Abstract
Recent evidence has shown significant roles of mitochondria-derived vesicles (MDVs) in mitochondrial quality control (MQC) system. Under mild stress condition, MDVs are formed to carry the malfunctioned mitochondrial components, such as mitochondrial DNA (mtDNA), peptides, proteins and lipids, to be eliminated to restore normal mitochondrial structure and functions. Under severe oxidative stress condition, mitochondrial dynamics (fission/fusion) and mitophagy are predominantly activated to rescue mitochondrial structure and functions. Additionally, MDVs generation can be also triggered as the major MQC machinery to cope with unhealthy mitochondria when mitophagy is unsuccessful for eliminating the damaged mitochondria or mitochondrial fission/fusion fail to recover the mitochondrial structure and functions. This review summarizes the current knowledge on MDVs and discuss their roles in physiologic and pathophysiologic conditions. In addition, the potential clinical relevance of MDVs in therapeutics and diagnostics of kidney stone disease (KSD) are emphasized.
Keywords: EVs, Extracellular vesicles, MDVs, Mitovesicles, Nephrolithiasis, Oxidative stress, Urolithiasis
Introduction
Vesicular transport is a regulatory mechanism in all living cells. The cell-derived vesicles are originated from various cellular organelles, including mitochondria. Several lines of evidence have demonstrated essential roles of mitochondria-derived vesicles (MDVs) in mitochondrial quality control (MQC) system [1–4]. This control system is crucial for mitochondrial homeostasis and cell survival regulation [5, 6]. As such, MDVs formation in the MQC system is recognized as the first-line and vital regulatory mechanism in both physiologic and pathologic conditions [2, 7]. Novel findings of MDVs generation during the past decade have amplified our understanding of non-mitophagy pathway for mitochondrial preservation and cell survival. Many lines of MDVs research have shown greater MDVs level in mild stress conditions than mitophagy, which is the canonical machinery for removing the damaged mitochondria [1, 8–11]. Recently, selective cargos of mitochondrial oxidized molecules, such as mitochondrial DNA (mtDNA), peptides, proteins and lipids, to be degraded by lysosomes have been shown [3, 12, 13]. Moreover, immune regulation by MDVs has been emphasized in several reports of inflammation-associated diseases [3, 12, 14]. As such, MDVs have gained a wide interest in many mitochondria-associated disorders/diseases, such as cancers [15, 16], aging [17–19], cardiovascular diseases [20, 21], and neurodegenerative disorders [22–24].
It is well known that kidney stone disease (KSD) is associated with oxidative stress and mitochondrial abnormalities in renal tissue [25–29]. Cellular mechanisms of mitochondrial dysfunction associated with kidney stone formation have been proposed [25]. For example, renal tubular inflammation and peroxidation of lipids and proteins in cell membranes induced by mitochondrial abnormalities can increase crystal deposition in the kidney [25]. Components of dead cells and fragmented organelles, including mitochondria, also serve as the sources for stone nidus (core component) formation [25]. Additionally, the damaged mitochondria can promote renal interstitial inflammation that further enhances development and formation of the Randall’s plaque, which is one of the common pathologies serving as the nidus for calcium oxalate (CaOx) kidney stone [25]. Therefore, preserving mitochondrial functions has been proposed as one of the preventive strategies against KSD [25].
In addition to the whole mitochondria and their fragments, several lines of recent evidence have implicated the involvement of intracellular and extracellular MDVs in kidney stone formation. This review therefore summarizes the current knowledge on roles of MDVs, particularly in KSD.
Overview of MDVs
The evolutionary origin of mitochondria is from archaebacteria that ordinarily transport vesicles in order to communicate with other living microorganisms, escape from host immune systems, and eliminate self-damaged materials [30, 31]. Thus, MDVs formation has been proposed as the ancient homeostatic process in living cells at mitochondrial level under physiologic and mild stress conditions [21, 32]. Although removal of the damaged mitochondria or mitochondrial contents by autophagy in the MQC system for cell homeostasis has been extensively studied [5, 6], several mechanisms of mitochondrial reinforcement and repair remain unclear. Hence, recent concepts of micromitophagy [33, 34], MDVs formation [1, 8, 10, 22], and mitophagy-independent machinery [35–37] have been emerged to explain mitochondrial stability [8], prevention of cell death [37] and tissue repair [38, 39].
The intracellular vesicles that contain mitochondrial components have been recognized as mitochondrial vesicles or MDVs [40]. They are the nanoscale vesicles (approximately 70–150 nm in diameter) surrounded by single or double membranes, i.e., outer mitochondrial membrane (OMM) and/or inner mitochondrial membrane (IMM) [7, 11, 22]. MDVs are also the specific cargos for mitochondrial nucleic acids (DNA and RNA) [3, 21, 41–45], proteins [3, 22, 46, 47], lipids [7, 32, 37], fragmented mitochondria [5, 48] and/or other mitochondrial components [49–51]. Previous studies have shown that MDVs play major roles in intracellular interactions of the parental mitochondria with lysosomes [44, 52], endosomes [7, 44], and peroxisomes [22, 53]. Additional reports have demonstrated intercellular roles of MDVs in removing malfunctioned part of mitochondria [3, 44, 54], transferring functional MDVs to communicate with the target cells that require more energy [55–57] and regulating immune response [58, 59].
MDVs are known as the key component of the first-line secure process in the MQC system, and their possible roles entirely differ from mitochondrial dynamics (fission/fusion) and mitophagy [1, 4, 5, 10]. Additionally, the number of MDVs is increased by mild stress or early stage of mitochondrial dysfunction [21]. Two main types of MDVs have been recognized in the MQC system, including steady-state MDVs [32, 60] and stress-induced MDVs [8, 39], both of which can be characterized by their specific markers. Translocase of outer mitochondrial membrane 20 (TOMM20), an OMM protein, is mostly found in steady-state MDVs (TOMM+-MDVs) [32], whereas pyruvate dehydrogenase (PDH) is predominantly found in oxidative stress-triggered MDVs (PDH+-MDVs) [61]. Unveiling the MDVs formation and their functional roles would make the image of mitochondria-related intracellular and intercellular communications much clearer.
Biogenesis of MDVs
Previously, mitochondrial membrane blebbing and mitophagy-related machinery had been proposed as the possible mechanisms for MDVs formation [7]. However, later evidence has clearly shown that MDVs are independent of mitochondrial dynamics and mitophagy [5, 40]. One of the newly proposed mechanisms for MDVs biogenesis is via PINK1 (phosphatase and tensin homolog-induced kinase 1)/Parkin (an E3 ubiquitin protein ligase containing ubiquitin-like domain at N-terminus)-dependent, but DRP1 (dynamin related protein 1)-independent process [7, 52, 61, 62]. In mild stress condition or slight mitochondrial damage, mitochondrial membrane curvature is initiated followed by PINK1 accumulation [8, 10, 40]. Parkin is then recruited at OMM, and the MDVs are scissored and released by an unclear mechanism [7, 8, 10, 40, 52]. The involvement of DRP1 in MDVs generation has been excluded as MDVs can be formed even when DRP1 is knocked down [40].
By contrast, several investigations have shown that MDVs can be formed in PINK1-deficient cells [4, 7, 62, 63]. Recent proteome study has documented a new molecular model of MDVs biogenesis in resting stage that depends on the microtubule-associated motor proteins, MIRO1 and MIRO2 (MIRO1/2), and DRP1-dependent mechanism for cutting and releasing MDVs from parental mitochondria, whereas Parkin and PINK1 are not involved in this mediated pathway [32]. MDVs formation begins at steady-state by mitochondrial membrane protrusion after MIRO1/2 formation followed by recruitment of DRP1 by 49- and 51-kDa mitochondrial dynamics DRP1 receptor protein (MiD49 and MiD51, respectively) or mitochondrial fission factor (MFF) [32]. To complete MDVs construction, DRP1 then catalyzes the cutting of thin membrane tube to release MDVs that can be delivered to their specific targets. However, further elucidations for precise mechanism are needed as this group of the investigators have previously demonstrated that DRP1 silencing does not affect MDVs formation [21, 52, 61, 64] (in contrast to their own recent findings). They have described that the contradictory results were due to dissimilar gene knockout technique in each work. DRP1 was > 95% silencing in the prior study by simple molecular technique but was completely deleted by a more effective method, namely clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) system, in a recent work [32]. Thus, MDVs can be formed in an incomplete DRP1-knockdown condition. Nevertheless, they have also suggested that the steady-state MDVs formation does not require Parkin and PINK1, which may be needed for generation and regulation of MDVs formation during oxidative stress and inflammatory conditions [48, 65]. Moreover, the dynamicity of MDVs formation may be also affected by techniques of detection, isolation, and diverse states of diseases or study models. Therefore, future studies on MDVs should clearly provide sufficient details of methodology and conditioning used in each study for clarification. And more extensive investigations are required for further elucidations of the precise mechanism(s) of MDVs biogenesis.
Classification and subtypes of MDVs
Most of the investigations on MDVs have been done inside the cells with their inter-organellar interactions [8, 38, 40]. However, MDVs are considerably diverse. Immuno-labelling together with high-resolution electron microscopy [4, 21, 57, 60, 66], proteomics and lipidomic profiling [22, 32, 46, 67] can enhance the study of MDVs. Currently, intracellular MDVs can be discriminated from other intracellular vesicles by using their specific markers, including OMM, IMM, mitochondrial matrix proteins and mtDNA [2, 7].
In addition to the intracellular MDVs, increasing evidence of extracellular MDVs has been documented. The secretion of extracellular MDVs has been suggested to be associated with endolysosomal and multivesicular body (MVB) formation, a mechanism that is similar to secretion of extracellular vesicles (EVs) [7, 11, 40, 46, 47, 68–73]. In general, EVs are classified based-on their diameter, biogenesis mechanism and specific protein markers. These EVs commonly include exosomes, microvesicles (MVs) and apoptotic bodies (ABs) [74, 75]. ABs are macrovesicles that are secreted from apoptotic cells during cell death by apoptotic mechanism [76]. The size of ABs extremely differs from that of MDVs. However, diameters of MVs and exosomes are approximately 100–1000 nm [77, 78] and 20–200 nm [79, 80], respectively, which overlap with that of MDVs (50–150 nm) [60]. As such, MVs can be discriminated from MDVs by their MVB-independent secretory mechanism [81, 82]. Nevertheless, exosomal secretion is MVB-dependent [83, 84] similar to that of MDVs [7, 40, 46, 68]. Thus, extracellular MDVs can be discriminated from exosomes by using corresponding specific markers. To discriminate the isolated MDVs from EVs by their differential size, high-resolution nanoparticle tracking analysis (NTA) is the method of choice [85–87]. Excluding MVs and exosomes with size overlapping that of MDVs would require high-resolution isolation and specific detection of mitochondrial components such as IMM, OMM, mitochondrial matrix proteins and mtDNA [7, 40, 46, 68, 88–90].
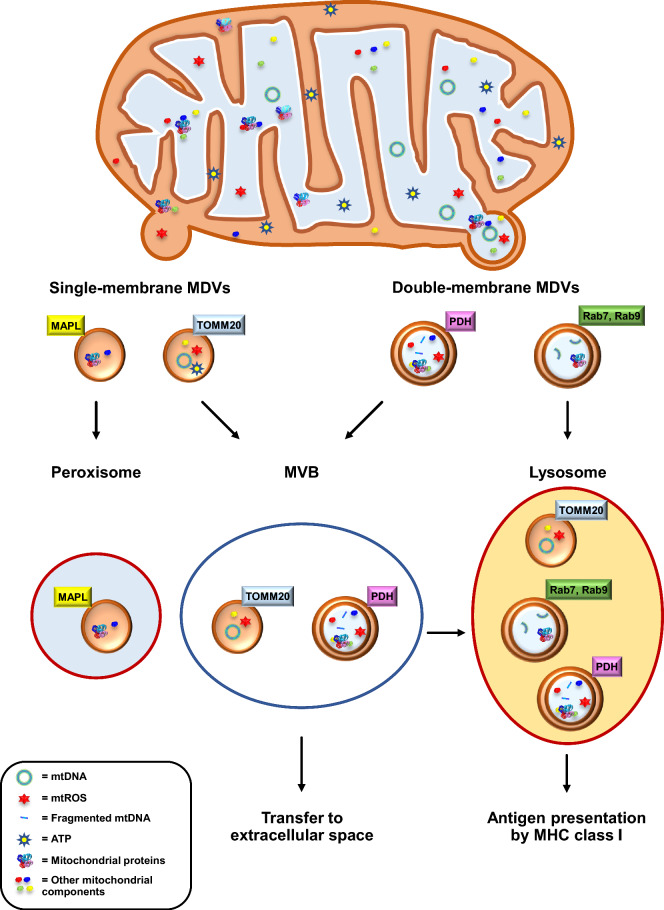
Packaging of MDVs is a complex mechanism associated with their diverse functions and destinations. Therefore, MDVs subtypes may be classified based on their specific contents and targets (Fig. 1). For example, MDVs containing mitochondria-anchored protein ligase (MAPL) are transported to peroxisomes [64, 91]. Similarly, MDVs containing peroxisomal biogenesis factor 3 (Pex3) and peroxisomal biogenesis factor 14 (Pex14) play crucial roles in peroxisomal biogenesis [92, 93]. Although MDVs containing Pex3/Pex14 or MAPL share the same targets, each of them functions differently.
Fig. 1.
Classification and subtypes of MDVs. MDVs can be classified based-on their membranes and specific cargos. The single-membrane MDVs contain outer mitochondrial membrane (OMM) proteins, whereas double-membrane MDVs contain OMM and inner mitochondrial membrane (IMM) proteins as well as mitochondrial matrix proteins. Based on these different cargos, there are specific protein markers for subtype classification. Mitochondria-anchored protein ligase (MAPL) and translocase of outer mitochondrial membrane 20 (TOMM20) are the common markers for single-membrane MDVs. Peroxisome is the terminal of MALP+-MDVs, while TOMM+-MDVs are excreted by multivesicular body (MVB) process like exosomes. Pyruvate dehydrogenase (PDH) are the specific protein marker for double-membrane MDVs, which are excreted by the MVB process. Moreover, MDVs formation in the presence of Rab7 (a small GTPase that monitors vesicular transport to late endosomes and lysosomes) and Rab9 can mediate antigen presentation via MHC class I
Additionally, MDVs can be classified based on the cellular status, including steady-state MDVs and stress-induced MDVs, which are the two distinct subtypes of MDVs widely investigated in several disease models [8, 32, 39, 60]. The steady-state MDVs are typically demonstrated as TOMM+/PDH− MDVs, whereas TOMM−/PDH+ MDVs (stress-induced MDVs) are predominantly found during oxidative stress [32, 61]. The biogenesis of the steady-state MDVs is PINK1/Parkin-independent, in contrast to that of the stress-induced MDVs as discussed above [52, 61]. After biogenesis, both TOMM+/PDH− and TOMM−/PDH+ MDVs carry the damaged mitochondrial components and transfer them to lysosomes for degradation to maintain mitochondrial structure and functions.
Another subtype of extracellular nanovesicles that correlate with MDVs has been recently isolated by high-resolution density gradient separation and termed as “mitovesicles” [68, 70]. Their size is approximately 6 nm and differs from other subtypes of MDVs or EVs. Mitovesicles are small double-membrane EVs that contain proteins involved in catabolic pathway, energy production and pro-fission process, but lack of proteins involved in biosynthesis, transport and pro-fusion process [19, 68]. Mechanisms of mitovesicles formation and release to extracellular space are not specified at this stage, but has been postulated to fuse with MVB before being secreted from the cells [68]. Moreover, mitovesicles serve as the functional vesicles based on the inside mitochondrial components [19, 68].
Although several subtypes of MDVs have been reported, their molecular machineries and biogenesis remain not well understood. Hence, specific cargos, functions, targets and subtypes of MDVs still require further elucidations for clarification.
Roles of MDVs in physiology and pathophysiology
Under physiologic state with mild stress, MDVs serve as a part of the crucial process in the MQC system to preserve mitochondrial functions [2, 21, 91]. MDVs formation has been proposed as the first-line mitochondrial safety to remove damaged mitochondrial components prior to detrimental derangement of the entire mitochondria and cell death activation [1, 5, 8, 38, 40, 46]. In addition, the increase of MDVs is the finest compensatory mechanism of the MQC system, when mitophagy does not work to eliminate the impaired mitochondria [1, 37, 63]. Thereafter, biogenesis of mitochondrial proteins and lipids is activated to restore the mitochondrial functions [5, 7, 94]. MDVs are therefore considered as a novel potential therapeutic target for maintaining the MQC system and preventing mitochondrial dysfunction in normal and disease conditions. MDVs also get involved in communications between mitochondria and other intracellular organelles. They not only transport the damaged compartments to endolysosomes for degradation but also transfer proteins and lipids to peroxisomal activation and biogenesis [92, 93]. Moreover, mitochondrial components such as BCL-2 (B-cell lymphoma 2) protein [5, 39, 40, 68, 95] and mtDNA from healthy mitochondria [21, 96, 97] can be sent to unhealthy mitochondria to recover their structure and functions, resulting in prevention of cell death [42, 98, 99].
Under pathophysiologic conditions, MDVs are the important regulator for immune response and inflammation [65, 100, 101]. During injury, mtDNA is recognized as one of the damage-associated molecular patterns (DAMPs) that can trigger pro-inflammatory response after binding to intracellular Toll-like receptors or nucleotide-binding oligomerization domain-containing protein (NOD)-like receptors [102, 103]. Additionally, mitochondrial reactive oxygen species (mtROS) has been shown to regulate proinflammatory signaling by increasing nuclear factor kappa B (NF-κB) gene expression and involving in hypoxia-inducible factor 1α (HIF1α)-induced inflammasome formation through NLRP3 (NLR family pyrin domain containing 3) [95, 104, 105]. Previous studies have also found that mitochondrial DAMPs can be released into the circulation, are recognized by pattern recognition receptors (PRRs), and promote tissue and organ injury [3, 103, 106–108]. Moreover, mitochondrial DAMPs can mediate neutrophil migration and degranulation, thereby further enhancing cellular injury and tissue inflammation [3, 103]. Many lines of evidence have shown that MDVs inhibit such inflammatory response and down-stream cascades by transferring the damaged mitochondrial components to be degraded by lysosomes and then fuse with MVB [109, 110]. Mitochondrial DAMPs in MVB are then released out as extracellular MDVs, which can inhibit pro-inflammatory activation. Moreover, MDVs-mediated antigen presentation is crucial for regulating the immune system [40, 48, 65, 111]. MDVs formation in the presence of Rab7 (a small GTPase that monitors vesicular transport to late endosomes and lysosomes), Rab9 and SNX9 (sorting nexin 9) can mediate antigen presentation after breaking down inside lysosomes by proteasome to load these mitochondrial antigens onto MHC class I molecules within endoplasmic reticulum and then transfer them to the cell surface [40, 48, 65, 111]. Therefore, MDVs are the important regulator for development, activation, differentiation and survival of diverse immune cells, including T-lymphocytes and macrophages [40, 48, 101, 112, 113].
Furthermore, MDVs can enhance anti-microbial machineries [40]. Methicillin-resistant Staphylococcus aureus (MRSA) infection can induce formation of MDVs containing mtROS and mitochondrial enzyme, superoxide dismutase-2 (SOD2) [114, 115]. These MDVs are then delivered to bacteria-containing phagosomes, where SOD2 can settle hydrogen peroxide activation and bacterial eradication.
Roles of mitochondrial dysfunction in kidney stone formation
Accumulative evidence has shown the involvement of mitochondrial dysfunction and oxidative damage in KSD development [25–29, 116–119]. Mitochondria are highly abundant in renal tubular cells that require high energy for keeping their regular functions, including water reabsorption and solute transports [120–123]. Interestingly, mitochondria are enriched in epithelial cells lining renal tubular segments that have been proposed as the initial areas for kidney stone formation [124–128]. Besides, interactions of mitochondria with oxalate and CaOx crystals have been shown as the important mechanisms involved in the pathogenesis of KSD [25, 26, 116, 129–131]. Several studies have demonstrated that oxalate and/or CaOx crystals can alter mitochondrial activities and induce ROS overproduction, leading to mitochondrial dysfunction and oxidative stress [25, 26, 116, 131–134].
Mechanistically, oxidative stress-induced stimuli can activate ROS overproduction and induce mitochondrial damage [135–137]. The damaged mitochondria fail to keep membrane potential properties and, hence, release calcium ion, mtDNA, mtROS, mitochondrial matrix proteins, OMM and IMM into the cytoplasm [4, 5, 21, 35]. These mitochondrial components further induce cell death, inflammatory response and renal tubulointerstitial tissue injury [134, 138]. Such tubular cell injury has been reported to induce CaOx crystal adhesion onto the cells, leading to crystal retention inside the renal tissue that is one of the important mechanisms for kidney stone formation [134, 139–142]. Additionally, the adhered crystals can further grow and aggregate with the surrounding crystals, resulting in stone nidus formation [127, 129, 134, 141].
Additionally, the damaged mitochondria and other cellular and organellar fragments can directly bind to CaOx crystals and serve as the stone nidus for crystal nucleation, growth and aggregation, which further enhance kidney stone formation [134, 143, 144]. Moreover, the damaged mitochondria can trigger inflammatory cascade at renal interstitial area [117, 145] by recruiting numerous inflammatory cells into this area, leading to accumulation of various proinflammatory cytokines and tissue inflammation [116, 129]. Together with supersaturation of calcium phosphate, which is common in the renal interstitium, Randall’s plaque starts to form [126, 146, 147]. After erosion into the urinary space, where CaOx is frequently supersaturated, this plaque then serves as the nidus for CaOx stone to grow [25].
Potential roles of MDVs in KSD
Several recent studies have continuously shown significant roles of urinary EVs (uEVs) in KSD [148–152]. uEVs are involved in inflammatory response and elimination of CaOx crystals, and may also serve as the composition of the stone matrix [150, 152]. Also, recent clinical studies have identified specific subtypes of uEVs as the potential biomarkers in the urine of kidney stone patients compared with healthy subjects [149, 150, 153]. Furthermore, pattern of uEVs subtypes in females with KSD (but not those derived from non-stone females) is similar to that in males with or without KSD [154]. Although MDVs have not yet been examined directly in KSD, numerous mitochondrial proteins have been identified in these uEVs. According to recent proteome and lipidome studies of MDVs [22, 46, 68], a large number of mitochondrial proteins and lipids have been identified in both MDVs and EVs [155–158]. We have also compared all of the proteins identified in EVs based on Vesiclepedia database (http://www.microvesicles.org/) with those identified in mitochondria based on The Human Protein Atlas (https://www.proteinatlas.org/). Interestingly, 244 proteins are commonly found in both EVs and mitochondria (Table 1). These findings are consistent with the data observed in recent proteome studies of EVs [46, 73]. Therefore, MDVs are expected to play similar roles as of uEVs in KSD.
Table 1.
Summary of proteins that are found in both EVs (http://www.microvesicles.org/) and mitochondria (https://www.proteinatlas.org/)
| No. | Gene symbol | Uniprot ID | Protein name(s) |
|---|---|---|---|
| 1 | DECR1 | Q16698 | 2,4-dienoyl-CoA reductase [(3E)-enoyl-CoA-producing], mitochondrial (EC 1.3.1.124) (2,4-dienoyl-CoA reductase [NADPH]) (4-enoyl-CoA reductase [NADPH]) (Short chain dehydrogenase/reductase family 18C member 1) |
| 2 | MRPS14 | O60783 | 28S ribosomal protein S14, mitochondrial (MRP-S14) (S14mt) (Mitochondrial small ribosomal subunit protein uS14m) |
| 3 | MRPS18B | Q9Y676 | 28S ribosomal protein S18b, mitochondrial (MRP-S18-b) (Mrps18-b) (S18mt-b) (28S ribosomal protein S18-2, mitochondrial) (MRP-S18-2) (Mitochondrial small ribosomal subunit protein bS18b) (Mitochondrial small ribosomal subunit protein mS40) |
| 4 | MRPS23 | Q9Y3D9 | 28S ribosomal protein S23, mitochondrial (MRP-S23) (S23mt) (Mitochondrial small ribosomal subunit protein mS23) |
| 5 | MRPS26 | Q9BYN8 | 28S ribosomal protein S26, mitochondrial (MRP-S26) (S26mt) (28S ribosomal protein S13, mitochondrial) (MRP-S13) (S13mt) (Mitochondrial small ribosomal subunit protein mS26) |
| 6 | MRPS27 | Q92552 | 28S ribosomal protein S27, mitochondrial (MRP-S27) (S27mt) (Mitochondrial ribosomal protein S27) (Mitochondrial small ribosomal subunit protein mS27) |
| 7 | DAP3 | P51398 | 28S ribosomal protein S29, mitochondrial (MRP-S29) (S29mt) (Death-associated protein 3) (DAP-3) (Ionizing radiation resistance conferring protein) (Mitochondrial small ribosomal subunit protein mS29) |
| 8 | MRPS31 | Q92665 | 28S ribosomal protein S31, mitochondrial (MRP-S31) (S31mt) (Imogen 38) (Mitochondrial small ribosomal subunit protein mS31) |
| 9 | MRPS35 | P82673 | 28S ribosomal protein S35, mitochondrial (MRP-S35) (S35mt) (28S ribosomal protein S28, mitochondrial) (MRP-S28) (S28mt) (Mitochondrial small ribosomal subunit protein mS35) |
| 10 | BCKDHB | P21953 | 2-oxoisovalerate dehydrogenase subunit beta, mitochondrial (EC 1.2.4.4) (Branched-chain alpha-keto acid dehydrogenase E1 component beta chain) (BCKDE1B) (BCKDH E1-beta) |
| 11 | MRPL2 | Q5T653 | 39S ribosomal protein L2, mitochondrial (L2mt) (MRP-L2) (Mitochondrial large ribosomal subunit protein uL2m) |
| 12 | MRPL21 | Q7Z2W9 | 39S ribosomal protein L21, mitochondrial (L21mt) (MRP-L21) (Mitochondrial large ribosomal subunit protein bL21m) |
| 13 | MRPL23 | Q16540 | 39S ribosomal protein L23, mitochondrial (L23mt) (MRP-L23) (L23 mitochondrial-related protein) (Mitochondrial large ribosomal subunit protein uL23m) (Ribosomal protein L23-like) |
| 14 | MRPL36 | Q9P0J6 | 39S ribosomal protein L36, mitochondrial (L36mt) (MRP-L36) (BRCA1-interacting protein 1) (Mitochondrial large ribosomal subunit protein bL36m) |
| 15 | MRPL40 | Q9NQ50 | 39S ribosomal protein L40, mitochondrial (L40mt) (MRP-L40) (Mitochondrial large ribosomal subunit protein mL40) (Nuclear localization signal-containing protein deleted in velocardiofacial syndrome) (Up-regulated in metastasis) |
| 16 | MRPL43 | Q8N983 | 39S ribosomal protein L43, mitochondrial (L43mt) (MRP-L43) (Mitochondrial large ribosomal subunit protein mL43) (Mitochondrial ribosomal protein bMRP36a) |
| 17 | MRPL44 | Q9H9J2 | 39S ribosomal protein L44, mitochondrial (L44mt) (MRP-L44) (EC 3.1.26.-) (Mitochondrial large ribosomal subunit protein mL44) |
| 18 | MRPL46 | Q9H2W6 | 39S ribosomal protein L46, mitochondrial (L46mt) (MRP-L46) (Mitochondrial large ribosomal subunit protein mL46) (P2ECSL) |
| 19 | MRPL52 | Q86TS9 | 39S ribosomal protein L52, mitochondrial (L52mt) (MRP-L52) (Mitochondrial large ribosomal subunit protein mL52) |
| 20 | MPST | P25325 | 3-mercaptopyruvate sulfurtransferase (MST) (EC 2.8.1.2) |
| 21 | HPDL | Q96IR7 | 4-hydroxyphenylpyruvate dioxygenase-like protein (HPD-like protein) (EC 1.13.-.-) (Glyoxalase domain-containing protein 1) |
| 22 | NT5DC3 | Q86UY8 | 5′-nucleotidase domain-containing protein 3 (EC 3.1.3.-) (GRP94-neighboring nucleotidase) |
| 23 | RPL7L1 | Q6DKI1 | 60S ribosomal protein L7-like 1 (Large ribosomal subunit protein uL30-like 1) |
| 24 | ADAMTS16 | Q8TE57 | A disintegrin and metalloproteinase with thrombospondin motifs 16 (ADAM-TS 16) (ADAM-TS16) (ADAMTS-16) (EC 3.4.24.-) |
| 25 | SMPDL3A | Q92484 | Acid sphingomyelinase-like phosphodiesterase 3a (ASM-like phosphodiesterase 3a) (EC 3.1.4.-) |
| 26 | NDUFAB1 | O14561 | Acyl carrier protein, mitochondrial (ACP) (CI-SDAP) (NADH-ubiquinone oxidoreductase 9.6 kDa subunit) |
| 27 | AGK | Q53H12 | Acylglycerol kinase, mitochondrial (hAGK) (EC 2.7.1.107) (EC 2.7.1.138) (EC 2.7.1.94) (Multiple substrate lipid kinase) (HsMuLK) (MuLK) (Multi-substrate lipid kinase) |
| 28 | FAHD1 | Q6P587 | Acylpyruvase FAHD1, mitochondrial (EC 3.7.1.5) (Fumarylacetoacetate hydrolase domain-containing protein 1) (FAH domain-containing protein 1) (Oxaloacetate decarboxylase) (OAA decarboxylase) (EC 4.1.1.112) (YisK-like protein) |
| 29 | NUDT9 | Q9BW91 | ADP-ribose pyrophosphatase, mitochondrial (EC 3.6.1.13) (ADP-ribose diphosphatase) (ADP-ribose phosphohydrolase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (Nucleoside diphosphate-linked moiety X motif 9) (Nudix motif 9) |
| 30 | AARS2 | Q5JTZ9 | Alanine–tRNA ligase, mitochondrial (EC 6.1.1.7) (Alanyl-tRNA synthetase) (AlaRS) |
| 31 | AARS2 | Q5JTZ9 | Alanine–tRNA ligase, mitochondrial |
| 32 | ALDH1B1 | P30837 | Aldehyde dehydrogenase X, mitochondrial (EC 1.2.1.3) (Aldehyde dehydrogenase 5) (Aldehyde dehydrogenase family 1 member B1) |
| 33 | ALDH7A1 | P49419 | Alpha-aminoadipic semialdehyde dehydrogenase (Alpha-AASA dehydrogenase) (EC 1.2.1.31) (Aldehyde dehydrogenase family 7 member A1) (EC 1.2.1.3) (Antiquitin-1) (Betaine aldehyde dehydrogenase) (EC 1.2.1.8) (Delta1-piperideine-6-carboxylate dehydrogenase) (P6c dehydrogenase) |
| 34 | AASS | Q9UDR5 | Alpha-aminoadipic semialdehyde synthase, mitochondrial (LKR/SDH) [Includes: Lysine ketoglutarate reductase (LKR) (LOR) (EC 1.5.1.8); Saccharopine dehydrogenase (SDH) (EC 1.5.1.9)] |
| 35 | MAOA | P21397 | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A) |
| 36 | ADGB | Q8N7X0 | Androglobin (Calpain-7-like protein) |
| 37 | ANKRD34B | A5PLL1 | Ankyrin repeat domain-containing protein 34B |
| 38 | ARMCX1 | Q9P291 | Armadillo repeat-containing X-linked protein 1 (ARM protein lost in epithelial cancers on chromosome X 1) (Protein ALEX1) |
| 39 | ARMCX2 | Q7L311 | Armadillo repeat-containing X-linked protein 2 (ARM protein lost in epithelial cancers on chromosome X 2) (Protein ALEX2) |
| 40 | DARS2 | Q6PI48 | Aspartate–tRNA ligase, mitochondrial (EC 6.1.1.12) (Aspartyl-tRNA synthetase) (AspRS) |
| 41 | ATP5MF | P56134 | ATP synthase subunit f, mitochondrial (ATP synthase membrane subunit f) |
| 42 | PFKL | P17858 | ATP-dependent 6-phosphofructokinase, liver type (ATP-PFK) (PFK-L) (EC 2.7.1.11) (6-phosphofructokinase type B) (Phosphofructo-1-kinase isozyme B) (PFK-B) (Phosphohexokinase) |
| 43 | CLPX | O76031 | ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial |
| 44 | DHX30 | Q7L2E3 | ATP-dependent RNA helicase DHX30 (EC 3.6.4.13) (DEAH box protein 30) |
| 45 | YME1L1 | Q96TA2 | ATP-dependent zinc metalloprotease YME1L1 (EC 3.4.24.-) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin-associated metalloprotease) (PAMP) (YME1-like protein 1) |
| 46 | ATRNL1 | Q5VV63 | Attractin-like protein 1 |
| 47 | AURKAIP1 | Q9NWT8 | Aurora kinase A-interacting protein (AURKA-interacting protein) (28S ribosomal protein S38, mitochondrial) (MRP-S38) (Mitochondrial small ribosomal subunit protein mS38) |
| 48 | CD72 | P21854 | B-cell differentiation antigen CD72 (Lyb-2) (CD antigen CD72) |
| 49 | BOLA3 | Q53S33 | BolA-like protein 3 |
| 50 | BDNF | P23560 | Brain-derived neurotrophic factor (BDNF) (Abrineurin) [Cleaved into: BDNF precursor form (ProBDNF)] |
| 51 | BRI3BP | Q8WY22 | BRI3-binding protein (I3-binding protein) (Cervical cancer 1 proto-oncogene-binding protein KG19) (HCCRBP-1) |
| 52 | KCTD6 | Q8NC69 | BTB/POZ domain-containing protein KCTD6 (KCASH3 protein) (Potassium channel tetramerization domain-containing protein 6) |
| 53 | CALHM2 | Q9HA72 | Calcium homeostasis modulator protein 2 (Protein FAM26B) |
| 54 | CASQ1 | P31415 | Calsequestrin-1 (Calmitine) (Calsequestrin, skeletal muscle isoform) |
| 55 | CPT2 | P23786 | Carnitine O-palmitoyltransferase 2, mitochondrial (EC 2.3.1.21) (Carnitine palmitoyltransferase II) (CPT II) |
| 56 | CASP3 | P42574 | Caspase-3 (CASP-3) (EC 3.4.22.56) (Apopain) (Cysteine protease CPP32) (CPP-32) (Protein Yama) (SREBP cleavage activity 1) (SCA-1) [Cleaved into: Caspase-3 subunit p17; Caspase-3 subunit p12] |
| 57 | SLC44A1 | Q8WWI5 | Choline transporter-like protein 1 (CDw92) (Solute carrier family 44 member 1) (CD antigen CD92) |
| 58 | CBX6 | O95503 | Chromobox protein homolog 6 |
| 59 | C21orf2 | O43822 | Cilia- and flagella-associated protein 410 (C21orf-HUMF09G8.5) (Leucine-rich repeat-containing protein 76) (YF5/A2) |
| 60 | CNKSR3 | Q6P9H4 | Connector enhancer of kinase suppressor of ras 3 (Connector enhancer of KSR 3) (CNK homolog protein 3) (CNK3) (CNKSR family member 3) (Maguin-like protein) |
| 61 | ATG4D | Q86TL0 | Cysteine protease ATG4D (EC 3.4.22.-) (AUT-like 4 cysteine endopeptidase) (Autophagin-4) (Autophagy-related cysteine endopeptidase 4) (Autophagy-related protein 4 homolog D) [Cleaved into: Cysteine protease ATG4D, mitochondrial] |
| 62 | COX7A2L | O14548 | Cytochrome c oxidase subunit 7A-related protein, mitochondrial (COX7a-related protein) (Cytochrome c oxidase subunit VIIa-related protein) (EB1) |
| 63 | CYC1 | P08574 | Cytochrome c1, heme protein, mitochondrial (EC 7.1.1.8) (Complex III subunit 4) (Complex III subunit IV) (Cytochrome b-c1 complex subunit 4) (Ubiquinol-cytochrome-c reductase complex cytochrome c1 subunit) (Cytochrome c-1) |
| 64 | DYNC2H1 | Q8NCM8 | Cytoplasmic dynein 2 heavy chain 1 (Cytoplasmic dynein 2 heavy chain) (Dynein cytoplasmic heavy chain 2) (Dynein heavy chain 11) (hDHC11) (Dynein heavy chain isotype 1B) |
| 65 | DCAF15 | Q66K64 | DDB1- and CUL4-associated factor 15 |
| 66 | DHRS2 | Q13268 | Dehydrogenase/reductase SDR family member 2, mitochondrial (EC 1.1.1.-) (Dicarbonyl reductase HEP27) (Protein D) (Short chain dehydrogenase/reductase family 25C member 1) |
| 67 | DHRS7 | Q9Y394 | Dehydrogenase/reductase SDR family member 7 (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase 4) (retSDR4) (Short chain dehydrogenase/reductase family 34C member 1) |
| 68 | DEPTOR | Q8TB45 | DEP domain-containing mTOR-interacting protein (DEP domain-containing protein 6) |
| 69 | DIABLO | Q9NR28 | Diablo homolog, mitochondrial |
| 70 | DIABLO | Q9NR28 | Diablo homolog, mitochondrial (Direct IAP-binding protein with low pI) (Second mitochondria-derived activator of caspase) (Smac) |
| 71 | DLD | P09622 | Dihydrolipoyl dehydrogenase, mitochondrial (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein) |
| 72 | DLST | P36957 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial (EC 2.3.1.61) (2-oxoglutarate dehydrogenase complex component E2) (OGDC-E2) (Dihydrolipoamide succinyltransferase component of 2-oxoglutarate dehydrogenase complex) (E2K) |
| 73 | DHODH | Q02127 | Dihydroorotate dehydrogenase (quinone), mitochondrial (DHOdehase) (EC 1.3.5.2) (Dihydroorotate oxidase) |
| 74 | DNAJA3 | Q96EY1 | DnaJ homolog subfamily A member 3, mitochondrial (DnaJ protein Tid-1) (hTid-1) (Hepatocellular carcinoma-associated antigen 57) (Tumorous imaginal discs protein Tid56 homolog) |
| 75 | DNLZ | Q5SXM8 | DNL-type zinc finger protein (Hsp70-escort protein 1) (HEP1) (mtHsp70-escort protein) |
| 76 | DMRTA2 | Q96SC8 | Doublesex- and mab-3-related transcription factor A2 (Doublesex- and mab-3-related transcription factor 5) |
| 77 | OPA1 | O60313 | Dynamin-like 120 kDa protein, mitochondrial (EC 3.6.5.5) (Optic atrophy protein 1) [Cleaved into: Dynamin-like 120 kDa protein, form S1] |
| 78 | RNF115 | Q9Y4L5 | E3 ubiquitin-protein ligase RNF115 (EC 2.3.2.27) (RING finger protein 115) (RING-type E3 ubiquitin transferase RNF115) (Rabring 7) (Zinc finger protein 364) |
| 79 | SIAH1 | Q8IUQ4 | E3 ubiquitin-protein ligase SIAH1 (EC 2.3.2.27) (RING-type E3 ubiquitin transferase SIAH1) (Seven in absentia homolog 1) (Siah-1) (Siah-1a) |
| 80 | EML6 | Q6ZMW3 | Echinoderm microtubule-associated protein-like 6 (EMAP-6) (Echinoderm microtubule-associated protein-like 5-like) |
| 81 | GFM1 | Q96RP9 | Elongation factor G, mitochondrial (EF-Gmt) (Elongation factor G 1, mitochondrial) (mEF-G 1) (Elongation factor G1) (hEFG1) |
| 82 | TSFM | P43897 | Elongation factor Ts, mitochondrial (EF-Ts) (EF-TsMt) |
| 83 | ECI2 | O75521 | Enoyl-CoA delta isomerase 2 (EC 5.3.3.8) (DRS-1) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (Diazepam-binding inhibitor-related protein 1) (DBI-related protein 1) (Dodecenoyl-CoA isomerase) (Hepatocellular carcinoma-associated antigen 88) (Peroxisomal 3,2-trans-enoyl-CoA isomerase) (pECI) (Renal carcinoma antigen NY-REN-1) |
| 84 | EIF4E2 | O60573 | Eukaryotic translation initiation factor 4E type 2 (eIF-4E type 2) (eIF4E type 2) (Eukaryotic translation initiation factor 4E homologous protein) (Eukaryotic translation initiation factor 4E-like 3) (eIF4E-like protein 4E-LP) (mRNA cap-binding protein 4EHP) (h4EHP) (mRNA cap-binding protein type 3) |
| 85 | SLC1A3 | P43003 | Excitatory amino acid transporter 1 (Sodium-dependent glutamate/aspartate transporter 1) (GLAST-1) (Solute carrier family 1 member 3) |
| 86 | EXOC3 | O60645 | Exocyst complex component 3 (Exocyst complex component Sec6) |
| 87 | EXD2 | Q9NVH0 | Exonuclease 3'-5' domain-containing protein 2 (EC 3.1.11.1) (3'-5' exoribonuclease EXD2) (EC 3.1.13.-) (Exonuclease 3'-5' domain-like-containing protein 2) |
| 88 | FASTK | Q14296 | Fas-activated serine/threonine kinase (FAST kinase) (EC 2.7.11.8) |
| 89 | FSTL4 | Q6MZW2 | Follistatin-related protein 4 (Follistatin-like protein 4) |
| 90 | FOXN4 | Q96NZ1 | Forkhead box protein N4 |
| 91 | FXN | Q16595 | Frataxin, mitochondrial (EC 1.16.3.1) (Friedreich ataxia protein) (Fxn) [Cleaved into: Frataxin intermediate form (i-FXN); Frataxin(56–210) (m56-FXN); Frataxin(78–210) (d-FXN) (m78-FXN); Frataxin mature form (Frataxin(81–210)) (m81-FXN)] |
| 92 | LGALS2 | P05162 | Galectin-2 (Gal-2) (Beta-galactoside-binding lectin L-14-II) (HL14) (Lactose-binding lectin 2) (S-Lac lectin 2) |
| 93 | GDAP1 | Q8TB36 | Ganglioside-induced differentiation-associated protein 1 (GDAP1) |
| 94 | GSE1 | Q14687 | Genetic suppressor element 1 |
| 95 | FP565260.6 | A0A0B4J2D5 | Glutamine amidotransferase-like class 1 domain-containing protein 3B, mitochondrial (Keio novel protein-I) (KNP-I) (Protein GT335) (Protein HES1) |
| 96 | GLDC | P23378 | Glycine dehydrogenase (decarboxylating), mitochondrial (EC 1.4.4.2) (Glycine cleavage system P protein) (Glycine decarboxylase) (Glycine dehydrogenase (aminomethyl-transferring)) |
| 97 | GADD45GIP1 | Q8TAE8 | Growth arrest and DNA damage-inducible proteins-interacting protein 1 (39S ribosomal protein L59, mitochondrial) (MRP-L59) (CKII beta-associating protein) (CR6-interacting factor 1) (CRIF1) (Mitochondrial large ribosomal subunit protein mL64) (Papillomavirus L2-interacting nuclear protein 1) (PLINP) (PLINP-1) (p53-responsive gene 6 protein) |
| 98 | AC093155.3 | Q7LGA3 | Heparan sulfate 2-O-sulfotransferase 1 |
| 99 | HS2ST1 | Q7LGA3 | Heparan sulfate 2-O-sulfotransferase 1 (2-O-sulfotransferase) (2OST) (EC 2.8.2.-) |
| 100 | HHIPL2 | Q6UWX4 | HHIP-like protein 2 |
| 101 | HIGD1A | Q9Y241 | HIG1 domain family member 1A, mitochondrial (Hypoxia-inducible gene 1 protein) (RCF1 homolog A) (RCF1a) |
| 102 | HIGD2A | Q9BW72 | HIG1 domain family member 2A, mitochondrial (RCF1 homolog B) (RCF1b) |
| 103 | HINT3 | Q9NQE9 | Histidine triad nucleotide-binding protein 3 (HINT-3) (EC 3.-.-.-) |
| 104 | NSD3 | Q9BZ95 | Histone-lysine N-methyltransferase NSD3 (EC 2.1.1.370) (EC 2.1.1.371) (Nuclear SET domain-containing protein 3) (Protein whistle) (WHSC1-like 1 isoform 9 with methyltransferase activity to lysine) (Wolf-Hirschhorn syndrome candidate 1-like protein 1) (WHSC1-like protein 1) |
| 105 | HCFC1 | P51610 | Host cell factor 1 (HCF) (HCF-1) (C1 factor) (CFF) (VCAF) (VP16 accessory protein) [Cleaved into: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6] |
| 106 | HSDL1 | Q3SXM5 | Inactive hydroxysteroid dehydrogenase-like protein 1 (Short chain dehydrogenase/reductase family 12C member 3) |
| 107 | PLD5 | Q8N7P1 | Inactive phospholipase D5 (Inactive PLD 5) (Inactive choline phosphatase 5) (Inactive phosphatidylcholine-hydrolyzing phospholipase D5) (PLDc) |
| 108 | ITGB5 | P18084 | Integrin beta-5 |
| 109 | ICAM3 | P32942 | Intercellular adhesion molecule 3 (ICAM-3) (CDw50) (ICAM-R) (CD antigen CD50) |
| 110 | ILF3 | Q12906 | Interleukin enhancer-binding factor 3 (Double-stranded RNA-binding protein 76) (DRBP76) (M-phase phosphoprotein 4) (MPP4) (Nuclear factor associated with dsRNA) (NFAR) (Nuclear factor of activated T-cells 90 kDa) (NF-AT-90) (Translational control protein 80) (TCP80) |
| 111 | IFT27 | Q9BW83 | Intraflagellar transport protein 27 homolog (Putative GTP-binding protein RAY-like) (Rab-like protein 4) |
| 112 | ISOC2 | Q96AB3 | Isochorismatase domain-containing protein 2 |
| 113 | IDH3G | P51553 | Isocitrate dehydrogenase [NAD] subunit gamma, mitochondrial (Isocitric dehydrogenase subunit gamma) (NAD( +)-specific ICDH subunit gamma) |
| 114 | IVD | P26440 | Isovaleryl-CoA dehydrogenase, mitochondrial (IVD) (EC 1.3.8.4) (Butyryl-CoA dehydrogenase) (EC 1.3.8.1) |
| 115 | KLHL29 | Q96CT2 | Kelch-like protein 29 (Kelch repeat and BTB domain-containing protein 9) |
| 116 | KLC4 | Q9NSK0 | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8) |
| 117 | LRRD1 | A4D1F6 | Leucine-rich repeat and death domain-containing protein 1 |
| 118 | LIAS | O43766 | Lipoyl synthase, mitochondrial (EC 2.8.1.8) (Lipoate synthase) (LS) (Lip-syn) (Lipoic acid synthase) |
| 119 | LONP1 | P36776 | Lon protease homolog, mitochondrial (EC 3.4.21.53) (LONHs) (Lon protease-like protein) (LONP) (Mitochondrial ATP-dependent protease Lon) (Serine protease 15) |
| 120 | ACSL5 | Q9ULC5 | Long-chain-fatty-acid–CoA ligase 5 (EC 6.2.1.3) (Arachidonate–CoA ligase) (EC 6.2.1.15) (Long-chain acyl-CoA synthetase 5) (LACS 5) |
| 121 | LRP12 | Q9Y561 | Low-density lipoprotein receptor-related protein 12 (LDLR-related protein 12) (LRP-12) (Suppressor of tumorigenicity 7 protein) |
| 122 | LRP4 | O75096 | Low-density lipoprotein receptor-related protein 4 (LRP-4) (Multiple epidermal growth factor-like domains 7) |
| 123 | MFSD12 | Q6NUT3 | Major facilitator superfamily domain-containing protein 12 |
| 124 | XK | P51811 | Membrane transport protein XK (Kell complex 37 kDa component) (Kx antigen) (XK-related protein 1) |
| 125 | MBLAC2 | Q68D91 | Metallo-beta-lactamase domain-containing protein 2 (EC 3.-.-.-) |
| 126 | MTX2 | O75431 | Metaxin-2 (Mitochondrial outer membrane import complex protein 2) |
| 127 | C19orf70 | Q5XKP0 | MICOS complex subunit MIC13 (Protein P117) |
| 128 | APOO | Q9BUR5 | MICOS complex subunit MIC26 (Apolipoprotein O) (MICOS complex subunit MIC23) (Protein FAM121B) |
| 129 | MGST1 | P10620 | Microsomal glutathione S-transferase 1 (Microsomal GST-1) (EC 2.5.1.18) (Microsomal GST-I) |
| 130 | SLC25A10 | Q9UBX3 | Mitochondrial dicarboxylate carrier (Solute carrier family 25 member 10) |
| 131 | SLC25A22 | Q9H936 | Mitochondrial glutamate carrier 1 (GC-1) (Glutamate/H( +) symporter 1) (Solute carrier family 25 member 22) |
| 132 | SLC25A18 | Q9H1K4 | Mitochondrial glutamate carrier 2 (GC-2) (Glutamate/H( +) symporter 2) (Solute carrier family 25 member 18) |
| 133 | TIMM13 | Q9Y5L4 | Mitochondrial import inner membrane translocase subunit Tim13 |
| 134 | PAM16 | Q9Y3D7 | Mitochondrial import inner membrane translocase subunit TIM16 (Mitochondria-associated granulocyte macrophage CSF-signaling molecule) (Presequence translocated-associated motor subunit PAM16) |
| 135 | TIMM50 | Q3ZCQ8 | Mitochondrial import inner membrane translocase subunit TIM50 |
| 136 | TOMM40 | O96008 | Mitochondrial import receptor subunit TOM40 homolog (Protein Haymaker) (Translocase of outer membrane 40 kDa subunit homolog) (p38.5) |
| 137 | CCDC51 | Q96ER9 | Mitochondrial potassium channel (MITOK) (Coiled-coil domain-containing protein 51) |
| 138 | ABCB8 | Q9NUT2 | Mitochondrial potassium channel ATP-binding subunit (ATP-binding cassette sub-family B member 8, mitochondrial) (ABCB8) (Mitochondrial ATP-binding cassette 1) (M-ABC1) (Mitochondrial sulfonylurea-receptor) (MITOSUR) |
| 139 | KIAA0391 | O15091 | Mitochondrial ribonuclease P catalytic subunit (EC 3.1.26.5) (Mitochondrial ribonuclease P protein 3) (Mitochondrial RNase P protein 3) (Protein only RNase P catalytic subunit) |
| 140 | SLC25A37 | Q9NYZ2 | Mitoferrin-1 (Mitochondrial iron transporter 1) (Mitochondrial solute carrier protein) (Solute carrier family 25 member 37) |
| 141 | MOCOS | Q96EN8 | Molybdenum cofactor sulfurase (MCS) (MOS) (MoCo sulfurase) (hMCS) (EC 2.8.1.9) (Molybdenum cofactor sulfurtransferase) |
| 142 | MORN1 | Q5T089 | MORN repeat-containing protein 1 |
| 143 | MYL3 | P08590 | Myosin light chain 3 (Cardiac myosin light chain 1) (CMLC1) (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular myosin alkali light chain) (Ventricular myosin light chain 1) (VLCl) (Ventricular/slow twitch myosin alkali light chain) (MLC-lV/sb) |
| 144 | B3GNT4 | Q9C0J1 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 4 (EC 2.4.1.149) (UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4) (BGnT-4) (Beta-1,3-Gn-T4) (Beta-1,3-N-acetylglucosaminyltransferase 4) (Beta3Gn-T4) |
| 145 | NNT | Q13423 | NAD(P) transhydrogenase, mitochondrial (EC 7.1.1.1) (Nicotinamide nucleotide transhydrogenase) (Pyridine nucleotide transhydrogenase) |
| 146 | NDUFA12 | Q9UI09 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 (13 kDa differentiation-associated protein) (Complex I-B17.2) (CI-B17.2) (CIB17.2) (NADH-ubiquinone oxidoreductase subunit B17.2) |
| 147 | NDUFA9 | Q16795 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial (Complex I-39kD) (CI-39kD) (NADH-ubiquinone oxidoreductase 39 kDa subunit) |
| 148 | NDUFB1 | O75438 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1 (Complex I-MNLL) (CI-MNLL) (NADH-ubiquinone oxidoreductase MNLL subunit) |
| 149 | NDUFB4 | O95168 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4 (Complex I-B15) (CI-B15) (NADH-ubiquinone oxidoreductase B15 subunit) |
| 150 | NDUFB5 | O43674 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5, mitochondrial (Complex I-SGDH) (CI-SGDH) (NADH-ubiquinone oxidoreductase SGDH subunit) |
| 151 | NDUFV1 | P49821 | NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial (EC 7.1.1.2) (Complex I-51kD) (CI-51kD) (NADH dehydrogenase flavoprotein 1) (NADH-ubiquinone oxidoreductase 51 kDa subunit) |
| 152 | NDUFV2 | P19404 | NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial (EC 7.1.1.2) (NADH-ubiquinone oxidoreductase 24 kDa subunit) |
| 153 | NDUFS3 | O75489 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial (EC 7.1.1.2) (Complex I-30kD) (CI-30kD) (NADH-ubiquinone oxidoreductase 30 kDa subunit) |
| 154 | CYB5R1 | Q9UHQ9 | NADH-cytochrome b5 reductase 1 (b5R.1) (EC 1.6.2.2) (Humb5R2) (NAD(P)H:quinone oxidoreductase type 3 polypeptide A2) |
| 155 | SLC11A1 | P49279 | Natural resistance-associated macrophage protein 1 (NRAMP 1) (Solute carrier family 11 member 1) |
| 156 | NGRN | Q9NPE2 | Neugrin (Mesenchymal stem cell protein DSC92) (Neurite outgrowth-associated protein) (Spinal cord-derived protein FI58G) |
| 157 | NGDN | Q8NEJ9 | Neuroguidin (Centromere accumulated nuclear protein 1) (CANu1) (EIF4E-binding protein) |
| 158 | SLC3A1 | Q07837 | Neutral and basic amino acid transport protein rBAT (NBAT) (D2h) (Solute carrier family 3 member 1) (b(0, +)-type amino acid transport protein) |
| 159 | NLRX1 | Q86UT6 | NLR family member X1 (Caterpiller protein 11.3) (CLR11.3) (Nucleotide-binding oligomerization domain protein 26) (Nucleotide-binding oligomerization domain protein 5) (Nucleotide-binding oligomerization domain protein 9) |
| 160 | NACC2 | Q96BF6 | Nucleus accumbens-associated protein 2 (NAC-2) (BTB/POZ domain-containing protein 14A) (Repressor with BTB domain and BEN domain) |
| 161 | REXO2 | Q9Y3B8 | Oligoribonuclease, mitochondrial (EC 3.1.-.-) (RNA exonuclease 2 homolog) (Small fragment nuclease) |
| 162 | OAT | P04181 | Ornithine aminotransferase, mitochondrial (EC 2.6.1.13) (Ornithine delta-aminotransferase) (Ornithine–oxo-acid aminotransferase) [Cleaved into: Ornithine aminotransferase, hepatic form; Ornithine aminotransferase, renal form] |
| 163 | PAX9 | P55771 | Paired box protein Pax-9 |
| 164 | Q9HBH1 | Peptide deformylase, mitochondrial (EC 3.5.1.88) (Polypeptide deformylase) | |
| 165 | FKBP8 | Q14318 | Peptidyl-prolyl cis–trans isomerase FKBP8 (PPIase FKBP8) (EC 5.2.1.8) (38 kDa FK506-binding protein) (38 kDa FKBP) (FKBP-38) (hFKBP38) (FK506-binding protein 8) (FKBP-8) (FKBPR38) (Rotamase) |
| 166 | MRPL58 | Q14197 | Peptidyl-tRNA hydrolase ICT1, mitochondrial (EC 3.1.1.29) (39S ribosomal protein L58, mitochondrial) (MRP-L58) (Digestion substraction 1) (DS-1) (Immature colon carcinoma transcript 1 protein) (Mitochondrial large ribosomal subunit protein mL62) |
| 167 | GPX4 | P36969 | Phospholipid hydroperoxide glutathione peroxidase (PHGPx) (EC 1.11.1.12) (Glutathione peroxidase 4) (GPx-4) (GSHPx-4) |
| 168 | PIWIL4 | Q7Z3Z4 | Piwi-like protein 4 |
| 169 | PCBP3 | P57721 | Poly(rC)-binding protein 3 (Alpha-CP3) (PCBP3-overlapping transcript) (PCBP3-overlapping transcript 1) |
| 170 | PNPT1 | Q8TCS8 | Polyribonucleotide nucleotidyltransferase 1, mitochondrial (EC 2.7.7.8) (3'-5' RNA exonuclease OLD35) (PNPase old-35) (Polynucleotide phosphorylase 1) (PNPase 1) (Polynucleotide phosphorylase-like protein) |
| 171 | KCNH3 | Q9ULD8 | Potassium voltage-gated channel subfamily H member 3 (Brain-specific eag-like channel 1) (BEC1) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Voltage-gated potassium channel subunit Kv12.2) |
| 172 | PCYOX1L | Q8NBM8 | Prenylcysteine oxidase-like (EC 1.8.3.-) |
| 173 | DPY19L2 | Q6NUT2 | Probable C-mannosyltransferase DPY19L2 (EC 2.4.1.-) (Dpy-19-like protein 2) (Protein dpy-19 homolog 2) |
| 174 | CARS2 | Q9HA77 | Probable cysteine–tRNA ligase, mitochondrial (EC 6.1.1.16) (Cysteinyl-tRNA synthetase) (CysRS) |
| 175 | EARS2 | Q5JPH6 | Probable glutamate–tRNA ligase, mitochondrial (EC 6.1.1.17) (Glutamyl-tRNA synthetase) (GluRS) |
| 176 | LARS2 | Q15031 | Probable leucine–tRNA ligase, mitochondrial (EC 6.1.1.4) (Leucyl-tRNA synthetase) (LeuRS) |
| 177 | PSTPIP2 | Q9H939 | Proline-serine-threonine phosphatase-interacting protein 2 (PEST phosphatase-interacting protein 2) |
| 178 | PSMG4 | Q5JS54 | Proteasome assembly chaperone 4 (PAC-4) (hPAC4) |
| 179 | SELENOO | Q9BVL4 | Protein adenylyltransferase SelO, mitochondrial (EC 2.7.7.-) (EC 2.7.7.n1) (Selenoprotein O) (SelO) |
| 180 | ATOH1 | Q92858 | Protein atonal homolog 1 (Class A basic helix-loop-helix protein 14) (bHLHa14) (Helix-loop-helix protein hATH-1) (hATH1) |
| 181 | FAM171B | Q6P995 | Protein FAM171B |
| 182 | FAM181B | A6NEQ2 | Protein FAM181B |
| 183 | FAM234A | Q9H0X4 | Protein FAM234A (Protein ITFG3) |
| 184 | FAM83F | Q8NEG4 | Protein FAM83F |
| 185 | JARID2 | Q92833 | Protein Jumonji (Jumonji/ARID domain-containing protein 2) |
| 186 | CCDC58 | Q4VC31 | Protein MIX23 (Coiled-coil domain-containing protein 58) |
| 187 | SCO2 | O43819 | Protein SCO2 homolog, mitochondrial |
| 188 | SCO2 | O43819 | Protein SCO2 homolog, mitochondrial |
| 189 | PCMTD2 | Q9NV79 | Protein-l-isoaspartate O-methyltransferase domain-containing protein 2 |
| 190 | PPOX | P50336 | Protoporphyrinogen oxidase (PPO) (EC 1.3.3.4) |
| 191 | PDK3 | Q15120 | Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 3, mitochondrial (EC 2.7.11.2) (Pyruvate dehydrogenase kinase isoform 3 |
| 192 | PDHB | P11177 | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial (PDHE1-B) (EC 1.2.4.1) |
| 193 | RAB38 | P57729 | Ras-related protein Rab-38 (Melanoma antigen NY-MEL-1) |
| 194 | RIMS3 | Q9UJD0 | Regulating synaptic membrane exocytosis protein 3 (Nim3) (RIM3 gamma) (Rab-3-interacting molecule 3) (RIM 3) |
| 195 | RMDN3 | Q96TC7 | Regulator of microtubule dynamics protein 3 (RMD-3) (hRMD-3) (Cerebral protein 10) (Protein FAM82A2) (Protein FAM82C) (Protein tyrosine phosphatase-interacting protein 51) (TCPTP-interacting protein 51) |
| 196 | RTKN | Q9BST9 | Rhotekin |
| 197 | RPS6KA6 | Q9UK32 | Ribosomal protein S6 kinase alpha-6 (S6K-alpha-6) (EC 2.7.11.1) (90 kDa ribosomal protein S6 kinase 6) (p90-RSK 6) (p90RSK6) (Ribosomal S6 kinase 4) (RSK-4) (pp90RSK4) |
| 198 | GFM2 | Q969S9 | Ribosome-releasing factor 2, mitochondrial (RRF2mt) (Elongation factor G 2, mitochondrial) (EF-G2mt) (mEF-G 2) (Elongation factor G2) (hEFG2) |
| 199 | SHMT2 | P34897 | Serine hydroxymethyltransferase, mitochondrial (SHMT) (EC 2.1.2.1) (Glycine hydroxymethyltransferase) (Serine methylase) |
| 200 | ANKRD44 | Q8N8A2 | Serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B (PP6-ARS-B) (Serine/threonine-protein phosphatase 6 regulatory subunit ARS-B) (Ankyrin repeat domain-containing protein 44) |
| 201 | ANKRD52 | Q8NB46 | Serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit C (PP6-ARS-C) (Serine/threonine-protein phosphatase 6 regulatory subunit ARS-C) (Ankyrin repeat domain-containing protein 52) |
| 202 | SARS2 | Q9NP81 | Serine–tRNA ligase, mitochondrial (EC 6.1.1.11) (SerRSmt) (Seryl-tRNA synthetase) (SerRS) (Seryl-tRNA(Ser/Sec) synthetase) |
| 203 | DHRS3 | O75911 | Short-chain dehydrogenase/reductase 3 (EC 1.1.1.300) (DD83.1) (Retinal short-chain dehydrogenase/reductase 1) (retSDR1) (Retinol dehydrogenase 17) (Short chain dehydrogenase/reductase family 16C member 1) |
| 204 | ACADS | P16219 | Short-chain specific acyl-CoA dehydrogenase, mitochondrial (SCAD) (EC 1.3.8.1) (Butyryl-CoA dehydrogenase) |
| 205 | SLC6A13 | Q9NSD5 | Sodium- and chloride-dependent GABA transporter 2 (GAT-2) (Solute carrier family 6 member 13) |
| 206 | SPATA2L | Q8IUW3 | Spermatogenesis-associated protein 2-like protein (SPATA2-like protein) |
| 207 | CYP27A1 | Q02318 | Sterol 26-hydroxylase, mitochondrial (EC 1.14.15.15) (5-beta-cholestane-3-alpha,7-alpha,12-alpha-triol 26-hydroxylase) (Cytochrome P-450C27/25) (Cytochrome P450 27) (Sterol 27-hydroxylase) (Vitamin D(3) 25-hydroxylase) |
| 208 | SDHA | P31040 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial (EC 1.3.5.1) (Flavoprotein subunit of complex II) (Fp) |
| 209 | SDHB | P21912 | Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial (EC 1.3.5.1) (Iron-sulfur subunit of complex II) (Ip) |
| 210 | SUCLG1 | P53597 | Succinate–CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial (EC 6.2.1.4) (EC 6.2.1.5) (Succinyl-CoA synthetase subunit alpha) (SCS-alpha) |
| 211 | SUCLG2 | Q96I99 | Succinate–CoA ligase [GDP-forming] subunit beta, mitochondrial (EC 6.2.1.4) (GTP-specific succinyl-CoA synthetase subunit beta) (G-SCS) (GTPSCS) (Succinyl-CoA synthetase beta-G chain) (SCS-betaG) |
| 212 | SDC3 | O75056 | Syndecan-3 (SYND3) |
| 213 | TTC27 | Q6P3X3 | Tetratricopeptide repeat protein 27 (TPR repeat protein 27) |
| 214 | TTC28 | Q96AY4 | Tetratricopeptide repeat protein 28 (TPR repeat protein 28) (TPR repeat-containing big gene cloned at Keio) |
| 215 | TTC9B | Q8N6N2 | Tetratricopeptide repeat protein 9B (TPR repeat protein 9B) |
| 216 | TRAF6 | Q9Y4K3 | TNF receptor-associated factor 6 (EC 2.3.2.27) (E3 ubiquitin-protein ligase TRAF6) (Interleukin-1 signal transducer) (RING finger protein 85) (RING-type E3 ubiquitin transferase TRAF6) |
| 217 | TLR2 | O60603 | Toll-like receptor 2 (EC 3.2.2.6) (Toll/interleukin-1 receptor-like protein 4) (CD antigen CD282) |
| 218 | TRABD | Q9H4I3 | TraB domain-containing protein (Protein TTG2) |
| 219 | ENY2 | Q9NPA8 | Transcription and mRNA export factor ENY2 (Enhancer of yellow 2 transcription factor homolog) |
| 220 | TFAP4 | Q01664 | Transcription factor AP-4 (Activating enhancer-binding protein 4) (Class C basic helix-loop-helix protein 41) (bHLHc41) |
| 221 | GUF1 | Q8N442 | Translation factor GUF1, mitochondrial (EC 3.6.5.-) (Elongation factor 4 homolog) (EF-4) (GTPase GUF1) (Ribosomal back-translocase) |
| 222 | MTIF3 | Q9H2K0 | Translation initiation factor IF-3, mitochondrial (IF-3(Mt)) (IF-3Mt) (IF3(mt)) (IF3mt) |
| 223 | TMEM141 | Q96I45 | Transmembrane protein 141 |
| 224 | TMEM70 | Q9BUB7 | Transmembrane protein 70, mitochondrial |
| 225 | TMPPE | Q6ZT21 | Transmembrane protein with metallophosphoesterase domain (EC 3.1.-.-) |
| 226 | TNRC18 | O15417 | Trinucleotide repeat-containing gene 18 protein (Long CAG trinucleotide repeat-containing gene 79 protein) |
| 227 | TRMT10C | Q7L0Y3 | tRNA methyltransferase 10 homolog C (HBV pre-S2 trans-regulated protein 2) (Mitochondrial ribonuclease P protein 1) (Mitochondrial RNase P protein 1) (RNA (guanine-9-)-methyltransferase domain-containing protein 1) (Renal carcinoma antigen NY-REN-49) (mRNA methyladenosine-N(1)-methyltransferase) (EC 2.1.1.-) (tRNA (adenine(9)-N(1))-methyltransferase) (EC 2.1.1.218) (tRNA (guanine(9)-N(1))-methyltransferase) (EC 2.1.1.221) |
| 228 | PUS1 | Q9Y606 | tRNA pseudouridine synthase A (EC 5.4.99.12) (tRNA pseudouridine(38–40) synthase) (tRNA pseudouridylate synthase I) (tRNA-uridine isomerase I) |
| 229 | TNFRSF19 | Q9NS68 | Tumor necrosis factor receptor superfamily member 19 (TRADE) (Toxicity and JNK inducer) |
| 230 | YARS2 | Q9Y2Z4 | Tyrosine–tRNA ligase, mitochondrial (EC 6.1.1.1) (Tyrosyl-tRNA synthetase) (TyrRS) |
| 231 | OTULIN | Q96BN8 | Ubiquitin thioesterase otulin (EC 3.4.19.12) (Deubiquitinating enzyme otulin) (OTU domain-containing deubiquitinase with linear linkage specificity) (Ubiquitin thioesterase Gumby) |
| 232 | UBE3C | Q15386 | Ubiquitin-protein ligase E3C (EC 2.3.2.26) (HECT-type ubiquitin transferase E3C) (HectH2) |
| 233 | CMPK2 | Q5EBM0 | UMP-CMP kinase 2, mitochondrial (EC 2.7.4.14) (Nucleoside-diphosphate kinase) (EC 2.7.4.6) |
| 234 | C7orf31 | Q8N865 | Uncharacterized protein C7orf31 |
| 235 | KIAA0930 | Q6ICG6 | Uncharacterized protein KIAA0930 |
| 236 | VASN | Q6EMK4 | Vasorin (Protein slit-like 2) |
| 237 | ACADVL | P49748 | Very long-chain specific acyl-CoA dehydrogenase, mitochondrial (VLCAD) (EC 1.3.8.9) |
| 238 | VWA5B1 | Q5TIE3 | von Willebrand factor A domain-containing protein 5B1 |
| 239 | XPNPEP3 | Q9NQH7 | Xaa-Pro aminopeptidase 3 (X-Pro aminopeptidase 3) (EC 3.4.11.9) (Aminopeptidase P3) (APP3) |
| 240 | ZCCHC24 | Q8N2G6 | Zinc finger CCHC domain-containing protein 24 |
| 241 | ZNF428 | Q96B54 | Zinc finger protein 428 (Enzyme-like protein PIT13) |
| 242 | ZNF625 | Q96I27 | Zinc finger protein 625 |
| 243 | ZNF782 | Q6ZMW2 | Zinc finger protein 782 |
| 244 | HKR1 | P10072 | Zinc finger protein 875 (Krueppel-related zinc finger protein 1) (Protein HKR1) |
Remarkably, both EVs and mitochondria are acknowledged to be the crucial players in kidney stone formation [25, 130, 131, 148, 159–161]. As such, using anti-oxidants and/or other means of preservation of mitochondrial functions are expected to be one of the ideal strategies for KSD prevention [129, 162–166]. Although mitochondrial dynamics and mitophagy have been investigated and proposed as the main processes in the MQC system in many diseases [167–170], their roles in KSD remain underinvestigated [171]. Interestingly, MDVs have been demonstrated as the novel key player in the MQC system that is the main mechanism for mitochondrial homeostasis and mitochondrial stress response in several diseases, including kidney disorders [5, 171]. Recent studies of MDVs have demonstrated that MDVs can reduce inflammatory response and preserve healthy mitochondria in mild stress, leading to reduction of tissue injury [169–172].
The beneficial roles of MDVs are mediated via the MQC system to place a limit on mitochondrial dysfunction under the normal and mild stress conditions [8, 10, 40]. Also, they are the substitutable machineries to replace the other impaired processes in the MQC system such as mitochondrial dynamics and mitophagy [1, 8, 10, 40, 46]. Thus, the damaged mitochondrial components induced by oxidative stress, including oxidized mtDNA, proteins and lipids, are eradicated from the unhealthy mitochondria by MDVs to restore the healthy mitochondria inside the cells [3, 7, 22, 100, 173]. These processes can further reduce oxidative stress and prevent cell death. Besides, MDVs can remove the excessive mtROS and other proinflammatory molecules that tend to trigger proinflammatory signaling and cytokine production [21, 39, 100]. Therefore, MDVs formation is considered as the rapid and foremost protective response to prevent mitochondrial dysfunction, cell death and tissue inflammation/injury under the oxidative stress condition.
MDVs carry not only the damaged mitochondrial components but also the healthy mitochondrial compartments that can be transferred and released to the unhealthy mitochondria for maintaining cellular functions and survival. Previous studies have demonstrated that MDVs can transport functional mtDNA, mitochondrial matrix, IMM, OMM and fragmented mitochondria to other malfunctioned mitochondria inside the same cell or outside (adjacent cells) [41, 174–179]. Recently, the in vitro synthesis of MDVs has been developed and applied for reduction of cell apoptosis [2, 40, 73]. In the study of myocardial ischemic/hypoxic injury, administration of exogenous (synthetic) MDVs has been demonstrated to serve as the new and effective therapeutic strategy [39, 57]. Most of previous studies have suggested that both intracellular and extracellular MDVs have the protective roles against mitochondrial damage, oxidative stress and tissue/organ injury. Although the clear evidence for the beneficial roles of MDVs in KSD prevention is not currently available, we propose that MDVs would also play such protective role to cope with mitochondrial dysfunction and oxidative stress that are common in KSD (Fig. 2). Therefore, MDVs may serve as the novel therapeutic target to prevent KSD related to mitochondrial dysfunction and oxidative stress as described above.
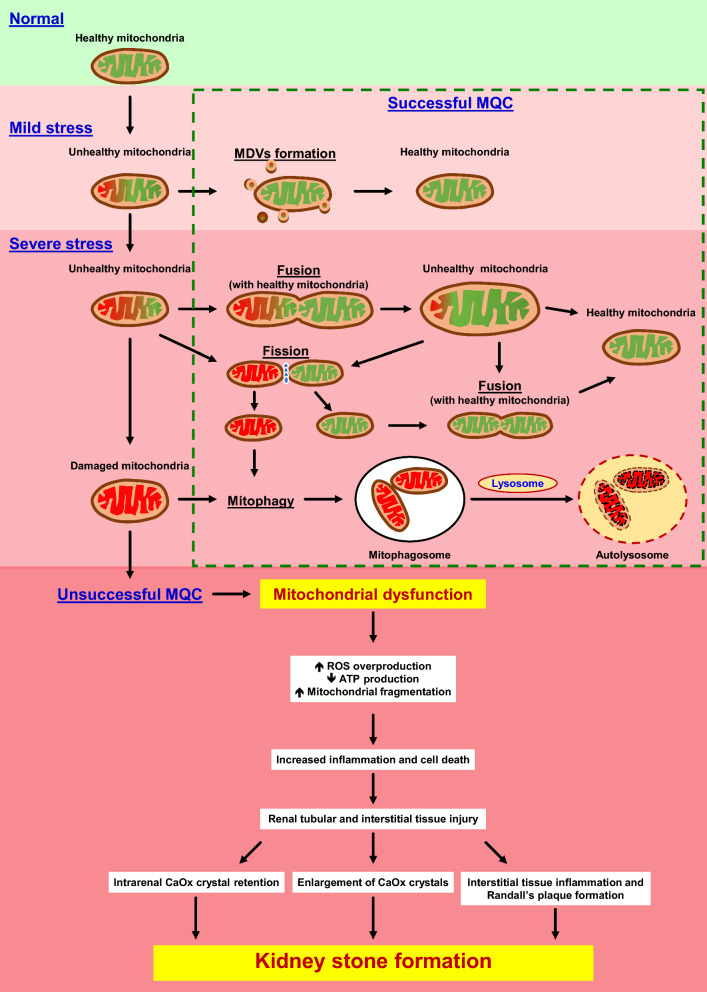
Fig. 2.
Roles of MDVs and MQC system in KSD. At early stage of oxidative stress with mild mitochondrial damage, MDVs (as a part of the MQC system) are formed to eliminate the malfunctioned mitochondrial components. Under severe oxidative stress condition, mitochondrial dynamics (fission/fusion) and mitophagy are predominantly activated to rescue mitochondrial structure and functions. When the MQC system is overwhelmed by extremely severe oxidative stress, mitochondrial dysfunction occurs, leading to ROS overproduction, mitochondrial degradation, inflammation, cell death, and renal tubulointerstitial injury. All these detrimental derangements lead to CaOx crystal deposition, growth, aggregation, nidus formation, Randall’s plaque development and, finally, kidney stone formation
Recently, uEVs serve as the important source for biomarker discovery in several kidney and non-kidney diseases [180–182]. In KSD, a recent proteome study of urinary exosomes has demonstrated greater levels of proteins in S100A family (S100A8, S100A9 and S100A12) in urinary exosomes derived from stone patients compared with those from healthy individuals [183]. Therefore, these exosomal S100A proteins may serve as the biomarkers for KSD. As MDVs share similar proteome and lipidome profiles with EVs, another important role of MDVs in diagnostics and prognostics of KSD should be more extensively investigated.
Conclusions and perspectives
MDVs are one of the most significant players in the MQC system to preserve mitochondrial structure and functions in normal and mild oxidative stress conditions [1, 4, 5, 10]. MDVs are also involved in various diseases, particularly cardiovascular diseases [20, 21] and neurodegenerative disorders [22–24]. In the kidney, the abundance of mitochondria per cell and their functions are critical for maintaining renal tubular cell functions along the nephron. During oxidative stress, mitochondrial dysfunction and tubulointerstitial inflammation occur and induce kidney stone formation [25, 116, 184]. Therefore, mitochondria are the key player in KSD development. The MQC system serves as the central machinery for mitochondrial homeostasis to prevent cell death and tissue injury [5, 6, 172]. MDVs, as the essential compartment of the MQC system [1–4], play the protective roles to rescue the malfunctioned mitochondria during mild stress to preserve their normal structure and functions (Fig. 2).
At early stage of oxidative stress with mild mitochondrial damage, MDVs formation is the rapid and effective process for preserving mitochondrial functions. Under severe oxidative stress condition, mitochondrial dynamics (fission/fusion) and mitophagy are predominantly activated to rescue mitochondrial structure and functions [185–188]. Additionally, MDVs generation can be also triggered as the major MQC machinery to cope with unhealthy mitochondria when mitophagy is unsuccessful for eliminating the damaged mitochondria or mitochondrial fission/fusion fails to recover the mitochondrial structure and functions [1, 8, 10, 40]. When the MQC system is overwhelmed by extremely severe oxidative stress, mitochondrial dysfunction occurs, leading to ROS overproduction, mitochondrial degradation, inflammation, cell death, and renal tubulointerstitial injury. All these detrimental derangements lead to CaOx crystal deposition, growth, aggregation, nidus formation, Randall’s plaque development and, finally, kidney stone formation [25, 129, 146, 147] (Fig. 2).
Nevertheless, the current knowledge on roles of MDVs under physiologic and pathophysiologic conditions remains incomplete. Several advanced methods/techniques have been continuously developed to further clarify the MDVs biology and functions, such as MDVs formation mechanisms, subtypes, specific contents, targets, and diagnostic/therapeutic potential [68, 75, 78, 189]. As MDVs seem to be more dynamic than we initially anticipated, isolation and purification of MDVs also need further development to obtain the specific subtype(s) of the purified MDVs. Differential ultracentrifugation is the primary method for MDVs isolation but still requires further improvement for better yield and higher purity [68, 75]. After isolation, characterizations can be done by morphological examination using high-resolution electron microscopy [19, 68]. To validate MDVs subtypes, proteome and lipidome studies should be performed followed by immunodetection [19, 68].
Recent evidence has demonstrated the therapeutic potential of MDVs in several diseases, including Parkinson’s disease [190], Down syndrome [68], Alzheimer’s disease [191], and myocardial ischemia [39, 192]. Interestingly, the synthetic MDVs have been successfully generated in vitro [39, 193–196]. These synthetic (exogenous) MDVs can be produced by activating the isolated mitochondria by chemical reaction, energy regenerating system, or mild stress-inducing reagents [39, 193]. This technique is therefore promising for further characterizations of MDVs and for developing MDVs-based therapeutic strategies in various diseases, including KSD.
In addition to the therapeutic/preventive potential, MDVs also have a promising role in diagnostics/prognostics of KSD. Future studies on biomarker discovery for KSD should focus on MDVs and their specific types. For example, at an initial phase of kidney stone development with mild stress condition or slight tissue injury, mtDNA and mtROS can be excreted through PDH+-MDVs and transferred to blood circulation and/or urine. Therefore, identification of urinary PDH+-MDVs containing mtDNA or mitochondrial proteins, together with evidence of supersaturation of crystalline compounds in the urine would yield an early biomarker for KSD.
Acknowledgements
Not applicable.
Author contributions
All authors (SC and VT) drafted the manuscript, read and approved the final manuscript, and are responsible for all aspects of the manuscript.
Funding
This study was supported by National Research Council of Thailand (NRCT) and Mahidol University (grant no. N42A650369).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and are also available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare NO competing interests.
Footnotes
The original version of this article was revised: incorrect Funding note. It was: This study was supported by Mahidol University research grant (MU’s Strategic Research Fund). It should be: This study was supported by National Research Council of Thailand (NRCT) and Mahidol University (grant no. N42A650369).
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
7/11/2023
A Correction to this paper has been published: 10.1186/s12967-023-04299-w
References
- 1.Towers CG, Wodetzki DK, Thorburn J, Smith KR, Caino MC, Thorburn A. Mitochondrial-derived vesicles compensate for loss of LC3-mediated mitophagy. Dev Cell. 2021;56(2029–42):e5. doi: 10.1016/j.devcel.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amari L, Germain M. Mitochondrial extracellular vesicles—origins and roles. Front Mol Neurosci. 2021;14:767219. doi: 10.3389/fnmol.2021.767219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Todkar K, Chikhi L, Desjardins V, El-Mortada F, Pepin G, Germain M. Selective packaging of mitochondrial proteins into extracellular vesicles prevents the release of mitochondrial DAMPs. Nat Commun. 2021;12:1971. doi: 10.1038/s41467-021-21984-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choong CJ, Okuno T, Ikenaka K, Baba K, Hayakawa H, Koike M, et al. Alternative mitochondrial quality control mediated by extracellular release. Autophagy. 2021;17:2962–2974. doi: 10.1080/15548627.2020.1848130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng MYW, Wai T, Simonsen A. Quality control of the mitochondrion. Dev Cell. 2021;56:881–905. doi: 10.1016/j.devcel.2021.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Picca A, Calvani R, Coelho-Junior HJ, Marzetti E. Mitophagy: at the heart of mitochondrial quality control in cardiac aging and frailty. Exp Gerontol. 2021;153:111508. doi: 10.1016/j.exger.2021.111508. [DOI] [PubMed] [Google Scholar]
- 7.Sugiura A, McLelland GL, Fon EA, McBride HM. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 2014;33:2142–2156. doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng T, Xie Y, Sheng H, Wang C, Lian Y, Xie N. Mitochondrial-derived vesicles: gatekeepers of mitochondrial response to oxidative stress. Free Radic Biol Med. 2022;188:185–193. doi: 10.1016/j.freeradbiomed.2022.06.233. [DOI] [PubMed] [Google Scholar]
- 9.Towers CG. Mitochondrial homeostasis is maintained in the absence of autophagy. Mol Cell Oncol. 2021;8:1984162. doi: 10.1080/23723556.2021.1984162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mondal P, Towers C. Beyond mitophagy: mitochondrial-derived vesicles can get the job done! Autophagy. 2022;18:449–451. doi: 10.1080/15548627.2021.1999562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts RF, Tang MY, Fon EA, Durcan TM. Defending the mitochondria: the pathways of mitophagy and mitochondrial-derived vesicles. Int J Biochem Cell Biol. 2016;79:427–436. doi: 10.1016/j.biocel.2016.07.020. [DOI] [PubMed] [Google Scholar]
- 12.Long G, Gong R, Wang Q, Zhang D, Huang C. Role of released mitochondrial DNA in acute lung injury. Front Immunol. 2022;13:973089. doi: 10.3389/fimmu.2022.973089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li YJ, Liu RP, Ding MN, Zheng Q, Wu JZ, Xue XY, et al. Tetramethylpyrazine prevents liver fibrotic injury in mice by targeting hepatocyte-derived and mitochondrial DNA-enriched extracellular vesicles. Acta Pharmacol Sin. 2022;43:2026–2041. doi: 10.1038/s41401-021-00843-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen M, Fan X, Shen Y, Wang X, Wu R, Wang Y, et al. Myeloid-derived suppressor cells ameliorate liver mitochondrial damage to protect against autoimmune hepatitis by releasing small extracellular vesicles. Int Immunopharmacol. 2022;114:109540. doi: 10.1016/j.intimp.2022.109540. [DOI] [PubMed] [Google Scholar]
- 15.Vikramdeo KS, Anand S, Khan MA, Khushman M, Heslin MJ, Singh S, et al. Detection of mitochondrial DNA mutations in circulating mitochondria-originated extracellular vesicles for potential diagnostic applications in pancreatic adenocarcinoma. Sci Rep. 2022;12:18455. doi: 10.1038/s41598-022-22006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poillet-Perez L, White E. MDVs to the rescue: how autophagy-deficient cancer cells adapt to defective mitophagy. Dev Cell. 2021;56:2010–2012. doi: 10.1016/j.devcel.2021.06.022. [DOI] [PubMed] [Google Scholar]
- 17.Sen A, Kallabis S, Gaedke F, Jungst C, Boix J, Nuchel J, et al. Mitochondrial membrane proteins and VPS35 orchestrate selective removal of mtDNA. Nat Commun. 2022;13:6704. doi: 10.1038/s41467-022-34205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leduc-Gaudet JP, Hussain SN, Gouspillou G. Parkin: a potential target to promote healthy ageing. J Physiol. 2022;600:3405–3421. doi: 10.1113/JP282567. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y, Perez-Gonzalez R, Miller C, Kurz M, D'Acunzo P, Goulbourne CN, et al. Sex Differentially alters secretion of brain extracellular vesicles during aging: a potential mechanism for maintaining brain homeostasis. Neurochem Res. 2022;47:3428–3439. doi: 10.1007/s11064-022-03701-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Opstad IS, Godtliebsen G, Strohl F, Myrmel T, Ahluwalia BS, Agarwal K, et al. Three-dimensional structured illumination microscopy data of mitochondria and lysosomes in cardiomyoblasts under normal and galactose-adapted conditions. Sci Data. 2022;9:98. doi: 10.1038/s41597-022-01207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cadete VJ, Deschenes S, Cuillerier A, Brisebois F, Sugiura A, Vincent A, et al. Formation of mitochondrial-derived vesicles is an active and physiologically relevant mitochondrial quality control process in the cardiac system. J Physiol. 2016;594:5343–5362. doi: 10.1113/JP272703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts RF, Bayne AN, Goiran T, Levesque D, Boisvert FM, Trempe JF, et al. Proteomic profiling of mitochondrial-derived vesicles in brain reveals enrichment of respiratory complex sub-assemblies and small TIM chaperones. J Proteome Res. 2021;20:506–517. doi: 10.1021/acs.jproteome.0c00506. [DOI] [PubMed] [Google Scholar]
- 23.Thorne NJ, Tumbarello DA. The relationship of alpha-synuclein to mitochondrial dynamics and quality control. Front Mol Neurosci. 2022;15:947191. doi: 10.3389/fnmol.2022.947191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Acunzo P, Kim Y, Ungania JM, Perez-Gonzalez R, Goulbourne CN, Levy E. Isolation of mitochondria-derived mitovesicles and subpopulations of microvesicles and exosomes from brain tissues. Nat Protoc. 2022;17:2517–2549. doi: 10.1038/s41596-022-00719-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaiyarit S, Thongboonkerd V. Mitochondrial dysfunction and kidney stone disease. Front Physiol. 2020;11:566506. doi: 10.3389/fphys.2020.566506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaiyarit S, Thongboonkerd V. Changes in mitochondrial proteome of renal tubular cells induced by calcium oxalate monohydrate crystal adhesion and internalization are related to mitochondrial dysfunction. J Proteome Res. 2012;11:3269–3280. doi: 10.1021/pr300018c. [DOI] [PubMed] [Google Scholar]
- 27.Sharma M, Naura AS, Singla SK. A deleterious interplay between endoplasmic reticulum stress and its functional linkage to mitochondria in nephrolithiasis. Free Radic Biol Med. 2021;168:70–80. doi: 10.1016/j.freeradbiomed.2021.03.031. [DOI] [PubMed] [Google Scholar]
- 28.Deng J, Yu B, Chang Z, Wu S, Li G, Chen W, et al. Cerium oxide-based nanozyme suppresses kidney calcium oxalate crystal depositions via reversing hyperoxaluria-induced oxidative stress damage. J Nanobiotechnol. 2022;20:516. doi: 10.1186/s12951-022-01726-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmatjan B, Ruotian L, Rahman A, Bin M, Heng D, Yi H, et al. Klotho inhibits the formation of calcium oxalate stones by regulating the Keap1-Nrf2-ARE signaling pathway. Int Urol Nephrol. 2022;55:263. doi: 10.1007/s11255-022-03398-9. [DOI] [PubMed] [Google Scholar]
- 30.Lewis WH, Ettema TJG. A microbial marriage reminiscent of mitochondrial evolution. Nature. 2021;591:375–376. doi: 10.1038/d41586-021-00500-6. [DOI] [PubMed] [Google Scholar]
- 31.Burki F. Mitochondrial evolution: going, going. Gone Curr Biol. 2016;26:R410–R412. doi: 10.1016/j.cub.2016.04.032. [DOI] [PubMed] [Google Scholar]
- 32.Konig T, Nolte H, Aaltonen MJ, Tatsuta T, Krols M, Stroh T, et al. MIROs and DRP1 drive mitochondrial-derived vesicle biogenesis and promote quality control. Nat Cell Biol. 2021;23:1271–1286. doi: 10.1038/s41556-021-00798-4. [DOI] [PubMed] [Google Scholar]
- 33.Lemasters JJ, Zhong Z. Mitophagy in hepatocytes: types, initiators and role in adaptive ethanol metabolism☆. Liver Res. 2018;2:125–132. doi: 10.1016/j.livres.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemasters JJ. Variants of mitochondrial autophagy: types 1 and 2 mitophagy and micromitophagy (Type 3) Redox Biol. 2014;2:749–754. doi: 10.1016/j.redox.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bao F, Zhou L, Zhou R, Huang Q, Chen J, Zeng S, et al. Mitolysosome exocytosis, a mitophagy-independent mitochondrial quality control in flunarizine-induced parkinsonism-like symptoms. Sci Adv. 2022;8:eabk2376. doi: 10.1126/sciadv.abk2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu CT. Multiple pathways for mitophagy: a neurodegenerative conundrum for Parkinson's disease. Neurosci Lett. 2019;697:66–71. doi: 10.1016/j.neulet.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teresak P, Lapao A, Subic N, Boya P, Elazar Z, Simonsen A. Regulation of PRKN-independent mitophagy. Autophagy. 2022;18:24–39. doi: 10.1080/15548627.2021.1888244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picca A, Guerra F, Calvani R, Romano R, Coelho-Junior HJ, Bucci C, et al. Mitochondrial-derived vesicles in skeletal muscle remodeling and adaptation. Semin Cell Dev Biol. 2022;143:37. doi: 10.1016/j.semcdb.2022.03.023. [DOI] [PubMed] [Google Scholar]
- 39.Li B, Zhao H, Wu Y, Zhu Y, Zhang J, Yang G, et al. Mitochondrial-derived vesicles protect cardiomyocytes against hypoxic damage. Front Cell Dev Biol. 2020;8:214. doi: 10.3389/fcell.2020.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popov LD. Mitochondrial-derived vesicles: recent insights. J Cell Mol Med. 2022;26:3323–3328. doi: 10.1111/jcmm.17391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takenaga K, Koshikawa N, Nagase H. Intercellular transfer of mitochondrial DNA carrying metastasis-enhancing pathogenic mutations from high- to low-metastatic tumor cells and stromal cells via extracellular vesicles. BMC Mol Cell Biol. 2021;22:52. doi: 10.1186/s12860-021-00391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lazo S, Noren Hooten N, Green J, Eitan E, Mode NA, Liu QR, et al. Mitochondrial DNA in extracellular vesicles declines with age. Aging Cell. 2021;20:e13283. doi: 10.1111/acel.13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, Weidling I, Koppel S, Menta B, Perez Ortiz J, Kalani A, et al. Detection of mitochondria-pertinent components in exosomes. Mitochondrion. 2020;55:100–110. doi: 10.1016/j.mito.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLelland GL, Lee SA, McBride HM, Fon EA. Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J Cell Biol. 2016;214:275–291. doi: 10.1083/jcb.201603105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, Berkowicz A, King K, Menta B, Gabrielli AP, Novikova L, et al. Pharmacologic enrichment of exosome yields and mitochondrial cargo. Mitochondrion. 2022;64:136–144. doi: 10.1016/j.mito.2022.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vasam G, Nadeau R, Cadete VJJ, Lavallee-Adam M, Menzies KJ, Burelle Y. Proteomics characterization of mitochondrial-derived vesicles under oxidative stress. FASEB J. 2021;35:e21278. doi: 10.1096/fj.202002151R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryan TA, Phillips EO, Collier CL, Jb Robinson A, Routledge D, Wood RE, et al. Tollip coordinates Parkin-dependent trafficking of mitochondrial-derived vesicles. EMBO J. 2020;39:e102539. doi: 10.15252/embj.2019102539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts RF, Fon EA. Presenting mitochondrial antigens: PINK1, Parkin and MDVs steal the show. Cell Res. 2016;26:1180–1181. doi: 10.1038/cr.2016.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manickam DS. Delivery of mitochondria via extracellular vesicles—a new horizon in drug delivery. J Control Release. 2022;343:400–407. doi: 10.1016/j.jconrel.2022.01.045. [DOI] [PubMed] [Google Scholar]
- 50.Al Amir Dache Z, Otandault A, Tanos R, Pastor B, Meddeb R, Sanchez C, et al. Blood contains circulating cell-free respiratory competent mitochondria. FASEB J. 2020;34:3616–3630. doi: 10.1096/fj.201901917RR. [DOI] [PubMed] [Google Scholar]
- 51.Velarde F, Ezquerra S, Delbruyere X, Caicedo A, Hidalgo Y, Khoury M. Mesenchymal stem cell-mediated transfer of mitochondria: mechanisms and functional impact. Cell Mol Life Sci. 2022;79:177. doi: 10.1007/s00018-022-04207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soubannier V, McLelland GL, Zunino R, Braschi E, Rippstein P, Fon EA, et al. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol. 2012;22:135–141. doi: 10.1016/j.cub.2011.11.057. [DOI] [PubMed] [Google Scholar]
- 53.Mohanty A, Zunino R, Soubannier V, Dilipkumar S. A new functional role of mitochondria-anchored protein ligase in peroxisome morphology in mammalian cells. J Cell Biochem. 2021;122:1686–1700. doi: 10.1002/jcb.30114. [DOI] [PubMed] [Google Scholar]
- 54.Rosina M, Ceci V, Turchi R, Chuan L, Borcherding N, Sciarretta F, et al. Ejection of damaged mitochondria and their removal by macrophages ensure efficient thermogenesis in brown adipose tissue. Cell Metab. 2022;34(533–48):e12. doi: 10.1016/j.cmet.2022.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D'Souza A, Burch A, Dave KM, Sreeram A, Reynolds MJ, Dobbins DX, et al. Microvesicles transfer mitochondria and increase mitochondrial function in brain endothelial cells. J Control Release. 2021;338:505–526. doi: 10.1016/j.jconrel.2021.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dave KM, Zhao W, Hoover C, D’Souza A, Manickam DS. Extracellular vesicles derived from a human brain endothelial cell line increase cellular ATP levels. AAPS PharmSciTech. 2021;22:18. doi: 10.1208/s12249-020-01892-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ikeda G, Santoso MR, Tada Y, Li AM, Vaskova E, Jung JH, et al. Mitochondria-rich extracellular vesicles from autologous stem cell-derived cardiomyocytes restore energetics of ischemic myocardium. J Am Coll Cardiol. 2021;77:1073–1088. doi: 10.1016/j.jacc.2020.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morrison TJ, Jackson MV, Cunningham EK, Kissenpfennig A, McAuley DF, O'Kane CM, et al. Mesenchymal stromal cells modulate macrophages in clinically relevant lung injury models by extracellular vesicle mitochondrial transfer. Am J Respir Crit Care Med. 2017;196:1275–1286. doi: 10.1164/rccm.201701-0170OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mao J, Li C, Wu F, She Z, Luo S, Chen X, et al. MSC-EVs transferring mitochondria and related components: a new hope for the treatment of kidney disease. Front Immunol. 2022;13:978571. doi: 10.3389/fimmu.2022.978571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao PJ, Eren E, Petralia RS, Gu JW, Wang YX, Kapogiannis D. Mitochondrial protrusions in neuronal cells. iScience. 2020;23:101514. doi: 10.1016/j.isci.2020.101514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Howard M, Erickson J, Cuba Z, Kim S, Zhou W, Gade P, et al. A secretory form of Parkin-independent mitophagy contributes to the repertoire of extracellular vesicles released into the tumour interstitial fluid in vivo. J Extracell Vesicles. 2022;11:e12244. doi: 10.1002/jev2.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramirez A, Old W, Selwood DL, Liu X. Cannabidiol activates PINK1-Parkin-dependent mitophagy and mitochondrial-derived vesicles. Eur J Cell Biol. 2022;101:151185. doi: 10.1016/j.ejcb.2021.151185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neuspiel M, Schauss AC, Braschi E, Zunino R, Rippstein P, Rachubinski RA, et al. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18:102–108. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 65.Matheoud D, Sugiura A, Bellemare-Pelletier A, Laplante A, Rondeau C, Chemali M, et al. Parkinson's disease-related proteins PINK1 and Parkin repress mitochondrial antigen presentation. Cell. 2016;166:314–327. doi: 10.1016/j.cell.2016.05.039. [DOI] [PubMed] [Google Scholar]
- 66.Dussouchaud A, Jacob J, Secq C, Verbavatz JM, Moras M, Larghero J, et al. Transmission electron microscopy to follow ultrastructural modifications of erythroblasts upon ex vivo human erythropoiesis. Front Physiol. 2021;12:791691. doi: 10.3389/fphys.2021.791691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pergande MR, Kang C, George D, Sutter PA, Crocker SJ, Cologna SM, et al. Lipidomic analysis identifies age-disease-related changes and potential new biomarkers in brain-derived extracellular vesicles from metachromatic leukodystrophy mice. Lipids Health Dis. 2022;21:32. doi: 10.1186/s12944-022-01644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.D'Acunzo P, Perez-Gonzalez R, Kim Y, Hargash T, Miller C, Alldred MJ, et al. Mitovesicles are a novel population of extracellular vesicles of mitochondrial origin altered in Down syndrome. Sci Adv. 2021;7:eabe5085. doi: 10.1126/sciadv.abe5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Picca A, Guerra F, Calvani R, Coelho-Junior HJ, Landi F, Bernabei R, et al. Extracellular vesicles and damage-associated molecular patterns: a Pandora's box in health and disease. Front Immunol. 2020;11:601740. doi: 10.3389/fimmu.2020.601740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.D'Acunzo P, Perez-Gonzalez R, Kim Y, Hargash T, Miller C, Alldred MJ, et al. Mitovesicles are a novel population of extracellular vesicles of mitochondrial origin altered in Down syndrome. Sci Adv. 2021;7:eabe5085. doi: 10.1126/sciadv.abe5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Picca A, Guerra F, Calvani R, Bucci C, Lo Monaco MR, Bentivoglio AR, et al. Mitochondrial dysfunction and aging: insights from the analysis of extracellular vesicles. Int J Mol Sci. 2019;20:805. doi: 10.3390/ijms20040805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Picca A, Guerra F, Calvani R, Marini F, Biancolillo A, Landi G, et al. Mitochondrial signatures in circulating extracellular vesicles of older adults with Parkinson's disease: results from the EXosomes in PArkiNson's Disease (EXPAND) Study. J Clin Med. 2020;9:504. doi: 10.3390/jcm9020504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zorova LD, Kovalchuk SI, Popkov VA, Chernikov VP, Zharikova AA, Khutornenko AA, et al. Do extracellular vesicles derived from mesenchymal stem cells contain functional mitochondria? Int J Mol Sci. 2022;23:7408. doi: 10.3390/ijms23137408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bagci C, Sever-Bahcekapili M, Belder N, Bennett APS, Erdener SE, Dalkara T. Overview of extracellular vesicle characterization techniques and introduction to combined reflectance and fluorescence confocal microscopy to distinguish extracellular vesicle subpopulations. Neurophotonics. 2022;9:021903. doi: 10.1117/1.NPh.9.2.021903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao D, Tao W, Li S, Chen Y, Sun Y, He Z, et al. Apoptotic body-mediated intercellular delivery for enhanced drug penetration and whole tumor destruction. Sci Adv. 2021;7:eabg0880. doi: 10.1126/sciadv.abg0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu L, Awoyemi AA, Fahy KE, Thapa P, Borchers C, Wu BY, et al. Keratinocyte-derived microvesicle particles mediate ultraviolet B radiation-induced systemic immunosuppression. J Clin Invest. 2021;131:e144963. doi: 10.1172/JCI144963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lovisolo F, Carton F, Gino S, Migliario M, Reno F. Photobiomodulation induces microvesicle release in human keratinocytes: PI3 kinase-dependent pathway role. Lasers Med Sci. 2022;37:479–487. doi: 10.1007/s10103-021-03285-2. [DOI] [PubMed] [Google Scholar]
- 79.Liu SL, Sun P, Li Y, Liu SS, Lu Y. Exosomes as critical mediators of cell-to-cell communication in cancer pathogenesis and their potential clinical application. Transl Cancer Res. 2019;8:298–311. doi: 10.21037/tcr.2019.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Noonin C, Thongboonkerd V. Exosome-inflammasome crosstalk and their roles in inflammatory responses. Theranostics. 2021;11:4436–4451. doi: 10.7150/thno.54004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rezaie J, Akbari A, Rahbarghazi R. Inhibition of extracellular vesicle biogenesis in tumor cells: a possible way to reduce tumorigenesis. Cell Biochem Funct. 2022;40:248–262. doi: 10.1002/cbf.3695. [DOI] [PubMed] [Google Scholar]
- 82.Schwager SC, Reinhart-King CA. Mechanobiology of microvesicle release, uptake, and microvesicle-mediated activation. Curr Top Membr. 2020;86:255–278. doi: 10.1016/bs.ctm.2020.08.004. [DOI] [PubMed] [Google Scholar]
- 83.Garner RT, Solfest JS, Nie Y, Kuang S, Stout J, Gavin TP. Multivesicular body and exosome pathway responses to acute exercise. Exp Physiol. 2020;105:511–521. doi: 10.1113/EP088017. [DOI] [PubMed] [Google Scholar]
- 84.Kanemoto S, Nitani R, Murakami T, Kaneko M, Asada R, Matsuhisa K, et al. Multivesicular body formation enhancement and exosome release during endoplasmic reticulum stress. Biochem Biophys Res Commun. 2016;480:166–172. doi: 10.1016/j.bbrc.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 85.Carnino JM, Lee H, Jin Y. Isolation and characterization of extracellular vesicles from Broncho-alveolar lavage fluid: a review and comparison of different methods. Respir Res. 2019;20:240. doi: 10.1186/s12931-019-1210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bano R, Ahmad F, Mohsin M. A perspective on the isolation and characterization of extracellular vesicles from different biofluids. RSC Adv. 2021;11:19598–19615. doi: 10.1039/D1RA01576A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang F, Cerione RA, Antonyak MA. Isolation and characterization of extracellular vesicles produced by cell lines. STAR Protoc. 2021;2:100295. doi: 10.1016/j.xpro.2021.100295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Opstad IS, Godtliebsen G, Ahluwalia BS, Myrmel T, Agarwal K, Birgisdottir AB. Mitochondrial dynamics and quantification of mitochondria-derived vesicles in cardiomyoblasts using structured illumination microscopy. J Biophotonics. 2022;15:e202100305. doi: 10.1002/jbio.202100305. [DOI] [PubMed] [Google Scholar]
- 89.Caielli S, Athale S, Domic B, Murat E, Chandra M, Banchereau R, et al. Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J Exp Med. 2016;213:697–713. doi: 10.1084/jem.20151876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sliter DA, Martinez J, Hao L, Chen X, Sun N, Fischer TD, et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018;561:258–262. doi: 10.1038/s41586-018-0448-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 91.Braschi E, Goyon V, Zunino R, Mohanty A, Xu L, McBride HM. Vps35 mediates vesicle transport between the mitochondria and peroxisomes. Curr Biol. 2010;20:1310–1315. doi: 10.1016/j.cub.2010.05.066. [DOI] [PubMed] [Google Scholar]
- 92.Rucktaschel R, Halbach A, Girzalsky W, Rottensteiner H, Erdmann R. De novo synthesis of peroxisomes upon mitochondrial targeting of Pex3p. Eur J Cell Biol. 2010;89:947–954. doi: 10.1016/j.ejcb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 93.Sugiura A, Mattie S, Prudent J, McBride HM. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature. 2017;542:251–254. doi: 10.1038/nature21375. [DOI] [PubMed] [Google Scholar]
- 94.Cadete VJJ, Vasam G, Menzies KJ, Burelle Y. Mitochondrial quality control in the cardiac system: an integrative view. Biochim Biophys Acta Mol Basis Dis. 2019;1865:782–796. doi: 10.1016/j.bbadis.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 95.Zhang Y, Tan J, Miao Y, Zhang Q. The effect of extracellular vesicles on the regulation of mitochondria under hypoxia. Cell Death Dis. 2021;12:358. doi: 10.1038/s41419-021-03640-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tarnopolsky MA, Kerkhof J, Stuart A, Bujak A, Nilsson MI, Hettinga B, et al. Bone marrow-derived mitochondrial DNA has limited capacity for inter-tissue transfer in vivo. FASEB J. 2020;34:9297–9306. doi: 10.1096/fj.202000463R. [DOI] [PubMed] [Google Scholar]
- 97.Hayakawa K, Esposito E, Wang X, Terasaki Y, Liu Y, Xing C, et al. Transfer of mitochondria from astrocytes to neurons after stroke. Nature. 2016;535:551–555. doi: 10.1038/nature18928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Peruzzotti-Jametti L, Bernstock JD, Willis CM, Manferrari G, Rogall R, Fernandez-Vizarra E, et al. Neural stem cells traffic functional mitochondria via extracellular vesicles. PLoS Biol. 2021;19:e3001166. doi: 10.1371/journal.pbio.3001166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sreekumar PG, Ishikawa K, Spee C, Mehta HH, Wan J, Yen K, et al. The mitochondrial-derived peptide humanin protects RPE cells from oxidative stress, senescence, and mitochondrial dysfunction. Invest Ophthalmol Vis Sci. 2016;57:1238–1253. doi: 10.1167/iovs.15-17053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ryan TA, Tumbarello DA. A central role for mitochondrial-derived vesicles in the innate immune response: implications for Parkinson's disease. Neural Regen Res. 2021;16:1779–1780. doi: 10.4103/1673-5374.306074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zeng X, Li X, Zhang Y, Cao C, Zhou Q. IL6 induces mtDNA leakage to affect the immune escape of endometrial carcinoma via cGAS-STING. J Immunol Res. 2022;2022:3815853. doi: 10.1155/2022/3815853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Picca A, Lezza AMS, Leeuwenburgh C, Pesce V, Calvani R, Bossola M, et al. Circulating mitochondrial DNA at the crossroads of mitochondrial dysfunction and inflammation during aging and muscle wasting disorders. Rejuvenation Res. 2018;21:350–359. doi: 10.1089/rej.2017.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Heid ME, Keyel PA, Kamga C, Shiva S, Watkins SC, Salter RD. Mitochondrial reactive oxygen species induces NLRP3-dependent lysosomal damage and inflammasome activation. J Immunol. 2013;191:5230–5238. doi: 10.4049/jimmunol.1301490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bulua AC, Simon A, Maddipati R, Pelletier M, Park H, Kim KY, et al. Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS) J Exp Med. 2011;208:519–533. doi: 10.1084/jem.20102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hauser CJ, Otterbein LE. Danger signals from mitochondrial DAMPS in trauma and post-injury sepsis. Eur J Trauma Emerg Surg. 2018;44:317–324. doi: 10.1007/s00068-018-0963-2. [DOI] [PubMed] [Google Scholar]
- 107.Dela Cruz CS, Kang MJ. Mitochondrial dysfunction and damage associated molecular patterns (DAMPs) in chronic inflammatory diseases. Mitochondrion. 2018;41:37–44. doi: 10.1016/j.mito.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Simmons JD, Lee YL, Mulekar S, Kuck JL, Brevard SB, Gonzalez RP, et al. Elevated levels of plasma mitochondrial DNA DAMPs are linked to clinical outcome in severely injured human subjects. Ann Surg. 2013;258:591–596. doi: 10.1097/SLA.0b013e3182a4ea46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mazumder S, Bindu S, De R, Debsharma S, Pramanik S, Bandyopadhyay U. Emerging role of mitochondrial DAMPs, aberrant mitochondrial dynamics and anomalous mitophagy in gut mucosal pathogenesis. Life Sci. 2022;305:120753. doi: 10.1016/j.lfs.2022.120753. [DOI] [PubMed] [Google Scholar]
- 110.Garg M, Johri S, Chakraborty K. Immunomodulatory role of mitochondrial DAMPs: a missing link in pathology? FEBS J. 2022. [DOI] [PubMed]
- 111.Xu Y, Shen J, Ran Z. Emerging views of mitophagy in immunity and autoimmune diseases. Autophagy. 2020;16:3–17. doi: 10.1080/15548627.2019.1603547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Torralba D, Baixauli F, Villarroya-Beltri C, Fernandez-Delgado I, Latorre-Pellicer A, Acin-Perez R, et al. Priming of dendritic cells by DNA-containing extracellular vesicles from activated T cells through antigen-driven contacts. Nat Commun. 2018;9:2658. doi: 10.1038/s41467-018-05077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ko JH, Kim HJ, Jeong HJ, Lee HJ, Oh JY. Mesenchymal stem and stromal cells harness macrophage-derived amphiregulin to maintain tissue homeostasis. Cell Rep. 2020;30(3806–20):e6. doi: 10.1016/j.celrep.2020.02.062. [DOI] [PubMed] [Google Scholar]
- 114.Abuaita BH, Schultz TL, O'Riordan MX. Mitochondria-derived vesicles deliver antimicrobial reactive oxygen species to control phagosome-localized Staphylococcus aureus. Cell Host Microbe. 2018;24(625–36):e5. doi: 10.1016/j.chom.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hussain SS, Kashatus DF. MDVs: spare the SOD and spoil the bug. Cell Host Microbe. 2018;24:616–618. doi: 10.1016/j.chom.2018.10.013. [DOI] [PubMed] [Google Scholar]
- 116.Patel M, Yarlagadda V, Adedoyin O, Saini V, Assimos DG, Holmes RP, et al. Oxalate induces mitochondrial dysfunction and disrupts redox homeostasis in a human monocyte derived cell line. Redox Biol. 2018;15:207–215. doi: 10.1016/j.redox.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Williams J, Holmes RP, Assimos DG, Mitchell T. Monocyte mitochondrial function in calcium oxalate stone formers. Urology. 2016;93:224–226. doi: 10.1016/j.urology.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Veena CK, Josephine A, Preetha SP, Rajesh NG, Varalakshmi P. Mitochondrial dysfunction in an animal model of hyperoxaluria: a prophylactic approach with fucoidan. Eur J Pharmacol. 2008;579:330–336. doi: 10.1016/j.ejphar.2007.09.044. [DOI] [PubMed] [Google Scholar]
- 119.Cao LC, Honeyman TW, Cooney R, Kennington L, Scheid CR, Jonassen JA. Mitochondrial dysfunction is a primary event in renal cell oxalate toxicity. Kidney Int. 2004;66:1890–1900. doi: 10.1111/j.1523-1755.2004.00963.x. [DOI] [PubMed] [Google Scholar]
- 120.Forbes JM. Mitochondria-power players in kidney function? Trends Endocrinol Metab. 2016;27:441–442. doi: 10.1016/j.tem.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 121.Forbes MS, Thornhill BA, Galarreta CI, Chevalier RL. A population of mitochondrion-rich cells in the pars recta of mouse kidney. Cell Tissue Res. 2016;363:791–803. doi: 10.1007/s00441-015-2273-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Flemming NB, Gallo LA, Ward MS, Forbes JM. Tapping into mitochondria to find novel targets for diabetes complications. Curr Drug Targets. 2016;17:1341–1349. doi: 10.2174/1389450116666150727114410. [DOI] [PubMed] [Google Scholar]
- 123.Konari N, Nagaishi K, Kikuchi S, Fujimiya M. Mitochondria transfer from mesenchymal stem cells structurally and functionally repairs renal proximal tubular epithelial cells in diabetic nephropathy in vivo. Sci Rep. 2019;9:5184. doi: 10.1038/s41598-019-40163-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mulay SR, Shi C, Ma X, Anders HJ. Novel insights into crystal-induced kidney injury. Kidney Dis (Basel) 2018;4:49–57. doi: 10.1159/000487671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wiener SV, Chen L, Shimotake AR, Kang M, Stoller ML, Ho SP. Novel insights into renal mineralization and stone formation through advanced imaging modalities. Connect Tissue Res. 2018;59:102–110. doi: 10.1080/03008207.2017.1409219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wiener SV, Ho SP, Stoller ML. Beginnings of nephrolithiasis: insights into the past, present and future of Randall's plaque formation research. Curr Opin Nephrol Hypertens. 2018;27:236–242. doi: 10.1097/MNH.0000000000000414. [DOI] [PubMed] [Google Scholar]
- 127.Khan SR. Histological aspects of the "fixed-particle" model of stone formation: animal studies. Urolithiasis. 2017;45:75–87. doi: 10.1007/s00240-016-0949-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hsi RS, Ramaswamy K, Ho SP, Stoller ML. The origins of urinary stone disease: upstream mineral formations initiate downstream Randall's plaque. BJU Int. 2017;119:177–184. doi: 10.1111/bju.13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dominguez-Gutierrez PR, Kwenda EP, Khan SR, Canales BK. Immunotherapy for stone disease. Curr Opin Urol. 2020;30:183–189. doi: 10.1097/MOU.0000000000000729. [DOI] [PubMed] [Google Scholar]
- 130.Peerapen P, Thongboonkerd V. Kidney stone proteomics: an update and perspectives. Expert Rev Proteomics. 2021;18:557–569. doi: 10.1080/14789450.2021.1962301. [DOI] [PubMed] [Google Scholar]
- 131.Peerapen P, Chaiyarit S, Thongboonkerd V. Protein network analysis and functional studies of calcium oxalate crystal-induced cytotoxicity in renal tubular epithelial cells. Proteomics. 2018;18:e1800008. doi: 10.1002/pmic.201800008. [DOI] [PubMed] [Google Scholar]
- 132.Vinaiphat A, Aluksanasuwan S, Manissorn J, Sutthimethakorn S, Thongboonkerd V. Response of renal tubular cells to differential types and doses of calcium oxalate crystals: integrative proteome network analysis and functional investigations. Proteomics. 2017;17:1700192. doi: 10.1002/pmic.201700192. [DOI] [PubMed] [Google Scholar]
- 133.Qian X, Wu W, Hu H, Yu X, Wang S, Zhu J, et al. The role of reactive oxygen species derived from different NADPH oxidase isoforms and mitochondria in oxalate-induced oxidative stress and cell injury. Urolithiasis. 2022;50:149–158. doi: 10.1007/s00240-022-01309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Joshi S, Khan SR. Opportunities for future therapeutic interventions for hyperoxaluria: targeting oxidative stress. Expert Opin Ther Targets. 2019;23:379–391. doi: 10.1080/14728222.2019.1599359. [DOI] [PubMed] [Google Scholar]
- 135.Zhang L, Du Z, He L, Liang W, Liu K, Gong S. ROS-induced oxidative damage and mitochondrial dysfunction mediated by inhibition of SIRT3 in cultured cochlear cells. Neural Plast. 2022;2022:5567174. doi: 10.1155/2022/5567174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang HW, Zhang Y, Tan PP, Jia LS, Chen Y, Zhou BH. Mitochondrial respiratory chain dysfunction mediated by ROS is a primary point of fluoride-induced damage in Hepa1-6 cells. Environ Pollut. 2019;255:113359. doi: 10.1016/j.envpol.2019.113359. [DOI] [PubMed] [Google Scholar]
- 137.Jin X, Xue B, Zhou Q, Su R, Li Z. Mitochondrial damage mediated by ROS incurs bronchial epithelial cell apoptosis upon ambient PM(2.5) exposure. J Toxicol Sci. 2018;43:101–111. doi: 10.2131/jts.43.101. [DOI] [PubMed] [Google Scholar]
- 138.Duann P, Lin PH. Mitochondria damage and kidney disease. Adv Exp Med Biol. 2017;982:529–551. doi: 10.1007/978-3-319-55330-6_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Thongboonkerd V. Proteomics of crystal-cell interactions: a model for kidney stone research. Cells. 2019;8:1076. doi: 10.3390/cells8091076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fong-ngern K, Vinaiphat A, Thongboonkerd V. Microvillar injury in renal tubular epithelial cells induced by calcium oxalate crystal and the protective role of epigallocatechin-3-gallate. FASEB J. 2017;31:120–131. doi: 10.1096/fj.201600543r. [DOI] [PubMed] [Google Scholar]
- 141.Khan SR. Reactive oxygen species, inflammation and calcium oxalate nephrolithiasis. Transl Androl Urol. 2014;3:256–276. doi: 10.3978/j.issn.2223-4683.2014.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Khamchun S, Thongboonkerd V. Cell cycle shift from G0/G1 to S and G2/M phases is responsible for increased adhesion of calcium oxalate crystals on repairing renal tubular cells at injured site. Cell Death Discov. 2018;4:106. doi: 10.1038/s41420-018-0123-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Govindaraj A, Selvam R. Increased calcium oxalate crystal nucleation and aggregation by peroxidized protein of human kidney stone matrix and renal cells. Urol Res. 2001;29:194–198. doi: 10.1007/s002400100177. [DOI] [PubMed] [Google Scholar]
- 144.Govindaraj A, Selvam R. An oxalate-binding protein with crystal growth promoter activity from human kidney stone matrix. BJU Int. 2002;90:336–344. doi: 10.1046/j.1464-410X.2002.02849.x. [DOI] [PubMed] [Google Scholar]
- 145.Yoodee S, Noonin C, Sueksakit K, Kanlaya R, Chaiyarit S, Peerapen P, et al. Effects of secretome derived from macrophages exposed to calcium oxalate crystals on renal fibroblast activation. Commun Biol. 2021;4:959. doi: 10.1038/s42003-021-02479-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Khan SR, Canales BK, Dominguez-Gutierrez PR. Randall's plaque and calcium oxalate stone formation: role for immunity and inflammation. Nat Rev Nephrol. 2021;17:417–433. doi: 10.1038/s41581-020-00392-1. [DOI] [PubMed] [Google Scholar]
- 147.O'Kell AL, Lovett AC, Canales BK, Gower LB, Khan SR. Development of a two-stage model system to investigate the mineralization mechanisms involved in idiopathic stone formation: stage 2 in vivo studies of stone growth on biomimetic Randall's plaque. Urolithiasis. 2019;47:335–346. doi: 10.1007/s00240-018-1079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Thongboonkerd V. Roles for exosome in various kidney diseases and disorders. Front Pharmacol. 2019;10:1655. doi: 10.3389/fphar.2019.01655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chirackal RS, Jayachandran M, Wang X, Edeh S, Haskic Z, Perinpam M, et al. Urinary extracellular vesicle-associated MCP-1 and NGAL derived from specific nephron segments differ between calcium oxalate stone formers and controls. Am J Physiol Renal Physiol. 2019;317:F1475–F1482. doi: 10.1152/ajprenal.00515.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang J, Kumar S, Jayachandran M, Herrera Hernandez LP, Wang S, Wilson EM, et al. Excretion of urine extracellular vesicles bearing markers of activated immune cells and calcium/phosphorus physiology differ between calcium kidney stone formers and non-stone formers. BMC Nephrol. 2021;22:204. doi: 10.1186/s12882-021-02417-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.He Z, Guan X, Liu Y, Tao Z, Liu Q, Wu J, et al. Alteration of exosomes secreted from renal tubular epithelial cells exposed to high-concentration oxalate. Oncotarget. 2017;8:92635–92642. doi: 10.18632/oncotarget.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yang Y, Wang Q, Xun Y, Li C, Wang S. The preliminary exploration of what role miRNAs derived from urinary exosomes play in kidney stone formation. Urology. 2022;166:104–110. doi: 10.1016/j.urology.2022.05.019. [DOI] [PubMed] [Google Scholar]
- 153.Jayachandran M, Yuzhakov SV, Kumar S, Larson NB, Enders FT, Milliner DS, et al. Specific populations of urinary extracellular vesicles and proteins differentiate type 1 primary hyperoxaluria patients without and with nephrocalcinosis or kidney stones. Orphanet J Rare Dis. 2020;15:319. doi: 10.1186/s13023-020-01607-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jayachandran M, Lugo G, Heiling H, Miller VM, Rule AD, Lieske JC. Extracellular vesicles in urine of women with but not without kidney stones manifest patterns similar to men: a case control study. Biol Sex Differ. 2015;6:2. doi: 10.1186/s13293-015-0021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013;13:1554–1571. doi: 10.1002/pmic.201200329. [DOI] [PubMed] [Google Scholar]
- 156.Stephens OR, Grant D, Frimel M, Wanner N, Yin M, Willard B, et al. Characterization and origins of cell-free mitochondria in healthy murine and human blood. Mitochondrion. 2020;54:102–112. doi: 10.1016/j.mito.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lischnig A, Bergqvist M, Ochiya T, Lasser C. Quantitative proteomics identifies proteins enriched in large and small extracellular vesicles. Mol Cell Proteomics. 2022;21:100273. doi: 10.1016/j.mcpro.2022.100273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Haraszti RA, Didiot MC, Sapp E, Leszyk J, Shaffer SA, Rockwell HE, et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J Extracell Vesicles. 2016;5:32570. doi: 10.3402/jev.v5.32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Thongboonkerd V, Kanlaya R. The divergent roles of exosomes in kidney diseases: pathogenesis, diagnostics, prognostics and therapeutics. Int J Biochem Cell Biol. 2022;149:106262. doi: 10.1016/j.biocel.2022.106262. [DOI] [PubMed] [Google Scholar]
- 160.Singhto N, Thongboonkerd V. Exosomes derived from calcium oxalate-exposed macrophages enhance IL-8 production from renal cells, neutrophil migration and crystal invasion through extracellular matrix. J Proteomics. 2018;185:64–76. doi: 10.1016/j.jprot.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 161.Singhto N, Kanlaya R, Nilnumkhum A, Thongboonkerd V. Roles of macrophage exosomes in immune response to calcium oxalate monohydrate crystals. Front Immunol. 2018;9:316. doi: 10.3389/fimmu.2018.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yu L, Gan X, Bai Y, An R. CREB1 protects against the renal injury in a rat model of kidney stone disease and calcium oxalate monohydrate crystals-induced injury in NRK-52E cells. Toxicol Appl Pharmacol. 2021;413:115394. doi: 10.1016/j.taap.2021.115394. [DOI] [PubMed] [Google Scholar]
- 163.Song Q, He Z, Li B, Liu J, Liu L, Liao W, et al. Melatonin inhibits oxalate-induced endoplasmic reticulum stress and apoptosis in HK-2 cells by activating the AMPK pathway. Cell Cycle. 2020;19:2600–2610. doi: 10.1080/15384101.2020.1810401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Albert A, Tiwari V, Paul E, Ganesan D, Ayyavu M, Kujur R, et al. Expression of heterologous oxalate decarboxylase in HEK293 cells confers protection against oxalate induced oxidative stress as a therapeutic approach for calcium oxalate stone disease. J Enzyme Inhib Med Chem. 2017;32:426–433. doi: 10.1080/14756366.2016.1256884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chhiber N, Kaur T, Singla S. Rottlerin a polyphenolic compound from the fruits of Mallotus phillipensis (Lam.) MullArg., impedes oxalate/calcium oxalate induced pathways of oxidative stress in male wistar rats. Phytomedicine. 2016;23:989–997. doi: 10.1016/j.phymed.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 166.Patel AB, Robertson WG, Choong S, Hothersall JS. Heat-shock protein 25 ameliorates calcium oxalate crystal-mediated oxidative stress in renal epithelial cells. BJU Int. 2006;98:1094–1099. doi: 10.1111/j.1464-410X.2006.06478.x. [DOI] [PubMed] [Google Scholar]
- 167.Tian H, Chen X, Liao J, Yang T, Cheng S, Mei Z, et al. Mitochondrial quality control in stroke: from the mechanisms to therapeutic potentials. J Cell Mol Med. 2022;26:1000–1012. doi: 10.1111/jcmm.17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tang Y, Huang Y, Wan Z, Zhou B, Wu Z. Mitochondrial quality control links two seemingly unrelated neurodegenerative diseases. Autophagy. 2022;18:2495–2497. doi: 10.1080/15548627.2022.2094605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sygitowicz G, Sitkiewicz D. Mitochondrial quality control: the role in cardiac injury. Front Biosci (Landmark Ed) 2022;27:96. doi: 10.31083/j.fbl2703096. [DOI] [PubMed] [Google Scholar]
- 170.Liu D, Cai ZJ, Yang YT, Lu WH, Pan LY, Xiao WF, et al. Mitochondrial quality control in cartilage damage and osteoarthritis: new insights and potential therapeutic targets. Osteoarthritis Cartil. 2022;30:395–405. doi: 10.1016/j.joca.2021.10.009. [DOI] [PubMed] [Google Scholar]
- 171.Tang C, Cai J, Yin XM, Weinberg JM, Venkatachalam MA, Dong Z. Mitochondrial quality control in kidney injury and repair. Nat Rev Nephrol. 2021;17:299–318. doi: 10.1038/s41581-020-00369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Eldeeb MA, Thomas RA, Ragheb MA, Fallahi A, Fon EA. Mitochondrial quality control in health and in Parkinson's disease. Physiol Rev. 2022;102:1721–1755. doi: 10.1152/physrev.00041.2021. [DOI] [PubMed] [Google Scholar]
- 173.Marzetti E, Guerra F, Calvani R, Marini F, Biancolillo A, Gervasoni J, et al. Circulating mitochondrial-derived vesicles, inflammatory biomarkers and amino acids in older adults with physical frailty and sarcopenia: a preliminary biosphere multi-marker study using sequential and orthogonalized covariance selection—linear discriminant analysis. Front Cell Dev Biol. 2020;8:564417. doi: 10.3389/fcell.2020.564417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dansako H, Ueda Y, Satoh S, Kato N. Extracellular vesicles activate ATM-Chk2 signaling pathway through the intercellular transfer of mitochondrial DNA in HBV-infected human hepatocytes. FASEB J. 2021;35:e21680. doi: 10.1096/fj.202002678R. [DOI] [PubMed] [Google Scholar]
- 175.Lou E. A ticket to ride: the implications of direct intercellular communication via tunneling nanotubes in peritoneal and other invasive malignancies. Front Oncol. 2020;10:559548. doi: 10.3389/fonc.2020.559548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jiang D, Chen FX, Zhou H, Lu YY, Tan H, Yu SJ, et al. Bioenergetic crosstalk between mesenchymal stem cells and various ocular cells through the intercellular trafficking of mitochondria. Theranostics. 2020;10:7260–7272. doi: 10.7150/thno.46332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Laberge A, Arif S, Moulin VJ. Microvesicles: intercellular messengers in cutaneous wound healing. J Cell Physiol. 2018;233:5550–5563. doi: 10.1002/jcp.26426. [DOI] [PubMed] [Google Scholar]
- 178.Sinclair KA, Yerkovich ST, Hopkins PM, Chambers DC. Characterization of intercellular communication and mitochondrial donation by mesenchymal stromal cells derived from the human lung. Stem Cell Res Ther. 2016;7:91. doi: 10.1186/s13287-016-0354-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ahmad T, Mukherjee S, Pattnaik B, Kumar M, Singh S, Kumar M, et al. Miro1 regulates intercellular mitochondrial transport & enhances mesenchymal stem cell rescue efficacy. EMBO J. 2014;33:994–1010. doi: 10.1002/embj.201386030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.van Heugten MH, Hoorn EJ, Fenton RA. Urinary extracellular vesicles: does cargo reflect tissue? Curr Opin Nephrol Hypertens. 2022;31:464–470. doi: 10.1097/MNH.0000000000000822. [DOI] [PubMed] [Google Scholar]
- 181.Reithmair M, Lindemann A, Mussack V, Pfaffl MW. Isolation and characterization of urinary extracellular vesicles for MicroRNA biomarker signature development with reference to MISEV compliance. Methods Mol Biol. 2022;2504:113–133. doi: 10.1007/978-1-0716-2341-1_9. [DOI] [PubMed] [Google Scholar]
- 182.Park S, Moon HY. Urinary extracellular vesicle as a potential biomarker of exercise-induced fatigue in young adult males. Eur J Appl Physiol. 2022;122:2175–2188. doi: 10.1007/s00421-022-04995-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wang Q, Sun Y, Yang Y, Li C, Zhang J, Wang S. Quantitative proteomic analysis of urinary exosomes in kidney stone patients. Transl Androl Urol. 2020;9:1572–1584. doi: 10.21037/tau-20-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ceban E, Banov P, Galescu A, Botnari V. Oxidative stress and antioxidant status in patients with complicated urolithiasis. J Med Life. 2016;9:259–262. [PMC free article] [PubMed] [Google Scholar]
- 185.Zhong Y, Sun D, Yao Y, Liu Q, Guo T, Wang X, et al. Autophagy and mitochondrial dynamics contribute to the protective effect of diosgenin against 3-MCPD induced kidney injury. Chem Biol Interact. 2022;355:109850. doi: 10.1016/j.cbi.2022.109850. [DOI] [PubMed] [Google Scholar]
- 186.Visavadiya NP, Pena GS, Khamoui AV. Mitochondrial dynamics and quality control are altered in a hepatic cell culture model of cancer cachexia. Mol Cell Biochem. 2021;476:23–34. doi: 10.1007/s11010-020-03882-9. [DOI] [PubMed] [Google Scholar]
- 187.Geto Z, Molla MD, Challa F, Belay Y, Getahun T. Mitochondrial dynamic dysfunction as a main triggering factor for inflammation associated chronic non-communicable diseases. J Inflamm Res. 2020;13:97–107. doi: 10.2147/JIR.S232009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Li S, Zhang J, Liu C, Wang Q, Yan J, Hui L, et al. The role of mitophagy in regulating cell death. Oxid Med Cell Longev. 2021;2021:6617256. doi: 10.1155/2021/6617256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Willms E, Cabanas C, Mager I, Wood MJA, Vader P. Extracellular vesicle heterogeneity: subpopulations, isolation techniques, and diverse functions in cancer progression. Front Immunol. 2018;9:738. doi: 10.3389/fimmu.2018.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ge P, Dawson VL, Dawson TM. PINK1 and Parkin mitochondrial quality control: a source of regional vulnerability in Parkinson's disease. Mol Neurodegener. 2020;15:20. doi: 10.1186/s13024-020-00367-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lazar SV, Mor S, Wang D, Goldbloom-Helzner L, Clark K, Hao D, et al. Engineering extracellular vesicles for Alzheimer's disease: an emerging cell-free approach for earlier diagnosis and treatment. WIREs Mech Dis. 2022;14:e1541. doi: 10.1002/wsbm.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fan H, He Z, Huang H, Zhuang H, Liu H, Liu X, et al. Mitochondrial quality control in cardiomyocytes: a critical role in the progression of cardiovascular diseases. Front Physiol. 2020;11:252. doi: 10.3389/fphys.2020.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Soubannier V, Rippstein P, Kaufman BA, Shoubridge EA, McBride HM. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS ONE. 2012;7:e52830. doi: 10.1371/journal.pone.0052830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhao X, Xu H, Li Y, Liu Y, Li X, Zhou W, et al. Silica nanoparticles perturbed mitochondrial dynamics and induced myocardial apoptosis via PKA-DRP1-mitochondrial fission signaling. Sci Total Environ. 2022;842:156854. doi: 10.1016/j.scitotenv.2022.156854. [DOI] [PubMed] [Google Scholar]
- 195.Wang ZH, Chen L, Li W, Chen L, Wang YP. Mitochondria transfer and transplantation in human health and diseases. Mitochondrion. 2022;65:80–87. doi: 10.1016/j.mito.2022.05.002. [DOI] [PubMed] [Google Scholar]
- 196.Wang R, Wang X, Zhang Y, Zhao H, Cui J, Li J, et al. Emerging prospects of extracellular vesicles for brain disease theranostics. J Control Release. 2022;341:844–868. doi: 10.1016/j.jconrel.2021.12.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and are also available from the corresponding author on reasonable request.