Abstract
Recent innovations in single-cell technologies have opened up exciting possibilities for profiling the omics of individual cells. Minimally invasive analysis tools that probe and remove the contents of living cells enable cells to remain in their standard microenvironment with little impact on their viability. This negates the requirement of lysing cells to access their contents, an advancement from previous single-cell manipulation methods. These novel methods have the potential to be used for dynamic studies on single cells, with many already providing high intracellular spatial resolution. In this article, we highlight key technological advances that aim to remove the contents of living cells for downstream analysis. Recent applications of these techniques are reviewed, along with their current limitations. We also propose recommendations for expanding the scope of these technologies to achieve comprehensive single-cell tracking in the future, anticipating the discovery of subcellular mechanisms and novel therapeutic targets and treatments, ultimately transforming the fields of spatial transcriptomics and personalised medicine.
Recent advances and applications in subcellular sampling in single-cell systems are reviewed, along with their promise and current limitations.
Introduction
Understanding the molecular basis of individual cells is essential for elucidating genetic, functional or compositional heterogeneity of tissues and organs for accurate design of disease models and patient responses to specific therapies.1 Imaging techniques such as RNAscope™,2 Multi Omic Single-scan Assay with Integrated Combinatorial Analysis (MOSAICA)3 and multiplexed error-robust fluorescent in situ hybridisation (MERFISH)4 can provide information on the contents of fixed cells, providing a ‘snapshot’ of the molecular profiles of single cells. Developments in single-cell imaging techniques such as synchrotron imaging,5–7 live cell imaging8 and single-molecule tracking9 have provided further insight into the subcellular dynamics of living cells. To fully understand the molecular basis of single cells, however, it is essential to extract the target molecules from individual cells before their analysis. This can aid the discovery of novel transcripts, proteoforms, and post-transcriptional and -translational modifications. Hitherto, microfluidic platforms incorporating various single-cell manipulation techniques such as microwell-based docking,10 electrokinetic single-cell focusing,11 fluorescence-activated cell sorting12 and optical tweezers,13,14 were widely employed for high-throughput chemical analysis of single cells. Although easy to implement, these methods require removing the target cell from their natural surroundings and, in most cases, their lysis in regulated environments. This leads to post-lysis analyte modifications and the loss of contextual information.
To overcome these limitations, the insertion of minimally invasive sampling devices based on atomic force microscopy (AFM) tips and micro/nanopipettes were proposed.15 For instance, the functionalised AFM tip-based selective mRNA profiling methods provided a non-destructive way to analyse specific gene expressions in single living cells while maintaining viability.16 A similar method, employing fluid force microscopy, extended the use of this platform to minimally invasive intracellular fluid extraction for tuneable aspiration of intracellular fluids for single-cell analysis.17,18 Actis et al. reported on a nanopipette-based biopsy platform for continuous intracellular sampling,19 which, when used in conjunction with matrix-assisted laser desorption/ionisation-mass spectrometry20 and scanning ion conductance microscopy,19 could aid single cell protein/nucleic acid analysis with spatial and temporal control. Both fluid force microscopy and nanopipette-based extraction strategies involve the withdrawal of cytoplasmic fluid from the cells. If this fluid exceeds femtoliter volumes, this can impact the survival of the cell. Further, the very low concentration (<pM) of some analyte molecules makes subsequent in vitro analysis challenging.
Minimally invasive single-cell sampling techniques that can concentrate and extract cellular components while maintaining cellular integrity have been developed as an alternative. Nadappuram et al. reported on the development of a dielectrophoretic nanotweezer for non-destructive subcellular concentration (trapping) and extraction of biomolecules.21 These nanotweezers employ dielectrophoresis (DEP) to trap molecules subjected to a nonuniform electric field22,23 and are composed of two individually addressable nanoelectrodes separated by a nanoscale septum to generate the relatively high electric field gradient required for trapping and extraction of different molecules. The capability of these nanotweezers for minimally invasive single-cell extraction of molecules and organelles under physiological environment was demonstrated by extracting genomic DNA, RNA and single mitochondrion from human osteosarcoma (U2OS) cells, human pulmonary artery endothelial (HPAE) cells and the axon of mouse primary hippocampal neurons, respectively.
Single-cell analysis technologies have greatly advanced the genomics field and underpin one of the most ambitious genomics projects post the sequencing of the human genome: the ‘Human Cell Atlas’, which aims to create a reference map of all human cells.15,24 Much of the state-of-the-art research in the field has been reviewed previously. This review focuses predominantly on single-cell analysis platforms that can manipulate and sample from individual cells with minimum perturbations, including those based on nanostraw structures, micro/nanopipettes, AFM and dielectrophoretic nanotweezers. These methods hold great promise for the non-destructive extraction of cellular components with spatiotemporal control and could significantly improve both clinical and non-clinical investigations into cellular processes with single-cell resolution.
1. Pipettes
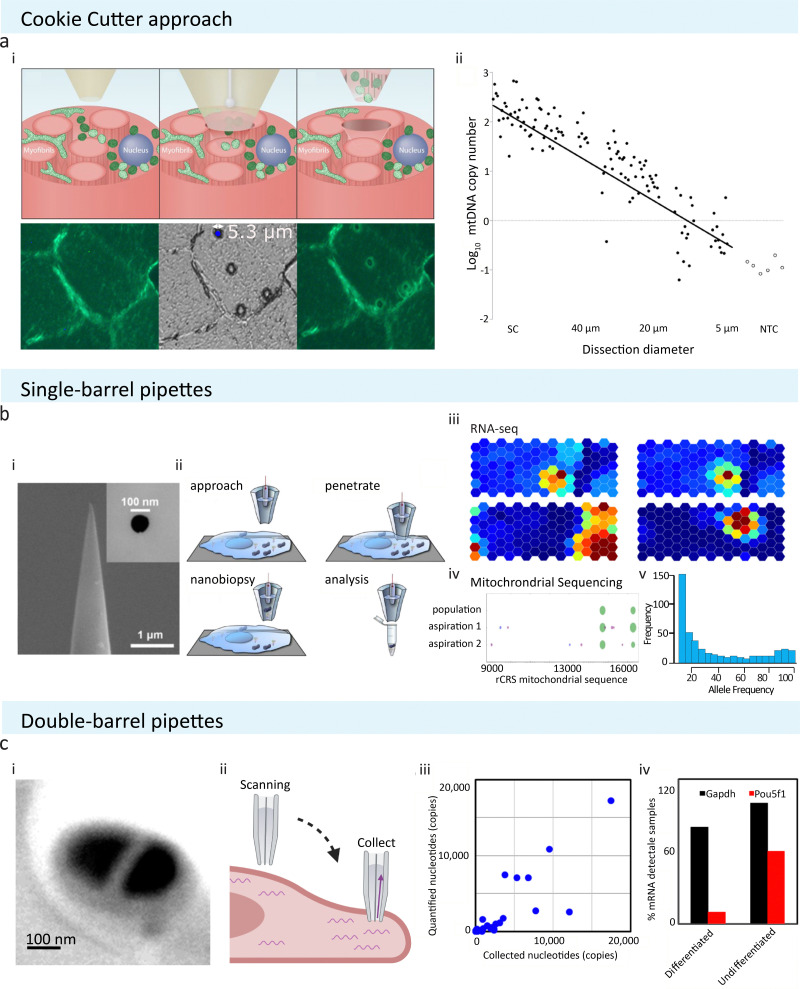
Pipettes are typically fine, hollow needles pulled from borosilicate glass or quartz capillaries. Nanopipettes can be pulled with controllable pore sizes over a wide range of diameters (usually 50 nm–5 μm) and are classified as micro or nanopipettes if they have a pore diameter above or below 100 nm, respectively. Different approaches in extracting localised cellular samples using micro and nanopipettes have been reported, such as electrowetting combined with scanning ion conductance microscopy (SICM) and aspiration by negative pressure. These pipettes can be either single or double-barrelled (Fig. 1(b)-i and (c)-i, respectively) and are commercially available or can be fabricated from capillaries using a laser puller.
Fig. 1. Schematic of pipette-based cellular sampling technologies and downstream analysis. (a) The ‘cookie cutter’ approach from Bury et al. extracts cellular content by plunging through tissue sections. (i) Localised cellular samples are removed alongside with the micropipette after puncturing. (ii) The number of samples obtained is depends on the pore size used; with a pipette of 5 μm in size, a single mitochondrion could be extracted. Reproduced from ref. 25. Copyright 2022, Springer Nature. (b) Single-barrel pipettes from Actis et al. could be used to extract cellular samples with or without SICM. (i) SEM image of a typical single barrel with tip size of 100 nm or below are commonly utilised in extracting localised cellular samples. Reprinted (adapted) with permission from ref. 19. Copyright 2014, American Chemical Society. (ii) A schematic showing the experimental procedure of nanobiopsy when combined with SICM to obtain cellular topography. (iii) and (iv) Extracted samples have been analysed using RNA-seq, mitochondrial sequencing, or RT-qPCR to study cell-to-cell variability or single cell mitochondrial heterogeneity. Reproduced with permission from ref. 19, 28 (Copyright 2018, Elsevier), and 35 (Copyright 2017, Elsevier). (c) Double-barrel pipettes used in combination with SICM to obtain cellular contents at precise locations. (i) SEM image shows a typical dual barrel pipette applied for SICM-nanobiopsy. (ii) During SICM-single-cell biopsy, one of the two barrels of the pipette was used to acquire cell topography via SICM, and then sample extraction was done by exerting negative pressure to aspirate cellular fluid at the desired cellular location. (iii) The amount of transcripts detectable via RT-qPCR is proportional to the amount of samples collected. (iv) The dual barrel pipettes have been used to obtain subcellular amounts of samples from stem cells and study the differentiation status of stem cells by analysing differentiation marker Pou5f1 in mouse embryonic stem cells. Reprinted (adapted) from ref. 29. Copyright 2016, American Chemical Society. The schematic was created with BioRender.com.
One method of extracting cytosolic content is by puncturing tissue sections using micropipettes to extract localised samples, namely the ‘cookie cutter’ approach (Fig. 1(a)).25 This technique allows for rapid, simple sample extraction without requiring complex probe modification. However, due to the large probe size (5 μm diameter) of the micropipettes (Fig. 1(a)-ii), this technique could be more invasive on living cells compared to other similar technologies discussed in later sections.
Another common way of aspirating cytoplasmic fluid is by using electrowetted nanopipettes. Electrowetting is a process in which the nanopipette is filled with an organic solvent, 1,2-dichloroethane, followed by the application of an electric field to alter the interfacial tension, allowing for cytoplasmic fluid to be aspirated into the pipette via interfacial electric stress (Fig. 1(b)-ii).19,26,27 This is often combined with the use of SICM, allowing for precise positioning of the nanopipettes by providing high optical resolution (Fig. 1(c)).19 In brief, the authors filled the nanopipette with an organic, hydrophobic solution of dichloroethane (DCE). When the filled nanopipette was inserted into a cell, a voltage was applied leading to altered surface tension of DCE. This allowed the movement of the aqueous cellular solution to flow in or out of the nanopipette tip. The flow was dictated by either a negative or positive voltage being applied during sample aspiration and collection. The collected samples could then be subjected to genomic or transcriptomic analysis such as quantitative reverse transcription polymerase chain reaction (RT-qPCR) or next-generation sequencing (NGS) such as RNA sequencing (RNA-seq) and whole mitochondrial genomic sequencing (Fig. 1(b)iii–v and (c)-iii, iv). This technique has been used to identify novel targets localised in the neurites of neurons.28 A dual barrel nanopipette system is another way of utilising SICM to increase the precision and resolution of sample capturing.29 Of the two barrels, one is filled with an aqueous solution for SICM imaging and the other with an organic solution (for electrowetting) for cytosol aspiration and follow-up downstream RT-qPCR analysis. The authors validated the efficacy and efficiency of the system on MCF-7 cells. They observed the aspiration of cytosol into the SICM pipette under a video microscope and investigated the extracted cytosolic samples with RT-qPCR. They then applied this method to mouse embryonic stem cells, either differentiated or undifferentiated, where they punctured the cells to collect cytosol and used RT-qPCR (Fig. 1(c)-iv) to analyse the Pou5f1 gene (undifferentiation marker) and GAPDH (housekeeping gene). This validated using dual barrel system to study mRNA localisation in single cells. The one disadvantage of this technique is that the authors found that it was necessary to penetrate the cells at least 3 μm underneath the cellular membrane for successful extraction of the cytosol, perhaps making the application of this technique on thin, flat cell types impractical. It should be noted that in some cases, unfortunately, electrowetting-based aspiration leads to increased cell toxicity,30 and this limitation should be considered when designing aspiration experiments.
Once a localised cellular samples are obtained, there are several ways for their characterisation. In most cases, samples are analysed by RT-qPCR of RNAs of interest from the extracted samples at a given cellular location. Saha-Shah et al. introduced the application of matrix-assisted laser desorption/ionisation-mass spectrometry (MALDI-MS).20 Nanopipettes were used for the collection of cellular fluids by controlled pressure actuation from onion cells, fruit fly larvae, and mouse brain tissue sections. The collected samples were then analysed by MALDI-MS, and the reported results were consistent with previously published data using other MS methods.31–34 Nanopipettes have shown further advantages in their <1 μm capturing precision and simplicity using liquid aspiration by suction. Like other cellular fluid aspiration methods, the major limitation of the technique lies with the geometry of the nanopipettes. This is not only limited to the diameter of the nanopore opening but also the shape of the pipette, such as the length of the shank. In this study, the authors have shown that the length of the shank directly influences the relationship between the pressure applied and the volume of cellular content aspirated.
Although cytosolic fluid extraction is often the main interest in many scientific fields, nanopipette-based subcellular sampling could also extract organelles such as mitochondria. Morris et al. demonstrated mitochondria extraction from neurons to evaluate the level of heteroplasmy in inbred mice while showing that specific genetic variants are preserved across generations in both murine and human models.35 Mitochondria were first fluorescently labelled in vitro using a MitoTracker dye. A motorised micropipette allowed the extraction of the cellular content, including the mitochondrion of interest, by applying negative pressure. The collected sample was lysed by protease K, and the mtDNA was PCR amplified before subjecting to whole genome sequencing and variant analysis (Fig. 1(b)-iv, v).
The benefit of utilising nanopipettes for aspirating cellular samples is that the pore or tip size of nanopipettes can be small and controllable to facilitate precisely localised cellular sampling.25,35 There are also plenty of ways of operating these fine capillaries in a highly precise manner, such as in conjunction with micromanipulators,20 making it one of the most suitable and widely adapted methods across a range of single-cell techniques. In summary, nanopipettes have been used to extract subcellular content in a range of different samples, from tissue sections to single cells and from the extraction of subcellular cytoplasmic fluids to single organelles. This provided a wide range of applications in various biological studies such as stem cell research and genomics analysis.29,35 There are, however, some limitations to nanopipette-based probes that need further addressing, such as increased cell toxicity due to electrowetting and non-specific aspiration of cytoplasmic fluids, which may require sample purification steps and makes the detection of low abundance molecules difficult.
2. Nanostraws
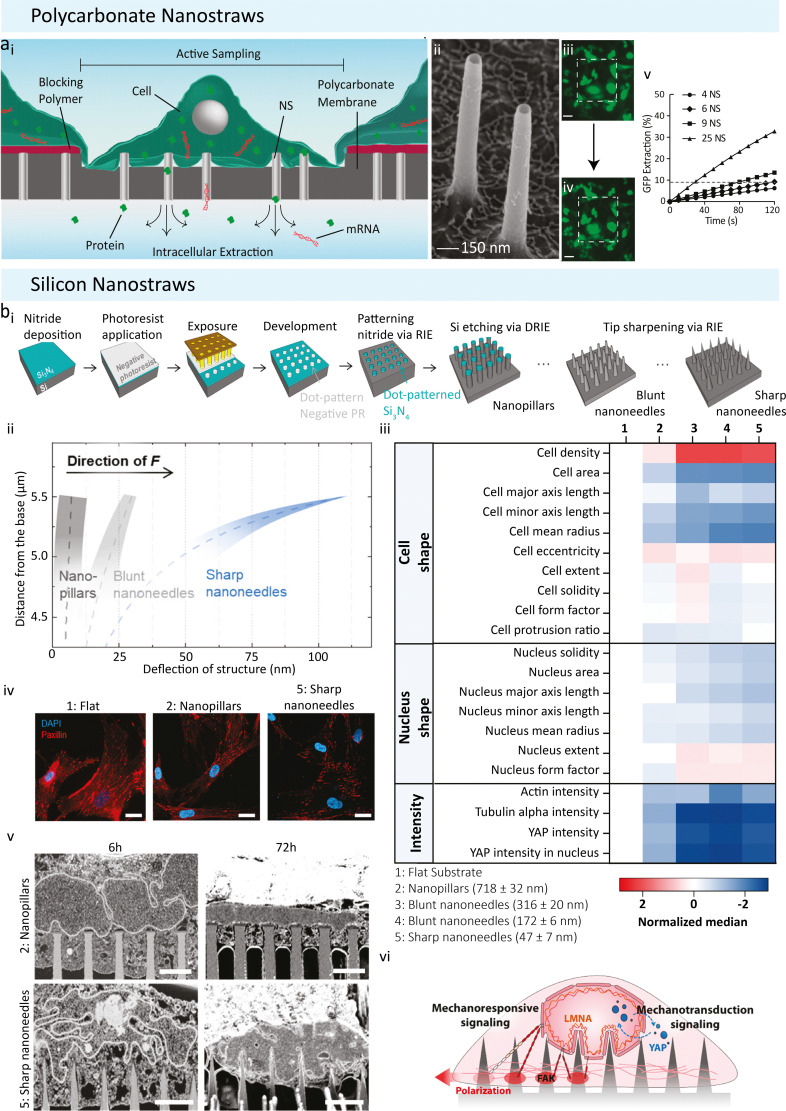
An alternative non-destructive approach to the nanopipettes is the nanostraw (NS) platform. They are highly effective in penetrating the cell membrane and providing direct fluidic intracellular access in live cells.36–41 NSs are hollow tube-like structures (diameters between 20 to 700 nm, length 1 to 3 μm) that are embedded in a cell culture-compatible membrane. There are two types of NS; polycarbonate-based (Fig. 2(a)) and silicon-based (Fig. 2(b)) ones. The polycarbonate NSs are fabricated from track-etched polycarbonate membranes, widely used for cell culture applications.36 Various methods for fabricating these types of NSs have been reported previously.37,41 These NSs are typically 150 nm in diameter and 1–3 μm long (Fig. 2(a)-ii). They are mounted onto the bottom of glass cylinders 2 to 5 mm in diameter, fitting into a 48 or 96-well plate for cell cultures. A thin layer of Al2O3 is deposited on the polycarbonate membrane, including the track-etched tracks, which in turn become the NSs walls through careful etching.36 The other type of NS is fabricated from silicon wafers using a top-down photolithography approach (Fig. 2(b)-i).38 This allows the fabrication of finely tuneable pillars with diameters ranging from 20 nm to 700 nm.38 These silicon NSs are 5 μm long and arranged in uniform square arrays with 2 μm spacing, making them suited for a variety of biological applications and allowing for a deeper cell penetration if required.
Fig. 2. Nanostraw use adapted from Cao et al. and Seong et al. (a) Polycarbonate nanostraws adapted from Cao et al. (i) Polycarbonate nanostraws allow intracellular species within the cell to diffuse through the NS and into the extraction buffer below the membrane. The size of the sampling region can be defined lithographically so that only the cells that grow in the active regions are sampled. (ii) Tilted view (45°) SEM image of the 150 nm diameter NS. (iii) and (iv) Fluorescent microscopy images of GFP of a culture of 26 cells on a 200 × 200 μm NS sampling region (white dashed squares, scale bar = 50 μm). (iii) GFP-expressing CHO cells before sampling. (iv) GFP-expressing CHO cells immediately after sampling. Locally diminished GFP intensities (dark spots) were observed in the cells after sampling, corresponding to the locations where GFP was removed from the cells. Brightness was increased to highlight the spots. (v) The percentage of the cell's initial GFP that diffuses into the extraction buffer as a function of time and the number of NS (the dashed line indicates the GFP extraction level after 2 min of diffusion from six NS). Reproduced from ref. 36. Copyright 2017, National Academy of Sciences. (B) Size-Tunable silicon based Nanostraws reported by Seong et al. (i) Schematic detailing the photolithography and dry silicon etching processes. Silicon nitride was deposited as a hard mask onto a silicon wafer, dot arrays of photoresist were patterned on the nitride layer by photolithography, silicon nitride, unprotected by photoresist was reactive ion etched (RIE), nanopillar arrays were produced via deep reactive ion etching (DRIE), and sharpened into nanoneedle arrays. (ii) Deflection of nanopillars (Dtip = 718 ± 32 nm), blunt nanoneedles (Dtip = 316 ± 20 nm), and sharp nanoneedles (Dtip = 47 ± 7 nm) when 300 nN of traction force (F) was imposed at the apex of each structure. Note that only the upper 1.5 μm of tip deflection is shown, as this is the region in which the deflection profiles differ the most. (iii) Normalized heatmap showing the change in population median on different substrates for a selected range of parameters. Note: data shown here are transformed and normalized to flat substrates; hence ‘1’ is blank (white). (iv) Representative confocal immunofluorescence images of paxillin-stained focal adhesions in hMSCs on different structures after 24 h of culture, scale bars = 25 μm. (v) Visualization of nuclear membrane–structure interfaces using FIB-SEM. FIB-SEM images show the extent of plasma membrane and nuclear envelope deformation after 6 h and 72 h of culture on nanopillars (Dtip = 718 ± 32 nm) and sharp nanoneedles (Dtip = 47 ± 7 nm), respectively (scale bars = 2 μm). Reproduced from ref. 38. Copyright 2020, American Chemical Society.
Both NSs have been employed for live subcellular sampling.36,38–40,42,43 A unique advantage of the NS platform is that cells of interest can be cultured directly on the NS-embedded substrate (Fig. 2(a)-i–iv). This allows high throughput sampling of intracellular proteins and cytoplasmic mRNA for both single-cell and bulk-cell extractions (20/30 cells).36 NSs with a 100 nm or smaller diameter could penetrate the cell membrane spontaneously,36,38 keeping a direct channel to extract intracellular biomolecules36,39,44–46 and work as an effective delivery mechanism.40,47–49 NSs with larger diameters require the application of a short electric pulse (10–35 V) for 2–5 minutes of cytoplasmic access. The pulses cause membrane poration and provide temporary fluidic access.36 Interestingly, the biological effect of NSs on cells depends on their diameter.36,38 The effect of NSs size on cell extractions and viability was explored by Seong et al., who tested four diameters (718 nm, 316 nm, 172 nm, 47 nm) and used immunofluorescent microscopy images and extracted morphological features of 100 000 individual human mesenchymal stem cells (hMSC) (Fig. 2(b)-iv).38 Cell body elongation was promoted, and a decrease in the number of cell protrusions was seen with decreasing tip diameter. The authors also found that sharper NSs guide cell alignment and polarisation by activated actomyosin contractility. Through the variation of tip diameters, one could directly influence cell morphology, polarisation and even gene expression in hMSCs (Fig. 2(b)-iii–v).38
NSs have more applications than just the extraction of biomolecules and delivery into cells. Another study by Palankar et al. combined NS arrays with atomic force microscopy to probe the mechanodynamics of human pluripotent stem cell-derived cardiomyocytes. In this study, the 3D NSs were used to guide subcellular cytoskeletal modifications of cardiomyocytes which ultimately led to changes in beating rates, calcium dynamics of the cells, and their stiffness.50 NS arrays of various geometries were found to have widespread applications in mechanoregulating cell functions51 such as underpinning of cell migration,52–54 the interplay between force and focal adhesion maturation,55,56 and the affiliation of nuclear deformations with the mechanotransduction process.57,58
NSs are highly versatile for multiple biological applications, including subcellular biomolecule sampling. Nevertheless, the technique is far more invasive than previously thought. The works of Seong et al. and Palankar et al. highlighted that NSs affect cellular morphology,30 gene expression,32 cellular alignment37 and poration of cell membranes;42 thereby suggesting that the platform is not best suited for continuous longitudinal subcellular sampling as it can affect the genetic expression profiles and morphology of the cells.
3. Atomic force microscopy-based FluidFM
Cellular sampling methods based on AFM have been developed by utilising the AFM tip's multifunctionality: its ability to scan an aqueous surface for high-resolution imaging of living cells under physiological conditions, combined with its ability to probe cells at the subcellular level to extract their contents or deliver exogenous substances such as drugs, nanoparticles and organelles.17,59 Osada et al. first demonstrated the use of AFM tips for cellular sampling from rat fibroblast-like VNOf90 cells and mouse osteoblast-like MC3T3-E1 cells maintained in their standard cultured environment.60 mRNAs were isolated via non-specific contact with the tip inserted into the cell and detected using RT-PCR. The same group further developed this to study the localisation and distribution of beta-actin mRNA in live rat VNOf90 cells via the detection of mRNA on the AFM tip using RT-PCR followed by qPCR.61 mRNAs were extracted from 4–9 different locations within the same cell, and the extraction was confirmed by quantifying beta-actin mRNAs using RT-qPCR. mRNAs were detected at these loci under two cellular conditions: cells in a quiescent state and cells stimulated by fetal bovine serum (FBS). Using this method, the authors found that at one subcellular location, the peripheral region from the nucleus, there was a larger number of positive results in FBS-stimulated cells (5 out of 8 cells in stimulated cells vs. 1 out of 8 in quiescent cells). Based on cellular morphology, the authors demonstrated that cells remained intact and viable after sampling. As this method was based on non-specific adsorption onto the tip, it is limited by how much mRNA can be captured at the tip, where it is unlikely that lower abundant mRNAs would be able to be detected sufficiently using AFM. Nevertheless, this was the first method of its kind to detect mRNAs inside living cells without removing them from their physiological environment or showing significant damage to the cell. Later developments into AFM-based subcellular extraction have been shown, including dielectrophoretic AFM extraction62 and the development of FluidFM,63 which have revealed exciting possibilities for subcellular sampling. In particular, FluidFM, as discussed in this section, has been employed for subcellular sampling, organelle transplantation from one cell to another, and sequential measurements from the same live cell (Live-seq).
3.1. FluidFM sampling
The use of standard AFM to manipulate or deliver molecules into cells is limited due to mechanical indentation or scraping of the AFM probe onto the sample.63 Methods based on the adsorption of biological material onto the tip are also limited by the minute amount of sample that can be obtained for downstream analysis. There may be additional difficulties with dispensing or biomolecule release from the probe. FluidFM is a technique first reported by Meister et al.63 that addresses some of the problems with AFM probing and has biological applications in subcellular sampling,18,64–67 controlled delivery or injection into cells,63,66–68 pick and place and patterning of cells,69–71 cellular adhesion interactions and surface properties,72–78 cellular electrophysiology79 and single-cell sensing.80 The current review focuses on extractions of cellular components from living cells, but other extensive reviews are available for applications outside this area.17,59,81,82
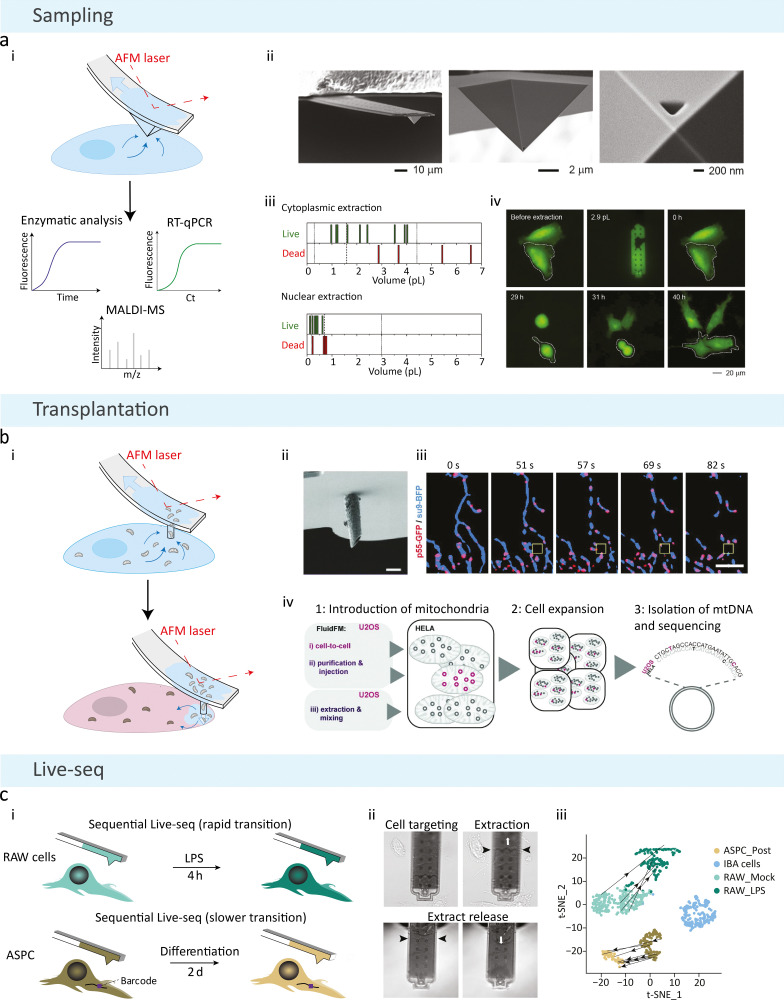
The FluidFM technique combines AFM with a pressure-driven micro-channelled silicon nitride probe for fluid-controlled extractions from single cells (Fig. 3(a)).18 This is achieved by incorporating a micro-sized channel into the AFM cantilever with a nanosized tip aperture, forming a continuous fluidic channel.63 For single-cell sampling, the standard probe is made up of a pyramidal tip with a 400 nm diameter triangular aperture,18 where the size of the aperture can be altered for precise control over extraction volumes and components of interest.66,67 The probe can also be fabricated in a variety of other sizes and tip shapes depending on the application.81 For subcellular sampling, the tip of the probe is lowered into a cell so that the aperture is completely inserted, a negative pressure is then applied, which causes the channel to fill with intracellular fluid, and the tip is withdrawn from the cell for dispensing and molecular analysis (Fig. 3(a)).18 FluidFM facilitates: (1) real-time monitoring of cells throughout the extraction procedure via an inverted optical microscope; (2) force-controlled positioning and insertion of the tip into the cell, monitored by a standard AFM laser; (3) femto- to pico-litre volumes17 to be removed from single cells, controlled by the pressure-driven microchannel and size of the aperture; and (4) controlled local dispensing of components using force-controlled feedback.
Fig. 3. Schematic of FluidFM methodologies. (a) Basic FluidFM sampling consists of an AFM cantilever with a continuous pressure-driven channel. (i) A pyramidal tip with a triangular aperture is inserted into a cell, and negative pressure is applied to aspirate the contents of the cell for downstream analysis. (ii) Scanning electron micrographs of the FluidFM probe for cellular sampling. (iii) Cell viability as a function of FluidFM extraction volumes from the cytoplasm and nucleus, where each line represents one count. The dashed line indicates the median, dotted lines represent the minimum and maximum native volumes. (iv) Fluorescent time-lapse imaging of an extracted cell (2.9 pL removed from the cytoplasm using FluidFM), marked with a dashed line. The extracted cell behaved similarly to the adjacent non-extracted cell, where they became round and divided to produce daughter cells at the same time. Reproduced with permission from ref. 18. Copyright 2016, Elsevier. (b) Mitochondrial transplantation from one cell to another is achieved with a cylindrical probe and a larger aperture size. (i) Following aspiration of mitochondria from the host cell, the probe is inserted into a new cell, and positive pressure is applied to inject the host's mitochondria. (ii) Scanning electron microscopy image of the cylindrical probe used for combining extraction and injection of mitochondria (scale bar = 2 μm). (iii) Time-lapse imaging of mitochondria and mitochondrial nucleoid extraction, showing an overlay of the mitochondrial matrix (su9-BFP) and mitochondrial nucleoids (p55-GFP), where the yellow box illustrates the position of the cantilever (scale bar = 5 μm). (iv) Procedure for quantifying mtDNA uptake and maintenance after transplantation, illustrating two FluidFM transplantation methods (cell-to-cell transplantation and injection of purified mitochondria), and the control performed for non-specific uptake (mixing with extracted donor cell mitochondria). Reproduced from ref. 66. Copyright 2022, PLOS. (c) FluidFM extraction is combined with Smartseq2 technology to form Live-seq: sequential transcriptomic profiling of live single cells. (i) Sequential sampling was performed in a rapid (top) and slow (bottom) cell state transition, where a unique barcode was used in the 3′ untranslated region of a green fluorescent protein reporter to identify the same cell over longer time periods. (ii) Images of the Live-seq sampling procedure using FluidFM, where the black arrows show the level of buffer and extract inside the probe, and the white arrows represent the under- or over-pressure applied. (iii) Direct trajectories of sequentially sampled cells from projections of Live-seq data. Reproduced from ref. 64. Copyright 2022, Springer Nature.
Guillaume-Gentil et al. first achieved successful sampling from single cells using FluidFM in 2016, demonstrating nuclear and cytoplasmic extractions from HeLa cells using a triangular aperture FluidFM system (Fig. 3(a)-ii).18 Extraction volumes up to 4 pl and 0.6 pl in the cytoplasm and nucleus, respectively, could be taken from cells with 82% and 86% cell viability after a single extraction (Fig. 3(a)-iii). The authors highlight that these volumes represented at least 90% of the total cytoplasmic volume and 20% of the total nuclear volume of the largest native volumes measured. Extraction volumes larger than this resulted in cell death. Despite this, the high extraction volumes showed that the cells could withstand a large loss of their native cytoplasmic volumes after the extraction procedure. The authors assessed cell viability up to 5 days after extraction and showed that extracted cells remained viable and behaved similarly to non-extracted cells, including undergoing cell division (Fig. 3(a)-iv). This was a key finding in portraying the low invasiveness of the technique, showing that removing these relatively large volumes did not affect the cells’ normal behaviour in culture. However, extracting 90% of the cytoplasmic volume may affect the microenvironment of some cell types and could lead to morphological changes, which may alter gene expression programs in cells.83 In addition to successful extraction, the aspirated volumes from the nucleus and cytoplasm were sufficient for transcript detection by RT-qPCR, corresponding to 0.01 pg of total cellular RNA. The authors also performed enzymatic assays on the cytoplasmic samples to demonstrate the integrity of extracted proteins and successfully measure apoptosis following treatment with staurosporine, a protein kinase inhibitor. The authors attempted to extract mitochondria with FluidFM, but this was unsuccessful; they concluded that this might be due to the small size of the probe aperture for isolating organelles. Overall, the detection and quantification of mRNA and protein enzymatic activities highlight the potential of FluidFM to be used in single-cell transcriptomics and proteomics, with the potential to tune the extraction volumes and tip apertures for high-resolution subcellular compartment analysis.
The same group showed that FluidFM could be expanded to single-cell metabolomics: 20 metabolites from the cytoplasm of live HeLa cells were detected from FluidFM extractions using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF-MS).65 0.8–2.7 μl volumes were taken from cells, and picolitre volumes were spotted onto a microarray substrate for MALDI-TOF-MS measurements. This work expanded the capabilities of single-cell metabolomic studies, where cells could be maintained in their standard microenvironment and not destroyed during the process. This, combined with high-throughput sensitive analytical techniques like MALDI-MS, enables simple and sensitive detection of metabolites in living cells and shows the potential for achieving metabolic profiling of cells over time.
More recently, the technology has been applied to fungal cells, where the same group showed that the FluidFM probe could penetrate through the cell wall and membrane of cells.67 Sampling from an injection into fungal cells has been relatively unexplored: the authors account for this lack of success due to difficulties working with rigid cell walls, small cell sizes, and immobilising this weakly-adhering cell type. Their success in working with fungal cells was achieved by spatially constraining yeast cells (Saccharomyces cerevisiae and Schizosaccharomyces pombe) on microstructured substrates to minimise displacement when the tip was inserted. Dimorphic yeast (Candida albicans) and multicellular fungal (Coprinopsis cinerea) cells were sufficiently adhered for sampling and injection when used on glass or polystyrene. In their previous work, forces up to 200 nN were used to insert into mammalian cells and inject into cell nuclei effectively.68 To penetrate the cell walls and membranes of cells, however, the authors required forces up to 2 μN to be used. They modified the length and thickness of the standard FluidFM probe to increase its stiffness so that these higher forces could be attained. The aperture size was also reduced to 200 nm to account for the smaller cell size and ensure that the apex was fully inside the cell. Extraction of cytoplasmic fluid from hyphal compartments of fungal cells was achieved, but the extraction procedure inhibited the growth of the targeted hypha, and vacuoles in the compartment were observed after extraction. This shows that the tool may be too invasive to extract from these small cell types without damage and is likely to induce physiological changes in the cell. The authors, therefore, evaluated its use for inducing fungal defence responses. Injection of labelled protein into the cells was also demonstrated, which did not affect cell viability. Overall, this work expands the applicability of single timepoint single-cell sampling to fungal cells and highlights the adaptability of the technology based on cell type and size. The FluidFM tip, however, limited the sampling and injection sites achievable in these cells, since the tip was too large for extraction from or injection into the nuclei of fungal cells. Despite the smaller aperture size on the modified probe, the extraction process appears to be too intrusive on fungal cells to continue their normal function post-extraction, so it may not be suitable for real-time transcriptomic or proteomic analysis in living fungal cells in its current state.
3.2. FluidFM-based organelle transplantation
Recently, mitochondrial isolation using FluidFM has been achieved by tuning the aperture size and probe geometry to allow the aspiration of both single and multiple mitochondria from cells.66 Two probes were used to extract mitochondria successfully: pyramidal probes with a larger aperture size than the standard FluidFM probe (1.2 μm2) and a new slanted cylindrical probe with a larger aperture size (1.6 μm2). This enabled whole mitochondria and parts of the mitochondrial network to be aspirated from the cells without compromising cell viability.
A significant advancement of the technology was achieved in this work by combining mitochondrial isolation with transplantation into a host cell using the newly developed cylindrical probe (Fig. 3(b)). Injection into a new cell was achieved via the application of positive pressure. Isolation of purified bulk mitochondria into a host cell was shown as a high throughput method as the same cantilever could be used to inject mitochondria into multiple host cells serially. The alternative methodology was the cell-to-cell approach, where mitochondria from a donor HeLa cell were isolated via FluidFM aspiration, followed by approaching and inserting the cantilever into a receiver cell (U20S) and injecting its contents. Perfluorooctane was used to prefill the inside of the cantilever to limit the extraction and injection volumes; extraction volumes of approximately 0.8 pl and injection volumes between 0.5 pl to 2 pl were achieved, where the flow resistance of the fluorocarbon prevented higher injection volumes.
The authors demonstrated an impressive 95% success rate of mitochondrial transplants into the cytosol of living cells, with 80% viability of the receiver cells after injection. For the injection of bulk purified mitochondria into host cells, a 46% viability of host cells was observed with the same transplant success rate. The authors attribute this lower viability to the mitochondrial quality being poorer after the purification steps; more time is spent outside the cell, leading to membrane rupture of some mitochondria, potentially leading to apoptosis in the host cell from cytochrome c leakage. The development of the new cylindrical probe shows that FluidFM can be an efficient tool for extraction, manipulation and transplantation of the contents of single cells, where, despite having a lower throughput, transplantation of mitochondria between two cells was shown to be particularly effective for inflicting minimal damage on the host cell.
In the same study, the dynamics of mitochondria following transplantation were monitored in real-time. Interestingly, mitochondria from both methods allowed for the donor cell's mitochondria to fuse to the host's mitochondrial network in 18 out of 22 cells, observed by differential fluorescent labelling of donor and host mitochondria, where the remaining four cells showed donor mitochondrial degradation. The authors found that fusion into the host's mitochondrial network occurred within 30 minutes for most cells. Not only was successful transplantation achieved between two different cell lines, but mitochondria from HeLa cells also successfully fused to primary cell (human endothelial keratinocytes, HEKa) networks 20 minutes after injection. Overall, three outcomes were observed following transplantation: (1) fusion of donor mitochondria into the mitochondrial network of the host in most cases, exhibiting a morphological transition to mitochondrial tubules rather than discrete spheres; (2) degradation or fragmentation of donor mitochondria inside the host cell; or (3) mitochondria that maintain their original shape and donor fluorescence, have not fused to the host's network and are not subjected to degradation. Time-lapse imaging was used to monitor the transplanted mitochondria and their behaviour over time (Fig. 3(b)-iii). They showed that mitochondria were accepted into host cells, with similar acceptance rates even when donor mitochondria were damaged from double-stranded break formation, dysfunctional oxidative phosphorylation proteins, or a loss of membrane potential. This method, therefore, allows for the successful integration of mitochondria into different cell types and has provided novel findings on the potential for healthy cells to survive or adapt to damaged mitochondria.
Long-term tracking of transplanted mitochondria (Fig. 3(b)-iv) was also achieved in this work, where up to 2.5% of mtDNA from donor U2OS cells was detected in HeLa host cells 14 days after transplantation, during which time the host cells would have expanded in culture. Controls were performed to show that this is not achievable from the non-specific uptake of mitochondria mixed inside the culture medium. This demonstrated that ‘foreign’ mtDNA could be incorporated and replicated in new cell types. This work expands the potential of subcellular sampling technologies, where the transplantation benefit could have wide implications in understanding mitochondrial quality control for maintaining cell homeostasis. This could also have great potential in mitochondrial regeneration therapies, where impaired mitochondria have been shown to be critical features of many diseases such as cancer,84 stroke,85 and neurodegenerative diseases.86,87 This new method fills a technological gap in subcellular sampling technologies and opens new possibilities for studying mitochondrial replacement therapies and mtDNA selection and expansion.
3.3. Single-cell transcriptome profiling: Live-seq
Recently, Chen et al. published a seminal paper demonstrating Live-seq, a technique that allows temporal measurements of a cell's transcriptome to be obtained using FluidFM technology (Fig. 3(c)).64 Live-seq combines FluidFM cytoplasmic extractions with Smart-seq2,88 a highly sensitive RNA-seq method for detecting low amounts of RNA at the picogram level. A 400 nm diameter triangular FluidFM aperture was used, and the standard FluidFM extraction procedure was optimised to minimise RNA degradation and sample loss (Fig. 3(c)-ii). This was achieved by: reducing the time taken during the extraction; performing extractions at room temperature; pre-loading the probe with sampling buffer containing RNAase inhibitors; and releasing the extract into a microlitre droplet containing reagents for downstream RNA-seq. The sensitivity of Smart-seq2 was also optimised to detect 1 pg of total RNA.64 An average volume of 1.1 pl was taken from 3 distinct cell types: IBA cells, primary mouse adipose stem and progenitor cells (ASPCs) and two macrophage-like RAW264.7 cell lines. Distinct cell types and states could be distinguished using the expression profiles obtained from Live-seq. Overall, cells were viable after single cytoplasmic extractions, including continuous cell growth, normal cell division, expected responses to external stimuli, and their ability to differentiate.
Cell viability after performing a single extraction from the 3 cell types was found to be between 85–89% and not to scale with the extraction volumes taken from cells. Time-dependent microscopy was used to monitor the growth dynamics of the smallest cell type studied, RAW cells. Interestingly, despite 40–70% of the total cell volume being taken during the extraction, RAW cells were shown to recover these volumes within 100–320 minutes, followed by continuing the same growth pattern as shown in controls. This shows that cells may withstand the loss of a large portion of their cytoplasm and recover to normal function. However, the time taken to recover the cell volume may delay cell growth and likely incur changes in gene expression until the volume is fully recovered. Therefore, the cells extracted using this method may not accurately represent a non-extracted cell, and the delay time may always need to be considered when studying cells over time. Some transcriptomic changes detected using single-cell RNA-seq were shown at one hour and four hours after Live-seq sampling compared to non-extracted controls, where 12 genes were shown to be significantly differentially expressed between Live-seq and control samples. In its current state, however, Live-seq may not be suitable for short-term transcriptomic studies due to the risk of the extraction procedure inducing stress and changes in gene expression in response to the loss of cytoplasmic volume.
In the same work, sequential sampling from the same living cell was achieved at two distinct time points using time-lapse fluorescent cell tracking. Two models were used to detect changes in cell expression (Fig. 3(c)-i): (1) application of lipopolysaccharide (LPS) stimulation in RAW cells to detect a fast molecular change 4 hours post-stimulation, (2) adipogenic differentiation of ASPCs to detect a slower change after two days. For the former treatment, 4 cells were successfully sequentially extracted, which reflected only around 40% of samples that passed the transcriptome quality control criteria based on the number of genes detected, mitochondrial read ratio, and unique mapped rates. For the latter condition, a unique barcode was used in a region of the green fluorescent protein expressed in the cell to ensure that the correct cell was sequentially extracted. 44 cells were sampled using this method, but only 8 pairs of profiles passed the quality control measures. Despite the relatively low efficiencies for both time-dependent studies, the Live-seq data obtained from the high-quality cells mapped correctly to their cell state clusters and showed an expected transition after each model based on single-cell RNA-seq data (Fig. 3(c)-iii). Using Live-seq, they discovered that Nfkbia and Gsn expression levels contributed to LPS phenotypic heterogeneity and validated this with conventional RNA-seq data. After differentiation, ASPCs were monitored for seven days after the second extraction and notably demonstrated no drop in viability compared to controls (95% viability of 44 sampled cells vs. 93% viability of control unsampled cells). They also maintained their adipogenic differentiated state over this period, highlighting that the cells maintained their functionality after two extractions from the same cell.
With the development of Live-seq, the technology has become one of the leading techniques for subcellular sampling, demonstrating that sequential measurements of the same cell are possible and that the technique is sensitive enough not only for detecting mRNA using RT-qPCR, but for sequencing the contents of single cells. FluidFM-based Live-seq has greatly expanded the scope of single-cell transcriptomics, demonstrating that cells do not need to be lysed to access their transcriptome via RNA-seq. Given that seemingly similar cells may respond differently to external stimuli, this technique enables the ground state of a particular cell to be accessed and tracked in order to fully understand the impact of the response. This has the potential to widen our knowledge of the true molecular mechanisms at the single-cell and subcellular level and understand the heterogeneity of responses in seemingly similar cell populations. Although large proportions of cells’ cytoplasmic volumes are taken using this technique, the robustness of cells to recover from this loss, and how the volumes taken are not correlated with cell viability, is impressive and may negate the invasiveness of the technique for monitoring the phenotypic and molecular dynamics of cells over longer time frames. On the other hand, because a large proportion of the cytoplasm is removed, the spatial resolution is more limited than other techniques that can sample from precise subcellular locations. Nevertheless, as the authors state, the development of lower input single-cell RNA-seq methods such as Smart-seq389 may mean that the volumes extracted could be decreased and the spatial precision improved if lower RNA inputs can be detected. Overall, Live-seq proves a fascinating method to monitor and map transcriptomic dynamics in the same cell over time, and the scope of the technique could have significant implications in understanding cellular dynamics and relating phenotypes to distinct transcriptomic profiles, contributing to the development of the single-cell transcriptomic atlas.
4. Dielectrophoretic systems
Dielectrophoresis (DEP) was first introduced in 1951 by Pohl90,91 and was coined to describe the motion of nonpolar dielectric particles resulting from interactions with a nonuniform electric field. Typically, a DEP system requires two individually addressable electrodes separated by an insulating material. Applying an alternating current (AC) across these two electrodes creates an electric field gradient around them, which could exert an attractive force on polarisable objects near the field. The magnitude of this attractive force can be modulated by increasing the voltage across the electrodes, decreasing the gap between electrodes, and altering the frequency of the individually applied voltages. The following equation can describe this:FDEP = 2πεmε0r3Re[CM(f)]∇E2RMSwhere εm and ε0 are the complex permittivities of the medium and vacuum, r is the radius of the particle, E refers to the electric field Re[CM(f)] is the real part of the Clausius-Mossotti factor.92
DEP-based techniques are relatively inexpensive and simple to perform, requiring only a pair of closely spaced electrodes and a function generator.22,93–99 Over the years, DEP has been employed in many forms of microfluidic disease detection systems. Based on the differences in the dielectric properties of cells, properties such as membrane conductance and membrane capacitance have been seen in different physiological and environmental states. However, this review section will focus on DEP methods developed for subcellular biomolecular capture and extraction. We will discuss two main DEP-based systems: AFM probe comprising a Dielectrophoretic Nanotweezer (DENT)62 and nanopipette-based DEP nanotweezers.21 These probes have a marked difference compared to previously mentioned techniques, as they avoid the need for electrowetting and non-specific aspiration of cytoplasmic fluid.
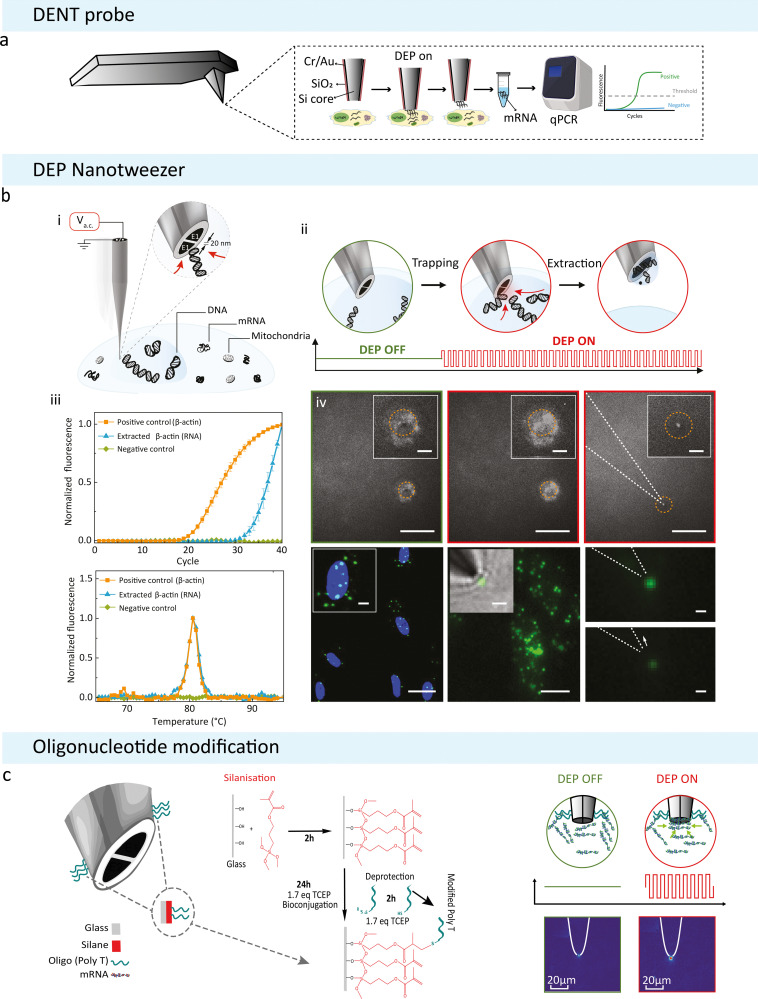
DENT was initially introduced as a modification of the classic AFM. In 2009, Nawarathna et al. modified the standard AFM probe and employed DEP to selectively target and capture cytoplasmic mRNA with minimal impact on cell viability (Fig. 4(a)). This modification utilises a nanoprobe comprising a closely distanced silicon core and a metal alloy layer (Cr/Au), which serve as the inner and outer electrodes.62 The application of AC voltage between the two electrodes (120 kHz, 5 V peak-to-peak for 60–75 s) generates an electric field with a dielectrophoretic force strong enough to polarise subcellular components and organelles to become induced dipoles that are attracted to and captured at the probe tip.100 This system was used to perform extractions of nucleic acids from the cytoplasm of living cells for further genetic analysis. The integration of DENT to the classical AFM probes enables one to precisely position the nanoprobe with nanometre precision within the cytoplasm of a living cell.62 Further, the surface modifications of the DENT probes by immobilising gene-specific primers complementary to the mRNA of interest using standard biotin-streptavidin chemistry increased the collection efficiency.62 Following this work, Li et al. used target-specific short mRNA capture primers for Rat β-actin, GAPDH, and neu on modified AFM probes using biotin–streptavidin chemistry. The results show that for each target, there was a 100-fold increase in capture when DEP was applied.16
Fig. 4. DEP based subcellular sampling techniques. (a) Schematic of a modified AFM probe with DENT. Created with BioRender.com. (b) DEP Nanotweezer schematic showing how DNA can be extracted from a live cell: (i) application of an a.c. voltage on the nanotweezer generates a highly localized electric field gradient suitable for targeted molecular trapping in solution or inside a cell. (ii) Step-by-step schematics of a single cell biopsy. The tip was approached and then inserted into the cell nucleus; the application of an a.c. bias traps DNA molecules at the nanotweezer tip, and, in the final step, the nanotweezer along with the accumulated material was withdrawn from the cell. (iv) Biopsies were also performed in cells stained with a non-specific RNA dye (SYTO RNASelect). The accumulation of labelled mRNA during DEP capture results in an increase in fluorescence at the nanotweezer tip (left and middle). The mRNA can still be seen at the tip once extracted from the cell (right, scale bars = 20 μm, insets = 5 μm). (v) Targeted mRNA trapping and extraction was performed by labelling, via in situ hybridization, of individual ETS-1 mRNA molecules with fluorescein isothiocyanate (green dots) (left). A high-resolution fluorescence image of individual ETS-1 mRNA molecules (middle) along with a superimposed bright-field image is shown (inset). The application of the a.c. voltage resulted in trapping of mRNA at the nanotweezer tip (top right), which was then pulled away by the subsequent withdrawal of the nanotweezers, causing a drop in the fluorescence signal (bottom right). Scale bars: left, 25 μm (inset, 5 μm); middle, 10 μm (inset, 2 μm); right, 1 μm. Reproduced from ref. 21. (c) Updated nanotweezer system with its glass surface modified to contain an RNA Trap Poly Thymine Oligomer sequence on the tip. Modifying the surface of the glass with a broadly binding oligomer was attempted to increase the overall amount of mRNA that can be held by a single trapping event. The modification was done using click chemistry to bind the oligomer to the glass surface depicted on the left. On the right is a diagrammatic representation of the modified nanotweezers trapping free-flowing mRNA in solution. Alexa Fluor 488 labelled mRNA was extracted from MCF-7 cancer cells and diluted in 100 mM KCl. The corresponding heatmaps show the tips of the nanotweezers before and after DEP was induced.
To increase the analytical throughput of the DENT AFM system, the same group integrated it with microfluidics in 2017.16 DENT AFM nanoprobes could penetrate a very thinly sealed microfluidic chip, approximately 1 μm, and isolate mRNAs from the cytoplasm of HeLa cells and circulating tumour cells. mRNA isolation with qPCR was used to compare the isolation efficiency at varying strengths of applied AC voltages (1.1 V, 1.5 V, 1.9 V peak-to-peak, 10 MHz), with greater voltages resulting in a higher number of mRNA transcripts captured.
A recent development in using DEP for subcellular capture and extractions is the DEP Nanotweezers developed by Nadappuram et al. in 2019 (Fig. 4(b)-i, ii).21 The nanotweezers were fabricated using nanopipettes made from double-barrelled quartz capillaries via laser pulling, and the nanoelectrodes were formed at the tips using pyrolytic carbon deposition. The fabrication process allows for a simple way to decrease the gap between the electrodes to sub-20 nm, which in turn allows the generation of large electric field gradients (∇|E|2 ≈ 1028 V2 m−3). The generated field gradient was two orders of magnitude higher than metal-based DEP platforms,22,23,101 and higher than previously reported single-cell screening systems based on DEP,62 which enabled the trapping of single molecules in physiological ionic strengths. The nanotweezers could capture and extract single DNA molecules directly from the nucleus, transcription factors from the cytoplasm (Fig. 4(b)-iii, iv) and even single organelles such as mitochondria. This technique provided the capability to sample subcellular biomolecules with high spatial resolution and minimal impact on cell and organelle viability.21 In the same group, the nanotweezer has been applied to study gene expression and mitochondrial patterns in primary neurons. Single mitochondria and mRNA have been isolated from individual neurons without compromising cell viability. From these extracted samples, mRNAs have been detected by RT-qPCR. Regions of the mtDNA from individual mitochondria have also been successfully sequenced.
Recently, surface modifications were carried out on the outer quartz glass surface of the nanotweezer to improve selectivity to nucleic acids and maximise capture efficiency.102 The nanotweezer tip was functionalised with a 20-base polyT oligonucleotide sequence using click chemistry. Thymine was chosen as a broad-spectrum recognition sequence to take advantage of the naturally occurring polyadenine tail on cytoplasmic mRNA (Fig. 4(c)).102 This strategy resolves the issue of sample retention when the DEP field is switched off during nanotweezer withdrawal from the sample and also increases the amount of mRNA that could be captured and extracted from both solutions at physiological conditions and from living cells.102 In addition, it was possible to collect a continuous expression profile over a period of 28 days of living MCF-7 cells.102 The cells tolerated weekly sampling and were kept alive over the full experimental timeframe. A future avenue to explore would be to create primer-specific oligonucleotide sequences for low copy and hard to detect mRNAs, for example, making a nanotweezer selective for ESR1, which according to RNAscope data, was found with very low confidence in the cytoplasm of MCF-7 cells.102
The nanotweezers in their functionalised state allowed us to achieve a truly continuous transcriptional profile for MCF-7 breast cancer cells. To our knowledge, this had not been done before; they allowed for continuous sampling of live cells, at the single cell level, over 28 days, totalling 18 biopsies per cell whilst not affecting cell viability. The potential to create single-cell transcriptional profiles with real temporal resolution in cells could lead to new insights into understanding diseases such as cancer and neurodegenerative diseases.
Conclusions
The development of single-cell analysis tools has led to ground-breaking discoveries of heterogeneous cell populations, novel cell types, mitochondrial heteroplasmy, subcellular mechanisms, and compartmentalised transcriptomics. Advances in technologies that probe and remove the contents of living cells have been shown to be particularly advantageous in the field compared to imaging-based techniques, as they minimise cellular alterations and allow cells to remain alive during the extraction procedure. This expands their potential for monitoring living cells over time. As opposed to using sequence-specific probes, they also allow for the contents of cells to be investigated using high-standard analysis tools such as mass spectrometry and next-generation sequencing and have the potential for a large number of targets to be analysed from the same cell. This could lead to new discoveries of proteoforms and post-transcriptional regulatory mechanisms in a variety of cell types and systems.
The intracellular extraction techniques discussed herein vary in size, throughput, cost, spatial resolution, and physiological impact on the cell (summarised in Table 1). Nanostraws are statically positioned inside single cells and have very high throughputs as individual cells can be cultured on top of multiple nanostraws, allowing for the parallel sampling of the same cell over multiple time points. However, cells need to be grown on specific substrates rather than a standard culture dish or cover glass, and the manufacturing process can be time-consuming and expensive due to the need for etching and photolithography. Other techniques based on micro- and nanopipettes or AFM probes can be positioned to specific subcellular regions of interest and controlled by micro- or nanomanipulators, providing as low as nanoscale resolution. These, however, lack throughput as only single serial extractions can be performed. Nanopipette-based systems are often integrated with SICM, which allows for precise and controlled positioning of the nanopipette inside a cell; the feedback mechanism ensures that appropriate insertion into the cell is achieved. Other pipette-based techniques, like the cookie cutter, expand nanoprobe methods to tissue section sampling, which could have applications in analysing clinical tissue samples or 3D cell cultures. FluidFM allows for force-controlled sub-picolitre volumes to be removed from cells for highly controlled sampling. More recently, the capabilities of FluidFM sampling have been greatly expanded to fungal cells, mitochondrial transplantation, and same-cell transcriptome profiling, pushing the scope of subcellular sampling. Nanopipette and FluidFM sampling, however, require the removal of cytoplasmic fluid from the cell, which raises questions regarding the viability of cells and the impact on cellular function after the extraction procedure. The nanotweezer is distinct in that it isolates polarisable molecules directly from specific regions of cells, negating the need to remove cytoplasmic fluid, which may otherwise trigger local changes or stress responses. The nanotweezer may therefore be less invasive than aspirating methods and allows for tighter control over the location of the desired material in the cell, increasing the spatial resolution. Even with the recent surface functionalisation of the nanotweezer, however, the amount of subcellular material extracted may still be limited for downstream analysis, such as RNA-seq. Nevertheless, as with all methods highlighted here, the development of lower input and highly sensitive next-generation sequencing methods may make investigating small quantities of biomolecules from spatially restricted cellular locations possible in the future.
Summary table of subcellular sampling technologies.
| Technique | Tip diameter | Cell types sampled | Biomolecules sampled | Analysis methods | No. of times same cell sampled | Throughput | Spatial precision | Cost | Cell viability | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Micro/nanopipette | 50 nm–5 μm | Neurons, drosophila larvae, onion cells, MCF-7, brain tissue | mRNA, metabolites, mitochondria | RT-qPCR, mtDNA sequencing, MALDI-MS | 1–2 | Low | High | Low | Moderate | 19, 20 and 25–35 |
| Nanostraw | 20–700 nm | hMSCs, cardiomyocytes | DNA, mRNA, proteins, metabolites | RT-qPCR, RNA-seq, ELISA, DNA sequencing | Up to 4 | High | Low | Moderate | Low | 36–48 |
| Fluid FM | 200 nm–1.1 μm pyramidal probe or 1.2 μm cylindrical probe | U2OS, HeLa, HeKa, ASPCs, IBA cells, macrophage-like cells, fungal cells | mRNA, proteins, metabolites, mitochondria | RT-qPCR, RNA-seq, enzymatic analysis, MALDI-MS | 1–2 | Moderate | High | High | Moderate to high | 18 and 63–67 |
| DENT | 300 nm | HeLa, fibroblasts, SK-BR-3, U937 | mRNA | RT-qPCR | 1 | Low | High | Moderate | N/A | 16 and 62 |
| Nanotweezers | 40–60 nm | U2OS, HPAECs, neurons, MCF-7 | DNA, mRNA, mitochondria | RT-qPCR, DNA sequencing | Up to 18 | Low | Very high | Low | High | 21 and 102 |
Since it is now known that seemingly similar cells in a population can still reveal high levels of heterogeneity, it is becoming more important to access the same single cell over time rather than obtaining multiple static measurements of different cells, which would only represent the average behaviour of a population. This could help us better understand cellular dynamics and, in doing so, gain a true representation of individual cell responses. Time-dependent serial sampling from the same cell has already been demonstrated using many of the sampling techniques described here (nanostraw, FluidFM and nanotweezer), with the nanotweezer notably demonstrating single-cell tracking and sampling over a 28 days period.
Further work into the capabilities of all these techniques for long-term studies of cells is required to fully achieve single-cell tracking, however. This includes assessments of: the number of times a single cell can be sampled without negatively affecting its viability; how long a cell can maintain normal function and stay alive after being sampled; the impact that sampling has on different cellular functions such as cell division, stress responses and energy production in both the short- and long-term; the possible impact that sampling has on a cell's omics and the timeframes at which this impact is observed; and the time required between each serial sampling procedure based on the effects on viability and function. It may be interesting to combine the single-cell sampling methods highlighted in this review with other live-cell imaging techniques for spatially resolved single-cell analysis. For sequential and consistent sampling of cells over time, automation of the techniques is also necessary, particularly for those with lower throughput. This is readily within reach with further development of these methods using machine learning. This could be used to increase the throughput of the techniques, as well as the spatial and temporal resolution of each sampling event. Machine learning could also be integrated in the downstream analysis of high-throughput sampling to give insight into proteomic and transcriptomic patterns that would otherwise be difficult to identify.
In terms of achieving temporal measurements within the same cell, there are uncertainties as to whether the samples would represent the entire cell using these technologies; there is a balance to be had between having minimal impact on the cell and sampling enough material to accurately represent the cell. These techniques may therefore be more suited to investigate precise locations of cells over time rather than single cells or the cytoplasm as a whole. This could be extremely valuable in spatial transcriptomics, where the nanometre resolution of these methods would be of great advantage for precisely profiling subcellular regions. For example, precise subcellular sampling could be used to investigate specific compartments of polarised cells, such as neurons, which could be used to discover novel localisation sites of protein synthesis machinery.103 This, in combination with live cell measurements over time, could help create a dynamic spatial transcriptome of a single cell. This could be used in various physiological conditions, such as after the application of a drug or in response to a stimulus and could be used to detect changes in specific locations of cells for compartmentalised mechanics. Of the techniques where both delivery and extraction are possible (nanostraws, nanopipettes and FluidFM), this objective could be further exploited through direct local delivery of biomolecules or organelles to specific compartments of a cell, followed by extracting localised cellular contents for precise intracellular tracking.
Many of these techniques are still in their infancy in regard to their biological applications. Thus far, these biological assessments have observed heterogeneity of cancer-associated genes over a 28 days period (nanotweezer),102 uncovered genes that contribute to a particular phenotype (Live-seq),64 revealed mitochondrial heteroplasmy in the same cell (micro/nanopipette),19,35 and evaluated mitochondrial quality control (FluidFM).66 However, further dynamic studies are necessary to answer key biological questions and demonstrate the full potential of the techniques, which, if successful, would have wide implications in disease tracking, cellular mechanics and treatment responses. This anticipates major contributions to the field of personalised medicine and the development of the single-cell atlas.
Conflicts of interest
There are no conflicts of interest to declare.
Supplementary Material
Acknowledgments
A. P. I. and J. B. E. acknowledge support from EPSRC grant EP/V049070/1, and Analytical Chemistry Trust Fund grant 600322/05. J. B. E. has also received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement no. 724300 and 875525). B. P. N. acknowledge support from the Analytical Chemistry Trust Fund and Community for Analytical Measurement Science fellowship (Ref. No. 600310/21/07). A. S. and A. M. C. acknowledge support from an EPSRC Centre for Doctoral Training Studentship from the Institute of Chemical Biology.
References
- Borland L. M. Kottegoda S. Phillips K. S. Allbritton N. L. Annu. Rev. Anal. Chem. 2008;1:191–227. doi: 10.1146/annurev.anchem.1.031207.113100. [DOI] [PubMed] [Google Scholar]
- Wang F. Flanagan J. Su N. Wang L. C. Bui S. Nielson A. Wu X. Vo H. T. Ma X. J. Luo Y. J. Mol. Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu T. Vallmitjana A. Gu J. La K. Xu Q. Flores J. Zimak J. Shiu J. Hosohama L. Wu J. Douglas C. Waterman M. L. Ganesan A. Hedde P. N. Gratton E. Zhao W. Nat. Commun. 2022;13:169. doi: 10.1038/s41467-021-27798-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. H. Boettiger A. N. Moffitt J. R. Wang S. Zhuang X. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. L. A. Fale P. L. V. Atharawi A. Wehbe K. Cinque G. Anal. Bioanal. Chem. 2018;410:6477–6487. doi: 10.1007/s00216-018-1245-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decelle J. Veronesi G. Gallet B. Stryhanyuk H. Benettoni P. Schmidt M. Tucoulou R. Passarelli M. Bohic S. Clode P. Musat N. Trends Cell Biol. 2020;30:173–188. doi: 10.1016/j.tcb.2019.12.007. [DOI] [PubMed] [Google Scholar]
- Mei L. Q. Zhang X. Yin W. Y. Dong X. H. Guo Z. Fu W. H. Su C. J. Gu Z. J. Zhao Y. L. Nanoscale. 2019;11:4767–4780. doi: 10.1039/C8NR10319D. [DOI] [PubMed] [Google Scholar]
- Braselmann E. Rathbun C. Richards E. M. Palmer A. E. Cell Chem. Biol. 2020;27:891–903. doi: 10.1016/j.chembiol.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T. Wu C. Curr. Opin. Genet. Dev. 2021;67:94–102. doi: 10.1016/j.gde.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breukers J., Struyfs C., Horta S., Thevissen K., Vanhoorelbeke K., Cammue B. P. and Lammertyn J., Handbook of Single-Cell Technologies, Springer, 2021, pp.311–341 [Google Scholar]
- Romero-Soto F. O. Polanco-Oliva M. I. Gallo-Villanueva R. C. Martinez-Chapa S. O. Perez-Gonzalez V. H. Electrophoresis. 2021;42:605–625. doi: 10.1002/elps.202000221. [DOI] [PubMed] [Google Scholar]
- Filby A. Carpenter A. E. N. Engl. J. Med. 2022;386:1755–1758. doi: 10.1056/NEJMcibr2200971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkin A. Schütze K. Dziedzic J. Euteneuer U. Schliwa M. Nature. 1990;348:346–348. doi: 10.1038/348346a0. [DOI] [PubMed] [Google Scholar]
- Ashkin A. Dziedzic J. M. Yamane T. Nature. 1987;330:769–771. doi: 10.1038/330769a0. [DOI] [PubMed] [Google Scholar]
- Actis P. Small Methods. 2018;2(3):1700300. doi: 10.1002/smtd.201700300. [DOI] [Google Scholar]
- Li X. Tao Y. L. Lee D. H. Wickramasinghe H. K. Lee A. P. Lab Chip. 2017;17:1635–1644. doi: 10.1039/C7LC00133A. [DOI] [PubMed] [Google Scholar]
- Guillaume-Gentil O. Potthoff E. Ossola D. Franz C. M. Zambelli T. Vorholt J. A. Trends Biotechnol. 2014;32:381–388. doi: 10.1016/j.tibtech.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Guillaume-Gentil O. Grindberg R. V. Kooger R. Dorwling-Carter L. Martinez V. Ossola D. Pilhofer M. Zambelli T. Vorholt J. A. Cell. 2016;166:506–516. doi: 10.1016/j.cell.2016.06.025. [DOI] [PubMed] [Google Scholar]
- Actis P. Maalouf M. M. Kim H. J. Lohith A. Vilozny B. Seger R. A. Pourmand N. ACS Nano. 2014;8:546–553. doi: 10.1021/nn405097u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha-Shah A. Weber A. E. Karty J. A. Ray S. J. Hieftje G. M. Baker L. A. Chem. Sci. 2015;6:3334–3341. doi: 10.1039/C5SC00668F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadappuram B. P. Cadinu P. Barik A. Ainscough A. Devine M. J. Kang M. Gonzalez-Garcia J. Kittler J. T. Willison K. R. Vilar R. Actis P. Wojciak-Stothard B. Oh S. H. Ivanov A. P. Edel J. B. Nat. Nanotechnol. 2019;14:80–88. doi: 10.1038/s41565-018-0315-8. [DOI] [PubMed] [Google Scholar]
- Barik A. Chen X. S. Oh S. H. Nano Lett. 2016;16:6317–6324. doi: 10.1021/acs.nanolett.6b02690. [DOI] [PubMed] [Google Scholar]
- Barik A. Zhang Y. Grassi R. Nadappuram B. P. Edel J. B. Low T. Koester S. J. Oh S.-H. Nat. Commun. 2017;8:1867. doi: 10.1038/s41467-017-01635-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A. Teichmann S. A. Lander E. S. Amit I. Benoist C. Birney E. Bodenmiller B. Campbell P. Carninci P. Clatworthy M. eLife. 2017;6:e27041. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bury A. G. Pyle A. Marcuccio F. Turnbull D. M. Vincent A. E. Hudson G. Actis P. Anal. Bioanal. Chem. 2022;414(18):5483–5492. doi: 10.1007/s00216-022-03944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforge F. O. Carpino J. Rotenberg S. A. Mirkin M. V. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11895–11900. doi: 10.1073/pnas.0705102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale S. E. C. Unwin P. R. Electrochem. Commun. 2008;10:723–726. doi: 10.1016/j.elecom.2008.02.023. [DOI] [Google Scholar]
- Toth E. N. Lohith A. Mondal M. Guo J. Fukamizu A. Pourmand N. J. Biol. Chem. 2018;293:4940–4951. doi: 10.1074/jbc.M117.800763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto Y. Takahashi Y. Zhou Y. S. Ito H. Ida H. Ino K. Matsue T. Shiku H. ACS Nano. 2016;10:6915–6922. doi: 10.1021/acsnano.6b02753. [DOI] [PubMed] [Google Scholar]
- Liang B. X. Zhong Y. Z. Wang B. Lin L. Liu J. Lin X. Huang Y. J. Hu M. J. Zhang B. L. Meng H. Jiang L. Jiang J. Y. Wu J. J. Zhang Y. T. Rong W. F. Yang X. F. Huang Z. L. Toxicol. Appl. Pharmacol. 2021;430:115728. doi: 10.1016/j.taap.2021.115728. [DOI] [PubMed] [Google Scholar]
- Niehoff A. C. Kettling H. Pirkl A. Chiang Y. N. Dreisewerd K. Yew J. Y. Anal. Chem. 2014;86:11086–11092. doi: 10.1021/ac503171f. [DOI] [PubMed] [Google Scholar]
- Stahl B. Linos A. Karas M. Hillenkamp F. Steup M. Anal. Biochem. 1997;246:195–204. doi: 10.1006/abio.1997.2011. [DOI] [PubMed] [Google Scholar]
- Palm W. Sampaio J. L. Brankatschk M. Carvalho M. Mahmoud A. Shevchenko A. Eaton S. PLoS Genet. 2012;8:e1002828. doi: 10.1371/journal.pgen.1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B. Nemes P. Nazarian J. Hathout Y. Hoffman E. P. Vertes A. Analyst. 2010;135:751–758. doi: 10.1039/B922854C. [DOI] [PubMed] [Google Scholar]
- Morris J. Na Y. J. Zhu H. Lee J. H. Giang H. Ulyanova A. V. Baltuch G. H. Brem S. Chen H. I. Kung D. K. Lucas T. H. O’Rourke D. M. Wolf J. A. Grady M. S. Sul J. Y. Kim J. Eberwine J. Cell Rep. 2017;21:2706–2713. doi: 10.1016/j.celrep.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. H. Hjort M. Chen H. D. Birey F. Leal-Ortiz S. A. Han C. M. Santiago J. G. Pasca S. P. Wu J. C. Melosh N. A. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E1866–E1874. doi: 10.1073/pnas.1615375114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G. Hu N. Xu A. M. Li X. L. Zhao Y. L. Xie X. Adv. Funct. Mater. 2020;30:1909890. doi: 10.1002/adfm.201909890. [DOI] [Google Scholar]
- Seong H. Higgins S. G. Penders J. Armstrong J. P. K. Crowder S. W. Moore A. C. Sero J. E. Becce M. Stevens M. M. ACS Nano. 2020;14:5371–5381. doi: 10.1021/acsnano.9b08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu A. M. Aalipour A. Leal-Ortiz S. Mekhdjian A. H. Xie X. Dunn A. R. Garner C. C. Melosh N. A. Nat. Commun. 2014;5:3613. doi: 10.1038/ncomms4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X. Xu A. M. Leal-Ortiz S. Cao Y. H. Garner C. C. Melosh N. A. ACS Nano. 2013;7:4351–4358. doi: 10.1021/nn400874a. [DOI] [PubMed] [Google Scholar]
- He G. Chen H. J. Liu D. Feng Y. P. Yang C. D. Hang T. Wu J. M. Cao Y. H. Xie X. Adv. Mater. Interfaces. 2018;5:1701535. doi: 10.1002/admi.201701535. [DOI] [Google Scholar]
- VanDersarl J. J. Xu A. M. Melosh N. A. Nano Lett. 2012;12:3881–3886. doi: 10.1021/nl204051v. [DOI] [PubMed] [Google Scholar]
- Chiappini C. Martinez J. O. De Rosa E. Almeida C. S. Tasciotti E. Stevens M. M. ACS Nano. 2015;9:5500–5509. doi: 10.1021/acsnano.5b01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G. Yang C. D. Hang T. Liu D. Chen H. J. Zhang A. H. Lin D. Wu J. M. Yang B. R. Xie X. ACS Sens. 2018;3:1675–1682. doi: 10.1021/acssensors.8b00367. [DOI] [PubMed] [Google Scholar]
- Mukherjee P. Nathamgari S. S. P. Kessler J. A. Espinosa H. D. ACS Nano. 2018;12:12118–12128. doi: 10.1021/acsnano.8b05473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappini C. Campagnolo P. Almeida C. S. Abbassi-Ghadi N. Chow L. W. Hanna G. B. Stevens M. M. Adv. Mater. 2015;27:5147–5152. doi: 10.1002/adma.201501304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. Chen H. Qiu R. Hanna M. Ma E. Hjort M. Zhang A. Lewis R. S. Wu J. C. Melosh N. A. Sci. Adv. 2018;4:29. doi: 10.1126/sciadv.aat8131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. H. Ma E. B. Cestellos-Blanco S. Zhang B. Qiu R. Y. Su Y. D. Doudna J. A. Yang P. D. Proc. Natl. Acad. Sci. U. S. A. 2019;116:7899–7904. doi: 10.1073/pnas.1818553116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. W. Shi Y. M. Miyamoto D. Nakazawa K. Miyake T. Sci. Rep. 2019;9:6806. doi: 10.1038/s41598-019-43340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palankar R. Glaubitz M. Martens U. Medvedev N. von der Ehe M. Felix S. B. Munzenberg M. Delcea M. Adv. Healthcare Mater. 2016;5:335–341. doi: 10.1002/adhm.201500740. [DOI] [PubMed] [Google Scholar]
- Schoen I. Hu W. Klotzsch E. Vogel V. Nano Lett. 2010;10:1823–1830. doi: 10.1021/nl100533c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Roure O. Saez A. Buguin A. Austin R. H. Chavrier P. Silberzan P. Ladoux B. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabodzey A. Alcaide P. Luscinskas F. W. Ladoux B. Biophys. J. 2008;95:1428–1438. doi: 10.1529/biophysj.107.119156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepat X. Wasserman M. R. Angelini T. E. Millet E. Weitz D. A. Butler J. P. Fredberg J. J. Nat. Phys. 2009;5:426–430. [Google Scholar]
- Balaban N. Q. Schwarz U. S. Riveline D. Goichberg P. Tzur G. Sabanay I. Mahalu D. Safran S. Bershadsky A. Addadi L. Geiger B. Nat. Cell Biol. 2001;3:466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- Paul R. Heil P. Spatz J. P. Schwarz U. S. Biophys. J. 2008;94:1470–1482. doi: 10.1529/biophysj.107.108688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl K. N. Booth-Gauthier E. A. Ladoux B. J. Biomech. 2010;43:2–8. doi: 10.1016/j.jbiomech.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Wang N. Tytell J. D. Ingber D. E. Nat. Rev. Mol. Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- Amarouch M. Y. El Hilaly J. Mazouzi D. Scanning. 2018:7801274. doi: 10.1155/2018/7801274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada T. Uehara H. Kim H. Ikai A. J. Nanobiotechnol. 2003;1:2. doi: 10.1186/1477-3155-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara H. Osada T. Ikai A. Ultramicroscopy. 2004;100:197–201. doi: 10.1016/j.ultramic.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Nawarathna D. Turan T. Wickramasinghe H. K. Appl. Phys. Lett. 2009;95:083117. doi: 10.1063/1.3213343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister A. Gabi M. Behr P. Studer P. Voros J. Niedermann P. Bitterli J. Polesel-Maris J. Liley M. Heinzelmann H. Zambelli T. Nano Lett. 2009;9:2501–2507. doi: 10.1021/nl901384x. [DOI] [PubMed] [Google Scholar]
- Chen W. Z. Guillaume-Gentil O. Rainer P. Y. Gabelein C. G. Saelens W. Gardeux V. Klaeger A. Dainese R. Zachara M. Zambelli T. Vorholt J. A. Deplancke B. Nature. 2022;608:733–740. doi: 10.1038/s41586-022-05046-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume-Gentil O. Rey T. Kiefer P. Ibanez A. J. Steinhoff R. Bronnimann R. Dorwling-Carter L. Zambelli T. Zenobi R. Vorholt J. A. Anal. Chem. 2017;89:5017–5023. doi: 10.1021/acs.analchem.7b00367. [DOI] [PubMed] [Google Scholar]
- Gabelein C. G. Feng Q. Sarajlic E. Zambelli T. Guillaume-Gentil O. Kornmann B. Vorholt J. A. PLoS Biol. 2022;20:e3001576. doi: 10.1371/journal.pbio.3001576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume-Gentil O. Gabelein C. G. Schmieder S. Martinez V. Zambelli T. Kunzler M. Vorholt J. A. Commun. Biol. 2022;5:180. doi: 10.1038/s42003-022-03127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume-Gentil O. Potthoff E. Ossola D. Dorig P. Zambelli T. Vorholt J. A. Small. 2013;9:1904–1907. doi: 10.1002/smll.201202276. [DOI] [PubMed] [Google Scholar]
- Martinez V. Forro C. Weydert S. Aebersold M. J. Dermutz H. Guillaume-Gentil O. Zambelli T. Voros J. Demko L. Lab Chip. 2016;16:1663–1674. doi: 10.1039/C5LC01466B. [DOI] [PubMed] [Google Scholar]
- Guillaume-Gentil O. Zambelli T. Vorholt J. A. Lab Chip. 2014;14:402–414. doi: 10.1039/C3LC51174J. [DOI] [PubMed] [Google Scholar]
- Dorig P. Stiefel P. Behr P. Sarajlic E. Bijl D. Gabi M. Voros J. Vorholt J. A. Zambelli T. Appl. Phys. Lett. 2010;97:023701. doi: 10.1063/1.3462979. [DOI] [Google Scholar]
- Yang J. S. Park J. Koehler M. Simpson J. Luque D. Rodriguez J. M. Alsteens D. Adv. NanoBiomed Res. 2021;1:2100077. doi: 10.1002/anbr.202100077. [DOI] [Google Scholar]
- Gunaratnam G. Dudek J. Jung P. Becker S. L. Jacobs K. Bischoff M. Hannig M. Microorganisms. 2021;9:2213. doi: 10.3390/microorganisms9112213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff E. Guillaume-Gentil O. Ossola D. Polesel-Maris J. LeibundGut-Landmann S. Zambelli T. Vorholt J. A. PLoS One. 2012;7:e52712. doi: 10.1371/journal.pone.0052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen N. Sarkar S. Hondroulis E. Sabhachandani P. Konry T. Talanta. 2017;174:409–413. doi: 10.1016/j.talanta.2017.06.038. [DOI] [PubMed] [Google Scholar]
- Sztilkovics M. Gerecsei T. Peter B. Saftics A. Kurunczi S. Szekacs I. Szabo B. Horvath R. Sci. Rep. 2020;10:61. doi: 10.1038/s41598-019-56898-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A. G. Kanyo N. Voros A. Szekacs I. Bonyar A. Horvath R. Sci. Rep. 2022;12:7747. doi: 10.1038/s41598-022-11770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelie-Guinlet M. Viela F. Viljoen A. Dehullu J. Dufrene Y. F. Curr. Opin. Biomed. Eng. 2019;12:1–7. doi: 10.1016/j.cobme.2019.08.001. [DOI] [Google Scholar]
- Ossola D. Amarouch M. Y. Behr P. Voros J. Abriel H. Zambelli T. Nano Lett. 2015;15:1743–1750. doi: 10.1021/nl504438z. [DOI] [PubMed] [Google Scholar]
- Schlotter T. Weaver S. Forro C. Momotenko D. Voros J. Zambelli T. Aramesh M. ACS Nano. 2020;14:12993–13003. doi: 10.1021/acsnano.0c04281. [DOI] [PubMed] [Google Scholar]
- Li M. Liu L. Q. Zambelli T. Nano Res. 2022;15:773–786. doi: 10.1007/s12274-021-3573-y. [DOI] [Google Scholar]
- Saha P. Duanis-Assaf T. Reches M. Adv. Mater. Interfaces. 2020;7:2001115. doi: 10.1002/admi.202001115. [DOI] [Google Scholar]
- Houthaeve G. De Smedt S. C. Braeckmans K. De Vos W. H. Nano Converg. 2022;9:6. doi: 10.1186/s40580-022-00298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod K. F. Annu. Rev. Cancer Biol. 2020;4:41–60. doi: 10.1146/annurev-cancerbio-030419-033405. [DOI] [Google Scholar]
- Hu Q. Lu J. F. Zhang X. H. Liu R. Yang S. H. J. Cereb. Blood Flow Metab. 2022;42:1748–1750. doi: 10.1177/0271678X221109685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh M. J. Chinnery P. F. Biochim. Biophys. Acta, Bioenerg. 2015;1847:1401–1411. doi: 10.1016/j.bbabio.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Johnson J. Mercado-Ayon E. Mercado-Ayon Y. Dong Y. N. Halawani S. Ngaba L. Lynch D. R. Arch. Biochem. Biophys. 2021;702:108698. doi: 10.1016/j.abb.2020.108698. [DOI] [PubMed] [Google Scholar]
- Picelli S. Bjorklund A. K. Faridani O. R. Sagasser S. Winberg G. Sandberg R. Nat. Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- Hagemann-Jensen M. Ziegenhain C. Chen P. Ramskold D. Hendriks G. J. Larsson A. J. M. Faridani O. R. Sandberg R. Nat. Biotechnol. 2020;38:708–714. doi: 10.1038/s41587-020-0497-0. [DOI] [PubMed] [Google Scholar]
- Pohl H. A., Dielectrophoresis: the behavior of neutral matter in nonuniform electric fields, Cambridge University Press, Cambridge, 1978 [Google Scholar]
- Pohl H. A. J. Appl. Phys. 1951;22:869. doi: 10.1063/1.1700065. [DOI] [Google Scholar]
- Cottet J. Fabregue O. Berger C. Buret F. Renaud P. Frenea-Robin M. Biophys. J. 2019;116:12–18. doi: 10.1016/j.bpj.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y. J. Gordon R. Nano Lett. 2012;12:402–406. doi: 10.1021/nl203719v. [DOI] [PubMed] [Google Scholar]
- Saleh A. A. E. Dionne J. A. Nano Lett. 2012;12:5581–5586. doi: 10.1021/nl302627c. [DOI] [PubMed] [Google Scholar]
- Wang K. Schonbrun E. Steinvurzel P. Crozier K. B. Nat. Commun. 2011;2:469. doi: 10.1038/ncomms1480. [DOI] [PubMed] [Google Scholar]
- Kang J. H. Kim K. Ee H. S. Lee Y. H. Yoon T. Y. Seo M. K. Park H. G. Nat. Commun. 2011;2:582. doi: 10.1038/ncomms1592. [DOI] [PubMed] [Google Scholar]
- Juan M. L. Gordon R. Pang Y. J. Eftekhari F. Quidant R. Nat. Phys. 2009;5:915–919. [Google Scholar]
- Xu H. X. Kall M. Phys. Rev. Lett. 2002;89:246802. doi: 10.1103/PhysRevLett.89.246802. [DOI] [PubMed] [Google Scholar]
- Novotny L. Bian R. X. Xie X. S. Phys. Rev. Lett. 1997;79:645–648. doi: 10.1103/PhysRevLett.79.645. [DOI] [Google Scholar]
- Ramos A. Morgan H. Green N. G. Castellanos A. J. Electrost. 1999;47:71–81. doi: 10.1016/S0304-3886(99)00031-5. [DOI] [Google Scholar]
- Freedman K. J. Otto L. M. Ivanov A. P. Barik A. Oh S. H. Edel J. B. Nat. Commun. 2016;7:10217. doi: 10.1038/ncomms10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteza Cabrejos A., Hong S., Nadappuram B. P., Magnani L., Ivanov A. and Edel J. B., Unpublished, 2023
- Holt C. E. Martin K. C. Schuman E. M. Nat. Struct. Mol. Biol. 2019;26:557–566. doi: 10.1038/s41594-019-0263-5. [DOI] [PubMed] [Google Scholar]