Abstract
Assembly of the functional complexes of the mitochondrial respiratory chain requires sophisticated and efficient regulatory mechanisms. In plants, the subunit composition and assembly factors involved in the biogenesis of cytochrome c oxidase (complex IV) are substantially less defined than in mammals and yeast. In this study, we cloned maize (Zea mays) Small kernel 11 (Smk11) via map-based cloning. Smk11 encodes a mitochondria-localized tetratricopeptide repeat protein. Disruption of Smk11 severely affected the assembly and activity of mitochondrial complex IV, leading to delayed plant growth and seed development. Protein interactions studies revealed that SMK11 might interact with four putative complex IV assembly factors, Inner membrane peptidase 1A (ZmIMP1A), MYB domain protein 3R3 (ZmMYB3R-3), cytochrome c oxidase 23 (ZmCOX23), and mitochondrial ferredoxin 1 (ZmMFDX1), among which ZmMFDX1 might interact with subunits ZmCOX6a and ZmCOX-X1; ZmMYB3R-3 might also interact with ZmCOX6a. The mutation of SMK11 perturbed the normal assembly of these subunits, leading to the inactivation of complex IV. The results of this study revealed that SMK11 serves as an accessory assembly factor required for the normal assembly of subunits into complex IV, which will accelerate the elucidation of the assembly of complex IV in plant mitochondria.
Tetratricopeptide-containing protein SMK11 plays critical roles in assembly of mitochondrial cytochrome c oxidase and is essential for seed development in maize.
Introduction
Mitochondria are indispensable cellular organelles present in nearly all aerobic eukaryotes where they are essential for multiple life processes. In the inner mitochondrial membrane, multiple-subunit protein complexes, complex I (NADH dehydrogenase), complex II (succinate dehydrogenase), complex III (cytochrome c reductase), and complex IV (cytochrome c oxidase) form the core of the mitochondrial electron transport chain and catalyze the multistep transfer of electrons from NADH and FADH2 to molecular oxygen, powering the production of ATP (Meyer et al. 2019). Mitochondria in plants contain a range of alternative electron transport pathways (alternative oxidases and NADH dehydrogenases) that are missing in mammals and partly in yeast. Such pathways bypass the energy-conserving electron transfer chain complexes and affect the redox environment of the latter (Rasmusson et al. 2008; Vanlerberghe, 2013; Møller et al. 2021).
Complex IV is the terminal electron acceptor of the oxidative phosphorylation (OXPHOS) system. It accepts single electron from reduced cytochrome c on the cytosolically oriented side of the inner membrane and eventually delivers four electrons to O2 on the matrix side of the membrane, to produce H2O. Dedicated investigations of plant complex IV are motivated by expected variations in the structure and assembly of complex IV between mammals, yeast, and plants caused by different functional contexts. Complex IV contains three core hydrophobic catalytic subunits cytochrome c oxidase 1 (COX1), COX2 and COX3 that are conserved in bacteria and that are encoded by the mitochondrial genome (mtDNA) in most eukaryotes. Additionally, there are more than ten nuclear-encoded subunits that likely act as protective structural scaffolds to stabilize the catalytic core (Pierron et al. 2012; Kolli et al. 2019). In plants, the cytochrome c oxidase consists of seven more or less conserved subunits (COX1, COX2, COX3, COX5b, COX6a and COX6b) that are also found in animals and fungi, and six potential plant-specific subunits named COX-X1, COX-X2, COX-X3, COX-X4, COX-X5, and COX-X6 (Millar et al. 2004; Klodmann et al. 2011; Senkler et al. 2017; Mansilla et al. 2018). In addition to these structural subunits, a large number of accessory proteins are required for the assembly and activity of complex IV, most of which have been identified in mammals and yeast, but not in plants (Ghezzi and Zeviani, 2012; Mansilla et al. 2018).
The assembly process of complex IV has been extensively studied in yeast and mammals, and the current evidence suggests that complex IV is assembled from linear and modular models, which are preassembled and incorporated into the catalytic core subunits orderly (Meyer et al. 2019). The first step of complex IV assembly in opisthokonts is the embeding of COX1 into the inner mitochondrial membrane (IMM) with the help of chaperones oxidase assembly protein 1 (OXA1), COX14, COX assembly factor 3 (COA3), MSS51, HSP70 in yeast, and C-X9-C motif containing 1 (CMC1) in human, to form the COX1 module (McStay et al. 2013; Bourens and Barrientos, 2017). Then, heme α synthesized by COX10 and COX15 is incorporated into COX1, and the assembly factor COA1 and the structural subunits COX5a, COX6a, and COX6b are added (Mansilla et al. 2018). After that, the CuB center is assembled into COX1 with the assistance of COX11, COX19, and COX17 (Bode et al. 2015). Subsequently, the insertion of COX2 in the IMM is conducted by OXA1, COX20, and COX18, and the CuA center is assembled and incorporated into COX2 with the release of COX18 to form the COX2 module (Bourens et al. 2014). In humans, the COX5b, COX6c, COX7b, COX7c, and COX8a are also assembled into the COX2 module, before the formation of the core COX1-COX2 module (Timón-Gómez et al. 2018). Finally, COX3 is incorporated into the complex with COX4, COX7 and COX13 in yeast, and COX6a, COX6b and COX7a in humans (Soto et al. 2012). The differences observed indicate that the biogenesis of complex IV has evolved to follow variations in mechanisms, necessitating specific investigations of different eukaryotes to reveal the factors involved.
According to the knowledge in yeast and humans, a total of 72 genes encoding putative complex IV-related proteins were predicted in Arabidopsis (Arabidopsis thaliana), among which, four accessory subunits (AtCOX5b, AtCOX5c, AtCOX6a and AtCOX6b) and six plant-specific structural subunits named COX-X1 to COX-X6 were included (Mansilla et al. 2018). However, their exact functions and how they are involved in the assembly process of complex IV remains unclear (Mansilla et al. 2018). To date, studies on the assembly of complex IV in plant have mainly focused on a restricted set of assembly factors that are involved in particular step of the total assembly process. AtCOX17 and AtCOX19, homologs of the yeast copper chaperones COX17 and COX19p, are involved in cytochrome c oxidase biogenesis in plants (Balandin and Castresana, 2002; Attallah et al. 2007). AtCOX10, a homolog of COX10p from yeast, is involved in synthesis of heme o (later converted to heme a) during complex IV biogenesis, and is essential for plant embryogenesis (Mansilla et al. 2015). Homolog of copper chaperone 1 (AtHCC1), a SCO1p homolog, plays a role in the copper insertion during complex IV assembly (Attallah et al. 2011). Disturbance of AtCOX11 expression was found to affect complex IV activity and many vital physiological processes, e.g. pollen germination and plant growth (Radin et al. 2015). AtOXA2b, a plant-specific tetratricopeptide repeat (TPR)-containing OXA protein, is crucial for proper membrane insertion of COX2 during biogenesis of complex IV and for plant growth (Kolli et al. 2019). Although AtOXA2b appears to function similarly to COX18 in humans, its function was shown to depend on the TPR domain, which is absent in COX18 (Kolli et al. 2019). Till now, the complex IV biogenesis mechanism in plant has less been characterized, as compared to that in yeast and humans, and knowledge is missing on multiple steps in the plant assembly process.
In the present study, we characterized a new mitochondrial-localized TPR-containing protein, small kernel 11 (SMK11), which is required for the assembly and activity of mitochondrial complex IV and seed development in maize (Zea mays). We further found that SMK11 may act as an accessory protein by interacting with four assembly factors Inner membrane peptidase 1A (ZmIMP1A), MYB domain protein 3R3 (ZmMYB3R-3), Cytochrome c oxidase 23 (ZmCOX23), and Mitochondrial ferredoxin 1 (ZmMFDX1) to mediate the assembly of subunits into complex IV. These findings will provide clues to clarify the assembly process of mitochondrial complex IV in plants.
Results
The smk11 mutant displays small kernels with delayed development
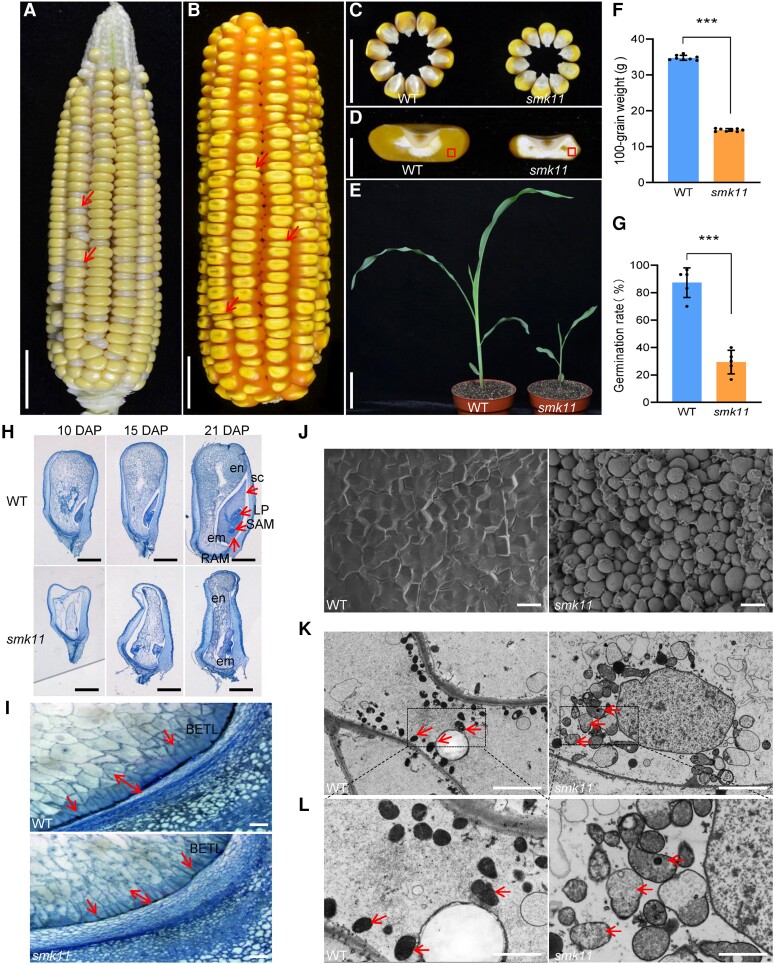
The smk11 mutant was isolated from a natural variation of a cross between inbred lines Chang7-2 and G390 during reproduction and exhibited a 3:1 segregation of wild-type (WT) (+/+ and smk11/+) and small kernel (smk11/smk11) phenotypes (2,715:875, χ2 = 0.72, P < 0.05), indicating that smk11 was a monogenic recessive mutant. Heterozygous mutant plants were crossed to inbred lines B73 and Mo17 and self-crossed to obtain F2 ears, respectively. The homozygous smk11 kernels could be clearly distinguished from segregating F2 ears at 18 days after pollination (DAP) with small and white endosperms compared with the WT (Fig. 1A;Supplemental Fig. S1). At 30 DAP, smk11 kernels were substantially smaller than the WT (Fig. 1B–D). At maturity, the 100-grain weight of smk11 mutants was nearly 58.0% less than that of the WT (p < 0.001) (Fig. 1F). We tested the effects of Smk11 mutation on seed viability and found that approximately 30.7% of the mutant seeds could germinate, but with a slower growth (Fig. 1G). At 18 days after planting, the viable smk11 seedlings displayed stunted growth and delayed development compared with the WT (Fig. 1E). These results suggest that the Smk11 mutation inhibits plant growth and seed development.
Figure 1.
Phenotypic features of maize smk11 mutant. A) A selfed ear segregates smk11 mutants at 18 DAP. Red arrows indicate the small kernels. B) A selfed ear segregates smk11 mutants at maturity. Arrows indicate the small kernels. C) Randomly selected mature smk11 and WT kernels from the segregated F2 population. D) Sections of mature WT and smk11 kernels. E) WT and smk11 mutant seedlings at 18 days after planting. F, G) Comparison of 100-grain weight (n = 8 ears) F) and germination rate (n = 5 ears) g) between WT and smk11 mutants. H) Longitudinal paraffin sections of smk11 and WT kernels at 10 DAP, 15 DAP and 21 DAP. I) The BETL cells on longitudinal paraffin sections of WT and smk11 mutant kernels at 15 DAP. Arrows indicate the BETL cells. J) Scanning electron microscopy of starch granules in the areas as boxed in Fig.1d of WT and smk11 endosperm. K) Mitochondrial ultrastructure of developing endosperms of WT and smk11 mutant at 15 DAP. Arrows indicate the Mitochondrion. L) Magnified view of mitochondria in WT and smk11 mutant. Arrows indicate the Mitochondrion. em, embryo; en, endosperm; LP, leaf primordia; RAM, root apical meristem; SAM, shoot apical meristem; sc, scutellum; BETL, BETL. Scale bars = 1 cm A-C) ; 5 mm D), 5 cm E), 1 mm H); 100 μm I); 100 μm J); 5 μm K), 2 μm L). Values are the average ± S.E. (*** P < 0.001, Student’s t test).
To investigate the embryo and endosperm developmental status of smk11 mutant, homozygous smk11 and WT kernels from the same segregating ear at 10, 15 and 21 DAP were sectioned and observed under microscopy. At 10 DAP, WT embryos were well developed to the coleoptilar stage with the endosperm filling the pericarp, whereas the smk11 embryos remained at the transition stage with an interspace between endosperm and pericarp (Fig. 1H). At 15 DAP, WT embryos had established the typical embryonic structures, including scutellum (sc), shoot apical meristem (SAM), root apical meristem (RAM), and leaf primordia (LP). In contrast, the smk11 embryos arrested at the transition stage showing a malformed embryo proper, and the endosperm developed slowly and closed to the pericarp (Fig. 1H). At 21 DAP, the embryos and endosperm of WT kernels were further enlarged, while the embryos of smk11 had gradually become larger but still with an abnormal morphology appearance. And until this stage, the endosperm of smk11 completely filled the entire pericarp (Fig. 1H).These results indicate that the mutation of Smk11 blocks maize embryogenesis and endosperm development, resulting in a small kernel phenotype (Fig. 1H).
Nutrients are transported from the maternal tissue into kernel endosperm through the basal endosperm transfer layer (BETL) cells. At 15 DAP, the BETL cells of WT kernels were darkly stained with thick and extensive cell wall ingrowths, whereas the smk11 BETL cells were less developed with reduced cell wall ingrowths, suggesting that the developmentally defective BETL cells in smk11 may reduce their kernel weights and result in small kernels (Fig. 1I). Scanning electron microscopy observation of outer region of mature endosperm in WT and smk11 kernels showed that the WT mature endosperm were tightly embedded in a proteinaceous matrix but were naked in smk11 mutant (Fig. 1J).
Positional cloning of Smk11 gene
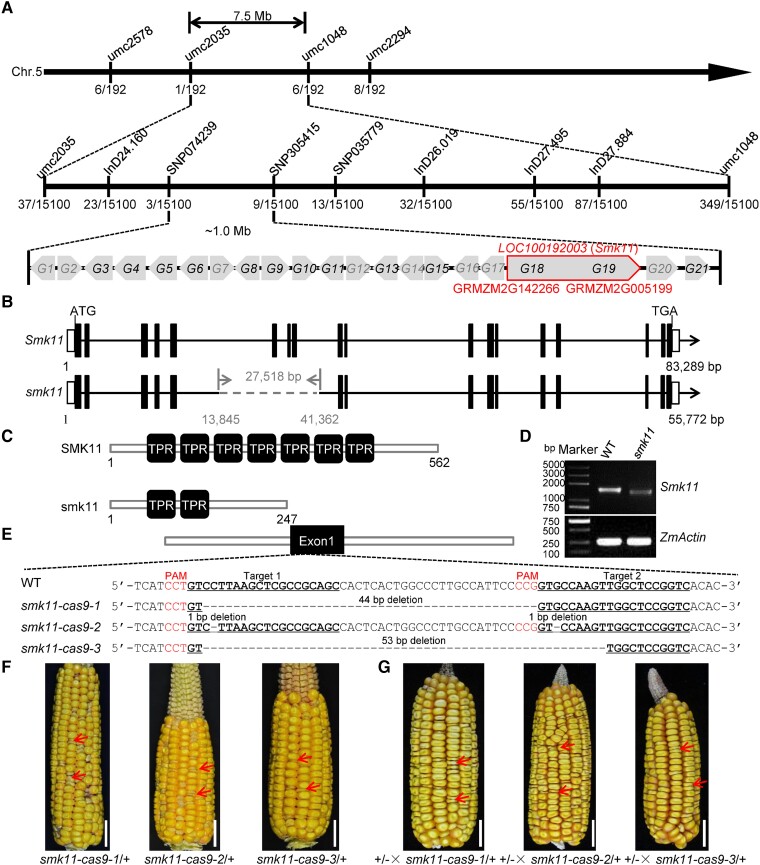
To identify the gene responsible for the mutant phenotype of smk11, genetic mapping of smk11 locus was carried out with an F2 population of B73 background. The smk11 mutant locus was initially mapped to a 7.5 Mb interval on the long arm of maize chromosome 5 by bulked segregant RNA-sequencing (BSR-seq) method. SSR (simple sequence repeat), Indel (insertion and deletion), and SNP (single nucleotide polymorphism) molecular markers were further developed by sequencing the genome sequences near the Smk11 locus in both parental lines for a fine mapping (Supplemental Table S1). Using an expanded population of 15,100 homozygous mutant kernels, the Smk11 gene was genetically narrowed down to a 1.0 Mb physical region between molecular markers SNP074239 and SNP305415 (Fig. 2A). According to the B73 reference genome (RefGen_v3), 21 predicted protein-coding genes (G1 to G21) were annotated, among which, eight genes (G1, G2, G7, G12, G14, G16, G17, and G20) could be excluded as candidates, because of their undetectable expression in kernels based on the MaizeGDB database (https://www.maizegdb.org/) and the published RNA-seq data (Supplemental Table S2). Among the remaining 13 genes, six of them (G3, G4, G5, G6, G8, and G13) had similar expression levels between WT and smk11 mutant, while four of them (G9, G10, G11, and G15) showed significantly upregulated expression and three genes (G18, G19, and G21) were significantly downregulated in smk11 mutant (Supplemental Fig. S2). Since the genome sequencing of G9, G10, G11, G15, and G21 revealed no sequence differences between WT and smk11 mutant, we focused on the candidate gene as G18 or G19. Surprisingly, G18 (GRMZM2G142266) and G19 (GRMZM2G005199), which are located adjacent to each other but approximately 29 Kb apart based on the MaizeGDB database (RefGen_v3), showed highly similar expression patterns. In addition, according to NCBI database (https://www.ncbi.nlm.nih.gov/), GRMZM2G142266 and GRMZM2G005199 were annotated as one gene, LOC100192003. To identify the authentic transcripts of GRMZM2G142266 and GRMZM2G005199, 3′-end sequence of GRMZM2G142266 was amplified from inbred line B73 kernels using 3′RACE (rapid amplification of cDNA ends) method, and the identified transcript was indeed highly consistent with the genome annotation of NCBI database, confirming that GRMZM2G142266 and GRMZM2G005199 are identical to LOC100192003 (Supplemental Fig. S3). We then performed whole-genome sequencing of the smk11 mutant using the Pacific Bioscience Sequel II platform and revealed a ∼27 Kb deletion (containing ∼25 Kb of intron 5, exon 6-8, and ∼0.7 Kb of intron 8) in LOC100192003 of smk11 mutant (Fig. 2B), which was confirmed by PCR amplification and sequencing using primer pairs crossing the flanking sequence of the deletion (Supplemental Fig. S4A–C). These results suggest that LOC100192003 was the strongest candidate gene for the Smk11.
Figure 2.
Map-based cloning and allelism test of Smk11. A) Fine-mapping of Smk11 using F2 populations of B73×smk11/+ with 15,000 individuals. The numbers under each bar indicate the number of recombinants. Twenty-one genes (G1 to G21) were annotated between two markers SNP074239 and SNP305415 based on the MaizeGDB database. Genes in gray indicate that they are not expressed in the embryo and endosperm. Genes in black indicate that they are expressed in the embryo and endosperm. Two genes (GRMZM2G142266 and GRMZM2G005199) were annotated as one gene LOC100192003 according to NCBI database. B) Diagram showing the gene structure of LOC100192003 (Smk11) and smk11. The lines represent introns, the black boxes represent exons, and the white boxes represent 5′ UTR and 3′ UTR. The gray dashed line stands for the deletion fragment in smk11. C) Schematic diagram of the maize SMK11 and smk11 protein structures. D) RT-PCR analysis of the expression of Smk11 in 15 DAP kernels of smk11 mutant and WT. The amplification product was normalized against ZmActin. E) The sequence in the LOC100192003 locus is targeted using CRISPR/Cas9. The two gRNA sequences (Target 1 and Target 2) are underlined, and the protospacer adjacent motif (PAM) are shown in red letter and the gray dashes represent deletions. Alignments of mutant sequences from three independent transgenic lines are indicated. F) Mature F2 ear of Cal × smk11-cas9-1, Cal × smk11-cas9-2, and Cal × smk11-cas9-1. Arrows indicate the mutant kernels. G) Allelism tests were performed using crosses between heterozygous smk11 (+/-) and heterozygous smk11-cas9-1 (smk11-cas9-1/+), smk11-cas9-2 (smk11-cas9-2/+), and smk11-cas9-3 (smk11-cas9-3/+). Arrows indicate the mutant kernels. Scale bars = 1 cm (F–G).
To investigate whether the Smk11 transcriptional level was affected in smk11 mutant, the full-length transcripts from WT and mutant kernels at 15 DAP were examined by RT-PCR. An expected band of ∼1.6 Kb was detected in the WT, whereas the corresponding band was undetectable in smk11; on the contrary, a relatively smaller band with a much lower transcript level was detected in smk11, compared with the complete Smk11 in WT, indicating a defective transcript of Smk11 in mutant (Fig. 2D).
Allelic confirmation and functional complementation of Smk11
To confirm that LOC100192003 was Smk11, targeted mutagenesis of LOC100192003 was performed using the CRISPR/Cas9 system. Eight independent transgenic lines were obtained, and three of them (smk11-cas9-1 to smk11-cas9-3) showed frameshift mutations caused by different types of deletions at the two target sequences, which were chosen for further experiments (Fig. 2E). T0 transgenic lines were crossed with their acceptor inbred line Cal, and then were self-crossed to generate T2 progenies. The kernels of T2 ears displayed a 3:1 segregation of WT and mutant (Supplemental Table S3), and the mutated kernels showed a similar small kernel phenotype to smk11 (Fig. 2F). Allelism tests between three independent knockout alleles and heterozygous smk11 revealed a 3:1 segregation of WT and smk kernel phenotypes on the F2 ears, further confirming that LOC100192003 is Smk11 (Fig. 2G).
Moreover, a transgenic functional complementation test was also performed by transforming the entire LOC100192003 coding sequence driven by Cauliflower mosaic virus (CaMV) 35S promoter into maize inbred line Zong31. Three independent transformants (OE1, OE3, and OE21) with different transcriptional levels of Smk11 were obtained (Supplemental Fig. S5C). The reciprocal cross tests between homozygous smk11/smk11 plants and OE1 plants were performed, and the F1 plants with heterozygous smk11/+ genotype and hemizygous transgenic locus were then self-pollinated and produced ears with 15:1 segregation of WT and small kernel, indicating that the smk11 mutant phenotype could be functionally complemented by the introduced Smk11 transgenic allele (Supplemental Fig. S5A, B, D; Supplemental Table S4). Taken together, these data strongly confirmed that LOC100192003 is Smk11.
Smk11 is constitutively expressed and encodes a mitochondrial-localized TPR domain-containing protein
The genomic data (https://www.ncbi.nlm.nih.gov/) revealed that Smk11 (LOC100192003) is 83,289 bp length, containing 18 exons and 17 introns (Fig. 2B). It is predicted to encode a 62.8 kDa protein comprising 562 amino acid residues. SMK11 was annotated as a tetratricopeptide repeat (TPR)-like superfamily protein, and the bioinformatics analysis (https://toolkit.tuebingen.mpg.de/tools/tprpred) predicted seven TPR motifs, which are composed of 34 amino acid residues in tandem repeating units (Fig. 2C). A neighbor-joining phylogenetic tree using the full-length SMK11 amino acid sequence and its potential homologous protein sequences from other species were constructed and indicated that SMK11 homologs are highly conserved in angiosperms (Supplemental Fig. S6A), especially with high similarity in the TPR domains (Supplemental Fig. S6B).
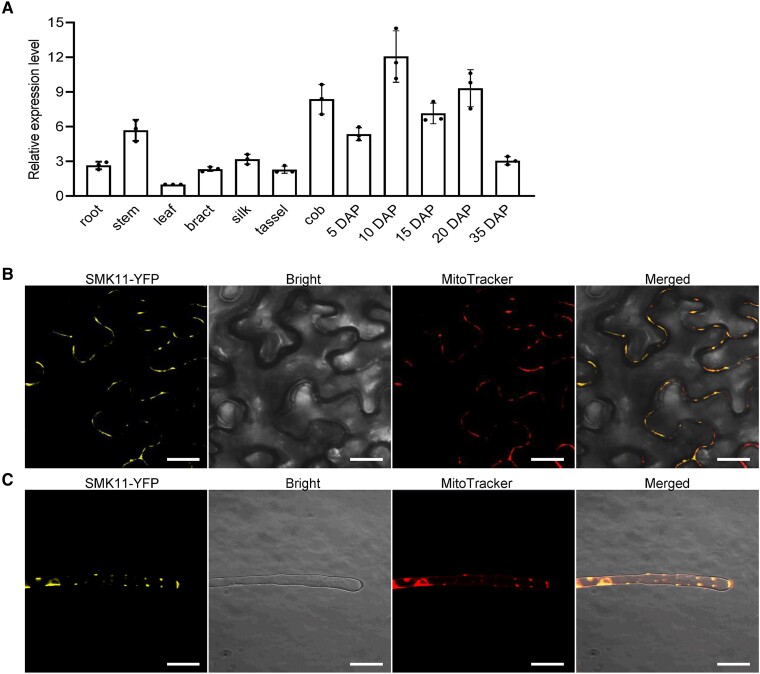
The spatiotemporal expression profile of Smk11 in various tissues of inbred line B73 was investigated by reverse transcription quantitative PCR (RT-qPCR). The results revealed that Smk11 is ubiquitously expressed in a broad range of tissues, including root, stem, leaf, bract, silk, tassel, cob and kernel at different development stages (Fig. 3A). To further characterize the function of SMK11, its subcellular localization was investigated in Nicotiana benthamiana leaf epidermis cells, Arabidopsis thaliana (Columbia) root hairs, and maize protoplasts. Fluorescent signals of p35S::SMK11-YFP showed co-localization with MitoTracker Red by confocal laser scanning microscopy observation, indicating that SMK11 is located in the mitochondria (Fig. 3B and C; Supplemental Fig. S7).
Figure 3.
Expression profile of Smk11 and subcellular localization of SMK11 protein. A) Expression profiles of Smk11 in various tissues and kernels at different development stages. ZmActin was used as an internal control. Data are the average ± S.E. of three biological replicates. B) Localization of SMK11-YFP in Nicotiana benthamiana mesophyll cells. MitoTracker Red was used to label mitochondria. C) Localization of SMK11-YFP in the root hairs of transgenic Arabidopsis thaliana. MitoTracker Red was used to label mitochondria. Scale bar = 20 µm (B, C).
Loss of function of Smk11 altered mitochondrial ultrastructure
Since Smk11 encodes a mitochondria-localized protein, we investigate the impact of the loss function of Smk11 on mitochondrial morphology. Ultra-thin sections of 15 DAP endosperm from smk11 and the WT kernels were observed by transmission electron microscopy (TEM). The result showed that the WT endosperm exhibited normal mitochondria with densely folded inner membranes, whereas the mitochondria of smk11 showed a poorly developed membrane system with large internal spaces and vaguely dissociated cristae structures (Fig. 1K and L), indicating that SMK11 is required for the proper structure and function of mitochondria during seed development.
Loss of function of SMK11 affects the assembly and activity of mitochondrial complex IV
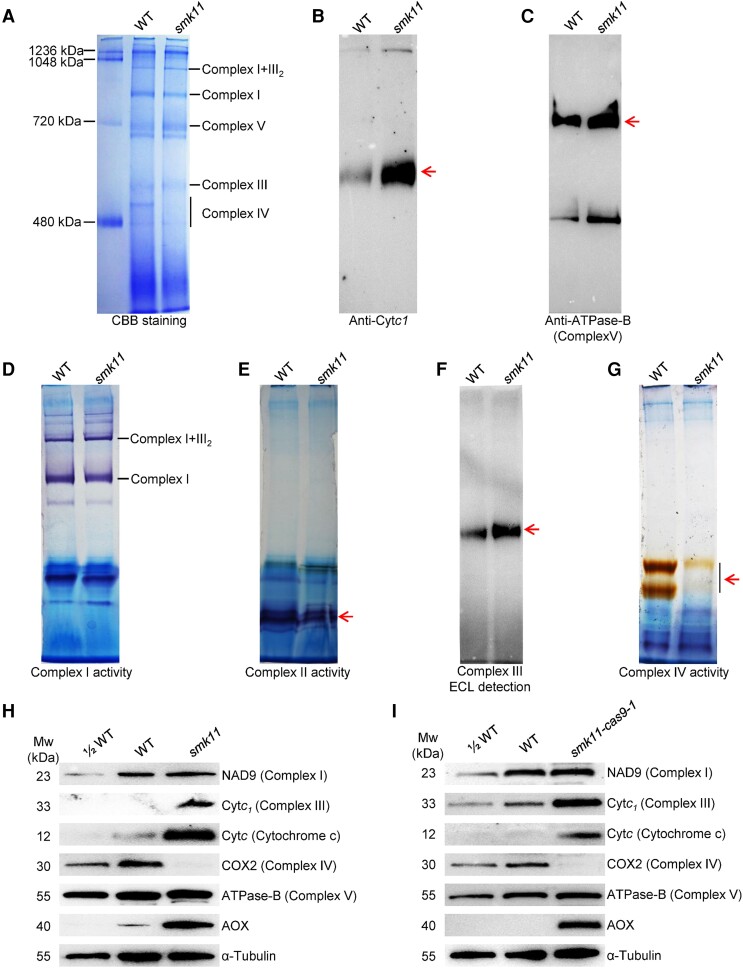
The mutation in Smk11 led to mitochondrial deformations, which promoted us to analyze whether the mutation of Smk11 affected the assembly and activity of mitochondrial complexes. Mitochondria from 15 DAP smk11 and WT kernels without pericarp were isolated, enriched and separated by BN-PAGE. Coomassie Brilliant Blue (CBB) staining showed comparable abundance of supercomplex I + III2, complex I, complex III and complex V between WT and smk11 mutant, however, the abundance of complex IV was strongly decreased in smk11 mutant (Fig. 4A). The in-gel activity staining of different complexes was also performed and the results showed that the activities of complex I and complex II had no substantial difference between WT and smk11 mutant (Fig. 4D and E), while the activity of complex IV was decreased obviously (Fig. 4G), which was consistent with the arrested assembly of complex IV. The peroxidase activity of the covalently bound heme of c-type cytochromes (Cytc1 and Cytc) standing for complex III was detected with a strong increase in smk11 mutant (Fig. 4F). Western blotting in denatured BN-PAGE was also performed using antibodies against Cytc1 (complex III) and ATPase-B (complex V) to further confirm the impact of the deficiency of SMK11 on the assembly of mitochondrial complexes. A remarkably increase of Cytc1 in smk11 mutant (Fig. 4B), yet an almost constant level of ATPase-B (Fig. 4C) between WT and smk11 were identified. We also investigated the assembly and activity of mitochondrial complexes in smk11-cas9-1 mutant, which showed consistent results, with substantial decrease in the abundance and activity of complex IV and an obviously increase in the abundance of complex III (Cytc1) (Supplemental Fig. S8A–C).
Figure 4.
Impact on mitochondrial complexes in smk11 kernels at 15 DAP. A) BN (blue native) gel was stained with CBB. The position of mitochondrial complexes is indicated. About 100 μg of mitochondrial protein was loaded in each lane. B) BN gels transferring to PVDF membranes were probed with anti-Cytc1 (a subunit of complex III) antibody. The band of Cytc1 was indicated in arrow. C) BN gels transferring to PVDF membranes were probed with anti-ATPase-B antibody (a subunit of complex V) antibody. The band of ATPase-B was indicated in arrow. D) BN gels were used for activity staining of complex I. E) BN gels were used for activity staining of complex II. The position of mitochondrial complex II was indicated in arrow. F) BN gels transferring to PVDF membranes were detected for peroxidase activity. The position of peroxidase was indicated in arrow. G) BN gels were used for activity staining of complex IV. The position of mitochondrial complex II was indicated in arrow. H, I) Western blot analysis with antibodies against NAD9 (subunit of complex I), Cytc1 (subunit of complex III), Cytc (cytochrome c), COXII (subunit of complex IV), ATPase-B (subunit of complex V) and AOX (alternative oxidase) in mitochondrial protein from smk11 and smk11-cas9-1 kernels at 15 DAP. α-Tubulin was used as a sample loading control.
To further investigate the potential effects of Smk11 mutation on the mitochondrial respiratory chain complexes, the steady-state levels of mitochondrial complex-related proteins were examined in both smk11 and smk11-cas9-1 mutants by western blot with specific antibodies against NAD9 (a subunit of complex I), Cytc1 (a subunit of complex III), Cytc (subunits of the cytochrome c maturation pathway), COX2 (a subunit of complex IV), and ATPase-B (a subunit of complex V). The results showed that the level of COX2 protein was dramatically reduced in both smk11 and smk11-cas9-1 mutants, compared with WT (Fig. 4H–I). In addition, the transcriptional levels of three mtDNA-encoded subunits of complex IV (COX1, COX2, and COX3) were also analyzed in smk11 mutant, with only a significant upregulation in COX2 (Supplemental Fig. S9A–C). The levels of Cytc1 and Cytc (cytochrome c, transferring electron from complex III to complex IV) proteins were remarkably accumulated in smk11 and smk11-cas9-1 mutants compared with the WT (Fig. 4H and I). In addition, no substantial change was detected in the levels of either NAD9 or ATPase-B proteins (Fig. 4H and I). These results indicate that the loss of function of SMK11 leads to failed assembly and activity of mitochondrial complex IV, but the protein accumulation of complex III was increased, which may imply a feedback regulatory mechanism in response to complex IV deficiency.
To further investigate the composition difference of mitochondrial complex IV between WT and smk11 mutant, the proteins at the position of complex IV in the BN-PAGE gel stained with CBB (Fig. 4A) were cut out for LC-MS/MS analysis. As a result, four subunits (ZmCOX6a, ZmCOX6b-1, ZmCOX-X1, and ZmCOX-X3) of complex IV were detected in WT, but were absent in the smk11 mutant (Table 1). In addition, the amount of ZmCOX-X2 and ZmCOX5b-3 subunits were dramatically decreased in smk11 mutant, indicating an association of SMK11 with these subunits of the complex IV (Table 1). Since antibodies to these subunits are not available, RT-qPCR was performed to detect the transcriptional levels of these genes, and the results showed that all genes were distinctly upregulated in the smk11 mutant (Supplemental Fig. S10). These results suggest that the mutation of SMK11 affected the proper assembly of complex IV in maize mitochondria.
Table 1.
Mass spectrometry analysis of mitochondrial complex IV in smk11 mutant at 15 DAP
| Gene ID | Description | Repeat I | Repeat II | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | smk11 | WT | smk11 | ||||||||||
| Coverage | Unique Peptides | PSM | Coverage | Unique Peptides | PSM | Coverage | Unique Peptides | PSM | Coverage | Unique Peptides | PSM | ||
| GRMZM2G145396 | ZmCOX-X2 | 38.46 | 3 | 6 | 11.54 | 1 | 1 | 44.87 | 4 | 7 | 14.82 | 1 | 1 |
| GRMZM2G134738 | ZmCOX5b-3 | 23.64 | 4 | 8 | 7.27 | 1 | 1 | 64.24 | 7 | 24 | 56.18 | 3 | 7 |
| GRMZM2G177432 | ZmCOX6a | 13.83 | 1 | 1 | — | — | — | 16.26 | 1 | 3 | — | — | — |
| GRMZM2G047512 | ZmCOX-X3 | 7.61 | 1 | 1 | — | — | — | 7.61 | 1 | 2 | — | — | — |
| GRMZM2G159691 | ZmCOX-X1 | 7.34 | 1 | 1 | — | — | — | 14.28 | 2 | 4 | — | — | — |
| GRMZM2G152925 | ZmCOX6b-1 | 5.81 | 1 | 1 | — | — | — | 15.70 | 3 | 3 | — | — | — |
The "-" in Table 1 indicates the proteins were unidentified in the LC-MS/MS analysis.
About 100 μg mitochondrial protein of WT and smk11 was loaded in each lane. BN-PAGE was performed and the gel was stained with CBB. The proteins at the position of mitochondrial complex IV were cut out and used for LC-MS/MS analysis by Shanghai Applied Protein Technology. Coverage represents the percentage of the protein sequence covered by identified peptides. Unique Peptides stand for the number of unique peptide sequences in the protein group. PSM (peptide spectrum matches) stands for the total number of identified peptide sequences for the protein, including those redundantly identified.
Alternative respiratory pathway was activated in smk11 mutant
The dysfunction of the cytochrome respiratory pathway usually leads to an upregulated expression of alternative oxidase pathway. The transcript and protein levels of Aox genes were analyzed in smk11 mutant. The RNA expression levels of Aox2 and Aox3 were upregulated in smk11 as compared to the WT, with about 64-fold and 8-fold increase, respectively (Supplemental Fig. S9D). Consistently, an obviously increased accumulation of AOX proteins in smk11 and smk11-cas9-1 mutants further confirmed the activation of the alternative oxidase pathway in response to complex IV deficiency (Fig. 4H and I). These results indicated that the respiratory function of smk11 mitochondria is severely perturbed because of the decrease in complex IV activity (Fig. 4G), which enhances the alternative pathway in the smk11 mutant.
SMK11 mediates the assembly and activity of complex IV
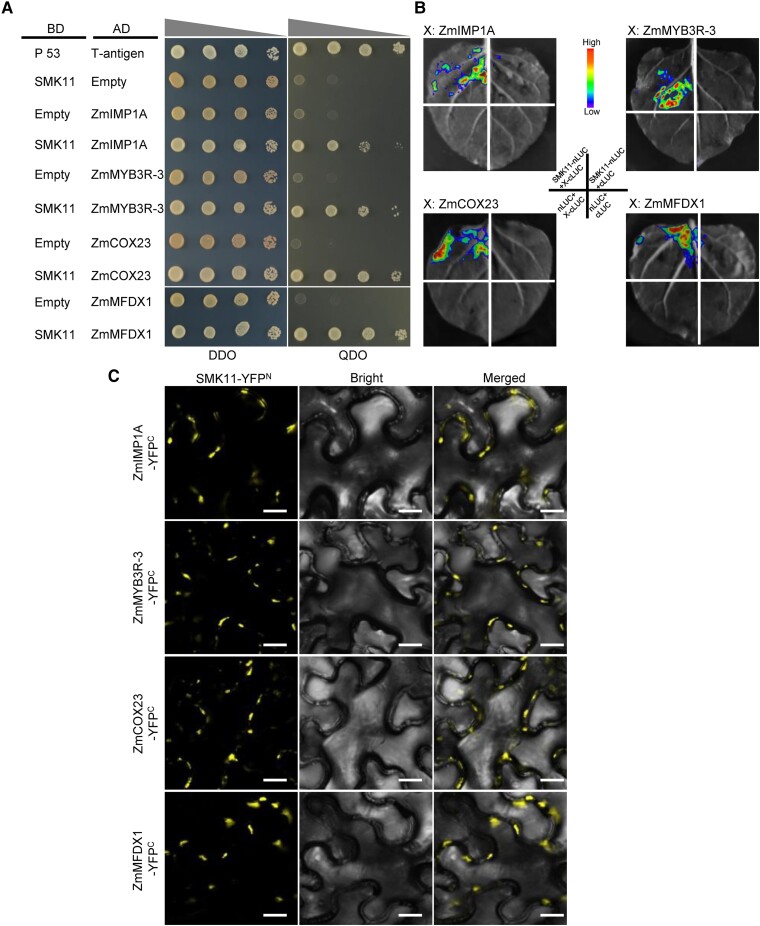
Since the amount of COX2, ZmCOX6a, ZmCOX6b-1, ZmCOX-X1, ZmCOX-X2, ZmCOX-X3, and ZmCOX5b-3 were decreased in smk11 mutant, we investigated potential protein interactions between SMK11 and these subunits by Y2H, respectively, yet did not find direct physical interaction (Supplemental Fig. S11). To further investigate the molecular mechanism by which SMK11 might be involved in the assembly of mitochondria complex IV, a cDNA library from embryo and endosperm of maize inbred line B73 at 15 DAP was screened. The Y2H assay results indicated that SMK11 may interact with four putative assembly factors of complex IV, including ZmIMP1A (GRMZM5G833660), ZmMYB3R-3 (GRMZM2G142938), ZmCOX23 (GRMZM2G031501), and ZmMFDX1 (Zm00001d037104) (Fig. 5A; Supplemental Table S5). The interactions of SMK11 with these four proteins were further verified by luciferase complementation image (LCI) assays and bimolecular fluorescence complementation (BiFC) assays. The co-expression of SMK11-nLUC/YFPN (N-terminal domains of luciferase/N-terminal of YFP) with ZmIMP1A-cLUC/YFPC (C-terminal domains of luciferase/C-terminal of YFP), ZmMYB3R-3-cLUC/YFPC, ZmCOX23-cLUC/YFPC, and ZmMFDX1-cLUC/YFPC, resulted in strong luciferase activity or YFP fluorescence signals, indicating that SMK11 can interact with them in Nicotiana benthamiana cells (Fig. 5B and C). In addition, in vitro pull-down assays were also performed. SMK11 was purified as fusion protein with maltose-binding protein (MBP) and His tags, and ZmIMP1A and ZmCOX23 were purified as fusion protein with MBP. The interactions between MBP-SMK11-His and MBP-ZmIMP1A, as well as MBP-SMK11-His and MBP-ZmCOX23 were investigated using His-mediated pull-down assays. Consistent with the observed interactions in the Y2H, LCI and BiFC assays, MBP-SMK11-His was able to bind to MBP-ZmIMP1A and MBP-ZmCOX23 in vitro, respectively (Supplemental Fig. S12). The MBP protein used as a negative control that was added to the binding reaction failed to be retained in the resin after high-stringency washes (Supplemental Fig. S12). Because ZmMYB3R-3 and ZmMFDX1 could not be prokaryotically expressed and purified, the His-mediated pull-down assays between SMK11 and ZmMYB3R-3 and ZmMFDX1 were not conducted. These results suggest that SMK11 may interact with these four putative assembly factors (ZmIMP1A, ZmMYB3R-3, ZmCOX23, and ZmMFDX1) of mitochondrial complex IV both in vitro and in vivo.
Figure 5.
Interactions between maize SMK11 and the assembly factors of complex IV were identified by Y2H, LUC, and BiFC assays. A) Yeast two-hybrid (Y2H) analysis between SMK11 and the four assembly factors (ZmCOX23, ZmMFDX1, ZmMYB3R-3, and ZmIMP1A) of complex IV. The Y2H Gold strain harboring the indicated bait and prey constructs were spotted on SD-Trp-Leu (DDO) and SD-Trp-Leu-His-Ade (QDO). The interaction between Human P53 and T-antigen was used as a positive control. AD, activating domain; BD, binding domain. B) LCI assays between SMK11 and the four assembly factors of complex IV. The specific combinations used for each interaction are indicated. The fluorescence signal intensity represents their interaction activities. C) BiFC assays between SMK11 and the four assembly factors of complex IV. Scale bar =50 μm.
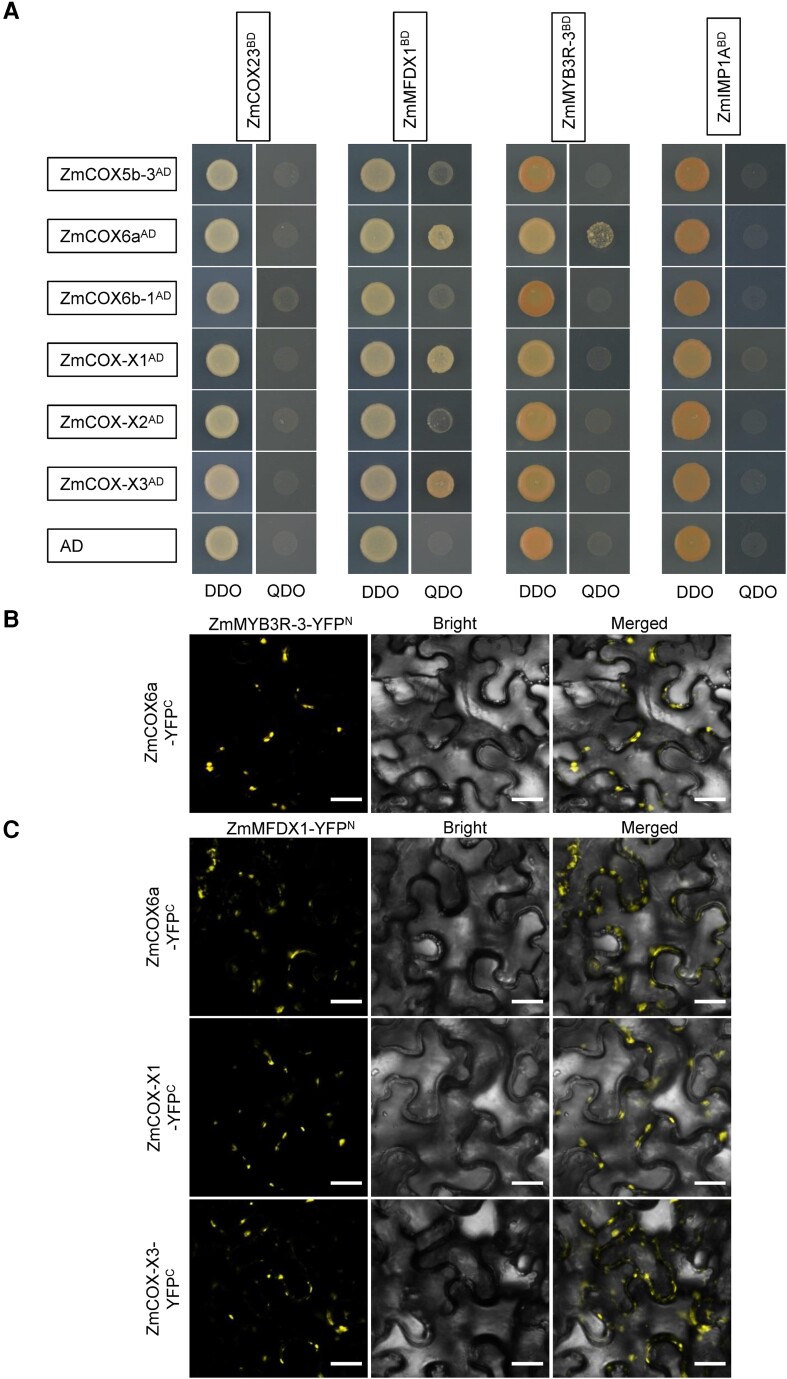
Because the mutation of Smk11 resulted in truncated SMK11 (mSMK11) with only two TPR domains (Fig. 2C), we performed Y2H assay to test whether this truncation affected the interactions with the four interacting proteins. The results showed that mSMK11 could not interact with these proteins, suggesting the possible specific function of SMK11 on the assembly of mitochondrial complex IV (Supplemental Fig. S13). In addition, the interactions between the four assembly factors (ZmIMP1A, ZmMYB3R-3, ZmCOX23, and ZmMFDX1) and the six subunits (ZmCOX5b-3, ZmCOX6a, ZmCOX6b-1, ZmCOX-X1, ZmCOX-X2, and ZmCOX-X3) of complex IV were also investigated by Y2H. As a result, ZmMFDX1 could interact with ZmCOX6a, ZmCOX-X1 and ZmCOX-X3; and ZmMYB3R-3 could interact with ZmCOX6a, which were verified by BiFC assays, suggesting an auxiliary function of SMK11 on the assembly of these subunits (Fig. 6A and C). These results strongly suggest that SMK11 play essential roles in the assembly of mitochondrial complex IV by affecting the assembly factors ZmIMP1A, ZmMYB3R-3, ZmCOX23, and ZmMFDX1.
Figure 6.
Interactions between maize assembly factors and subunits of complex IV were identified by Y2H assay and BiFC assay. A) Yeast two-hybrid (Y2H) analysis between the four assembly factors (ZmCOX23, ZmMFDX1, ZmMYB3R-3 and ZmIMP1A) and the four subunits (ZmCOX5b-3, ZmCOX6a, ZmCOX-X1, and ZmCOX-X2) of complex IV. The Y2H Gold strain harboring the indicated bait and prey constructs were spotted on SD-Trp-Leu (DDO) and SD-Trp-Leu-His-Ade (QDO). The interaction between the four assembly factors and AD was used as a negative control. AD, activating domain; BD, binding domain. B) BiFC assay between ZmMYB3R-3 and ZmCOX6a. C) BiFC assays between ZmMFDX1 and ZmCOX6a, ZmCOX-X1, as well as ZmCOX-X3. Scale bar =50 μm.
Discussion
The composition of plant mitochondrial complexes has been investigated using BN-PAGE and mass spectrometry, however, the exact subunit set of complex IV is currently not fully defined, as the status of some potential subunits are still under debate (Senkler et al. 2017). Therefore, most proteins related to complex IV structure and assembly in plants have been annotated by sequence homology to yeast and human counterparts (Mansilla et al. 2018). In this study, we cloned a mitochondria-localized TPR gene Smk11, and its disruption severely affects the assembly and activity of mitochondrial complex IV, which would lead to insufficient energy supply and affect plant growth and seed development. SMK11 thus constitutes a new complex IV biogenesis factor. Loss of function of SMK11 resulted in a dramatic reduction in COX2 protein accumulation (Fig. 4H and I), and a significant upregulation of COX2 gene (Supplemental Fig. S9B). However, the editing and splicing of three mitochondrial genes for complex IV subunits (COX1, COX2, and COX3) were not affected in the smk11 mutant (Supplemental Fig. S14). These results indicate that SMK11 serves as a key factor involved in the assembly of complex IV, rather than an RNA processing factor.
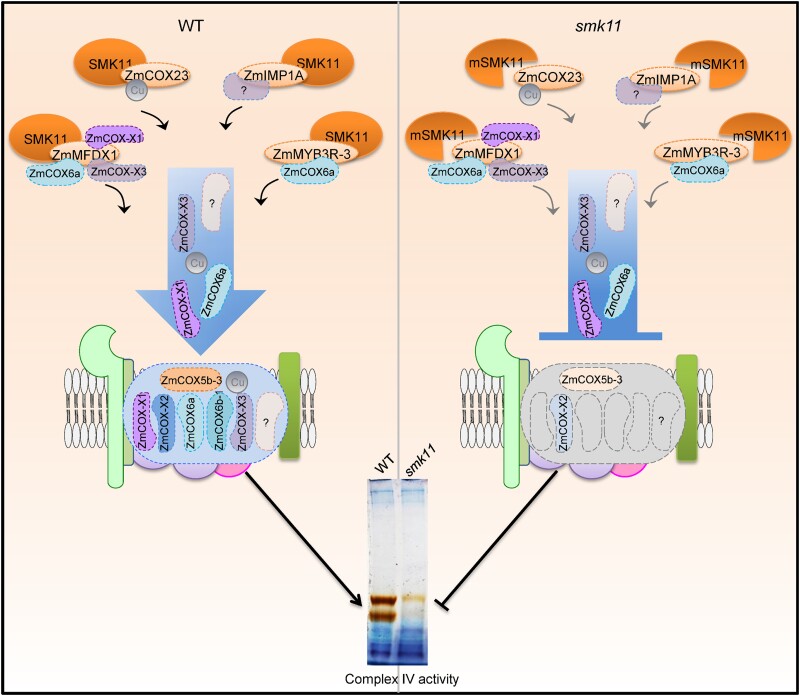
TPR-containing proteins preferably to bond target proteins (D’Andrea and Regan, 2003; Delannoy et al. 2007). It was reported that Arabidopsis thaliana OXA2b, which contains a TPR domain at the C-terminus, plays a crucial role in the complex IV assembly by interacting with COX2 (Kolli et al. 2019). Mouse TTC19 (tetra-tricopeptide repeat domain 19) is linked with complex III biogenesis by removing UQCRFS1(Bottani et al. 2017). Based on this, we presumed that SMK11 may play role in the biogenesis of mitochondrial complex IV in maize. We found that SMK11 may interact with four putative maize assembly factors of complex IV, including ZmIMP1A (putative ortholog of IMP1 from yeast, and IMMP1 from human), ZmMYB3R-3 (putative ortholog of IMP2 from yeast, and IMMP2 from human), ZmCOX23 (putative ortholog of COX23 from yeast and human), and ZmMFDX1 (putative ortholog of YAH1 from yeast, and FDX2 from human) (Fig. 5A–C). In addition, through LC-MS/MS analysis we identified the amount of several complex IV subunits including ZmCOX5b-3 (putative ortholog of COX4 from yeast, and COX5B from human), ZmCOX6a (putative ortholog of COX13 from yeast, and COX6A from human), ZmCOX-X1 (putative ortholog of AT5G27760), and ZmCOX-X2 (putative ortholog of AT1G01170), were dramatically decreased in the smk11 mutant (Table 1). We could not exclude the possibility that other assembly factors also interact with SMK11 and the incorporation of other subunits into complex IV were also affected due to the technology limitation. However, our results clearly suggest that SMK11 acts as an assembly factor, to assist the assembly of subunits into complex IV (Fig. 7). This function is consistent with the notion that the TPR domain primarily mediates protein–protein interactions (D'Andrea and Regan, 2003; Perez-Riba and Itzhaki, 2019) and provides a scaffold for integration of further protein factors into the assembly process for complex IV.
Figure 7.
A proposed working model of the role of SMK11 in the assembly of mitochondrial complex IV in maize. In the wild type (left), SMK11 interacts with ZmIMP1A, ZmMYB3R-3, ZmCOX23, and ZmMFDX1 to facilitate the assembly of the subunits including ZmCOX5b-3, ZmCOX6a, ZmCOX-X1, and ZmCOX-X2 into complex IV, guaranteeing the integrity and activity of complex IV. In the smk11 mutant (right), mutation of SMK11 (mSMK11) inhibits the interaction with the four assembly factors, leading to the disruption of assembly and activity of complex IV. The question marks stand for the proteins unidentified. The blot shown at the bottom is the same as that shown in Figure 4g.
Assembly factor YAH1 is a yeast mitochondrial ferredoxin and is essential for the electron transfer reactions catalyzing the conversion of heme o to heme α, which is the form incorporated into COX1. We found that ZmMFDX1 (the plant putative ortholog of YAH1) could interact with subunits ZmCOX6a, ZmCOX-X1, and ZmCOX-X3, which could not be detected by LC-MS/MS of complex IV from smk11 mitochondria, suggesting that ZmMFDX1 may play additional roles in the assembly of complex IV compared with YAH1, by directly incorporating subunits into complex IV. ZmCOX6a is the putative ortholog of COX12 that was found in the assembly intermediates containing COX3 and being involved in the formation of COX dimers as well as the incorporation of COX into supercomplexes (Mansilla et al. 2018; Timón-Gómez et al. 2018). ZmCOX6a interacts not only with ZmMFDX1 but also with ZmMYB3R-3, the putative ortholog of IMP2, which is required for the stability and activity of IMP1 peptidase (Mansilla et al. 2018). This indicates that the assembly process of ZmCOX6a into complex IV needs the help of several assembly factors. IMP1, putative ortholog of ZmIMP1A, is required for the maturation of mitochondrial proteins located in the intermembrane space (IMS) during the COX2 biogenesis. COX23, putative ortholog of ZmCOX23, is supposed to be involved in copper trafficking and insertion into the complex under assembly (Barros et al. 2004). We found that SMK11 could also interact with ZmIMP1A and ZmCOX23, however, we could not draw a firm conclusion as to how SMK11 plays a role in the maturation of mitochondrial proteins and copper assembly.
In a conclusion, we identified a mitochondria-localized TPR gene Smk11 by map-based cloning, and found that SMK11 might interact with four putative complex IV assembly factors ZmIMP1A, ZmMYB3R-3, ZmCOX23, and ZmMFDX1. The mutation of SMK11 perturbed the normal assembly of these subunits, leading to inactivation of complex IV. Our results suggest that SMK11 serves as an accessory assembly factor to mediate the normal assembly of subunits into complex IV, and could accelerate the elucidation of the assembly of complex IV in plant mitochondria.
Materials and methods
Plant materials
The maize (Zea mays) smk11 mutant was isolated from the progeny of a hybrid plant derived from a cross between inbred lines Chang7-2 and G390 during reproduction. Heterozygous smk11 plants were crossed into B73 and Mo17 genetic backgrounds to generate respective F1 plants, which were then self-pollinated to produce F2 segregated populations. Homozygous mutant kernels from F2 segregating ears in B73 background were used as the mapping population. Kernels of smk11 and the WT from Mo17 background were used for phenotypic observations, cytological analysis, as well as RNA and protein extraction. All plants were cultivated in the field in Beijing (summer) or Sanya (winter), China.
Nicotiana benthamiana and Arabidopsis thaliana (Columbia) plants were grown in a growth chamber under a 16/8 h day/night photoperiod at a temperature of 20-25 °C and 70% relative humidity.
Light microscopy, transmission electron microscopy, and scanning electron microscopy
For light microscopy analysis of embryo and endosperm, developing WT and smk11 mutant kernels at 10 DAP, 15 DAP, and 21 DAP were harvested from the same F2 heterozygous ears and cut along the longitudinal axis for paraffin section preparation. For light microscopy analysis of BETL cells, WT and smk11 mutant kernels at 15 DAP were dissected along the horizontal and longitudinal axes and fixed in 2.5% (v/v) glutaraldehyde for resin section preparation. The subsequent procedures were performed as described preciously (Fan et al. 2021). The images of paraffin- and resin-embedded sections were taken under a Nikon Ti microscope and an Olympus IX71 microscope, respectively.
For transmission electron microscopy analysis of endosperm, immature WT and smk11 kernels at 15 DAP from the same segregated ears were collected and treated according to a previously described method (Fan et al. 2021). The prepared samples were observed with a Hitachi 7700 transmission electron microscope.
For scanning electronic microscope analysis, mature WT and smk11 seeds from the same self-pollinated ear were rifted with a razor and immediately immersed in 2.5% (w/v) glutaraldehyde. The samples were critical-point dried and spray-coated with gold in an E-100 ion sputter, and then observed under a Hitachi S3400N scanning electron microscopy.
Genetic mapping of smk11 locus
The initial mapping of smk11 was performed by bulked segregant RNA-sequencing (BSR-seq) method with pooled RNA samples extracted from embryo and endosperm of WT and mutant kernels (30 kernels each sample) at 15 DAP from the same F2 segregated ears in B73 genetic background. The cDNA libraries were constructed following standard Illumina protocols and sequenced with an Illumina Novaseq System by Berry Genomics (Beijing, China). Two independent biological replicates with two different ears were performed. The smk11 locus was preliminarily mapped to a 7.5 Mb interval between two SSR markers umc2035 and umc1048 on chromosome 5. A population of 15,100 homozygous mutant kernels from B73 background, and the developed InDel (InD24.160, InD26.019, InD27.495 and InD27.884) and SNP (SNP074239, SNP305415 and SNP035779) molecular markers were further used to localize the Smk11 locus to a 1.0 Mb physical region. Primer sequences for the markers are listed in Supplemental Table S1.
For whole-genome sequencing, genomic DNA from maize homozygous smk11 mutant leaves at the three-leaf stage was extracted using DNeasy Plant Mini Kit (QiaGen). The integrity and quality of the DNA was examined with the Agilent 4200 Bioanalyzer. About 8 μg of genomic DNA was sheared and concentrated with AMPure PB magnetic beads. Each SMRT bell library was constructed with the Pacific Biosciences SMRT Bell Template Prep Kit 1.0. Sequencing was conducted on the Pacific Bioscience Sequel II platform and genome was assembled by Annoroad Gene Technology company.
Allelism test and complementation test
For allelism test, two 20 bp gRNA sequences (GTCCTTAAGCTCGCCGCAGC and GTGCCAAGTTGGCTCCGGTC) were designed in the first exon of Smk11 using CRISPR-P (http://crispr.hzau.edu.cn/CRISPR/) and cloned into the BsaI site of pBUE411 vector to construct the pBUE411-2gRNA-Smk11 vector (Xing et al. 2014). The vector was transferred into Agrobacterium tumefaciens strain LBA4401 and then transformed into maize immature embryos of inbred line Cal as described previously (Liu et al. 2015). Eight independent Cas9-edited knockout lines (named smk11-cas9-1 to smk11-cas9-8) were obtained, and three of them that contain frameshift mutations were chosen for further experiments.
For functional complementation test, the entire coding sequence of Smk11 was amplified from inbred line B73 cDNA and cloned into the BglII and BstEII sites of pCAMBIA3301 vector, and the resulting vector was transferred into Agrobacterium tumefaciens strain LBA4401. Agrobacterium-mediated maize transformation using immature embryos of inbred line Zong31 was performed as described above. The reciprocal cross tests between T1 positive transformation events and homozygous smk11/smk11 plants were conducted to generate F1 plants, which were then self-pollinated to validate the complementation.
RNA extraction, RT-PCR, and RT-qPCR
Maize tissues were collected and immediately frozen in liquid nitrogen, and then ground to a fine powder with a mortar and pestle. Total RNA was extracted with an RNAprep Pure Plant Kit (TianGen) according to the manufacturer’s instructions. About 1.5 μg of total RNA was reverse transcribed with HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme) using oligo d(T)20 primer.
For reverse transcription PCR (RT-PCR) analysis, the full-length transcripts of Smk11 and Aoxs in WT and mutant kernels at 15 DAP were amplified with primer pairs listed in Supplemental Table S1.
Reverse transcription quantitative PCR (RT-qPCR) was performed using ChamQ SYBR qPCR Master Mix (High ROX Premixed) (Vazyme) with an ABI 7300-sequence detection system (Applied Biosystems). The gene expression levels were assessed using the 2−△△Ct method with ZmActin as the internal control (Livak and Schmittgen, 2001). Statistically significant differences of expression levels of genes were calculated by Student’s t test with three independent biological replicates. All the primers used for RT-qPCR analysis are listed in Supplemental Table S1.
Subcellular localization
The full-length coding sequence without the stop codon of Smk11 was amplified from inbred line B73 cDNA and then cloned into the AhdI site of pGWC entry vector using pEASY®-Basic Seamless Cloning and Assembly Kit (TransGen). The correct entry plasmid was introduced into pEarleyGate101 (https://www.arabidopsis.org/index.jsp) plant expression vector through an LR site-specific recombination (Invitrogen). The resulting p35S::SMK11-YFP vector was transiently expressed in Nicotiana benthamiana leaf and stably transformed into Arabidopsis thaliana plants via the Agrobacterium tumefaciens strain GV3101 as previously described (Fan et al. 2021). The recombinant vector was also transiently transformed into maize etiolated mesophyll cell protoplasts by enzymatic hydrolysis as described previously (Ren et al. 2020). The YFP fluorescence was captured and imaged using an LSM 980 confocal microscope (Zeiss). Fluorescence of YFP and MitoTracker were detected with excitation at 514 and 579 nm, respectively.
Analysis of mitochondrial protein complexes
The mitochondria from approximately 15 g fresh kernels without pericarps of WT and smk11 kernels at 15 DAP were isolated and enriched according to the method in the previous report (Fan et al. 2021).
For complex activity analysis, about 100 μg of enriched mitochondrial proteins were solubilized according to the standard procedures of the NativePAGE Sample Prep Kit (Thermo Fisher Scientific) and separated by 3-13% blue native polyacrylamide gel electrophoresis (BN-PAGE) as described previously (Fan et al. 2021). The gel strips were stained by Coomassie Brilliant Blue R-250 and/or used for in-gel complex I, complex II, and complex IV activity assays as described previously (Wittig et al. 2007).
For BN-PAGE western blotting, the BN-PAGE gel strips were incubated in 50 mM Tris-HCl, pH 6.8, 8 M urea, 5% (w/v) SDS, 20% (v/v) glycerol, and 5% (v/v) β-mercaptoethanol for 1 h to denature the complexes and then transferred onto a PVDF membrane using a semi-dry blotter (Bio-Rad) for western blotting with antiserum against Cytc1, ATPase-B, and COX2, respectively (Sun et al. 2018).
For western blotting, 1/2 WT (12.5 μg), WT (25 μg), and smk11 (25 μg) denatured mitochondria were separated on a 12.5% SDS-PAGE gel, and then transferred onto a PVDF membrane. Antiserum against NAD9, Cytc1, Cytc, COX2, ATPase-B, AOX, and α-Tubulin (Agrisera) were used as primary antibodies with a 1:1,000 dilution, and goat anti-rabbit antibody conjugated to horseradish peroxidase was used as secondary antibody with a 1:5,000 dilution. Detection was carried out with the SuperSignalTM West Pico PLUS Chemiluminescent Substrate Kit (Thermo Fisher Scientific) according to the user manual.
Sequence analysis
The potential homologous proteins of SMK11 used for phylogenetic analysis were identified using the Basic Local Alignment Search Tool (BLAST) at National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) public databases. ClustalW and MEGA 5 softwares with default parameters were used for multiple sequence alignment and phylogenetic tree construction. Sequence data from this article can be found in the GenBank/EMBL databases under accession number NP_001130899.1 for SMK11. The other accession numbers can be found in Supplemental Fig. S6.
Yeast two-hybrid assay
The yeast library using the cDNA from embryo and endosperm of maize inbred line B73 at 15 DAP was constructed by Shanghai OE Biotech Co., Ltd. The full-length coding sequence of Smk11 was cloned into the EcoRI and BamHI sites of pGBKT7 plasmid to produce fusion bait protein with GAL4 BD domain, and then transformed into yeast strain Y2HGold. pGBKT7-SMK11 was used to screen the cDNA library according to the Matchmaker Gold Yeast Two-Hybrid System User Manual. The ORFs of SMK11 potentially interacting proteins ZmIMP1A (GRMZM5G833660), ZmMYB3R-3 (GRMZM2G142938), ZmCOX23 (GRMZM2G031501), and ZmMFDX1 (Zm00001d037104) were amplified from maize B73 cDNA using the primers listed in Supplemental Table S1 and inserted in frame into the EcoRI and BamHI sites of pGADT7 plasmid. pGBKT7-SMK11 and pGADT7-Preys were co-transformed into yeast strain Y2HGold using a Frozen-EZ Yeast Transformation II Kit (Zymo Research). The mutant coding sequence of Smk11 (mSmk11) was also cloned into the pGBKT7 plasmid to produce pGBKT7-mSMK11 fusion protein and co-transformed into yeast strain Y2HGold with four SMK11-interacting proteins. The resulting co-transformants were spotted on SD/-Leu/-Trp medium (DDO) and SD/-Ade/-His/-Leu/-Trp medium (QDO), respectively, with a dilution series. The interaction between pGBKT7-53 (Human P53) and pGADT7-T (T-antigen) was used as a positive control, and the interaction between pGADT7-empty and pGBKT7-SMK11 were used as negative control.
Luciferase complementation image assays
The full-length ORFs of Smk11 (without stop codon) as well as SMK11-interacting proteins ZmIMP1A, ZmMYB3R-3, ZmCOX23, and ZmMFDX1 were cloned into the KpnI and SalI sites of pCAMBIA1300-nLUC and the KpnI and BamHI sites of pCAMBIA1300-cLUC vectors with a ClonExpress Ultra One Step Cloning Kit (Vazyme Biotech co., Ltd), respectively. These expression vectors were transferred into Agrobacterium tumefaciens strain GV3101 and cultured to OD600 = 0.8, then centrifuged, and the pellets were re-suspended in infiltration buffer, respectively (Fan et al. 2021). Different combinations of nLUC and cLUC vectors were mixed equally and then infiltrated into Nicotiana benthamiana leaves using needleless syringes. After growing for 48 h in a growth chamber with a 16/8 h day/night photoperiod, the leaves were injected with 1 mM luciferin, and the resulting luciferase signals were captured using a Plant In Vivo Molecular Imaging System (LB985 NightSHADE, Berthold). All primers related to vector construction are listed in Supplementary Table S1.
Bimolecular fluorescence complementation assay
The full-length ORFs (without stop codon) of Smk11 and the four SMK11-interacting proteins ZmIMP1A, ZmMYB3R-3, ZmCOX23, and ZmMFDX1 were cloned into the gateway binary vector pEarleyGate201-YN and pEarleyGate202-YC by LR site-specific recombination to generate fusion p35S::SMK11-YFPN and p35S:: SMK11-interacting protein-YFPC vectors, respectively. The different combinations of YFPN and YFPC fusion constructs were co-transformed into Nicotiana benthamiana leaf as described in the luciferase (LUC) assay, and the YFP fluorescence was collected by an LSM980 Confocal Scanning Microscope (Zeiss). Relevant primer sequences are given in Supplementary Table S1. Fluorescence of YFP was detected with excitation at 514, and the master gains of YFP were between 690 and 730.
Protein purification and pull-down assays
The SMK11 coding sequence (containing the 6×His coding sequence at the C-terminus) was cloned into the BamHI site of pMAL-c2x expression vector to generate fusion construct with MBP and His tags. The coding sequences of ZmIMP1A and ZmCOX23 were cloned into the pMAL-c2x expression vector to generate fusion constructs with MBP tag. These constructs were transformed into TransB (DE3) strains of Escherichia coli. MBP-ZmIMP1A and MBP-ZmCOX23 proteins were purified using amylose resin (NEB), and MBP-SMK11-His was purified using Ni-NTA agarose (Qiagen) according to the manufacturers’ instructions.
For pull-down assays, equal volumes of the two interacting proteins as well as MBP protein were mixed, and the mixture was incubated with Ni-NTA agarose beads for 3 to 5 h at 4°C. After incubation, the beads were collected by simple centrifugation (600 g, 5 min) and then washed five times with washing buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, and 0.05% (v/v) Tween 20, pH = 8.0). The beads were then re-suspended with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, and 0.05% (v/v) Tween 20, pH = 8.0) to elute the proteins from beads. Finally, the bound proteins within 1×SDS loading buffer were heated at 99°C for 5 min and subjected to SDS-PAGE. Western blot was used to detect the targeted recombinant proteins with anti-MBP (NEB) antibody. The primers are listed in Supplementary Table S1.
BN-PAGE combined with mass spectrometry
To investigate the composition difference of mitochondrial complex IV between WT and smk11 mutant, the proteins at the position of complex IV in the BN-PAGE gel stained with CBB were cut out and used for LC-MS/MS analysis by Shanghai Applied Protein Technology.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: SMK11, NP_001130899.1; ZmIMP1A, NP_001147250.2; ZmMYB3R-3, XP_020400651.1; ZmCOX23, NP_001140638.1; ZmMFDX1, NP_001296807.1; ZmCOX-X2, NP_001151357.1; ZmCOX5b-3, NP_001144681.2; ZmCOX6a, NP_001142528.2; ZmCOX-X3, NP_001140914.1; ZmCOX-X1, NP_001152359.2; ZmCOX6b-1, NP_001131473.1.
Supplementary Material
Acknowledgments
We thank Dr. Baocai Tan at Shandong University for providing Cytc1 antibody. All authors have no conflicts of interest to declare.
Contributor Information
Zhenjing Ren, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China.
Kaijian Fan, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China.
Sihan Zhen, College of Agronomy and Biotechnology, China Agricultural University, Beijing 100094, China.
Jie Zhang, College of Agronomy and Biotechnology, China Agricultural University, Beijing 100094, China.
Yan Liu, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China.
Junjie Fu, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China.
Chunlai Qi, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China.
Qianhan Wei, College of Agronomy and Biotechnology, China Agricultural University, Beijing 100094, China.
Yao Du, College of Agronomy and Biotechnology, China Agricultural University, Beijing 100094, China.
Wurinile Tatar, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China.
Xiaofeng Zhang, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China.
Guoying Wang, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China.
Allan G Rasmusson, Department of Biology, Lund University, SE-22362 Lund, Sweden.
Jianhua Wang, College of Agronomy and Biotechnology, China Agricultural University, Beijing 100094, China.
Yunjun Liu, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . The homozygous smk11 plants could produce small and inviable progeny.
Supplemental Figure S2 . RT-qPCR analysis of 13 expressed genes in candidate interval.
Supplemental Figure S3 . The cDNA sequence of LOC100192003 in inbred line B73 obtained using 3′RACE.
Supplemental Figure S4 . The sequencing conducted on the Pacific Bioscience Sequel II platform was confirmed by PCR amplification.
Supplemental Figure S5 . Functional complementation test of smk11.
Supplemental Figure S6 . Phylogenetic relationships and amino acid alignment of SMK11 and its homologs.
Supplemental Figure S7 . Localization of SMK11 in maize protoplasts.
Supplemental Figure S8 . Impact on mitochondrial complexes in smk11-cas9-1 kernels at 15 DAP.
Supplemental Figure S9 . RT-qPCR analysis of COX1, COX2, COX3, as well as Aox1, Aox2, and Aox3 expression in smk11 kernels at 15 DAP.
Supplemental Figure S10 . RT-qPCR analysis of the expression of four assembly factors and six subunits of complex IV.
Supplemental Figure S11 . SMK11 does not interact with the seven subunits of complex IV.
Supplemental Figure S12 . Recombinant protein MBP-SMK11-His interacts with MBP-ZmIMP1A and MBP-ZmCOX23, as determined using in vitro His pull-down assays.
Supplemental Figure S13 . Mutation of SMK11 (mSMK11) could not interact with the four assembly factors of complex IV.
Supplemental Figure S14 . Sequence analysis of COX1, COX2 and COX3 transcripts in WT and smk11.
Supplemental Table S1 . Primers used in this study.
Supplemental Table S2 . The expression patterns of 21 candidate genes in a 1-Mb Interval.
Supplemental Table S3 . Segregating ratio of heterozygous smk11-cas9/+ ears.
Supplemental Table S4 . Complementation statistics (15:1) of smk11 mutation with Smk11 transgene.
Supplemental Table S5 . List of SMK11-interacting proteins identified by Y2H, LUC, and BiFC assays.
Author contributions
Yj.L., J.W., Z.R., and K.F. designed the experiments; Z.R., K.F., Y.L., C.Q., Q.W., Y.D., X. Z., and W.T. performed the experiments; Z.R., K.F., S.Z., J.Z., J.F., G.W., and Y.L. analyzed the data; Z.R., K.F., A.R. and Yj.L. wrote the article.
Funding
This work was supported by the National Natural Science Foundation of China (31871631) and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS).
References
- Attallah CV, Welchen E, Martin AP, Spinelli SV, Bonnard G, Palatnik JF, Gonzalez DH. Plants contain two SCO proteins that are differentially involved in cytochrome c oxidase function and copper and redox homeostasis. J Exp Bot. 2011:62(12):4281–4294. 10.1093/jxb/err138 [DOI] [PubMed] [Google Scholar]
- Attallah CV, Welchen E, Pujol C, Bonnard G, Gonzalez DH. Characterization of Arabidopsis thaliana genes encoding functional homologues of the yeast metal chaperone Cox19p, involved in cytochrome c oxidase biogenesis. Plant Mol Biol. 2007:65(3):343–355. 10.1007/s11103-007-9224-1 [DOI] [PubMed] [Google Scholar]
- Balandin T, Castresana C. AtCOX17, an Arabidopsis homolog of the yeast copper chaperone COX17. Plant Physiol. 2002:129(4):1852–1857. 10.1104/pp.010963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros MH, Johnson A, Tzagoloff A. COX23, a homologue of COX17, is required for cytochrome oxidase assembly. J Biol Chem. 2004:279(30):31943–31947. 10.1074/jbc.M405014200 [DOI] [PubMed] [Google Scholar]
- Bode M, Woellhaf MW, Bohnert M, van der Laan M, Sommer F, Jung M, Zimmermann R, Schroda M, Herrmann JM. Redox-regulated dynamic interplay between Cox19 and the copper-binding protein Cox11 in the intermembrane space of mitochondria facilitates biogenesis of cytochrome c oxidase. Mol Biol Cell. 2015:26(13):2385–2401. 10.1091/mbc.E14-11-1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottani E, Cerutti R, Harbour ME, Ravaglia S, Dogan SA, Giordano C, Fearnley IM, D'Amati G, Viscomi C, Fernandez-Vizarra E, et al. TTC19 plays a husbandry role on UQCRFS1 turnover in the biogenesis of mitochondrial respiratory complex III. Mol Cell. 2017:67(1):96–105. 10.1016/j.molcel.2017.06.001 [DOI] [PubMed] [Google Scholar]
- Bourens M, Barrientos A. A CMC1-knockout reveals translation-independent control of human mitochondrial complex IV biogenesis. EMBO Rep. 2017:18(3):477–494. 10.15252/embr.201643103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourens M, Boulet A, Leary SC, Barrientos A. Human COX20 cooperates with SCO1 and SCO2 to mature COX2 and promote the assembly of cytochrome c oxidase. Hum Mol Genet. 2014:23(11):2901–2913. 10.1093/hmg/ddu003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003:28(12):655–662. 10.1016/j.tibs.2003.10.007 [DOI] [PubMed] [Google Scholar]
- Delannoy E, Stanley WA, Bond CS, Small ID. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem Soc Trans. 2007:35(6):1643–1647. 10.1042/BST0351643 [DOI] [PubMed] [Google Scholar]
- Fan K, Ren Z, Zhang X, Liu Y, Fu J, Qi C, Tatar W, Rasmusson AG, Wang G, Liu Y. The pentatricopeptide repeat protein EMP603 is required for the splicing of mitochondrial Nad1 intron 2 and seed development in maize. J Exp Bot. 2021:72(20):6933–6948. 10.1093/jxb/erab339 [DOI] [PubMed] [Google Scholar]
- Ghezzi D, Zeviani M. Assembly factors of human mitochondrial respiratory chain complexes: physiology and pathophysiology. Adv Exp Med Biol. 2012:748:65–106. 10.1007/978-1-4614-3573-0_4 [DOI] [PubMed] [Google Scholar]
- Klodmann J, Senkler M, Rode C, Braun HP. Defining the protein complex proteome of plant mitochondria. Plant Physiol. 2011:157(2):587–598. 10.1104/pp.111.182352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolli R, Soll J, Carrie C. OXA2b is crucial for proper membrane insertion of COX2 during biogenesis of complex IV in plant mitochondria. Plant Physiol. 2019:179(2): 601–615. 10.1104/pp.18.01286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang Y, Liu Y, Lu W, Wang G. Metabolic effects of glyphosate on transgenic maize expressing a G2-EPSPS gene from Pseudomonas fluorescens. J Plant Biochem Biot. 2015:24(2): 233–241. 10.1007/s13562-014-0263-9 [DOI] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△ct method. Methods. 2001:25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mansilla N, Garcia L, Gonzalez DH, Welchen E. AtCOX10, a protein involved in haem o synthesis during cytochrome c oxidase biogenesis, is essential for plant embryogenesis and modulates the progression of senescence. J Exp Bot. 2015:66(21):6761–6775. 10.1093/jxb/erv381 [DOI] [PubMed] [Google Scholar]
- Mansilla N, Racca S, Gras D, Gonzalez DH, Welchen E. The complexity of mitochondrial complex IV: an update of cytochrome c oxidase biogenesis in plants. Int J Mol Sci. 2018:19(3): 662. 10.3390/ijms19030662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay GP, Su CH, Tzagoloff A. Modular assembly of yeast cytochrome oxidase. Mol Biol Cell. 2013:24(4):440–452. 10.1091/mbc.e12-10-0749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EH, Welchen E, Carrie C. Assembly of the complexes of the oxidative phosphorylation system in land plant mitochondria. Annu Rev Plant Biol. 2019:70(1):23–50. 10.1146/annurev-arplant-050718-100412 [DOI] [PubMed] [Google Scholar]
- Millar AH, Eubel H, Jänsch L, Kruft V, Heazlewood JL, Braun HP. Mitochondrial cytochrome c oxidase and succinate dehydrogenase complexes contain plant specific subunits. Plant Mol Biol. 2004:56(1):77–90. 10.1007/s11103-004-2316-2 [DOI] [PubMed] [Google Scholar]
- Møller IM, Rasmusson AG, Van Aken O. Plant mitochondria-past, present and future. Plant J. 2021:108(4):912–959. 10.1111/tpj.15495 [DOI] [PubMed] [Google Scholar]
- Perez-Riba A, Itzhaki LS. The tetratricopeptide-repeat motif is a versatile platform that enables diverse modes of molecular recognition. Curr Opin Struct Biol. 2019:54:43–49. 10.1016/j.sbi.2018.12.004 [DOI] [PubMed] [Google Scholar]
- Pierron D, Wildman DE, Hüttemann M, Markondapatnaikuni GC, Aras S, Grossman LI. Cytochrome c oxidase: evolution of control via nuclear subunit addition. Biochim Biophys Acta. 2012:1817(4):590–597. 10.1016/j.bbabio.2011.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radin I, Mansilla N, Rödel G, Steinebrunner I. The Arabidopsis COX11 homolog is essential for cytochrome c oxidase activity. Front Plant Sci. 2015:6:1091. 10.3389/fpls.2015.01091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson AG, Geisler DA, Møller IM. The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion. 2008:8(1):47–60. 10.1016/j.mito.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Ren RC, Yan XW, Zhao YJ, Wei YM, Lu X, Zang J, Wu JW, Zheng GM, Ding XH, Zhang XS, et al. The novel E-subgroup pentatricopeptide repeat protein DEK55 is responsible for RNA editing at multiple sites and for the splicing of nad1 and nad4 in maize. BMC Plant Biol. 2020:20(1):553. 10.1186/s12870-020-02765-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkler J, Senkler M, Eubel H, Hildebrandt T, Lengwenus C, Schertl P, Schwarzländer M, Wagner S, Wittig I, Braun HP. The mitochondrial complexome of Arabidopsis thaliana. Plant J. 2017:89(6):1079–1092. 10.1111/tpj.13448 [DOI] [PubMed] [Google Scholar]
- Soto IC, Fontanesi F, Liu J, Barrientos A. Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim Biophys Acta. 2012:1817(6):883–897. 10.1016/j.bbabio.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Zhang X, Shen Y, Wang H, Liu R, Wang X, Gao D, Yang Y, Liu Y, Tan B. The pentatricopeptide repeat protein EMPTY PERICARP8 is required for the splicing of three mitochondrial introns and seed development in maize. Plant J. 2018:95(5):919–932. 10.1111/tpj.14030 [DOI] [PubMed] [Google Scholar]
- Timón-Gómez A, Nývltová E, Abriata LA, Vila AJ, Hosler J, Barrientos A. Mitochondrial cytochrome c oxidase biogenesis: recent developments. Semin Cell Dev Biol. 2018:76:163–178. 10.1016/j.semcdb.2017.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC. Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int J Mol Sci. 2013:14(4):6805–6847. 10.3390/ijms14046805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I, Karas M, Schägger H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics. 2007:6(7):1215–1225. 10.1074/mcp.M700076-MCP200 [DOI] [PubMed] [Google Scholar]
- Xing H, Dong L, Wang Z, Zhang H, Han C, Liu B, Wang X, Chen Q. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014:14(1):327. 10.1186/s12870-014-0327-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.