mol Microbiol. 1997;25:819–825. doi: 10.1111/j.1365-2958.1997.mmi494.x. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez-Beato A R, Lopez R, Garcia J L. Molecular characterization of PcpA: a novel choline-binding protein of Streptococcus pneumoniae. FEMS Microbiol. 1998;164:207–214. doi: 10.1111/j.1574-6968.1998.tb13087.x. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez-Beato A R, Ronda C, Garcia J L. Tracking the evolution of the bacterial choline binding domain: molecular characterization of the Clostridium acetobutylicum NCIB 502 cspA gene. J Bacteriol. 1995;177:1098–1103. doi: 10.1128/jb.177.4.1098-1103.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneewind O, Fowler A, Faull K. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 23.Schuchat A, Robinson K, Wenger J, Harrison L, Farley M, Reingold A, Lefkowitz L, Perkins B. Bacterial meningitis in the United States. N Engl J Med. 1997;337:970–976. doi: 10.1056/NEJM199710023371404. [DOI] [PubMed] [Google Scholar]
- 24.Shiroza T, Ueda S, Kuramitsu H K. Sequence analysis of the gtfB gene from Streptococcus mutans. J Bacteriol. 1987;169:4263–4270. doi: 10.1128/jb.169.9.4263-4270.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talkington D F, Crimmins D L, Voellinger D C, Yother J, Briles D E. A 43-kilodalton pneumococcal surface protein, PspA: isolation, protective abilities, and structural analysis of the amino-terminal sequence. Infect Immun. 1991;59:1285–1289. doi: 10.1128/iai.59.4.1285-1289.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomasz A. Choline in the cell wall of a bacterium: novel type of polymer-linked choline in pneumococcus. Science. 1967;157:694–697. doi: 10.1126/science.157.3789.694. [DOI] [PubMed] [Google Scholar]
- 27.Tu A, Fulgham R, McCrory M, Briles D, Szalai A. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect Immun. 1999;67:4720–4724. doi: 10.1128/iai.67.9.4720-4724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuomanen E I, Austrian R, Masure H R. The pathogenesis of pneumococcal infection: correlation of clinical events with molecular mechanisms. N Engl J Med. 1995;332:1280–1284. doi: 10.1056/NEJM199505113321907. [DOI] [PubMed] [Google Scholar]
- 29.Ueda S, Shiroza T, Kuramitsu H K. Sequence analysis of the gtfC gene from Streptococcus mutans GS-5. Gene. 1988;69:101–109. doi: 10.1016/0378-1119(88)90382-4. [DOI] [PubMed] [Google Scholar]
- 30.Von Eichel-Streiber C, Sauerborn M. Clostridium difficile toxin A carries a C-terminal repetitive structure homologous to the carbohydrate binding region of streptococcal glycosyltransferases. Gene. 1990;96:107–113. doi: 10.1016/0378-1119(90)90348-u. [DOI] [PubMed] [Google Scholar]
- 31.Wani J, Gilbert J, Plaught A, Weiser J. Identification, cloning and sequencing of the immunoglobulin AI protease gene of Streptococcus pneumoniae. Infect Immun. 1996;64:2240–2245. doi: 10.1128/iai.64.10.3967-3974.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiser J N, Goldberg J B, Pan N, Wilson L, Virji M. The phosphorylcholine epitope undergoes phase variation on a 43-kilodalton protein in Pseudomonas aeruginosa and on pili of Neisseria meningitidis and Neisseria gonorrhoeae. Infect Immun. 1998;66:4263–4267. doi: 10.1128/iai.66.9.4263-4267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiser J N, Pan N, McGowan K L, Musher D, Martin A, Richards J. Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J Exp Med. 1998;187:631–640. doi: 10.1084/jem.187.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wren B W. A family of clostridial and streptococcal ligand-binding proteins with conserved C-terminal repeat sequences. Mol Microbiol. 1991;5:797–803. doi: 10.1111/j.1365-2958.1991.tb00752.x. [DOI] [PubMed] [Google Scholar]
- 35.Yother J, White J M. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J Bacteriol. 1994;176:2976–2985. doi: 10.1128/jb.176.10.2976-2985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]