Abstract
OBJECTIVE:
The contribution of genetic factors to the presence of an overactive bladder is recognized. This study aimed to (1) assemble and synthesize available data from studies assessing differential gene expression in patients with overactive bladder vs controls without overactive bladder and (2) determine possible correlations and functional pathways between genes.
DATA SOURCES:
We searched PubMed, Ovid or Medline, and Wiley Cochrane Central Register of Controlled Trials databases between January 1, 2000, and December 15, 2021.
STUDY ELIGIBILITY CRITERIA:
Studies were included if gene expression was detected and quantified using molecular approaches performed on human bladder tissue specimens directly and excluded if the gene expression analysis was carried out from blood and urine specimens alone.
METHODS:
A systematic review was completed to identify publications that reported differently expressed gene candidates among patients with overactive bladder vs healthy individuals. Gene networking connections and pathway analysis were performed employing Metascape software, where inputs were identified from our systematic review of differentially expressed genes in overactive bladder.
RESULTS:
A total of 9 studies were included in the final analysis and 11 genes were identified as being up-regulated (purinergic receptor P2X 2 [P2RX2], smoothelin [SMTN], growth-associated protein 43 [GAP43], transient receptor potential cation channel subfamily M member 8 [TRPM8], cadherin 11 [CDH1], gap junction protein gamma 1 [GJC1], cholinergic receptor muscarinic 2 [CHRM2], cholinergic receptor muscarinic 3 [CHRM3], and transient receptor potential cation channel subfamily V member 4 [TRPV4]) or down-regulated (purinergic receptor P2X 2 [P2RX3] and purinergic receptor P2X 5 [P2RX5]) in patients with overactive bladder. Gene network analysis showed that genes are involved in chemical synaptic transmission, smooth muscle contraction, blood circulation, and response to temperature stimulus. Network analysis demonstrated a significant genetic interaction between TRPV4, TRPM8, P2RX3, and PR2X2 genes.
CONCLUSION:
Outcomes of this systematic review highlighted potential biomarkers for treatment efficacy and have laid the groundwork for developing future gene therapies for overactive bladder in clinical settings.
Keywords: gene expression, genomics, overactive bladder, systematic review, urge urinary incontinence
Introduction
Overactive bladder (OAB) is a storage symptom syndrome characterized by urgency, with or without incontinence, in the absence of urinary tract infection or another obvious pathology.1 Recent reports indicate between 20% and 50% of women are affected by OAB in the United States alone.2,3 Patients with OAB often suffer social, sexual, occupational, and psychological effects, which severely affect their quality of life.4 The pathophysiology of OAB is multifaceted with various mechanisms. The influence of the cholinergic system, nitric oxide, and adrenergic mechanisms are crucial to regulating bladder filling and storage processes; moreover, up to 70% of OAB cases are considered idiopathic.5 Although there exist several treatment options for OAB, success rates are highly variable, likely reflecting the multifaceted nature of OAB phenotypes.6,7 Genetic factors are implicated in the development of OAB, but, to date, there has been limited evaluation of the genetic components underlying OAB.8-10
The urothelium directly communicates with suburothelial afferents acting as luminal sensors. Low pH, high potassium concentration in the urine, and increased osmolality can affect sensory nerves. Detrusor tissue from patients with idiopathic instability shows increased electrically evoked contractions but normal sensitivity to muscarinic agonists. The large conductance, voltage- and calcium-activated K+ channel, known as the big potassium (BK), is highly expressed on urinary bladder smooth muscle cells and regulates bladder detrusor muscle function, as has been demonstrated using gene transfer for the BK channel for the treatment of OAB. Gene expression profiling enables the evaluation of the gene expression patterns and can identify many potential drug targets for treatment and biomarkers for treatment efficacy.11-13 Comparative genomic studies between normal bladder and OAB-afflicted tissues may provide a more direct way to evaluate the OAB pathophysiology. The bladder’s unique suitability for instillation gene therapies makes the gene therapies superior because of the endurance of bladder architecture histologically indistinguishable from normal controls without any evidence of cystitis or systemic spread of the infection.14 The identification of OAB-associated genes may lead to groundbreaking biomarkers and novel gene therapy approaches.
Objective
This review aimed to summarize current evidence from studies evaluating differential gene expression levels between normal and OAB bladder tissues to determine genes associated with OAB and identify possible correlations and networks between genes and functional pathways.
Methods
Eligibility criteria, information sources, and search strategy
A systematic literature review was conducted to identify publications investigating genetic contributions in the development of OAB and assess potential targets in a prognostic and/or clinical therapeutic context. The search was performed in compliance with the Preferred Reporting Items for Systematic Reviews guidelines for systematic reviews.15 The study was preregistered in the international Open Science Framework to host our protocol (https://osf.io/knfud) and study data in a publicly available database.
A systematic search strategy was developed by an academic librarian (A.C.S.) at our institution to identify publications that identified differently expressed gene candidates among patients with OAB. The search was performed using PubMed, Ovid or Medline, and Wiley Cochrane Central Register of Controlled Trials. The search results were limited to research published between January 1, 2000, and December 15, 2021, to include as many pertinent articles as possible. The search was performed using Medical Subject Headings terms, such as “overactive bladder,” “urge urinary incontinence,” “detrusor overactivity,” “genetic,” “gene,” and “gene expression.” The comprehensive search strategies are available (Supplemental Table 1). Of note, 2 researchers (I.I. and P.M.) screened the generated library using Rayyan to discover pertinent studies that contributed to this review. Rayyan was used to accelerate the literature review process by operating machine learning technologies. It relies on initial input from the users in terms ofkey words and phrases,16 and users (I.I and P.M) confirmed whether to include or exclude the returned studies.
Study selection
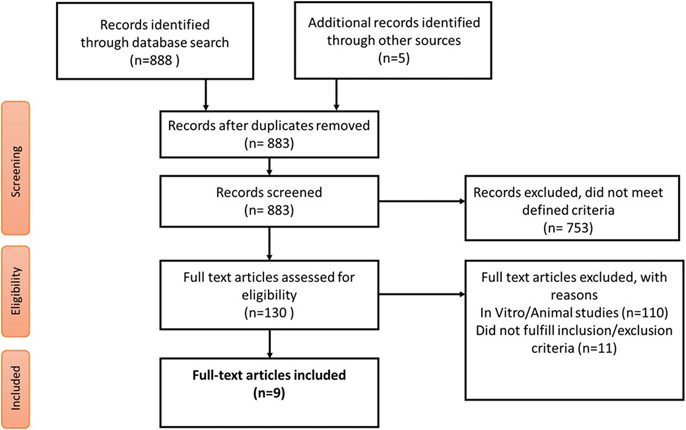
The inclusion criteria included primary research data examining gene expression analysis of patients with OAB vs controls without OAB. Studies were included if gene expression was detected and quantified using molecular techniques performed on bladder tissue specimens directly and excluded if the gene expression analysis was carried out from urine or blood specimens alone. In vitro and animal gene expression studies were excluded to clarify differentially expressed genes in humans alone. No other limitation was placed initially on data collection. All manuscript titles were screened for relevance, and eligible manuscripts were subjected to a full-text analysis. Figure 1 illustrates the overall manuscript selection approach in a flow diagram.
FIGURE 1. Flow diagram for study selection.
Other sources indicate a Google search.
Data extraction
Following selection, a standardized form was used, with parameters that included study characteristics, such as reference, study design, study population and age, gene selection process, and analytical method used. The primary outcomes of this study were to report examined significant genes and different analytical methods used across studies. Genes were selected and considered to be related to OAB if their expression was consistently corroborated through all statistical and analytical methods (reverse transcriptase-polymerase chain reaction, immunohistochemistry, etc.) used within the included study.
Gene network analysis
Database for Annotation, Visualization, and Integrated Discovery (DAVID),17 a web-accessible program, was employed for functional annotation of most significant genes. In addition to primary outcomes, GeneMANIA,18,19 a user-friendly, real-time, multiple association network integration algorithm, was employed to create gene set functional hypotheses. A list of the most significant differentially expressed genes was uploaded to the GeneMANIA server. The network of genes displaying co-expression, physical interactions, co-localization, shared protein domains, and predicted interactions was generated. The created network emphasizes genes and scores them based on comparative networks in the initial list.
Pathway analysis
In addition, for each given gene list, pathway and functional enrichment analyses have been carried out with the Gene Ontology Biological Processes and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway using Metascape 3.20 Protein-protein interaction (PPI) enrichment analysis was also carried out with the STRING, one of the most comprehensive PPI data sources. Only physical interactions in STRING (physical score of >0.132) were used. The resultant network contains the subset of proteins that form physical interactions with at least 1 other member in the list.
Assessment of risk of bias
The included studies were evaluated using the Newcastle-Ottawa Quality Assessment Scale21 and defined using the Agency for Healthcare Research and Quality (AHRQ) standards (Supplemental Table 2). In addition, the Joanna Briggs Institute (JBI) crucial appraisal tool31 was used to assess the relevance and results of the included manuscripts (Supplemental Table 3).
Results
Systematic review results
Study selection.
The literature search returned 130 studies, of which, only 9 met the inclusion criteria by presenting gene expression data from bladder tissue specimens that were quantified with molecular biology methods. These 9 studies were identified from the final literature search and included in the subsequent genetic analysis.
Study characteristics.
For the 9 included studies, genes were assessed and categorized as being either over- or underexpressed in patients with OAB vs normal patients. Genes with notable over- or underexpression were examined further for genetic associations, interactions between their coded proteins, and related pathways.
Risk of bias of included studies.
Overall, the included studies had fair to good quality as assessed by AHRQ standards (Supplemental Table 2). Included studies in the systematic review met the criteria of JBI critical appraisal checklist (Supplemental Table 3).
Synthesis of results between the studies.
A total of 9 genes were identified as overexpressed in OAB patients (purinergic receptor P2X 2 [P2RX2], smoothelin [SMTN], growth-associated protein 43 [GAP43], transient receptor potential cation channel subfamily M member 8 [TRPM8], cadherin 11 [CDH1], gap junction protein gamma 1 [GJC1], cholinergic receptor muscarinic 2 [CHRM2], cholinergic receptor muscarinic 3 [CHRM3], and transient receptor potential cation channel subfamily V member 4 [TRPV4]) and a total of 2 genes (purinergic receptor P2X [P2RX3] and purinergic receptor P2X 5 [P2RX5]) were reported with reduced expression. A summary of reported outcomes of the included studies can be found in Table 1.
TABLE 1.
Summary of reported outcomes in the studies included
| Reference | Study design | Study population and age (average/range) | Results Summary: gene selection process | Analytical Methods used |
|---|---|---|---|---|
| O’reilly et al,22 2002 | Case control | 20 female patients 35 to 75 y old (mean age, 51.8 y) diagnosed with detrusor instability. 20 age- and sex-matched controls 35 to 71 y old (mean age, 53.6 y) were recruited, in whom urodynamics showed a stable bladder. | Quantitative RT-PCR reveals significantly decreased P2X1, P2X4, and P2X7 expression but significantly increased P2X2 expression in idiopathic detrusor instability bladders. As only P2X2 expression is increased in idiopathic detrusor instability bladders, it strongly implies that a P2X1 or P2X2 heteromultimer is crucial for the changes in purinergic innervation in idiopathic detrusor instability bladders. | Immunohistochemistry Quantitative RT-PCR |
| Maake et al,23 2006 | Case control | Detrusor samples of 13 OABs (sensory urge and detrusor instability) were obtained before botulinum toxin injection and compared with those of 8 normally contractile, no obstructed bladders obtained during radical cystectomy. | In the smooth muscle of patients with detrusor instability and sensory urge, a significant 2.4- and 2.2-fold increase, respectively, in SMTN variant 1 messenger RNA was observed compared with that of normal controls. Analyses at the SMTN protein level confirmed significant up-regulation in these bladder dysfunctions by a factor of 2.3 and 1.8, respectively. No significant difference in SMTN expression was observed between detrusor instability and sensory urge. | Northern blot Quantitative RT-PCR Immunohistochemistry |
| Moore et al,24 2001 | Case control | Detrusor samples were taken from: controls, at cystectomy for cancer or cystoscopic biopsy for hematuria (n=22; age, 33–88 y), child bladder, at surgical correction of vesicoureteral reflux (n=21; age 4 mo to 2 y), and adults with detrusor instability at cystoscopy-cystodistension (n=18; age 30–81 y). | The lack of P2X3 and P2X5 may impair control of detrusor contractility and contribute to the pathophysiology of urge incontinence. | Immunohistochemistry |
| Schofield et al,25 2005 | Case control | A series of 18 women with urodynamically proven detrusor instability (median age, 62 y; range, 39–85 y), who were refractory to treatment, underwent cystoscopy and cold cup biopsy. Controls (n=26; median age, 65 y) were females without urgency or urge incontinence, undergoing cystoscopy for other indications. |
The increase in GAP43 with age and with previous cystitis history suggests that neuronal sprouting is important in some subsets of patients with idiopathic detrusor instability. | Immunohistochemistry |
| Mukerji et al,26 2006 | Case control | Bladder specimens obtained from patients with PBS (n=16), detrusor overactivity (n=14), and asymptomatic microscopic hematuria (controls, 17). |
There was marked increase of TRPM8-immunoreactive nerve fibers in IDO (P=.0249) and PBS (P<.0001) specimens, compared with controls. A significantly higher number of TRPM8-immunoreactive axons were also seen in the IDO (P=.0246) and PBS (P<.0001) groups. Urothelial TRPM8 and TRPM8-immunoreactive thick myelinated fibers seemed unchanged in IDO and PBS. | Immunohistochemistry |
| Roosen et al,27 2009 | Case control | 32 patients with OAB and refractory detrusor overactivity, and 8 controls without lower urinary tract symptoms underwent cystoscopic bladder biopsy. | Significant 2-fold up-regulation of cadherin-11 was found in the suburothelium of patients with OAB compared with that in controls (P=.018), whereas β-catenin was similar in the groups (P=.6). | Quantitative immunohistochemistry |
| Neuhaus et al,28 2005 | Case control | Control tissue samples were taken from the bladder dome, excluding the trigonal area, at cystectomies or as biopsies during transurethral tumor resections (5 women: mean age, 65 y; SD, 10.5; range, 54–81; 6 men: 73.5 y; SD, 5.7; range, 64–80. | Semiquantitative analyses showed significantly higher Cx43 expression in the detrusor muscle and a tendency to higher Cx45 expression in the suburothelial layer associated with urge symptoms, whereas Cx40 expression was unaffected. | Indirect immunofluorescence |
| Roberts et al,29 2020 | Case control | Samples from patients with no symptoms of OAB (non-OAB cohort: age, 72.0±2.3 y) and those from patients with symptoms of OAB (clinical diagnosis, frequency ≥8/d, urgency with or without urgency incontinence), diagnosed with IDO (OAB with no neurologic abnormality, OAB cohort: age, 60.0± 5.0 y) were collected. |
OABs exhibited greater TRPV4-induced ATP release with age dependence. These data provide the first evidence in humans for the key functional role of TRPV4 in urothelium with specific mechanisms and identify TRPV4 up-regulation in aging and OABs | Western blotting |
| Mukerji et al,30 2006 | Case control | Bladder specimens were obtained from patients with detrusor overactivity (n=12) and controls with asymptomatic microscopic hematuria (n=16). | Muscarinic receptor subtypes 2 and 3 immunoreactivity significantly correlated with the urgency score (P=.0002 and P=.0206, respectively) and muscarinic receptor subtype 2 immunoreactivity correlated with the frequency score (P=.0029). | Immunohistochemistry |
ATP, adenosine triphosphate; GAP43, growth-associated protein 43; IDO, idiopathic detrusor overactivity; OAB, overactive bladder; P2X, purinergic receptor P2X; PBS, painful bladder syndrome; RT-PCR, reverse transcriptase-polymerase chain reaction; SMTN, smoothelin; TRPM8, transient receptor potential cation channel subfamily M member 8; TRPV4, transient receptor potential cation channel subfamily V member 4.
Genetic analysis
Database for Annotation, Visualization, and Integrated Discovery functional analysis.
Functional analysis revealed that most up-regulated genes share common annotations or biology in the involvement of intrinsic components of the nuclear inner membrane, ligand-gated cation channel activity, nucleotide receptor activity, actin-mediated cell contraction, modulation of chemical synaptic transmission, calcium ion transmembrane transporter activity, complex of collagen trimers, cell-cell junction organization, acetylcholine receptor signaling, adenylate cyclase-inhibiting G protein–coupled receptor (GPCR) signaling, postsynaptic signal transduction, and G protein–coupled amine receptor activity. Moreover, the down-regulated genes share similar features in terms of regulation of cytosolic calcium ion concentration, protein autophosphorylation, inorganic cations import across the plasma membrane, extracellular ligand-gated ion channel activity, positive regulation of cytosolic calcium ion, and response to purine-containing compound (Table 2).
TABLE 2.
Genetic analysis (DAVID) of investigated significant genes
| DAVID symbol |
DAVID name | Results | Functions |
|---|---|---|---|
| Up-regulated genes | |||
| P2RX2 | Purinergic receptor P2X 2 | Up-regulated in OAB | Nuclear inner membrane Intrinsic component of nuclear inner membrane Ligand-gated cation channel activity Nucleotide receptor activity Ligand-gated channel activity |
| SMTN | Smoothelin | Up-regulated in OAB | Smooth muscle contraction Contractile fiber Actin-mediated cell contraction Actin filament-based movement Actin-myosin filament sliding |
| GAP43 | Growth-associated protein 43 | Up-regulated in OAB | Transmitter-gated ion channel activity Extracellular ligand-gated ion channel activity Neuron projection organization Neurotransmitter receptor activity Modulation of chemical synaptic transmission |
| TRPM8 | Transient receptor potential cation channel subfamily M member 8 | Up-regulated in OAB | Calcium ion transport Calcium ion transmembrane transporter activity Divalent inorganic cation transmembrane transporter Antimicrobial humoral response Ligand-gated cation channel activity |
| CDH11 | Cadherin 11 | Up-regulated in OAB | Cell-cell junction organization Extrinsic component of membrane Cell-cell junction Banded collagen fibril Complex of collagen trimers |
| GJC1 | Gap junction protein gamma 1 | Up-regulated in OAB | Regulation of heart contraction Regulation of blood circulation Cell-cell signaling involved in cardiac conduction Cardiac muscle cell contraction sinoatrial node cell to atrial cardiac muscle cell communication |
| CHRM2 | Cholinergic receptor muscarinic 2 | Up-regulated in OAB | Acetylcholine receptor signaling pathway Cellular response to acetylcholine Adenylate cyclase-inhibiting G protein—coupled receptor signaling pathway Postsynaptic signal transduction Serotonin receptor activity |
| CHRM3 | Cholinergic receptor muscarinic 2 | Up-regulated in OAB | Serotonin receptor activity G protein—coupled amine receptor activity Acetylcholine receptor signaling pathway Cellular response to acetylcholine Adenylate cyclase-inhibiting G protein |
| TRPV4 | Transient receptor potential cation channel subfamily V member 4 | Down-regulated in OAB | Regulation of cytosolic calcium ion concentration Voltage-gated potassium channel activity Cellular calcium ion homeostasis Protein autophosphorylation Inorganic cations import across plasma membrane |
| Down-regulated genes | |||
| P2RX3 | Purinergic receptor P2X 3 | Down-regulated in OAB | Nuclear inner membrane Intrinsic component of nuclear inner membrane Nucleotide receptor activity Excitatory extracellular ligand-gated ion channel activity Extracellular ligand-gated ion channel activity |
| P2RX5 | Purinergic receptor P2X 5 | Down-regulated in OAB | Extracellular ligand-gated ion channel activity Calcium ion transport into cytosol Cytosolic calcium ion transport Positive regulation of cytosolic calcium ion Response to purine-containing compound |
DAVID, Database for Annotation, Visualization, and Integrated Discovery; OAB, overactive bladder.
GeneMANIA network analysis.
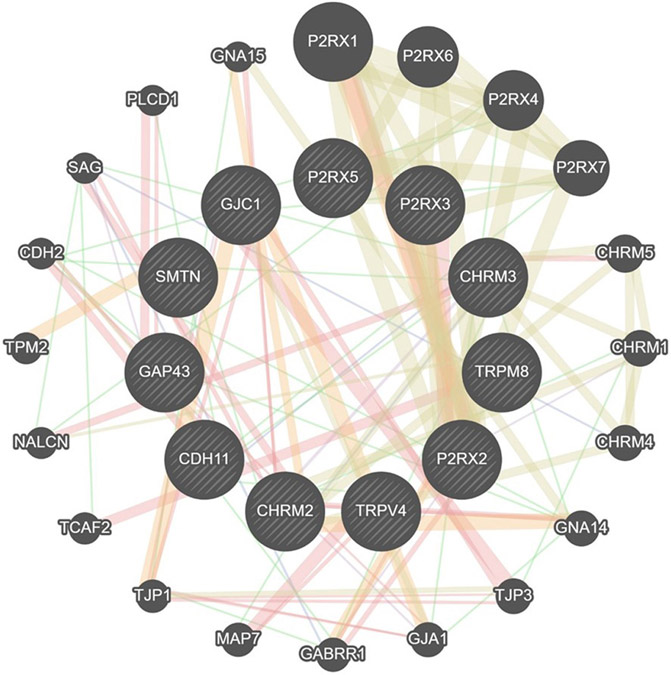
We ran GeneMANIA to analyze significantly overexpressed (P2RX2, SMTN, GAP43, TRPM8, CDH11, GJC1, CHRM2, CHRM3, and TRPV4) and underexpressed (P2RX3 and P2RX5) genes to further predict network connections. Interactions between genes are displayed as a network diagram. The color of the line connecting the genes indicates the type of communication (yellow: shared protein domains [47.23%]; orange: predicted communication [17.10%]; red: physical interactions [16.64%]; purple: co-expression [12.86%]; blue: co-localization [6.06%]) (Figure 2).
FIGURE 2. GeneMANIA interaction analysis for upregulated genes in OAB.
The most significant genes (P2RX2, SMTN, P2RX3, P2RX5, GAP43, TRPM8, CDH11, GJC1, TRPV4, CHRM2, CHRM3) are depicted in dash lines. The color of the line connecting the genes indicates the type of communication (purple, co-expression; red, physical interactions; Blue, co-localization; yellow, shared protein domains; orange, predicted communication; green, genetic interactions).
CDH2, cadherin 2; CDH11, cadherin 11; CHRM1, cholinergic receptor muscarinic 1; CHRM2, chinergic receptor muscarinic 2; CHRM3, cholinergic receptor muscarinic 3; CHRM4, cholinergic receptor muscarinic 4; CHRM5, cholinergic receptor muscarinic 5; GABRR1, gamma-aminobutyric acid receptor subunit rho-1; GAP43, growth-associated protein 43; GJA1, gap junction protein alpha 1; GJC1, gap junction protein gamma 1; GNA14, guanine nucleotide-binding protein subunit alpha-14; GNA15, guanine nucleotide-binding protein subunit alpha-15; NACLN, sodium leak channel, nonselective; MAP7, microtubule associated protein 7; P2RX, purinergic receptor P2X; PLCD1, phospholipase C delta 1; SAG, S-antigen visual arrestin; SMTN, smoothelin; TCAF2, TRPM8 channel associated factor 2; TJP1, tight junction protein 1; TJP3, tight junction protein 3; TPM2, tropomyosin 2; TRPM8, transient receptor potential cation channel subfamily M member 8; TRPV4, transient receptor potential cation channel subfamily V member 4.
Process enrichment analysis.
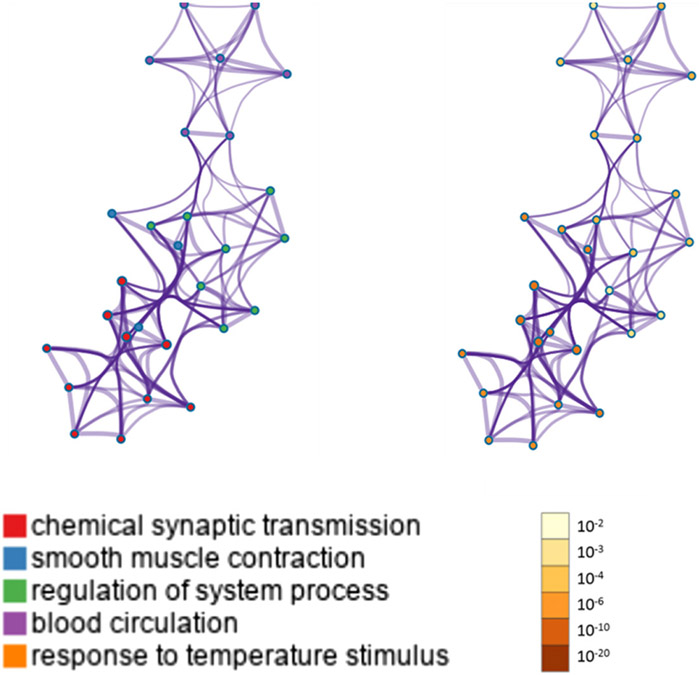
The most significant genes were found to be involved in chemical synaptic transmission, smooth muscle contraction, regulation of nervous system process, blood circulation, and response to temperature stimulus with the most statistically significant term within a cluster chosen to represent the cluster (Table 3).
TABLE 3.
Top 5 clusters with their representative enriched terms (1 per cluster) via genetic analysis method
| GO | Category | Description | Count | % | Log10(P) | Log10(q) |
|---|---|---|---|---|---|---|
| G0:0007268 | G0 Biological Processes | Chemical synaptic transmission | 6 | 54.55 | −8.67 | −4.75 |
| G0:0006939 | G0 Biological Processes | Smooth muscle contraction | 4 | 36.36 | −8.49 | −4.75 |
| G0:0044057 | G0 Biological Processes | Regulation of nervous system process | 5 | 45.45 | −5.99 | −2.99 |
| G0:0008015 | G0 Biological Processes | Blood circulation | 4 | 36.36 | −5.03 | −2.29 |
| G0:0009266 | G0 Biological Processes | Response to temperature stimulus | 3 | 27.27 | −4.59 | −1.94 |
“Count” is the number of genes in the user-provided lists with membership within the genetic analysis method. “%” is the percentage of all the user-provided genes that are found in the given ontology term (only input genes with at least 1 ontology term annotation are included in the calculation). “Log10(P)” is the P value in log base 10. “Log10(q)” is the multitest adjusted P value in log base 10.
GO, Gene Ontology.
Kyoto Encyclopedia of Genes and Genomes pathway analysis.
KEGG analyses reported significant pathways based on their log(P) values. The KEGG pathway analysis showed that the most significant genes were primarily involved in “chemical synaptic transmission,” “smooth muscle contraction,” “regulation of nervous system process,” “blood circulation,” and “response to temperature stimulus” pathways. Metascape displayed the top enrichment clusters for differentially expressed genes, which were discretely colored to encode P values of increasing statistical significance (Figure 3).
FIGURE 3. Metascape enrichment analysis for the included genes.
Network of enriched terms colored by cluster ID, where nodes sharing the same cluster ID are typically close to each other (threshold: 0.3 kappa score; similarity score of > 0.3).
ID, identification.
Protein-protein interaction network analysis.
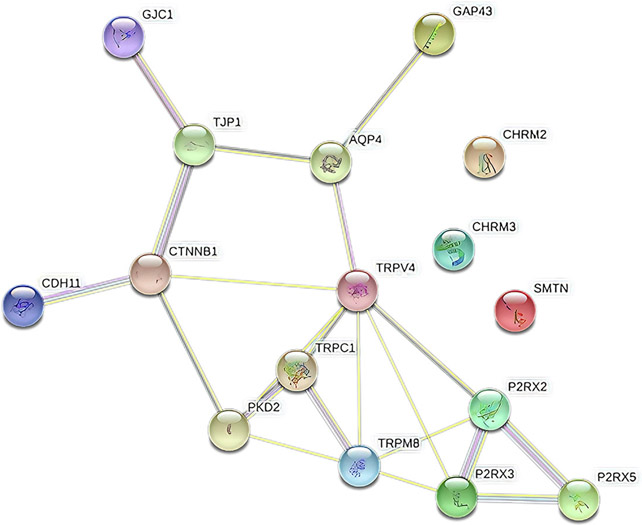
The network showed significantly more interactions between the TRPM8, TRPV4, P2RX3, and P2RX2 genes with a PPI enrichment P value of 3.41 × 10−5. Such an enrichment indicates the proteins, as a group, are at least partially biologically connected. The pathways between the constructed 4 genes were involved in “calcium signaling pathways” and “neuroactive ligand-receptor interaction” pathways (Figure 4). A schematic overview of up-regulated and down-regulated genes and their evidenced-based localization, functions, and connections can be found in Figure 5.
FIGURE 4. Protein-Protein Interaction Network based on studied genes.
The network showed significantly more interactions between the TRPM8, TRPV4, P2RX3, and PR2X2 genes (number of nodes: 16; number of edges: 21).
AQP4, aquaporin-4; CDH11, cadherin 11; CHRM2, chinergic receptor muscarinic 2; CHRM3, cholinergic receptor muscarinic 3; CTNNB1, catenin beta 1; GAP43, growth-associated protein 43; GJC1, gap junction protein gamma 1; P2RX, purinergic receptor P2X; PKD2, polycystin 2, transient receptor potential cation channel; SMTN, smoothelin; TJP1, tight junction protein 1; TRPC1, transient receptor potential cation channel subfamily C member 1; TRPM8, transient receptor potential cation channel subfamily M member 8; TRPV4, transient receptor potential cation channel subfamily V member 4.
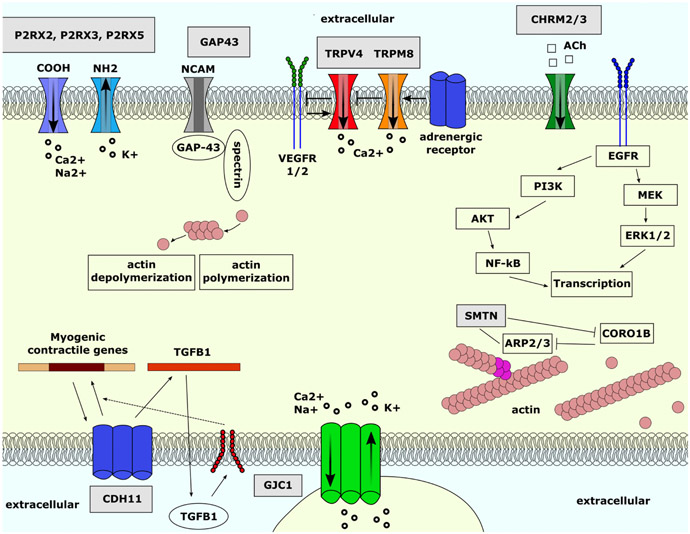
FIGURE 5. Overall summary of significant genes and their associated connections.
Ach, acetylcholine ARP2/3, actin-related protein 2/3; CDH11, cadherin 11; CHRM2, chinergic receptor muscarinic 2; CHRM3, cholinergic receptor muscarinic 3; CORO1B, coronin 1B; CTNNB1, catenin beta 1; EGFR, epidermal growth factor receptor; ERK1/2, extracellular signal-regulated kinase 1/2; GAP43, growth-associated protein 43; GJC1, gap junction protein gamma 1; NCAM, beural cell adhesion molecule; NF-κB, nuclear factor kappa B; P2RX, purinergic receptor P2X; PI3K, phosphoinositide 3-kinases; PKD2, polycystin 2, transient receptor potential cation channel; SMTN, smoothelin; TGBF1, transforming growth factor beta 1; TJP1, tight junction protein 1; TRPC1, transient receptor potential cation channel subfamily C member 1; TRPM8, transient receptor potential cation channel subfamily M member 8; TRPV4, transient receptor potential cation channel subfamily V member 4; VEGFR1, vascular endothelial growth factor receptor 1; VEGFR2, vascular endothelial growth factor receptor 2.
Comment
Principal findings
Up-regulated genes.
Purinergic receptor P2X 2.
The P2RX2 protein—coding gene located on chromosome 12 is of the purinoceptor family for adenosine triphosphate (ATP) and acts as a moderator for neuron to neuron and neuron to smooth muscle synaptic transmissions.32 P2RX2 has been implicated in other processes, including sound perception, neuron action potential, regulation of ion concentrations, and ischemia.33 P2RX2 overexpression has been associated with spinal injuries and hearing loss.34 The appropriate presence of P2RX2 is necessary to maintain the proper functioning of the bladder and up-regulation of P2RX2 is crucial for the changes in purinergic innervation in idiopathic detrusor instability bladders.22
Smoothelin.
The SMTN gene, located on chromosome 22, is responsible for coding proteins expressed only in smooth muscle cells.35 The SMTN gene is associated with the maintenance of the actin cytoskeleton and actin cytoskeletal dynamics.36 SMTN is involved in smooth muscle cell contraction, and the analysis of smooth muscle cells shows SMTN levels are significantly increased in patients with OAB.23
Growth-associated protein 43.
Growth-associated protein 43 is membrane bound, localized to the internal surface of the growth cone membrane, and is highly expressed during axonal growth and regeneration.37 GAP43 was up-regulated in degenerated primary afferent nerves during acute cystitis.38 Overexpression of GAP43 was reported in patients with OAB, indicating that GAP43 might be involved in the regulation of neuronal degeneration and regeneration during inflammatory states.25
Transient receptor potential cation channel subfamily M member 8.
TRPM8 has a crucial role in influencing transmembrane calcium ion transport, ligand-gated calcium channels, and thermogenesis.39 TRPM8 is typically activated in response to low temperature and is associated with dry eye disease40 and obesity41 in mouse studies. Overexpression of TRPM8 has been related to bladder activity, possibly through interactions with mechanosensitive C fibers.42 A recent rat study suggested that combination therapy of TRPM8 antagonist and β3-adrenoceptor agonist or anticholinergic agent can be a potential treatment option for obtaining additive effects compared with monotherapy for OAB.43 Marked increase of TRPM8-immunoreactive nerve fibers was detected in patients with OAB compared with controls without OAB.26
Cadherin 11.
Cadherins are cell surface glycoproteins that mediate Ca2+–dependent, hemophilic, cell-cell adhesion in epithelial tissues.44 The CDH11 gene is responsible for encoding a cadherin superfamily type II classical cadherin.45 Cadherin 11 up-regulation in suburothelial myofibroblasts in patients with overactive bladder may be significant in overactive bladder pathogenesis.27
Gap junction protein gamma 1.
Connexin proteins form gap junctions between adjacent cells to regulate cell-to-cell communications.46 GJC1 gene, also known as Cx45, is implicated in the initiation of inflammation through the activation of purinergic signaling pathways.47 GJC1 relates to extracellular ATP signaling in the urothelium of patients with bladder overactivity and urge incontinence. Both Cx43 and Cx45 are expressed at low levels in normal detrusor. The up-regulation of Cx45 in the myofibroblast cell layer supports the idea that alterations in sensory signaling are also involved in the pathogenesis of idiopathic OAB.28
Cholinergic receptor muscarinic 2.
The CHRM2 gene fits into the superfamily of GPCRs. Muscarinic acetylcholine receptors (M1–M5) activate a multitude of signaling pathways essential for controlling neuronal excitability and feedback management of acetylcholine release.48 Overexpression of CHRM2 was found in patients with persistent detrusor overactivity.49 Increased expression of the CHRM2 gene in myofibroblastlike cells suggests a potential role in pathophysiological mechanisms and the therapeutic effect of antimuscarinic agents in OAB.30-
Cholinergic receptor muscarinic 3.
CHRM3 gene is located on chromosome 1 and primarily responsible for smooth muscle contraction and glandular tissue secretion.48 Similar to CHRM2, increased expression of the CHRM3 gene has potential pathophysiological mechanisms in idiopathic OAB.30
Transient receptor potential cation channel subfamily V member 4.
TRPV4 is mainly expressed in bladder basal and intermediate urothelial cells and functions as a Ca2+ influx pathway activated by hypotonic cell swelling.50 TRPV4 mutation has also been identified in a spectrum of neuromuscular diseases that includes congenital distal spinal muscular atrophy, and hereditary motor and sensory neuropathy type IIC.51 Up-regulation of this gene was reported to be related to the pathogenesis of OAB.29
Down-regulated genes.Purinergic receptor P2X 3.
P2RX3 is 1 of 7 genes encoding the purinoceptor gene family and is expressed by sensory and autonomic neurons. Underexpression of P2RX3 resulted in a reduction in pain sensation and inflammation, suggesting that it plays an essential signaling role in the nervous system.52 Down-regulation of P2RX3 was associated with bladder hyporeflexia in 1 mouse study.53 P2RX3 deficiency may impair the control of detrusor contractility and contribute to the pathophysiology of urge incontinence. Therefore, antagonists to P2RX3 may have therapeutic potential in the treatment of disorders of urine storage and voiding, such as OAB.24
Purinergic receptor P2X 5.
P2RX5 receptor gene, located on chromosome 17, is also a ligand-gated ion channel. It is located on chromosome 17 and plays a crucial role in skeletal muscle tissue regeneration.54 The up-regulation of P2RX5 has been associated with T-cell activation, implying critical functionality in the immune system.55 Down-regulation of the gene has been correlated to detrusor instability and impaired bladder control.24
This systematic review identified a total of 11 genes with potential involvement in the pathogenesis of OAB. GeneMANIA analysis highlighted, in particular, P2RX genes as having “predicted communication” between them, indicating possible links in up- and down-regulation of genes in this family. The network analysis revealed further interactions between TRPV4, TRPM8, P2RX3, and P2RX2 genes. The pathways constructed between the 4 genes were involved in “calcium signaling pathways” and “neuroactive ligand-receptor interaction” (Figure 4). Opening voltage-dependent Ca2+ channels on the cell surface triggers bladder emptying contraction via a large influx of extracellular calcium. The activation of purinergic receptor genes was also implicated in bladder afferent hyperexcitability. Further exploration of this interaction network through molecular-level experimental studies may lead to understating the pathophysiology of the occurrence and development of OAB.
TRPV4 has been expressed mainly in bladder basal and intermediate urothelial cells and contributes to bladder filling detection. Reportedly, TRPV4 senses bladder urothelial cultured cell stretching, which is converted to ATP signals in the micturition reflex pathway during the storage phase. Up-regulation of TRPV4 might lead to a decreased sensation of bladder fullness via abnormal wall stretch-related signaling to afferent pathways and results in detrusor overactivity. An experimental study showed that intravesical activation of the cation channel TRPV4 improves bladder function in a rat model for detrusor underactivity.56 Conversely, TRPV4−/− mice exhibit an abnormal voiding pattern and decreased urothelial stretch-evoked ATP release.50 Currently, there is limited understanding of this difference between the 2 species. A study by Roberts et al29 showed that urothelial tissue in human OAB bladders spontaneously releases more ATP than cohort non-OAB bladders. Up-regulation of TRPV4 caused a greater quantity of stretch-induced ATP release in the mucosa and detrusor smooth muscle from aging bladders of patients with OAB. In addition, aging pig bladder mucosa exhibited greater TRPV4-induced ATP release with age dependence.29
Our analysis revealed that TRMP8 was expressed in patients with OAB as part of a response to the temperature stimulus pathway. TRPM8 is a cold-activated ion channel that plays a crucial role in detecting environmental temperatures and is targeted by antagonists that may be useful for reducing cold hypersensitivity resulting from nerve damage.57 It is noteworthy to mention that some people with OAB specifically report worsening symptoms with exposure to low temperatures, and this may represent a unique phenotype related to differential expression of TRMP8.58
In a previous animal model study, TRPM8 channel knockout mice had significantly less cold sensitivity.59 In addition, TRPM8 in the dorsal root ganglion might play a role in urinary urgency induced by cold sensation.60 Our study has highlighted further investigation of this gene as a new therapeutic opportunity for overactive bladder patients. In addition, this may lead to individualized treatments in patients with TRMP8 overexpression and could be a way to identify those patients with distinct phenotypes. Selective TRPM8 antagonists may provide valuable treatment for disorders related to the hyperactivity of bladder afferent nerves.
Comparison with existing literature
To date, most genomic studies61,62 were used from urine and blood samples, not actual bladder tissue specimens. Of note, 1 study that included 37 reports highlighted the prevalence of nerve growth factors in urine and serum samples collected from patients.61 Furthermore, they noted the prevalence of signal transduction pathways and, to a lesser extent, inflammatory responses associated with urgency urinary incontinence (UUI). Interestingly, the C-reactive protein was noted to be present in the serum of patients with UUI, but it was not significantly present in urine samples. In terms of tissue markers, alterations in the expression of GAP43, P2X (1—7), and TRPV1 were found to be implicated in cases of UUI,61 similar to the results of our findings.
Strength and limitations
The limitations of this study included the small sample size in some of the studies examined and the lack of information on the severity of OAB in the patients they analyzed. It should also be noted that we included studies where gene analysis was performed on bladder tissue specimens directly and excluded studies if the gene expression analysis was performed from urine and blood samples of patients vs healthy individuals.
Our initial aim was to run a meta-analysis based on publicly available Gene Expression Omnibus (GEO) data to categorize differentially expressed genes between controls without OAB and patients with OAB. GEO is a database repository of high-throughput gene expression data via hybridization arrays, chips, and microarrays. However, we could not find any available GEO datasets to perform a meta-analysis that could lead to robust findings for biomarker discovery studies. To the best of our knowledge, no optimized approach is established currently to run a meta-analysis using all molecular gene expression methods. Only microarray and RNA sequencing studies can be used to run meta-analysis using ImaGEO, ExAtlas, and Network Analyst web-based bioinformatics tools. Therefore, our gene expression analysis in the study was not an exhaustive representation of existing literature and might not be considered a deep analysis of available literature.
It is worth mentioning that any distinctions in gene expression level observed in patients with OAB vs controls without OAB are not necessarily involved in the development of OAB, which could be a consequence of the condition. Turning this into a potential biomarker or prediction tool might prove challenging, as it requires patients to give up bladder tissue. However, developing new treatments and tracking responses to treatment based on reported genes in this study could be a beneficial approach in the future. Despite these limitations, our systematic review highlighted differentially expressed genes in patients with OAB vs controls without OAB and emphasized several crucial genetic alterations as candidate targets from available reported literature.
Conclusions and implications
Further evaluation of the genes and pathways listed in our study would benefit researchers and clinicians. Our study did not conclude any clinical significance based on the in silico approach findings. Functional validation of potent trial targets and pathways needs to be further emphasized in vitro and using animal models. A better understanding of genes with poorer clinical outcomes might be prioritized for future targeting in clinical settings. Exploration of these specific pathways and gene interactions may be the key to unlocking the hidden mechanisms underlying the pathogenesis of OAB.
This systematic review identified differential expression of genes and gene-gene interactions among people with OAB, which may provide insight into distinct OAB phenotypes. Comprehensive experiments should be undertaken to evaluate the differential expression of these genes further. The outcomes of this systematic review may lay the groundwork for the development of future genes or other therapies and biomarkers for treatment efficacy of OAB in clinical practice.
Supplementary Material
AJOG at a Glance.
Why was this study conducted?
Although several studies have investigated genetic markers for overactive bladder (OAB) present in blood and urine, there is a distinct lack in the literature of tissue-level gene network analyses concerning OAB.
Key findings
Of note, 9 genes were found to be up-regulated (purinergic receptor P2X 2 [P2RX2], smoothelin [SMTN], growth-associated protein 43 [GAP43], transient receptor potential cation channel subfamily M member 8 [TRPM8], cadherin 11 [CDH1], gap junction protein gamma 1 [GJC1], cholinergic receptor muscarinic 2 [CHRM2], cholinergic receptor muscarinic 3 [CHRM3], and transient receptor potential cation channel subfamily V member 4 [TRPV4]) and 2 genes were down-regulated (purinergic receptor P2X 2 [P2RX3] and purinergic receptor P2X 5 [P2RX5]) in bladder tissues of patients with OAB.
What does this add to what is known?
This was a systematic review of genetic biomarkers associated with OAB at the tissue level. Study findings could offer a better means of assessing overactive bladder pathophysiology and provide direction for future analyses.
Footnotes
The authors report no conflict of interest.
This study received no financial support.
Contributor Information
Ilaha Isali, Division of Female Pelvic Medicine and Reconstructive Surgery, Department of Urology, University Hospitals, Cleveland Medical Center, Cleveland, OH.
Phillip McClellan, Division of Female Pelvic Medicine and Reconstructive Surgery, Department of Urology, University Hospitals, Cleveland Medical Center, Cleveland, OH.
Thomas R. Wong, Division of Female Pelvic Medicine and Reconstructive Surgery, Department of Urology, University Hospitals, Cleveland Medical Center, Cleveland, OH
Clara Sun, Division of Female Pelvic Medicine and Reconstructive Surgery, Department of Urology, University Hospitals, Cleveland Medical Center, Cleveland, OH.
Amber Catherine Stout, Core library, University Hospitals, Cleveland Medical Center, Cleveland, OH.
Fredrick R. Schumacher, Department of Population and Quantitative Health Sciences, Case Western Reserve University, Cleveland, OH.
Sarah Markt, Department of Population and Quantitative Health Sciences, Case Western Reserve University, Cleveland, OH.
Chen-Han Wilfred Wu, Division of Female Pelvic Medicine and Reconstructive Surgery, Department of Urology, University Hospitals, Cleveland Medical Center, Cleveland, OH; Department of Genetics and Genome Sciences, Case Western Reserve University, Cleveland, OH.
Kathryn L. Penney, Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, MA; Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA.
Sherif El-Nashar, Department of Obstetrics and Gynecology, Mayo Clinic, Jacksonville, FL.
Adonis Hijaz, Division of Female Pelvic Medicine and Reconstructive Surgery, Department of Urology, University Hospitals, Cleveland Medical Center, Cleveland, OH.
David Sheyn, Division of Female Pelvic Medicine and Reconstructive Surgery, Department of Urology, University Hospitals, Cleveland Medical Center, Cleveland, OH.
REFERENCES
- 1.Leron E, Weintraub AY, Mastrolia SA, Schwarzman P. Overactive bladder syndrome: evaluation and management. Curr Urol 2018;11:117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srikrishna S, Robinson D, Cardozo L, Vella M. Management of overactive bladder syndrome. Postgrad Med J 2007;83:481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coyne KS, Margolis MK, Kopp ZS, Kaplan SA. Racial differences in the prevalence of overactive bladder in the United States from the epidemiology of LUTS (EpiLUTS) study. Urology 2012;79:95–101. [DOI] [PubMed] [Google Scholar]
- 4.Abrams P, Kelleher CJ, Kerr LA, Rogers RG. Overactive bladder significantly affects quality of life. Am J Manag Care 2000;6:S580–90. [PubMed] [Google Scholar]
- 5.Tyagi P. Pathophysiology of the urothelium and detrusor. Can Urol Assoc J 2011;5(Suppl2):S128–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willis-Gray MG, Dieter AA, Geller EJ. Evaluation and management of overactive bladder: strategies for optimizing care. Res Rep Urol 2016;8:113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutchinson A, Nesbitt A, Joshi A, Clubb A, Perera M. Overactive bladder syndrome: management and treatment options. Aust J Gen Pract 2020;49:593–8. [DOI] [PubMed] [Google Scholar]
- 8.Çırakoğlu A, Fejzullahu A, Benli E, Yuce A, Ayyıldız A, Aynacıoğlu AŞ. Association between the Trp64Arg polymorphism of the ADRB3 gene and overactive bladder. Neurourol Urodyn 2021;40:1780–5. [DOI] [PubMed] [Google Scholar]
- 9.Andersson KE, Christ GJ, Davies KP, Rovner ES, Melman A. Gene therapy for overactive bladder: a review of BK-channel α-subunit gene transfer. Ther Clin Risk Manag 2021;17:589–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fırat E, Aybek Z, Akgün Ş, Küçüker K, Akıa H, Aybek H. Relation of ADRB3, GEF, ROCK2 gene polymorphisms to clinical findings in overactive bladder. World J Urol 2020;38:2571–5. [DOI] [PubMed] [Google Scholar]
- 11.Segundo-Val IS, Sanz-Lozano CS. Introduction to the gene expression analysis. Methods Mol Biol 2016;1434:29–43. [DOI] [PubMed] [Google Scholar]
- 12.Rovner E, Chai TC, Jacobs S, et al. Evaluating the safety and potential activity of URO-902 (hMaxi-K) gene transfer by intravesical instillation or direct injection into the bladder wall in female participants with idiopathic (non-neurogenic) overactive bladder syndrome and detrusor overactivity from two double-blind, imbalanced, placebo-controlled randomized phase 1 trials. Neurourol Urodyn 2020;39:744–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wein AJ. Re: evaluating the safety and potential activity of URO-902 (hMaxi-K) gene transfer by intravesical instillation or direct injection into the bladder wall in female participants with idiopathic (non-neurogenic) overactive bladder syndrome and detrusor overactivity from two double-blind, imbalanced, placebo-controlled randomized Phase 1 trials. J Urol 2020;204:884–5. [DOI] [PubMed] [Google Scholar]
- 14.Morris BD Jr, Drazan KE, Csete ME, et al. Adenoviral-mediated gene transfer to bladder in vivo. J Urol 1994;152:506–9. [DOI] [PubMed] [Google Scholar]
- 15.Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: the PRISMA statement. BMJ 2009;339:b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan-a web and mobile app for systematic reviews. Syst Rev 2016;5:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang DW, Sherman BT, Tan Q, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 2007;8:R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isali I, Mahran A, Khalifa AO, et al. Gene expression in stress urinary incontinence: a systematic review. Int Urogynecol J 2020;31:1–14. [DOI] [PubMed] [Google Scholar]
- 19.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol 2008;9(Suppl1):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo CK, Mertz D, Loeb M. Newcastle-Ottawa Scale: comparing reviewers’ to authors’ assessments. BMC Med Res Methodol 2014;14:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Reilly BA, Kosaka AH, Knight GF, et al. P2X receptors and their role in female idiopathic detrusor instability. J Urol 2002;167:157–64. [PubMed] [Google Scholar]
- 23.Maake C, Landman M, Wang X, Schmid DM, Ziegler U, John H. Expression of smoothelin in the normal and the overactive human bladder. J Urol 2006;175:1152–7. [DOI] [PubMed] [Google Scholar]
- 24.Moore KH, Ray FR, Barden JA. Loss of purinergic P2X(3) and P2X(5) receptor innervation in human detrusor from adults with urge incontinence. J Neurosci 2001;21:RC166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schofield EC, Clausen JA, Burcher E, Moore KH. GAP-43 immunoreactivity of sub-epithelial and detrusor muscle nerve fibres in patients with refractory idiopathic detrusor overactivity. Neurourol Urodyn 2005;24:325–33. [DOI] [PubMed] [Google Scholar]
- 26.Mukerji G, Yiangou Y, Corcoran SL, et al. Cool and menthol receptor TRPM8 in human urinary bladder disorders and clinical correlations. BMC Urol 2006;6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roosen A, Apostolidis A, Elneil S, et al. Cadherin-11 up-regulation in overactive bladder suburothelial myofibroblasts. J Urol 2009;182:190–5. [DOI] [PubMed] [Google Scholar]
- 28.Neuhaus J, Pfeiffer F, Wolburg H, Horn LC, Dorschner W. Alterations in connexin expression in the bladder of patients with urge symptoms. BJU Int 2005;96:670–6. [DOI] [PubMed] [Google Scholar]
- 29.Roberts MWG, Sui G, Wu R, et al. TRPV4 receptor as a functional sensory molecule in bladder urothelium: stretch-independent, tissue-specific actions and pathological implications. FASEB J 2020;34:263–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukerji G, Yiangou Y, Grogono J, et al. Localization of M2 and M3 muscarinic receptors in human bladder disorders and their clinical correlations. J Urol 2006;176:367–73. [DOI] [PubMed] [Google Scholar]
- 31.Munn Z, Barker TH, Moola S, et al. Methodological quality of case series studies: an introduction to the JBI critical appraisal tool. JBI Evid Synth 2020;18:2127–33. [DOI] [PubMed] [Google Scholar]
- 32.Brake AJ, Wagenbach MJ, Julius D. New structural motif for ligand-gated ion channels defined by an ionotropic ATP receptor. Nature 1994;371:519–23. [DOI] [PubMed] [Google Scholar]
- 33.North RA. Molecular physiology of P2X receptors. Physiol Rev 2002;82:1013–67. [DOI] [PubMed] [Google Scholar]
- 34.Yan D, Zhu Y, Walsh T, et al. Mutation of the ATP-gated P2X(2) receptor leads to progressive hearing loss and increased susceptibility to noise. Proc Natl Acad Sci U S A 2013;110:2228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinto SM, Manda SS, Kim MS, et al. Functional annotation of proteome encoded by human chromosome 22. J Proteome Res 2014;13:2749–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blaine J, Dylewski J. Regulation of the actin cytoskeleton in podocytes. Cells 2020;9:1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denny JB. Molecular mechanisms, biological actions, and neuropharmacology of the growth-associated protein GAP-43. Curr Neuropharmacol 2006;4:293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vizzard MA, Boyle MM. Increased expression of growth-associated protein (GAP-43) in lower urinary tract pathways following cyclophosphamide (CYP)-induced cystitis. Brain Res 1999;844:174–87. [DOI] [PubMed] [Google Scholar]
- 39.Sisco NJ, Helsell CVM, Van Horn WD. Competitive interactions between PIRT, the cold sensing ion channel TRPM8, and PIP2 suggest a mechanism for regulation. Sci Rep 2019;9:14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fakih D, Baudouin C, Réaux-Le Goazigo A, Mélik Parsadaniantz S. TRPM8: a therapeutic target for neuroinflammatory symptoms induced by severe dry eye disease. Int J Mol Sci 2020;21:8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang C, Zhai M, Yan D, et al. Dietary menthol-induced TRPM8 activation enhances WAT “browning” and ameliorates diet-induced obesity. Oncotarget 2017;8:75114–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merrill L, Gonzalez EJ, Girard BM, Vizzard MA. Receptors, channels, and signalling in the urothelial sensory system in the bladder. Nat Rev Urol 2016;13:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aizawa N, Fujimori Y, Nakanishi O, et al. Efficacy of the combination of KPR-5714, a novel transient receptor potential melastatin 8 (TRPM8) antagonist, and β3-adrenoceptor agonist or anticholinergic agent on bladder dysfunction in rats with bladder overactivity. Eur J Pharmacol 2021;899:173995. [DOI] [PubMed] [Google Scholar]
- 44.Mialhe A, Levacher G, Champelovier P, et al. Expression of E-, P-, N-cadherins and catenins in human bladder carcinoma cell lines. J Urol 2000;164:826–35. [DOI] [PubMed] [Google Scholar]
- 45.Frei JA, Niescier RF, Bridi MS, et al. Regulation of neural circuit development by Cadherin-11 provides implications for autism. eNeuro 2021;8. ENEURO.0066—21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dbouk HA, Mroue RM, El-Sabban ME, Talhouk RS. Connexins: a myriad of functions extending beyond assembly of gap junction channels. Cell Commun Signal 2009;7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu CY, Zhang WS, Zhang H, Cao Y, Zhou HY. The role of connexin-43 in the inflammatory process: a new potential therapy to influence keratitis. J Ophthalmol 2019;2019:9312827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gosso FM, de Geus EJ, Polderman TJ, Boomsma DI, Posthuma D, Heutink P. Exploring the functional role of the CHRM2 gene in human cognition: results from a dense genotyping and brain expression study. BMC Med Genet 2007;8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barbosa JABA, Reis ST, Nunes M, et al. The obstructed bladder: expression of collagen, matrix metalloproteinases, muscarinic receptors, and angiogenic and neurotrophic factors in patients with benign prostatic hyperplasia. Urology 2017;106:167–72. [DOI] [PubMed] [Google Scholar]
- 50.Gevaert T, Vriens J, Segal A, et al. Deletion of the transient receptor potential cation channel TRPV4 impairs murine bladder voiding. J Clin Invest 2007;117:3453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dai J, Cho TJ, Unger S, et al. TRPV4-pathy, a novel channelopathy affecting diverse systems. J Hum Genet 2010;55:400–2. [DOI] [PubMed] [Google Scholar]
- 52.Brederson JD, Jarvis MF. Homomeric and heteromeric P2X3 receptors in peripheral sensory neurons. Curr Opin Investig Drugs 2008;9:716–25. [PubMed] [Google Scholar]
- 53.Cockayne DA, Hamilton SG, Zhu QM, et al. Urinary bladder hyporeflexia and reduced pain-related behaviour in P2X3-deficient mice. Nature 2000;407:1011–5. [DOI] [PubMed] [Google Scholar]
- 54.Kim H, Walsh MC, Takegahara N, et al. The purinergic receptor P2X5 regulates inflammasome activity and hyper-multinucleation of murine osteoclasts. Sci Rep 2017;7:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abramowski P, Ogrodowczyk C, Martin R, Pongs O. A truncation variant of the cation channel P2RX5 is upregulated during T cell activation. PLoS One 2014;9:e104692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deruyver Y, Weyne E, Dewulf K, et al. Intravesical activation of the cation channel TRPV4 improves bladder function in a rat model for detrusor underactivity. Eur Urol 2018;74:336–45. [DOI] [PubMed] [Google Scholar]
- 57.Izquierdo C, Martín-Marténez M, Gómez-Monterrey I, González-Muñiz R. TRPM8 channels: advances in structural studies and pharmacological modulation. Int J Mol Sci 2021;22:8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tae BS, Park TY, Jeon BJ, et al. Seasonal variation of overactive bladder symptoms in female patients. Int Neurourol J 2019;23:334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dhaka A, Murray AN, Mathur J, Earley TJ, Petrus MJ, Patapoutian A. TRPM8 is required for cold sensation in mice. Neuron 2007;54:371–8. [DOI] [PubMed] [Google Scholar]
- 60.Shibata Y, Ugawa S, Imura M, et al. TRPM8-expressing dorsal root ganglion neurons project dichotomizing axons to both skin and bladder in rats. NeuroReport 2011;22:61–7. [DOI] [PubMed] [Google Scholar]
- 61.Post WM, Ruiz-Zapata AM, Grens H, et al. Genetic variants and expression changes in urgency urinary incontinence: a systematic review. Neurourol Urodyn 2020;39:2089–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Antunes-Lopes T, Carvalho-Barros S, Cruz CD, Cruz F, Martins-Silva C. Biomarkers in overactive bladder: a new objective and noninvasive tool? Adv Urol 2011;2011:382431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.