Abstract
Plant disease outbreaks pose significant risks to global food security and environmental sustainability worldwide, and result in the loss of primary productivity and biodiversity that negatively impact the environmental and socio-economic conditions of affected regions. Climate change further increases outbreak risks by altering pathogen evolution and host–pathogen interactions and facilitating the emergence of new pathogenic strains. Pathogen range can shift, increasing the spread of plant diseases in new areas. In this Review, we examine how plant disease pressures are likely to change under future climate scenarios and how these changes will relate to plant productivity in natural and agricultural ecosystems. We explore current and future impacts of climate change on pathogen biogeography, disease incidence and severity, and their effects on natural ecosystems, agriculture and food production. We propose that amendment of the current conceptual framework and incorporation of eco-evolutionary theories into research could improve our mechanistic understanding and prediction of pathogen spread in future climates, to mitigate the future risk of disease outbreaks. We highlight the need for a science–policy interface that works closely with relevant intergovernmental organizations to provide effective monitoring and management of plant disease under future climate scenarios, to ensure long-term food and nutrient security and sustainability of natural ecosystems.
Subject terms: Pathogens, Microbiome, Climate change, Plant sciences
In this Review, Singh et al. explore the impact of future climate scenarios on plant pathogen burden and biogeography, their interaction with the plant microbiome and the consequences on plant disease and productivity in different ecosystems. They propose different approaches to ensure long-term global food security.
Introduction
Increasing incidence and severity of plant disease outbreaks poses significant and growing risks to primary productivity, global food security and biodiversity loss for many vulnerable areas of the world1–7. These disease outbreaks cause yield and ecological losses. For example, the annual crop yield loss caused by pathogens (microorganisms that cause diseases and constrain host health and productivity) and pests alone is estimated at US$220 billion3–6, directly impacting food security, regional economies and other linked socio-economic aspects. This is further exacerbated by post-harvest loss caused by pathogenic microorganisms such as Penicillium spp. and Xanthomonas euvesicatoria1. Furthermore, climate change poses an increased risk of intensification of plant diseases, putting at risk the world’s food supply and natural plant biodiversity7–9. It is postulated that any potential yield gains in the next five decades will be offset by climate change-mediated altered disease pressure caused by known and emerging pathogens10. Similarly, the spread of pathogens linked to climate change is considered one of the main threats to forest health globally11. Therefore, improved knowledge of climate change impacts on the molecular, epidemiological and ecological interactions between pathogens, plants and the associated microbial communities is needed to develop climate-resilient agricultural and natural ecosystems4,6.
Plants are infected by a diverse range of pathogens, including bacteria, fungi, oomycetes, viruses and nematodes, that differ in their lifestyles (biotrophs, deriving nutrients from living cells, to necrotrophs, deriving nutrients from dead cells), infection strategies (intracellular or extracellular) and target plant tissues (for example, xylem, phloem, roots or leaves) (Supplementary Table 1). A key challenge to predicting plant diseases in space and time is to understand how these different pathogens interact with, and respond to, multiple drivers of disease (for example, other pathogens, host/vectors, commensal microorganisms and environment), and how they jointly respond to climate change. Theoretically, climate change may facilitate plant infection in multiple ways including by altering pathogen evolution, changing host–pathogen interactions and vector physiology, and facilitating the emergence of new strains of pathogens, which in turn can break down host-plant resistance7,12,13. Climate change can also result in the range shifts of pathogens and hosts, which would increase the spread of plant disease into new areas8,10,14,15. Yet we have limited knowledge of how different components of climate change (for example, temperature and rainfall fluctuation) and their interaction with anthropogenic activities impact plant pathogens in both agricultural and natural ecosystems. For example, the abundance of fungal soil-borne plant pathogens is likely to increase in most natural ecosystems under projected climate change scenarios, with significant but unquantified consequences for primary productivity globally14. Similarly, variation in relative humidity affects the abundance and infectivity of pathogens16.
Climate change will expectedly increase plant diseases in crops. Firstly, globalization and international trade have intensified movement of crop pathogens between continents in the past few decades17,18, increasing the risk of transmission from disease-prevalent to disease-free regions. Plant species or cultivars that have not coevolved with the introduced pathogen in the new geographical location are likely to foster pathogen prevalence and disease outbreaks. An example of trade and transport as drivers of pathogen emergence is wilt disease of banana, also known as Panama disease, caused by the soil-borne fungus Fusarium oxysporum f. sp. cubense, which likely originated in Southeast Asia and then spread globally during the twentieth century19. Secondly, climate and ecological changes and modern land management practices dominated by monocultures and high-density crops likely facilitated the emergence and adaptation of plant pathogens able to disseminate beyond their normal geographical ranges. For example, soybean and wheat are extensively grown in high-density monocultures, and their yields are compromised by a plethora of pests and pathogens. Soybean rust caused by the fungus Phakopsora pachyrhizi and wheat blotch caused by the fungus Zymoseptoria tritici are among the most destructive diseases on these crops, and yield losses of more than 50% have been documented during severe epidemics2,20. Despite the complexity of natural ecosystems (for example, biodiversity interactions), climate change and the linked emergence and evolution of pathogens pose similar challenges for wild plant communities and productivity21. For example, global warming-associated range expansion of Phytophthora cinnamomi could have significant negative impact on indigenous plant communities in many parts of the world22,23. A further increase in disease burden as a result of climate change could have devastating consequences for many plant species, food production and security, ecosystem sustainability and social conflicts.
This Review discusses how plant pathogen loads and disease pressure are likely to change under future climate scenarios. We explore current and future impacts of climate change and land use intensification on pathogen biogeography, on interactions between the plant microbiome and plant pathogens, and on plant disease incidence and severity, and their collective influence on agriculture and primary production. We analyse possible mechanisms by which pathogen invasion affects the plant microbiome, and how this knowledge might be harnessed to mitigate the risk of disease outbreaks, via improved disease surveillance, predictive modelling and effective sustainable management strategies8,12. Finally, we propose different approaches that combine pathogen monitoring and policy frameworks to ensure the long-term sustainability of global food security and environmental sustainability.
Climate change and plant diseases
Predicting the impacts of climate change on plant disease is complex and challenging, as multiple aspects of plants, pathogens and the environment are involved. These factors include the distribution and abundance of taxa (geographical range, niche preference), their fitness and virulence, abiotic interactions, plant–microorganism evolutionary processes, host and vector biology, and environmental conditions. For instance, many soil opportunistic pathogens can cause disease outbreaks when environmental conditions become favourable for pathogen replication and vulnerable hosts are available24,25. Supplementary Table 1 summarizes the responses of several plant pathogens, the damage they cause to the plant and their geographical distributions21. Climate change can also indirectly affect plant–pathogen interactions through alterations in the biochemical, physiological, ecological and evolutionary processes of the plant host and/or pathogen7,24,26,27 (Fig. 1). For example, prolonged drought causes water stress in forest trees, which results in increased susceptibility to infection by pathogens causing dieback disease from the genus Phytophthora, thus facilitating the occurrence of potentially new diseases28–30. Overall, the direct impacts of climate change are likely to vary depending on the pathogen, host identity and properties of biomes. Discussed in the following sections, there is limited but increasing evidence suggesting that climate change has a direct impact on pathogen virulence and disease development.
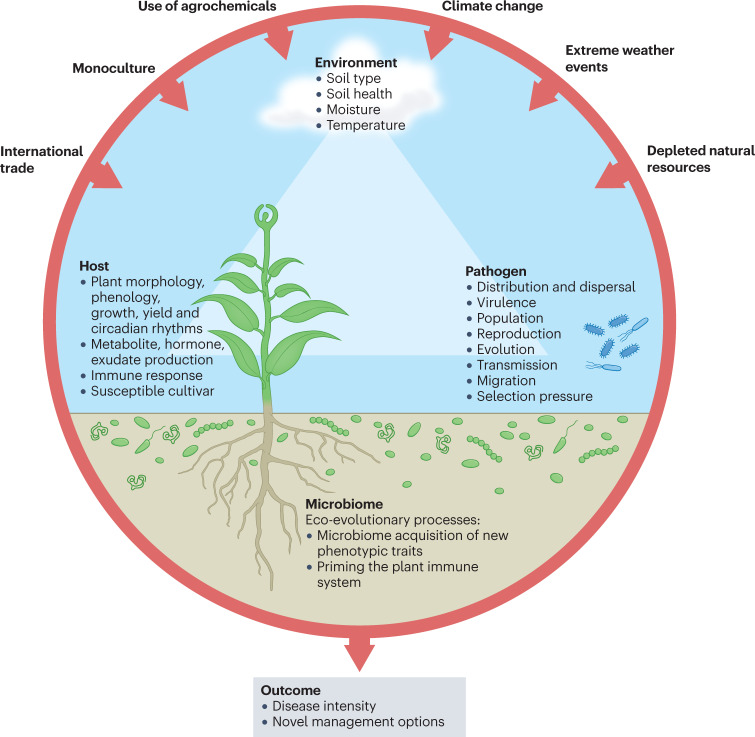
Fig. 1. A new angle in the disease triangle paradigm that considers the plant microbiome as a pivotal factor influencing plant disease.
Intimate interactions among the plant, the environment, the soil and plant microbiomes, and invading pathogens impact the outcome of infection processes, disease severity and productivity of the plant. Environmental change and human activities (for example, global commodity and climate change) drive pathogen evolution and have increased disease threats to global crops. Genetically uniform crop monocultures and high planting density in modern agriculture have accelerated the emergence of virulent pathogens capable of overcoming disease-resistant crop varieties and promote the pathogen’s population size and genetic variability. Similarly, overreliance on pesticides has also fostered rapid emergence of new strains of pathogens. Pathogen transmission and anthropogenic pathogen movement due to, for example, international trade spreads pathogens to places free of natural enemies, and allows exchange of genetic material via horizontal gene transfer, facilitating adaptation to local hosts. Depletion of natural resources and natural landscapes has caused deterioration of the agroecosystem diversity. Emerging evidence suggests that soil and plant microbiomes influence the three angles of the disease paradigm — the host, the pathogen and the environment — by either facilitating or supressing pathogen attacks, by affecting plant physiology and immune response, and providing a line of defence and manipulating environmental conditions. For example, in disease-suppressive soils, indigenous microbiomes can reduce disease incidence, even in the presence of a pathogen, a susceptible host and a conductive environment. Explicit consideration of the role of the microbiome can improve our mechanistic understanding of disease outbreaks, which may lead to more effective prediction, monitoring and management of disease outbreaks. Better land management practices can improve overall soil health by influencing the diversity and functions of soil microbial communities, and could potentially be used to steer microbiomes that suppress diseases.
Elevated temperature
Climate warming can significantly impact aspects of the population dynamics of pathogens, such as overwintering and survival, population growth rates or the number of generations of polycyclic species. For example, a reduced diurnal temperature decreases the latency period of the coffee leaf rust pathogen Hemileia vastatrix, promoting rust epidemics in Central America31. Warming temperatures shorten the pathogen’s incubation period, resulting in increased abundance of the pathogen over a growing season. Higher temperature (along with high humidity) is linked to enhanced disease severity of potato blight pathogen (Phytophthora infestans) and phoma stem canker of oilseed rape. Increased mean winter temperatures enhanced the infection rates of American chestnut by fungal Phytophthora spp. causing extreme tree death events in North America32. Finally, a 30-year study reported the link between early snowmelt and higher snow blight (Phacidium infestans) infection in pine trees33.
Changes in global temperatures can profoundly impact the occurrence of pathogens in agricultural and natural ecosystems, increasing the risk of exposure to new pests and pathogens. Global warming is projected to increase the abundance of many fungal soil-borne plant pathogens, with significant consequences for primary productivity14. Warming temperatures can result in the development of new strains of pathogens that are better adapted and more virulent7,12,13,19. The severity of Fusarium head blight of wheat is likely to increase due to the shift from the milder Fusarium culmorum that prefers cool and wet conditions to the more aggressive Fusarium graminearum that prefers warm and humid conditions34. Similarly, more aggressive and temperature-tolerant novel strains of Puccinia striiformis have replaced older strains and are causing major outbreaks of wheat rust in the United States, Australia and Europe35,36. Warming temperatures can increase the range of many pathogens that are currently limited by requirements for overwintering, such as wheat stem rust caused by Puccinia graminis f. sp. tritici37. On the other hand, over a period of 30 years with a steady rise in summer temperatures, local extinction of Triphragmium ulmarie, the rust pathogen that infects Filipendula ulmaria (meadowsweet), was observed38. Other pathogens, such as Phytophthora infestans, are predicted to be little impacted by warming temperatures due to their lower thermal preferences39.
The molecular basis for why plants are more susceptible to pathogens at high temperatures is not well understood13. However, elevated temperatures can suppress plant immunity, leading to increased pathogen infection24. In Arabidopsis, production of salicylic acid, a hormone critical to plant defence, is suppressed at high temperatures40 due to impaired activation of master immune transcription factors such as CBP60g (ref. 41). The CBP60g family transcription factors are widely conserved in plants42, and understanding their role in thermosensitive regulation of plant immunity provides clues for improved understanding of the warming effect on plant diseases. In rice, warm temperatures enhance expression of abscisic acid biosynthesis and responsive genes, and this is associated with increased susceptibility to bacterial blight disease; interestingly, suppression of the abscisic acid pathways was associated with resistance at elevated temperatures43. A recent study reported that induction of jasmonic acid biosynthesis and signalling genes by Magnaporthe oryzae results in enhanced susceptibility to rice blast disease in rice at warm temperatures44.
Elevated carbon dioxide
Varied disease incidence in conditions of increased carbon dioxide (CO2) concentrations suggests pathogen and host-dependent responses to CO2. Elevated CO2 levels increased the severity of powdery mildew on cucurbits caused by Sphaerotheca fuliginea45, as well as head blight and blotch on wheat caused by Fusarium spp. and Septoria tritici, respectively46, whereas the susceptibility of soybean towards the downy mildew pathogen Peronospora manshurica was reduced47. Similarly, changes in the leaf surface characteristics caused by elevated CO2 treatment enhanced rust disease of aspen trees48 but reduced the disease severity of brown spot disease of maple trees49. Atmospheric CO2 impacts plant immune responses and hormone levels that can influence plant–pathogen interactions. For example, increased basal expression of jasmonic acid-responsive genes under elevated CO2 enhanced resistance to the necrotrophic leaf pathogen Botrytis cinerea, but reduced the resistance to the hemi-biotrophic leaf pathogen Pseudomonas syringae pv. Tomato50. Reduction in the effectiveness of plant defence pathways under elevated CO2 increased the susceptibility of wheat against the two major pathogens Z. tritici and F. graminearum that cause S. tritici blotch and Fusarium head blight, respectively46. Elevated CO2 impacts tripartite biotic interactions between wheat, barley yellow dwarf virus (BYDV) and its aphid vector Rhopalosiphum padi. BYDV infection increased the aboveground nitrogen content of wheat growing under elevated CO2, as compared with non-infected plants, thereby reducing vector performance and phloem ingestion51. Elevated CO2 clearly influences the outcome of plant–pathogen interactions, but currently no unifying framework exists to understand and predict its effects and consequences.
Climate change-induced variability in water availability
Variations in relative humidity and soil moisture are among the main drivers of abundance and infectivity of plant pathogens, and therefore climate-induced changes in humidity will likely impact future plant disease outbreaks16. Many fungal diseases require high humidity for spore germination and infection of their host plants16. High humidity generally promotes the virulence of pathogens infecting aerial plant tissues. Infection rates by Sclerotinia sclerotiorum in lettuce52 and the stem rot pathogen Phytophthora sojae are higher under increased humidity53. Humidity-dependent expression of bacterial effectors that modify plant immune responses promotes establishment of P. syringae in the aqueous intercellular space (apoplast) of Arabidopsis leaves54. Higher humidity is also correlated with the increased production of the mycotoxin deoxynivalenol by F. graminearum, a pathogen infecting a range of grains, which results in significant economic losses and a reduction in food quality55,56. On the contrary, for M. oryzae, the causal agent of rice blast, and Streptomyces spp., causing bacterial scab in potato, lower moisture conditions increase pathogen numbers and disease severity57,58. Recent analyses suggest that an overall increase in relative humidity can increase the incidence of fungal-caused diseases in general16.
The impacts of drought on infection rates of pathogens and disease severity vary dramatically59. For example, diseases such as pea root rot (caused by Aphanomyces euteiches), onion white rot (Sclerotium cepivorum), wheat take-all (Gaeumannomyces graminis var. tritici), wheat crown rot (Fusarium spp.), brassica black leg (Leptosphaeria maculans) and grapevine black foot (Ilyonectria/Dactylonectria spp.) increase in severity with the increase in the length and frequency of drought. On the other hand, drought reduced the severity of kiwifruit sclerotinia rot (S. sclerotiorum) and radiata pine red needle cast (Phytophthora pluvialis)59. Similar results were reported for the bacterial pathogen Xylella fastidiosa of grape60. In general, necrotrophs will accelerate drought-induced tree mortality by depleting tree resources as a result of repair and compartmentalization processes, whereas biotroph-caused diseases are expected to be less severe in drought because of the strong connection between pathogen performance and tree nutritional status. However, if biotrophs are able to invade stressed trees, they are expected to cause more severe drought-dependent impacts on trees because they deplete carbohydrate reserves important for tree drought tolerance61.
Drought-mediated shifts in the direction and strength of plant–pathogen interactions across an aridity gradient can modify disease range expansions in response to climate change15. For example, drought and higher rates of tree mortality in arid regions accelerate the decline in pine blister rust at low elevations, whereas lower alternate host occurrence at high elevations dampens infection probabilities, even as the climatic conditions become more hospitable. Drought can also result in the emergence of new pathogens that can withstand harsh environmental conditions and take advantage of the changes in plant physiology in response to stress. For example, drought favours the infection of chickpea plant by the dry root rot fungal pathogen Macrophomina phaseolina62. Drought-induced lowering of plant basal immune responses increased potato yellow vein virus infection and yellow vein disease symptoms63. These changes further modify host–virus–vector (greenhouse whitefly) interactions resulting in enhanced horizontal transmission of the virus.
Other variables and future scenarios
Although we have a limited understanding on the combined effects of multiple environmental factors on plant–pathogen interactions, a few studies have demonstrated that the combined effects are more pronounced than individual effects64–66 and, in some cases, combinations of factors are required for outbreaks16,67. For example, an abnormally warm and humid pre-harvest season as a result of climate change was ascribed to the outbreak of M. oryzae triticum, the causal agent of wheat blast disease, in Bangladesh68. Similarly, high humidity and increased temperature promoted the disease incidence of B. cinerea in grape berries69. Altered climates (for example, increased temperature and soil moisture) can promote pathogen invasion and transmission across novel geographical and host ranges. In this respect, some fungal pathogens are more likely to spread in new regions of temperate and boreal biomes, as annual temperatures increase14, with disproportionally high negative impacts predicted on yield in Europe, China and some South American countries. Recent efforts have focused on understanding the global distribution of plant fungal pathogens under future climatic conditions14, but progress is limited by our knowledge of the current distribution of many important pathogens5,70 (Supplementary Table 1). Modelling combined with experimental data suggested that the prevalence of key soil-borne fungal pathogens belonging to Alternaria, Fusarium, Venturia and Phoma genera will likely increase under projected global warming14. Further, our re-analysis of published global survey data14,71 suggests that the relative abundance of some important soil-borne fungal taxa such as Penicillium spp., which damage fruit quality and production, are strongly associated with shifts in temperature and organic matter. Similarly, range expansion for Botryosphaeria dothidea and Neufusicoccum parvum resulting in more frequent and intensive disease outbreaks is predicted to be linked to climate change72. Conversely, the relative abundance of other soil-borne pathogens, such as the Oomycota taxa Phytophthora spp. and Pythium spp.70, may be highly sensitive to changes in soil pH associated with modifications in land use, and their distribution is likely to vary in response to climate change and land use intensification, with implications for food security worldwide (Fig. 2). Multiple environmental factors are known to interact with soil-borne pathogens, explaining complex patterns in the distribution of these microorganisms at a global scale (Fig. 2). Global shifts in the distribution of pathogens are concerning, as increasing evidence suggests that pathogens cause more damage in newly invaded regions and on new hosts than in their native region and hosts. As an example, ash dieback caused by the fungus Hymenoscyphus fraxineus causes minimal damage to ash native to Asia, where the pathogen originated, but has devastated European ash trees since invading Europe 30 years ago.
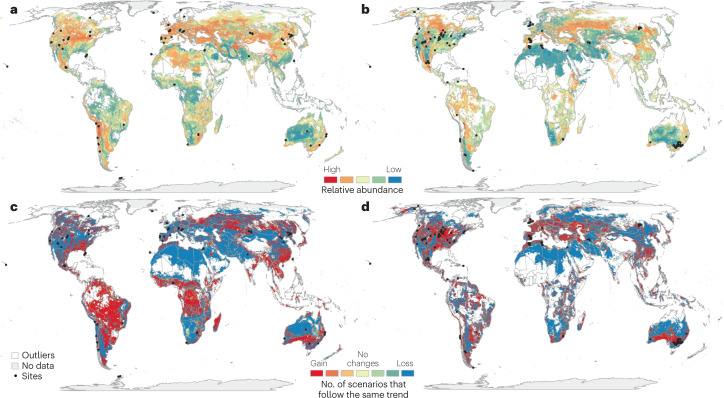
Fig. 2. Projected shifts in relative abundance of soil-borne pathogens from current to future climates.
a,b, Current relative abundance of soil-borne potential plant pathogens: Phytophthora spp. and Pythium spp. (panel a) and Penicillium sp. (panel b). c,d, The projected change in their abundance under predicted future climates (2050): Phytophthora spp. and Pythium spp. (panel c) and Penicillium sp. (panel d) (also see Supplementary Table 1). Previously developed models14 were implemented to project each map of the current and future relative abundance of plant pathogenic taxa worldwide. To implement these models, we performed exploratory correlation analyses to identify the most important factors associated with potential plant pathogen distributions from available data (see Supplementary Information). We used available data sets of climate variables, vegetation type, elevation and soil variables to identify the global distribution14,141. To perform projections of these pathogens in future climates, we used climatic and land use available data sets142–145. The prediction can be improved, as data from other locations will become available in the future. Areas of the projection away from the sampling points have been marked in white. The masking criterion was P < 0.01 to show the areas generated by the model in the projection that are closer to the sampling points (see Supplementary Information).
The interaction between climatic changes and evolutionary processes can also directly impact future pathogen outbreaks. Increases in frequency and intensities of extreme weather events can help spread pathogens to new locations, as for the case of soybean rust, which was introduced from Brazil into the United States by a hurricane2. Plant pathogens can also evolve to infect other plant species and/or become more virulent to overcome chemical and resistant cultivar control, or they may evolve into new pathogens by interspecific hybridization and extensive mutations. For example, a hybrid Phytophthora alni species that originated from the hybridization of Phytophthora uniformis and Phytophthora multiformis is the cause of severe decline of alder populations across Europe73. Overall, climate change will cause unknown shifts in the pathogen biology, host specificity and environmental favourability, making predictions challenging. Forecasting disease epidemic risks is also compounded by the interaction between agronomic practices (for example, agrochemicals, irrigation, plant variety), native and alternate host availability, connectivity and pathogen dispersal mechanisms (for example, airborne or vector-borne). Intensifying extreme weather events together with variations in land use and global trade are likely to further exacerbate pathogen transmission and disease incidence74. However, we have little knowledge about how these interactions among pathogen, biogeography, host and environment will influence disease management and efficacy of chemical, cultural and biological controls, key unknowns in securing food and environmental security for current and future generations.
Climate change, the plant microbiome and disease
Responses of the plant microbiome to climate change can also indirectly impact disease incidence. Plant microbiomes are likely impacted by climate change both via alterations in the starting inoculum from bulk soil or the rhizosphere75–77 and by host responses, which include changes in host physiology, morphology, exudation patterns and immune responses27 (Fig. 3). This is important because plant-associated microbiomes exert strong influences on host physiology, and contribute to the regulation of its metabolism, immune function and fitness in novel environments25,27,78, and play a key role in preventing the colonization and growth of pathogens. A range of mechanisms contribute to the control of plant pathogens by the plant microbiome, including the modulation of plant immune responses, competition with pathogens for resources and space, and/or production of antifungal effectors, lytic enzymes and secondary metabolites (including antibiotics, bacteriocins, toxins and siderophores)25. Plants can employ the ‘cry for help’ strategy that uses chemical stimuli for recruitment of beneficial microorganisms and traits from the soil, in order to enhance their capacity to combat pest-induced or pathogen-induced stresses79–81. The selective recruitment of beneficial microorganisms occurs through modulation of plant–microbiome signalling pathways78,79,82, altered root exudation patterns81,83,84 and/or production of volatiles85. The most well-studied example of microbiota-mediated disease protection is disease-suppressive soil, where active microbiota contribute to disease reduction, even in the presence of the pathogen, susceptible host and favourable environmental conditions80,86,87. Complex ecological interactions and communication between plants and pathogens, between plant microbiomes and pathogens, and between plants and their microbiomes define disease outcomes but the specific mechanisms of interaction and the identity of communication molecules remain unclear88.
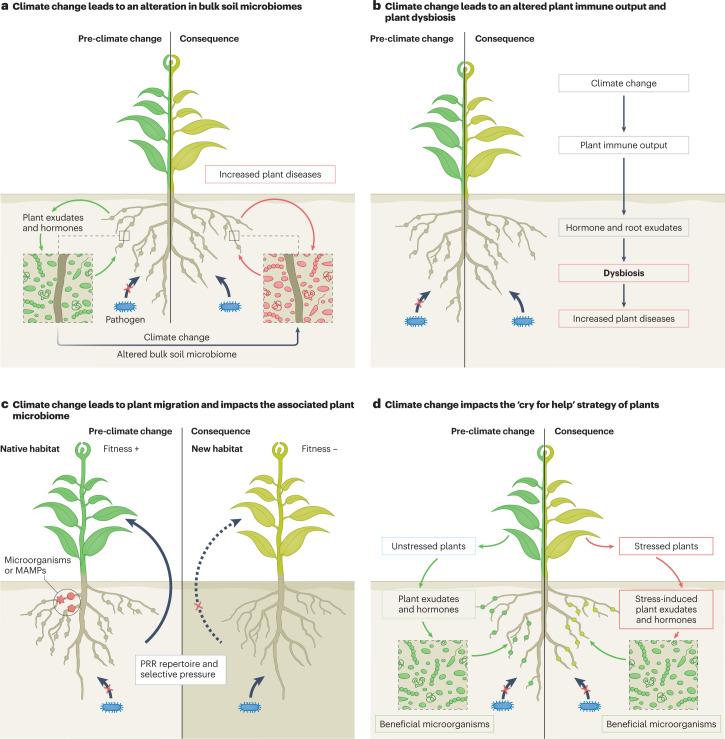
Fig. 3. Responses of plant microbiomes to novel climates and their consequences on disease occurrences.
Four scenarios are proposed for microbiome responses to climate change. Green plants symbolize the healthy state of plants prior to climate change, whereas yellow plants indicate the effect of climate change and pathogen infection on plants. a, Plants employ an array of mechanisms that depend on optimal immune response, root exudates and hormonal balance to assemble complex microbiomes. Plant-associated microbiomes in soil, particularly in the rhizosphere, provide the first line of defence against pathogens. Climate change will likely alter the structure of the microbial reservoir in the bulk soil. This together with changes in the plant immunity may alter the rhizosphere microbiome assembly and change the first line of defence, which would allow the pathogen to breach. b, Plants maintain homeostasis of associated (leaf, root, stem, endophytes) microbiomes via tight and complex regulation of immune systems. Climate change can alter plant physiology and immune response, such as the production of the root exudates, volatile organic carbons and phytohormones, which may constrain the ability of plants to recruit and assemble beneficial microbiomes and promote dysbiosis of the plant leading to diseases. c, Plant migration to new locations (niche range shift) may interrupt the plant immune system and its mutualistic coevolution with indigenous soil microbiomes that support healthy plant growth and disease tolerance. As such, plant migration may expose them to local pathogens to which they are susceptible, whereas in some cases, migrating plants will escape local pathogens. Similarly, niche range shift in pathogens (along with evolutionary processes) can make the pathogen more transmissible and virulent in new regions in the absence of an effective immune response of local plants and resistance by local microflora. d, The ‘cry for help’ strategy of the plant, which refers to the plant recruiting beneficial microorganisms when under pathogen attack, is also likely be altered under climate change. Climate change may constrain the abilities of plants to produce signal molecules (for example, root exudates, volatiles and so on) to attract beneficial microorganisms and/or shift microbial composition and traits, or their ability to respond to these signals. Climate change could also reduce the burden of pathogen attacks where a shift in microbiomes has either enriched beneficial microorganisms or primed the plant immune response. MAMP, microorganism-associated molecular pattern; PRR, pattern recognition receptor.
Climate change variables (elevated CO2, warming and drought) can also increase root exudation and alter exudate composition in both crops and trees to attract beneficial microorganisms, which ultimately support plant growth89–91. Pathogens can overcome the first line of plant defence either directly, via competition with native microbial communities, or indirectly, by inducing changes in plant biology and physiology (for example, root exudation)83. The xylem-colonizing vascular wilt fungal pathogen Verticillium dahliae exploits effector proteins with antibacterial properties to manipulate the plant microbiota and colonize the host92. Pathogens can also modulate the plant microbiome by altering plant defence responses. For example, immune suppression by the wheat fungal pathogen Z. tritici causes fluctuations in the leaf microbial communities that render the plant vulnerable to further infections93. If the pathogen manages to displace a highly interacting keystone microbial species that facilitates interactions in the community, the entire microbial network may collapse, resulting in severe impairment of plant performance94. Members of the plant microbiome can themselves facilitate pathogen progression in some cases via signalling, metabolic interactions and weakening of the host immune response95,96. For example, the causal agent of olive knot disease, Pseudomonas savastanoi pv. savastanoi, exchanges quorum sensing signals with the native non-pathogenic strains of Pantoea agglomerans (an epiphyte, grows on the surface of the plant) and Erwinia toletana (an endophyte, lives within the plant) for increased colonization and disease incidence97. In many cases, shifts in the structure and function of plant microbiota have been observed for various plant–pathogen complexes98–100. A pathogen can co-occur with a range of members of plant microbiota101; however, it is not yet clear whether observed changes in the microbial community composition contribute causally to pathogen colonization and disease.
As climate change can directly impact plant immune responses24,40,102, there is a possibility that plant microbial dysbiosis can facilitate pathogen invasion. For example, climate change-induced alteration in plant immune systems, which suppress pathogen invasion, can also negatively affect plant-microbiome assembly. For example, warming can increase or decrease microorganism-associated molecular pattern (MAMP)-triggered immunity and suppress plant effector-triggered immunity26,103,104. Similarly, production of salicylic acid is decreased under warming and drought. Climate-induced shifts in geographical distribution are suggested to have resulted in changes in the MAMP-induced selective pressure on pattern recognition receptor (PRR) repertoires, thereby impacting pathogen response, host fitness and the microbiome105. As the plant microbiome provides the first line of defence against pathogen invasion, changes in its structure and function can influence pathogen colonization. Plant immunity (including effector-triggered immunity, pattern-triggered immunity, RNA interference and defence hormone induction) has evolved not only to limit the growth of pathogens but also to maintain the homeostasis of the plant microbiome105. A recent study demonstrated the role of pattern-triggered immunity in preventing dysbiosis in the phyllosphere by regulating community structure and microbial abundances106. However, it remains to be evaluated whether plant immune-mediated alterations in the microbial community structure impact plant–pathogen interactions directly via shifting their overall response towards the incoming pathogen or indirectly via changes in plant fitness.
Although plant–microbiome–pathogen interactions will likely be altered in future climatic scenarios, we have limited knowledge to predict the overall directions and outcomes. We propose amendments to existing concepts (disease triangle and Koch’s postulates) and argue for explicit consideration of environmental and host microbiomes in disease concepts to obtain better mechanistic understanding of outbreaks for improved disease management. This can be further boosted by explicit inclusion of eco-evolutionary frameworks in future research, which will improve mechanistic knowledge and predictive models of pathogen invasion and disease outbreak (Box 1). For example, an experimental evolution study demonstrated that the relationship between a plant virus and its natural host can evolve from pathogenic to mutualistic under severe drought conditions107. As microbiomes are critical contributors to plant adaptation, we postulate that beneficial plant–microbiome interactions will evolve to maximize plant fitness against combined biotic and abiotic stresses under future climatic conditions27,108. For example, when exposed to soil-borne pathogens, the root-specific transcription factor MYB72 and the β-glucosidase BGLU42 regulate the synthesis and secretion of a coumarin molecule that inhibits pathogens but favours rhizobacteria, which induce systemic resistance109. Interestingly, coumarin accumulation is induced by osmotic and temperature stresses110, indicating a possible interplay of the plants’ ‘cry for help’ for selective recruitment of microbiota to tackle multiple stresses.
Box 1 Eco-evolutionary theories for prediction and management of plant diseases.
Utilizing invasion ecology theory can help predict the success of new and emerging pathogens in new environments. Despite some variability, experimental and observational studies on plant communities have demonstrated that invasion success is linked to high dispersal ability, growth rate and resource use efficiency of alien species and/or to indigenous communities with low species diversity, high frequency of disturbances and rapid changes in resources139. Pathogen invasions are widespread and, similar to plant invasion, include dispersal, establishment, growth and spread, and impacts140. Ultimately, the success of pathogen invasion is likely influenced by the composition and diversity of the indigenous soil and plant microbial communities, where abiotic feedback (for example, pH and resource availability) and biotic feedback (for example, microbial–microbial interactions or impact of viruses and invertebrates; see the figure) modulate the rate and stability of colonization. Employing invasion theoretical frameworks can thus help in better predicting successful invasion by new pathogens, while also having the potential to support the development of disease management approaches, for example by providing tools to identify biological controls of pathogens146. However, it is important to note that microorganisms do have some distinct characteristics in terms of their physiology, genetics and behaviour. For example, unlike plant competition, microbial competition occurs via indirect interactions regulated by antibiotics, rapid gene acquisition and quorum sensing147. Explicit consideration of distinct microbial (pathogen) characteristics in an empirical framework can provide critical advancement in the understanding of pathogen invasion.
Other mechanisms that can help pathogen colonization include frequent disturbances (for example, increasing extreme weather events) and evolutionary processes. Frequent disturbances often alter indigenous communities, creating new niches and, hence, promoting invasion by alien species or niche occupation by less dominant species140. This is likely facilitated by increases in resource availability and decreased competition, as the abundance of the indigenous community decreases following disturbance (niche availability)148. Niche (complementarity) theory can help in identifying the relative contribution of microbial physiology, alteration in the indigenous community and disturbance-induced niche availability to the success of pathogen invasions. In addition, evolutionary processes can promote rapid diversification of pathogens that can promote a higher rate of colonization. Short generation times, large diversities and population sizes, and horizontal gene transfer can result in invading pathogens and/or indigenous populations of soil and plant microbial communities being able to rapidly acquire new phenotypes (for example, antibiotic biosynthetic genes, fungicide resistance genes) with direct impacts on the rate and success of pathogen colonization147. A system-based approach that addresses key eco-evolutionary mechanisms (pathogen ability versus habitat properties versus disturbance-induced niche availability) and identifies their relative contributions, combined with appropriate modelling tools, can significantly advance the discipline. For example, the use of random forest and structural equation models can identify the relative contribution of different factors, whereas the use of spatial individual-based models that incorporate an adaptive process, diversification and emergent behaviour can significantly improve our ability to predict the rate and success of pathogen invasion under different climate and environmental settings149–151. This framework can also be used to predict the success of microbial inoculants for pathogen controls.
The figure shows that pathogen (or any other invading or introduced microorganism) colonization involves invasion (dispersal ability), followed by microbial establishment, which requires the microorganisms to overcome biotic and abiotic constraints of the invaded environment, and to find a niche and manipulate the environment for proliferation and spread140. Effective colonization will have functional consequences. Ecological theories suggest that four ecological processes (dispersal, selection, drift and diversification) determine the success of colonization of new habitats by invading microorganisms.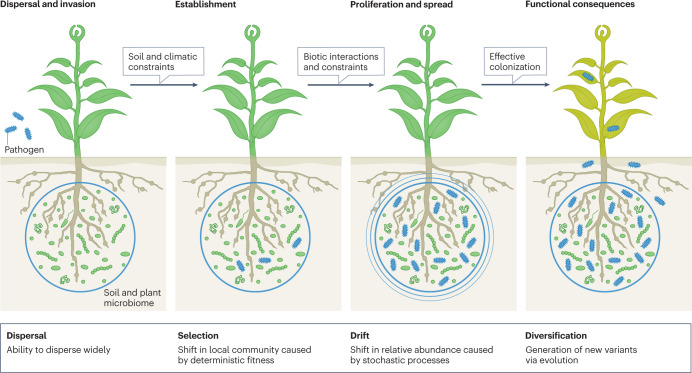
Paths forward
We propose three paths forward that address different but complementary disciplines of disease management.
Modern agriculture, pathogens and future mitigation strategies for sustainable land management
Pathogens are known to be highly sensitive to land management practices. For example, nitrogen and phosphorus fertilization consistently promote pathogenic over mutualistic fungi in grassland soils across four continents111. Conventional approaches to manage diseases rely on chemical fungicides and the use of disease-resistant varieties to control pathogens, but these methods have arguably reached their plateau, as in the case of soil-borne fungal pathogens which have increasingly become resistant to fungicides112. Currently, no effective chemical controls are available for diseases caused by widespread soil-borne pathogens such as Fusarium and Verticillium spp. in many crops113. Further, extensive use of chemical controls is increasingly discouraged due to both policy (for example, EU Green deals require 50% reduction in chemical pesticide use by 2030) and consumer demands, given their negative impacts on biodiversity including beneficial microorganisms, soil health and food quality (chemical residues), and ultimately human health114,115. Development of ecologically friendly chemicals needs prioritization, but these are currently expensive, and their development is time consuming. Also, there remains a risk: mid-term and long-term assessments have shown that chemicals initially considered environmentally friendly, such as organophosphorus and neonicotinoid, are in fact environmentally damaging.
The control of pathogens in natural ecosystems is addressed by different strategies at all levels. For example, maintaining species diversity in forest ecosystems could substantially reduce invasion of generalist plant pathogens116. Agroecosystems can adopt some of these approaches by promoting agrobiodiversity, intercropping and regular rotations of crops to improve resilience. However, cultural controls using rotations or fallow are economically challenging and are increasingly becoming ineffective, as pathogens evolve to be less sensitive to these practices. In the past, four or five non-cotton rotations were sufficient to reduce Fusarium and Verticillium wilt diseases in Australian cotton farms and allow profitable cotton production. Now, five to seven rotations are required for disease management117. Current strategies to address these problems include integrating disease resistance genes into crops by breeding (taking 10–20 years to release new crop-resistant cultivars) and through transgene cloning or gene editing (taking a few years but facing public and political concerns)118,119. In some cases, plant gene-mediated resistance can be overcome rapidly by evolving pathogens. The implementation of non-chemical tools such as biological control is complex, and outcomes are inconsistent120. Thus, fundamentally new approaches are needed to move beyond the current paradigm of disease management. Approaches that harness ecological and evolutionary interactions and other nature-based methods can provide future effective tools.
Technical innovations to monitor, manage and mitigate disease risks under global change
Current approaches to monitor, manage and mitigate disease risks are constrained by a focus on single-pathogen, single-crop and single-disease paradigms121. However, expanding studies that include other key aspects of disease, such as associated soil microbial and faunal communities and their interactions — a phytobiome approach122 — can provide better scientific knowledge to drive improvement of predictive and management tools123,124. Similarly, integrating the biology and ecology of vectors and their response to climate change can improve predictability and risk associated with vector-borne pathogens (Box 2). However, accomplishing this is not trivial and will require a transformative shift in our approaches to plant diseases. For example, traditional notions such as the disease triangle would benefit by including emerging ecological concepts in host–pathogen interactions, such as the role of soil and plant (also vector) microbiomes (Fig. 1). Similarly, certain diseases can be caused by multiple microorganisms (for example, tomato pith necrosis, grapevine decline diseases125) working in tandem and some of them cannot be isolated and re-introduced, limiting the effectiveness of the Koch postulates in establishing causative relationships between a microorganism and a disease125.
A transdisciplinary approach to understand pathogen biology and ecology from molecular to global scales is needed. Integrating available pathogen biology data with transport, trade, climate and geography can improve monitoring and predictive power of disease incidence. This can be further boosted by biochemical sensors, permanent observatories (airborne signals and vectors), satellite and remote sensing tools, artificial intelligence and the involvement of farmers and other volunteers to get an early report of disease, which can contribute towards effective surveillance126. Local data can be used to forecast the spread and severity of disease, and this can be upscaled to regional and global scales utilizing modelling and artificial intelligence tools (Box 2). These predictive tools would also contribute towards assessing the impact of disease on food production and other socio-economic indicators (jobs, income, mental health), thus supporting the development and implementation of effective mitigation tools5. In parallel, decision support systems based on systems biology approaches would be effective to manage disease risks, surveillance and forecasting. Such approaches should include precision agriculture, new eco-friendly chemicals and biologicals (for example, the microbiome and synthetic biology products), consideration of host genetic diversity and prediction of climate change on pathogens, host and vectors121. By integrating the impacts of climate change on agriculture and ecological sciences into predictive tools that also consider socio-economic information and international trade routes, better policies to effectively manage the risks of plant diseases could be developed.
A concerted effort involving technical innovations in microbiome, synthetic biology, precision agriculture and agroecological tools is needed to develop effective and sustainable solutions for plant disease managements (Box 3). Precision agricultural technologies based on drones, artificial intelligence and machine learning can identify disease early and assist effective and environmentally friendly targeted control measures, such as plant removal or precise dispersal of chemicals to plants. Recent advances in microbial (for example, microbiome engineering approaches), biochemical (volatiles and plant elicitors) and synthetic biology (synthetic microbial community (SynCom)) tools provide new pathways of disease management and reduced dependency on chemical controls127,128. In particular, there are emerging interests to harness plant and soil microbiomes to mitigate the negative consequences of climate change, varying from direct manipulation of microbiomes to indirect manipulation of their functions through changes in land management and farming practices, and the use of inoculants or biochemical products27. Beneficial plant microbiota can be harnessed to enhance plant fitness, reduce pathogen loads and prime plant defence signalling pathways129–131. Hence, microbial tools such as inoculants or microbiome engineering in situ promise optimized plant growth under the increasingly stressful conditions and pathogen attacks. Identifying beneficial seed and plant microbiota that can resist pathogen infection could, potentially, provide effective management tools as some of these microorganisms are inherited or actively recruited by the plant, and can hence improve the probability of successful host colonization and, ultimately, disease resistance132,133. Further, future plant breeding programmes should explicitly adopt a plant holobiont (host plus associate microbiomes) concept to ensure that new varieties can harness microbial symbionts to combat disease incidence134. Such tools can play an important role in mitigating deleterious consequences of pathogens on farm productivity and food availability under a climate change scenario135. However, systematic and coordinated studies are needed to advance understanding of the ecological and evolutionary processes that underpin interactions between plants, the associated microbiomes and pathogen invasions and how these are influenced by climate change, land management, agronomic practices and other ecosystem characteristics. This needs to be done with consideration of the climatic zone, crop and disease types, as well as management practices.
Box 2 Modelling future disease outbreaks.
There is emerging consensus that climate change will increase disease risk and pest impacts in many parts of the world10,152. Crop models that included crop phenology and pathogen development show that the impact of climate change on crop losses would be host–pathogen dependent39,153,154. For example, shifts in planting dates due to cooler seasons will reduce the global risk of potato late blight by the end of this century39. On the other hand, climate change-mediated early anthesis in wheat will cause an increased incidence of Fusarium ear blight disease in China153. Mathematical models that combine epidemiology with spatial components and population heterogeneities are powerful tools to quantify the likelihood of success of management practices applied to invading pests and pathogens155–158. These models provide critical information on the most effective control measures, time and suitability of the intervention, site selection and cost balance to realistically manage emerging pests and pathogens. Unfortunately, the lack of robust quantitative and standardized data hinders efforts to make meaningful predictions to infer climate change impacts on disease epidemiology across crops, agroecosystems and regions159. Furthermore, although extreme weather events are predicted to increase in frequency and intensity in the future160, due to the lack of robust data sets we have limited modelling capability to predict the impact of these events on disease incidence and severity.
Disease epidemiology and dynamics depend on many factors including the interplay of the pathogen with the host plant, microbial communities, the changing environment and more. Linking information across scales (from genomes to landscapes) to predict disease outcomes in a rapidly changing world is a significant challenge161. However, many connections at intermediate scales are viable with integrated application of new systems biology approaches and powerful analytical and modelling techniques. For example, ‘omics’ combined with robust physiological/morphological/symptom training data sets can be used for predicting different aspects of plant–pathogen interactions including gene regulatory networks, pathogen effector proteins, pathogen adaptive strategies and genes involved in plant–pathogen interactions under different climate change scenarios. Genome-scale network reconstructions can model intracellular metabolism to predict virulence and pathogen–host interactions under a range of environmental and physiological conditions162–164. Such models are now being used to provide detailed insights into the interactions between the invading pathogen and the host-associated microbiome to predict disease incidence and interventions165. Ecological modelling approaches can provide information on the successful colonization of invading pathogens under a complex host-associated microbiome. For example, ‘game theory’ and the ‘Lotka–Volterra model’ can be expanded to describe microbial interactions related to pathogen colonization or extinction to predict disease emergence166. Statistical models can provide information on the direct and indirect impacts of environmental and biotic variables on disease incidence167–169, whereas dynamic network models allow incorporation of several aspects of disease epidemiology including molecular and cellular reactions, plant–vector–pathogen interactions, species interactions in the microbiome as well as international trade and social networks (reviewed elsewhere170). Predictions of disease risks based on combinations of aerobiological models for inoculum transmission and crop-growth models offer a framework to quantify the impact of future climates on the risk of disease occurrence and spread171,172. However, most model development involves multidisciplinary knowledge integration, and the modelling approach should be transparent and flexible to allow users to select the level of details with which they would like to engage126. A model example is EMULSION, which couples a generic simulation engine to a domain-specific modelling language through structured text files, is readable by scientists from different fields (epidemiologists, biologists, economists) and allows those scientists to validate or revise assumptions at any stage of model development173.
Box 3 Technical innovations to better manage future disease outbreaks.
Advancing fundamental knowledge of pathogen, vector and host biology, ecology and evolution is a key step forward to unravel the complexities of disease incidence and severity and the socio-economic impacts. How these variables will respond to climate change and the integration of such knowledge with climate and weather data, international trade and early pathogen detection can provide effective tools for forecasting and monitoring. These data can form the essential part of new integrated models along with big data analytics and artificial intelligence to predict regions at most risk of future disease outbreaks.
Molecular profiling is the most reliable method to detect and track the spread of plant disease, Several methods are now available to rapidly extract DNA in as little as 1 min without using sophisticated laboratory instruments or expensive kits and chemicals174,175, and could be scaled up to improve disease tracking in situ. For example, smartphone-based diagnostic platforms are now available that perform isothermal nucleic acid amplification and are designed for rapid and inexpensive pathogen detection in plants175. Ultraportable platforms such as POCKET (point-of-care kit for the entire test176) are inexpensive and will enable a versatile sample-to-answer approach for pathogen diagnostics. Additionally, miniaturized sequencing platforms such as MinION or SmidgION will significantly improve portable DNA/RNA analysis. Using the hand-on sequencing platform in cost-efficient whole-genome sequencing technologies (US$1 per gigabase177) is critical to enable surveillance (identity of genotype and strain), thus expediting studies on the origin of outbreaks, tracking transmission and pathogen evolution.
Other methods, including hyperspectral imaging, volatile organic compound fingerprinting and remote sensing, have the potential to revolutionize the pathogen and disease surveillance sector. Hyperthermal and thermal imaging data sets provide unique signatures to differentiate the infection of olive trees by two different xylem-limited pathogens (Xylella fastidiosa and Verticillium dahliae)178. A smartphone-based volatile organic compound sensor can profile key plant metabolites at parts per million levels within 1 min, and have been used to detect Phytophthora infestans in both laboratory-inoculated and field-collected tomato leaves with more than 95% accuracy179. High-resolution imagery captured by remote sensing technologies (for example, satellites and drones) coupled with advanced machine learning approaches can capture subtle changes in plant chemistry and detect pathogens far earlier than symptoms become visible180,181. Drones are also useful to detect and monitor spores of plant pathogens182,183. Digital epidemiology, which translates and analyses information from media, newswires, official reports and crowdsourcing and disseminates the information through media (including websites, email lists and mobile alerts), should be integrated in tracking disease dynamics around the world184.
Data mining and big data analytics provide unique opportunities to advance our understanding of complex biological processes at a level of accuracy without precedence126. Integrating knowledge gathered through these approaches into collaborative international research programmes, such as the Centre for Agriculture and Bioscience International (CABI) Global Burden of Crop Loss initiative or the Group on Earth Observations Global Agricultural Monitoring GEOGLAM) Initiative’s Crop Monitor that collate diverse data sources to build predictive disease impact models, can potentially enable targeted responses to plant pathogens. Baseline data on the plant pathogen and microbial communities are required to generate global microbial maps and detect climate-mediated changes. Initiatives such as the Global Initiative of Crop Microbiome130 and the Soil Biodiversity Observatory Network185 are working towards profiling the soil/plant-associated microbiome globally.
Science–policy interface and social innovations
Plant diseases affect all four pillars of food security: access, availability, utilization and stability136. Additionally, plant diseases can significantly impact the sociopolitical stability of a region or country in the absence of an effective policy framework to monitor, manage and mitigate disease impacts. One of the best examples is Phytophthora blight of potato, which caused the Irish famine in 1859, triggering the death of two million people and mass migrations; this event was a devastating consequence of the lack of effective policies to manage disease-associated risks. Other examples include famine and death linked to brown spot disease in rice in Bengal, India, and the recent outbreak of coffee rust that caused a more than 50% loss in coffee productivity and led to hunger, poverty and mass migration from Central America5,126. Clearly, prioritizing evidence-based policy to monitor, manage and mitigate the impact of plant diseases is critical to maintain socio-economic well-being. For effective management of plant diseases, we therefore propose a knowledge hub and networks of science–policy interfaces.
Knowledge hub
Climate-induced range shifts and emerging pathogens threaten agricultural productivity, trade and access to international markets137. Effective surveillance, forecasting tools and policies would help mitigate these risks to food security and human well-being. The current monitoring of plant disease is coordinated mostly at the regional and national levels, with strong disparities between developing and developed countries138. These disparities and the lack of international coordination hamper a quick response to new emerging or fast-spreading diseases. A global approach, such as the recently proposed global surveillance system138, is urgently needed to continuously monitor and predict global hot spots of important plant diseases, and their socio-economic impacts. Such a system would allow real-time monitoring and quick response to mitigate risks of new emerging or fast-spreading diseases internationally, and could be modelled and upscaled by joining together existing regional surveillance systems with national and regional hubs (for example, USABlight.org for the monitoring of Phytophthora infestans). This network could help assemble, analyse and store data, and provide evidence and tools for monitoring and predicting disease progression and risks. This global information is critical to establish regulatory frameworks that include effective phytosanitary and quarantine rules for international trade.
Science–policy–society interfaces
To be effective, networks such as the global surveillance system would require the establishment of science–policy–society interfaces (SPSIs) operating at local, regional and global scales. If in continuous engagement with other organizations that are actively operating at the interface of science, policy and society, these SPSIs would provide assessment, monitoring, forecasting and provision of recommendations for policy actions that consider science, non-traditional knowledge (for example, indigenous and private sectors) and socio-economic conditions. Already active organizations such as the Consultative Group for International Agricultural Research (One CGIAR) would be in a strong position to coordinate a larger knowledge network, given the broad skill base and locations of their institutions in developing countries where new data need to be generated. With adequate mandate and resources, One CGIAR could coordinate periodic assessments, forecasting and monitoring of plant diseases, in partnership with the Food and Agriculture Organization of the United Nations (FAO). This could be boosted by harnessing expertise from other organizations and the global research community, such as the Intergovernmental Panel on Climate Change (IPCC) and the International Platform for Biodiversity and Ecosystem Services (IPBES), which act upon climate change and biodiversity loss, respectively. In fact, a formal association with these intergovernmental bodies including One Health will be mutually beneficial (Box 4), allowing the SPSIs to use their data, resources and models to predict disease incidence linked to climate change, biodiversity loss and consequences for plant and human health. Similar to the COVID-19 pandemic, in a globalized economy and trade market, plant pathogen transmission is difficult to control or restrict to a particular region. Thus, a global effort is essential to manage socio-economic risks associated with the likely increases in plant disease outbreaks, especially in low-income countries. Plant disease control in developing countries will not only reduce risk of transmission into those areas but also contribute towards food and job security for local communities, with the potential to reduce illegal migration and national or regional conflicts.
Box 4 Plant diseases as an integral part of the One Health concept.
The One Health concept consists of effective measures for surveillance, forecasting and mitigation of zoonotic diseases and can provide an important framework and infrastructure to better manage plant diseases74. Plant diseases and their control impact multiple dimensions of human health, including the availability and quality of food, food safety (mycotoxin-producing plant pathogens such as Aspergillus spp. and Fusarium spp.), vectors of human pathogens (Escherichia coli, Salmonella spp., Listeria monocytogenes in lettuce and other vegetables and fruits), antimicrobial resistance (transfer of antimicrobial resistance from food microbiomes to human gut microorganisms) and exposure to harmful chemical pesticides (for example, organophosphate chronic exposure and their carcinogenic and genotoxic effects)114,136. Explicit consideration of plant diseases in the One Health concept can provide an effective framework to simultaneously mitigate risks of human health that are directly linked to plant health (for example, human infection, agrochemicals) and spread of plant diseases. The One Health concept poses a transdisciplinary approach to identify epidemics of human disease based on biological and evolutionary aspects of the pathogen and host, habitat characteristics and connectivity and surveillance of wildlife reservoirs. Potential constraints, challenges and solutions for plant diseases are fundamentally similar to human and animal pathogens, and employing common approaches could help in identifying, predicting, monitoring and managing emerging plant diseases. Similarly, the One Health concept provides a holistic framework encompassing human and veterinary medicines, analytical practices, social and conservation science, environmental science and policymaking to manage the outbreak and spread of zoonotic diseases. A similar framework is needed for plant disease to manage increasing risks associated with climate change. In addition, the adoption of One Health data sharing can overcome some cultural, economic and political constraints in plant diseases and can be facilitated through capacity building and data repositories. A framework that allows incorporation of multiple potential pathways that can impact public health is critical to systematically and holistically manage risks associated with public health.
Inclusion of plant health into One Health can also further strengthen the monitoring and managing of human health risks associated with food safety (for example, human pathogens in agricultural produce)74. This will help streamline regulatory frameworks, improve surveillance measures and enable development of effective tools for human health. Inclusion of plant disease in One Health can also contribute towards capacity building in low-income countries through the integration of activities with other intergovernmental agencies including those that impact global biodiversity (International Platform for Biodiversity and Ecosystem Services (IPBES)) and climate change (Intergovernmental Panel on Climate Change (IPCC)). Overall, inclusion of plant disease (health) in the One Health concept will be mutually beneficial for human health outcomes as well ensuring food security and environmental sustainability74.
Conclusion and future perspectives
The current research landscape lacks some key fundamental knowledge to exploit emerging tools to manage disease risks. A holistic solution will require significant expansions of our current knowledge beyond disease monitoring and chemical controls. We need to improve our understanding of pathogen, vector and host biological, ecological and evolutionary responses to climate change. This should include identification of pathobiomes (group of microorganisms and invertebrates that help or hamper infection and disease progression) and their response to climate change. The use of existing ecological theories (for example, invasion theory; co-existing theory/network theory for microbiome coalescence) could provide a strong framework to study and predict pathogen transmission in new regions or to new hosts, and how they interact with host and soil microbiomes139,140. Similarly, the integration of evolutionary processes (for example, new phenotype acquisition by pathogens or indigenous microflora via horizontal gene transfer or mutations) could advance our fundamental knowledge on the mechanisms of pathogenicity (Box 1). We also need an improved understanding of plant phenology (the study of seasonal changes in plants) and disease interactions. Different plant species are vulnerable to pathogens at different stages of plant growth. For example, Fusarium and Verticillium pathogens mainly infect the host at early stages of growth to cause wilt disease117. Similarly, for many diseases (for example, powdery mildews of grapevine and strawberry), young leaves are more susceptible to pathogen infection than the mature leaves. Given that climate change will likely impact plant growth and phenology, we need to address how these changes might affect the susceptibility of plants to disease, in order to develop targeted strategies for disease management.
Measuring pathogen movement through air and water systems is needed to forecast pathogen loads as a result of climate change-driven changes in weather, wind direction and extreme weather events. Individual and interactive effects of climate factors such as temperature, precipitation and drought on disease manifestation and their interaction in different climatic zones should also be considered121. To achieve this, it is possible to use permanent observatories that monitor pollutants and microorganisms, together with drone technologies capable of sampling at 100 m above the ground. Better tools for disease surveillance and management are needed. Remote sensing and drones, enhanced sensor-based technologies (for example, analysis of metagenomes or volatiles) and population genomics along with data mining of social networks can be refined with more accurate data to improve disease surveillances. There is a need for improved modelling tools that integrate climate, weather, epidemiological and socio-economic models for prediction of future outbreaks and effective risk management. Importantly, most of the attention has been placed on diseases of commercial crops, yet the role of wild and native plants, which can act as alternate hosts or barriers to pathogen spread, in disease incidence remains underexplored. Climate change is likely to have impact on the range shift of wild plants, and how this will affect plant disease and epidemics is not known8. Similarly, we have very little understanding on how plant-associated microbiomes, which play a critical role in disease progression or restriction, will respond to climate change and the consequences for pathogen infections. Finally, to achieve effective monitoring and management of plant diseases, socio-economic aspects must be considered. The implementation of effective new computational information systems that support organizations in decision-making (detection or decision support systems) need to be user-friendly and accessible to small-holder farmers who may not be technology savvy. Easy-to-use tools with substantial training will be needed to implement new approaches that have better predictive power and more effective management advice. In this respect, mobile phone-based applications could be a useful tool as most people are familiar with their operation. The lack of adoption of new tools remains a major challenge, mostly because of poor predictive power. However, agricultural scientists could adopt an approach similar to weather forecasting, that is largely accepted by most stakeholders including farming communities. Overall, collaborations with socio-economic and behavioural sciences can contribute towards the development of strategies that encourage greater adoption of all these tools, such as subsidies and/or insurance to cover crop failure.
Available scientific evidence and simulation models suggest that plant disease pressures will significantly increase as climate change intensifies, thus negatively impacting food safety and the sustainability of natural ecosystems. As the magnitude and mechanisms of these impacts remain largely uncertain, effective monitoring and management of plant pathogens should be one of the highest priorities to minimize disease, ensure food safety and environmental sustainability, and promote better socio-economic outcomes. This is made difficult by scientific knowledge gaps around the ecological and evolutionary response of pathogens, hosts and vectors to climate change, and the transmission and emergence of new pathogens under increasing intensity and frequency of extreme weather events and international trade. We propose amendment of existing concepts (disease triangle and Koch’s postulates) and incorporation of eco-evolutionary theories to improve the mechanistic understanding and prediction of disease outbreaks under future climatic conditions and in new regions. Concerted efforts to integrate and harness emerging tools (for example, genomics, satellite, digital, big data, machine learning) for early detection, monitoring and prediction of disease outbreaks will enable sustainable management of disease from local to global scales. These are not trivial goals and will require coordinated research and policy actions from all levels of relevant organizations. We believe that the formation of a dedicated knowledge hub–SPSI in partnership with existing intergovernmental bodies in the context of One Health could help achieve those goals. To improve the detection, monitoring and management of plant pathogens in the face of changing climate, it will be key that important stakeholders (research funding providers, policymakers, intergovernmental agencies) worldwide commit to dedicate more resources to research and SPSIs.
Supplementary information
Acknowledgements
B.K.S. acknowledges funding from the Australian Research Council (DP210102081; DP230101448) for microbiome research. E.E. is supported by an Australian Research Council fellowship (DE210101822). E.G. acknowledges funding from Generalitat Valenciana and a European Social Fund grant (APOSTD/2021/188). M.D.-B. is supported by the Spanish Ministry of Science and Innovation (PID202-115813RA-100). P.T. and J.E.L.’s research is supported by the US National Science Foundation (no. 2120117). J.E.L. receives additional funding from the Foundation of Food and Agriculture (ICRC20-0000000084).
Author contributions
All authors contributed equally to all aspects of the manuscript.
Peer review
Peer review information
Nature Reviews Microbiology thanks Gabriele Berg, Hang-Wei Hu, Marcel van der Heijden and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41579-023-00900-7.
References
- 1.Tripathi, A. N., Tiwari, S. K. & Behera, T. K. in Postharvest Technology Ch. 5 (ed. Ahiduzzaman, M. D.) (IntechOpen, 2022).
- 2.Fones HN, et al. Threats to global food security from emerging fungal and oomycete crop pathogens. Nat. Food. 2020;1:332–342. doi: 10.1038/s43016-020-0075-0. [DOI] [PubMed] [Google Scholar]
- 3.Chakraborty S, Newton AC. Climate change, plant diseases and food security: an overview. Plant. Pathol. 2011;60:2–14. doi: 10.1111/j.1365-3059.2010.02411.x. [DOI] [Google Scholar]
- 4.Rohr JR, et al. Emerging human infectious diseases and the links to global food production. Nat. Sustain. 2019;2:445–456. doi: 10.1038/s41893-019-0293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ristaino JB, et al. The persistent threat of emerging plant disease pandemics to global food security. Proc. Natl Acad. Sci. USA. 2021 doi: 10.1073/pnas.2022239118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Dijk M, Morley T, Rau ML, Saghai Y. A meta-analysis of projected global food demand and population at risk of hunger for the period 2010–2050. Nat. Food. 2021;2:494–501. doi: 10.1038/s43016-021-00322-9. [DOI] [PubMed] [Google Scholar]
- 7.Velasquez AC, Castroverde CDM, He SY. Plant–pathogen warfare under changing climate conditions. Curr. Biol. 2018;28:R619–R634. doi: 10.1016/j.cub.2018.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burdon JJ, Zhan J. Climate change and disease in plant communities. PLoS Biol. 2020;18:e3000949. doi: 10.1371/journal.pbio.3000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muluneh MG. Impact of climate change on biodiversity and food security: a global perspective—a review article. Agric. Food Secur. 2021;10:36. doi: 10.1186/s40066-021-00318-5. [DOI] [Google Scholar]
- 10.Chaloner TM, Gurr SJ, Bebber DP. Plant pathogen infection risk tracks global crop yields under climate change. Nat. Clim. Change. 2021;11:710–715. doi: 10.1038/s41558-021-01104-8. [DOI] [Google Scholar]
- 11.Trumbore S, Brando P, Hartmann H. Forest health and global change. Science. 2015;349:814–818. doi: 10.1126/science.aac6759. [DOI] [PubMed] [Google Scholar]
- 12.Newbery F, Qi A, Fitt BDL. Modelling impacts of climate change on arable crop diseases: progress, challenges and applications. Curr. Opin. Plant. Biol. 2016;32:101–109. doi: 10.1016/j.pbi.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Cohen SP, Leach JE. High temperature-induced plant disease susceptibility: more than the sum of its parts. Curr. Opin. Plant Biol. 2020;56:235–241. doi: 10.1016/j.pbi.2020.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Delgado-Baquerizo M, et al. The proportion of soil-borne pathogens increases with warming at the global scale. Nat. Clim. Change. 2020;10:550–554. doi: 10.1038/s41558-020-0759-3. [DOI] [Google Scholar]
- 15.Dudney J, et al. Nonlinear shifts in infectious rust disease due to climate change. Nat. Commun. 2021;12:5102. doi: 10.1038/s41467-021-25182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romero F, et al. Humidity and high temperature are important for predicting fungal disease outbreaks worldwide. N. Phytol. 2022;234:1553–1556. doi: 10.1111/nph.17340. [DOI] [PubMed] [Google Scholar]
- 17.Brown JK, Hovmøller MS. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science. 2002;297:537–541. doi: 10.1126/science.1072678. [DOI] [PubMed] [Google Scholar]
- 18.Sikes BA, et al. Import volumes and biosecurity interventions shape the arrival rate of fungal pathogens. PLoS Biol. 2018;16:e2006025. doi: 10.1371/journal.pbio.2006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher MC, et al. Threats posed by the fungal kingdom to humans, wildlife, and agriculture. mBio. 2020 doi: 10.1128/mBio.00449-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goellner K, et al. Phakopsora pachyrhizi, the causal agent of Asian soybean rust. Mol. Plant Pathol. 2010;11:169–177. doi: 10.1111/j.1364-3703.2009.00589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeger MJ. The impact of climate change on disease in wild plant populations and communities. Plant Pathol. 2022;71:111–130. doi: 10.1111/ppa.13434. [DOI] [Google Scholar]
- 22.Thompson SE, Levin S, Rodriguez-Iturbe I. Rainfall and temperatures changes have confounding impacts on Phytophthora cinnamomi occurrence risk in the southwestern USA under climate change scenarios. Glob. Chang. Biol. 2014;20:1299–1312. doi: 10.1111/gcb.12463. [DOI] [PubMed] [Google Scholar]
- 23.Rigg JL, McDougall KL, Liew ECY. Susceptibility of nine alpine species to the root rot pathogens Phytophthora cinnamomi and P. cambivora. Australas. Plant Pathol. 2018;47:351–356. doi: 10.1007/s13313-018-0564-x. [DOI] [Google Scholar]
- 24.Cheng YT, Zhang L, He SY. Plant–microbe interactions facing environmental challenge. Cell Host Microbe. 2019;26:183–192. doi: 10.1016/j.chom.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK. Plant–microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 2020;18:607–621. doi: 10.1038/s41579-020-0412-1. [DOI] [PubMed] [Google Scholar]
- 26.Desaint H, et al. Fight hard or die trying: when plants face pathogens under heat stress. N. Phytol. 2021;229:712–734. doi: 10.1111/nph.16965. [DOI] [PubMed] [Google Scholar]
- 27.Trivedi P, Batista BD, Bazany KE, Singh BK. Plant–microbiome interactions under a changing world: responses, consequences and perspectives. N. Phytol. 2022;234:1951–1959. doi: 10.1111/nph.18016. [DOI] [PubMed] [Google Scholar]
- 28.Desprez-Loustau M-L, Marçais B, Nageleisen L-M, Piou D, Vannini A. Interactive effects of drought and pathogens in forest trees. Ann. Sci. 2006;63:597–612. doi: 10.1051/forest:2006040. [DOI] [Google Scholar]
- 29.Ryu M, Mishra RC, Jeon J, Lee SK, Bae H. Drought-induced susceptibility for Cenangium ferruginosum leads to progression of Cenangium-dieback disease in Pinus koraiensis. Sci. Rep. 2018;8:16368. doi: 10.1038/s41598-018-34318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hossain M, Veneklaas EJ, Hardy G, Poot P. Tree host–pathogen interactions as influenced by drought timing: linking physiological performance, biochemical defence and disease severity. Tree Physiol. 2019;39:6–18. doi: 10.1093/treephys/tpy113. [DOI] [PubMed] [Google Scholar]
- 31.Toniutti L, et al. Influence of environmental conditions and genetic background of Arabica coffee (C. arabica L) on leaf rust (Hemileia vastatrix) pathogenesis. Front. Plant. Sci. 2017;8:2025. doi: 10.3389/fpls.2017.02025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gustafson EJ, Miranda BR, Dreaden TJ, Pinchot CC, Jacobs DF. Beyond blight: Phytophthora root rot under climate change limits populations of reintroduced American chestnut. Ecosphere. 2022;13:18. doi: 10.1002/ecs2.3917. [DOI] [Google Scholar]
- 33.Barbeito I, Brücker RL, Rixen C, Bebi P. Snow fungi-induced mortality of Pinus cembra at the alpine treeline: evidence from plantations. Arct. Antarct. Alp. Res. 2013;45:455–470. doi: 10.1657/1938-4246-45.4.455. [DOI] [Google Scholar]
- 34.Parikka P, Hakala K, Tiilikkala K. Expected shifts in Fusarium species’ composition on cereal grain in Northern Europe due to climatic change. Food Addit. Contam. A. 2012;29:1543–1555. doi: 10.1080/19440049.2012.680613. [DOI] [PubMed] [Google Scholar]
- 35.Walter S, et al. Molecular markers for tracking the origin and worldwide distribution of invasive strains of Puccinia striiformis. Ecol. Evol. 2016;6:2790–2804. doi: 10.1002/ece3.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vidal T, et al. Success and failure of invasive races of plant pathogens: the case of Puccinia striiformis f. sp. tritici in France. Plant. Pathol. 2022 doi: 10.1111/ppa.13581. [DOI] [Google Scholar]
- 37.Ma L, et al. Effect of low temperature and wheat winter-hardiness on survival of Puccinia striiformis f. sp. tritici under controlled conditions. PLoS ONE. 2015;10:e0130691. doi: 10.1371/journal.pone.0130691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhan J, Ericson L, Burdon JJ. Climate change accelerates local disease extinction rates in a long-term wild host–pathogen association. Glob. Chang. Biol. 2018;24:3526–3536. doi: 10.1111/gcb.14111. [DOI] [PubMed] [Google Scholar]
- 39.Sparks AH, Forbes GA, Hijmans RJ, Garrett KA. Climate change may have limited effect on global risk of potato late blight. Glob. Chang. Biol. 2014;20:3621–3631. doi: 10.1111/gcb.12587. [DOI] [PubMed] [Google Scholar]
- 40.Castroverde CDM, Dina D. Temperature regulation of plant hormone signaling during stress and development. J. Exp. Bot. 2021 doi: 10.1093/jxb/erab257. [DOI] [PubMed] [Google Scholar]
- 41.Kim JH, et al. Increasing the resilience of plant immunity to a warming climate. Nature. 2022;607:339–344. doi: 10.1038/s41586-022-04902-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Q, Majsec K, Katagiri F. Pathogen‐driven coevolution across the CBP60 plant immune regulator subfamilies confers resilience on the regulator module. N. Phytol. 2022;233:479–495. doi: 10.1111/nph.17769. [DOI] [PubMed] [Google Scholar]
- 43.Cohen SP, et al. RNA-seq analysis reveals insight into enhanced rice Xa7-mediated bacterial blight resistance at high temperature. PLoS ONE. 2017;12:e0187625. doi: 10.1371/journal.pone.0187625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu J, et al. Warm temperature compromises JA-regulated basal resistance to enhance Magnaporthe oryzae infection in rice. Mol. Plant. 2022;15:723–739. doi: 10.1016/j.molp.2022.02.014. [DOI] [PubMed] [Google Scholar]
- 45.Khan MR, Rizvi TF. Effect of elevated levels of CO2 on powdery mildew development in five cucurbit species. Sci. Rep. 2020 doi: 10.1038/s41598-020-61790-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vary Z, Mullins E, McElwain JC, Doohan FM. The severity of wheat diseases increases when plants and pathogens are acclimatized to elevated carbon dioxide. Glob. Change Biol. 2015;21:2661–2669. doi: 10.1111/gcb.12899. [DOI] [PubMed] [Google Scholar]
- 47.Eastburn DM, Degennaro MM, Delucia EH, Dermody O, Mcelrone AJ. Elevated atmospheric carbon dioxide and ozone alter soybean diseases at SoyFACE. Glob. Change Biol. 2010;16:320–330. doi: 10.1111/j.1365-2486.2009.01978.x. [DOI] [Google Scholar]
- 48.Karnosky DF, et al. Interacting elevated CO2 and tropospheric O3 predisposes aspen (Populus tremuloides Michx.) to infection by rust (Melampsora medusae f. sp. tremuloidae) Glob. Change Biol. 2002;8:329–338. doi: 10.1046/j.1354-1013.2002.00479.x. [DOI] [Google Scholar]
- 49.Mcelrone AJ, Reid CD, Hoye KA, Hart E, Jackson RB. Elevated CO2 reduces disease incidence and severity of a red maple fungal pathogen via changes in host physiology and leaf chemistry. Glob. Change Biol. 2005;11:1828–1836. doi: 10.1111/j.1365-2486.2005.001015.x. [DOI] [Google Scholar]
- 50.Zhou Y, Van Leeuwen SK, Pieterse CMJ, Bakker PAHM, Van Wees SCM. Effect of atmospheric CO2 on plant defense against leaf and root pathogens of Arabidopsis. Eur. J. Plant Pathol. 2019;154:31–42. doi: 10.1007/s10658-019-01706-1. [DOI] [Google Scholar]
- 51.Trębicki P, et al. Virus infection mediates the effects of elevated CO2 on plants and vectors. Sci. Rep. 2016;6:22785. doi: 10.1038/srep22785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mamo BE, et al. Epidemiological characterization of lettuce drop (Sclerotinia spp.) and biophysical features of the host identify soft stem as a susceptibility factor. PhytoFrontiers. 2021;1:182–204. doi: 10.1094/PHYTOFR-12-20-0040-R. [DOI] [Google Scholar]
- 53.Tada T, Tanaka C, Katsube-Tanaka T, Shiraiwa T. Effects of wounding and relative humidity on the incidence of Phytophthora root and stem rot in soybean seedlings. Phsiol. Mol. Plant Pathol. 2021;116:101737. doi: 10.1016/j.pmpp.2021.101737. [DOI] [Google Scholar]
- 54.Xin X-F, et al. Bacteria establish an aqueous living space in plants crucial for virulence. Nature. 2016;539:524–529. doi: 10.1038/nature20166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andersen KF, Madden LV, Paul PA. Fusarium head blight development and deoxynivalenol accumulation in wheat as influenced by post-anthesis moisture patterns. Phytopathology. 2015;105:210–219. doi: 10.1094/PHYTO-04-14-0104-R. [DOI] [PubMed] [Google Scholar]
- 56.Qiu JB, Dong F, Yu MZ, Xu JH, Shi JR. Effect of preceding crop on Fusarium species and mycotoxin contamination of wheat grains. J. Sci. Food Agric. 2016;96:4536–4541. doi: 10.1002/jsfa.7670. [DOI] [PubMed] [Google Scholar]
- 57.Johansen TJ, Dees MW, Hermansen A. High soil moisture reduces common scab caused by Streptomyces turgidiscabies and Streptomyces europaeiscabiei in potato. Acta Agric. Scand. B Soil Plant Sci. 2015;65:193–198. [Google Scholar]
- 58.Bidzinski P, et al. Transcriptional basis of drought-induced susceptibility to the rice blast fungus Magnaporthe oryzae. Front. Plant Sci. 2016 doi: 10.3389/fpls.2016.01558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wakelin SA, et al. Climate change induced drought impacts on plant diseases in New Zealand. Australas. Plant Pathol. 2018;47:101–114. doi: 10.1007/s13313-018-0541-4. [DOI] [Google Scholar]
- 60.Choi H-K, Iandolino A, da Silva FG, Cook DR. Water deficit modulates the response of Vitis vinifera to the Pierce’s disease pathogen Xylella fastidiosa. Mol. Plant Microbe Interact. 2013;26:643–657. doi: 10.1094/MPMI-09-12-0217-R. [DOI] [PubMed] [Google Scholar]
- 61.Oliva J, Stenlid J, Martinez-Vilalta J. The effect of fungal pathogens on the water and carbon economy of trees: implications for drought-induced mortality. N. Phytol. 2014;203:1028–1035. doi: 10.1111/nph.12857. [DOI] [PubMed] [Google Scholar]
- 62.Rai A, Irulappan V, Muthappa S-K. Dry root rot of chickpea: a disease favored by drought. Plant Dis. 2021 doi: 10.1094/PDIS-07-21-1410-FE. [DOI] [PubMed] [Google Scholar]
- 63.Vasquez DF, et al. Drought as a modulator of plant–virus–vector interactions: effects on symptom expression, plant immunity and vector behaviour. Plant Pathol. 2022;71:1282–1292. doi: 10.1111/ppa.13554. [DOI] [Google Scholar]
- 64.Webb KM, et al. A benefit of high temperature: increased effectiveness of a rice bacterial blight disease resistance gene. N. Phytol. 2010;185:568–576. doi: 10.1111/j.1469-8137.2009.03076.x. [DOI] [PubMed] [Google Scholar]
- 65.Cohen SP, Leach JE. Abiotic and biotic stresses induce a core transcriptome response in rice. Sci. Rep. 2019;9:6273. doi: 10.1038/s41598-019-42731-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teshome DT, Zharare GE, Naidoo S. The threat of the combined effect of biotic and abiotic stress factors in forestry under a changing climate. Front. Plant Sci. 2020 doi: 10.3389/fpls.2020.601009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sewelam N, El-Shetehy M, Mauch F, Maurino VG. Combined abiotic stresses repress defense and cell wall metabolic genes and render plants more susceptible to pathogen infection. Plants. 2021 doi: 10.3390/plants10091946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Islam MT, Kim KH, Choi J. Wheat blast in Bangladesh: the current situation and future impacts. Plant Pathol. J. 2019;35:1–10. doi: 10.5423/PPJ.RW.08.2018.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ciliberti N, Fermaud M, Roudet J, Rossi V. Environmental conditions affect Botrytis cinerea infection of mature grape berries more than the strain or transposon genotype. Phytopathology. 2015;105:1090–1096. doi: 10.1094/PHYTO-10-14-0264-R. [DOI] [PubMed] [Google Scholar]
- 70.Juroszek P, von Tiedemann A. Linking plant disease models to climate change scenarios to project future risks of crop diseases: a review. J. Plant Dis. Prot. 2015;122:3–15. doi: 10.1007/BF03356525. [DOI] [Google Scholar]
- 71.Guerra CA, et al. Global hotspots for soil nature conservation. Nature. 2022;610:693–698. doi: 10.1038/s41586-022-05292-x. [DOI] [PubMed] [Google Scholar]
- 72.Batista E, Lopes A, Miranda P, Alves A. Can species distribution models be used for risk assessment analyses of fungal plant pathogens? A case study with three Botryosphaeriaceae species. Eur. J. Plant Pathol. 2022 doi: 10.1007/s10658-022-02587-7. [DOI] [Google Scholar]
- 73.Mizeriene G, et al. Patterns of genetic diversification in the invasive hybrid plant pathogen Phytophthora × alni and its parental species P. uniformis. Phytopathology. 2020;110:1959–1969. doi: 10.1094/PHYTO-12-19-0475-R. [DOI] [PubMed] [Google Scholar]
- 74.Morris CE, Géniaux G, Nédellec C, Sauvion N, Soubeyrand S. One Health concepts and challenges for surveillance, forecasting, and mitigation of plant disease beyond the traditional scope of crop production. Plant Pathol. 2022;71:86–97. doi: 10.1111/ppa.13446. [DOI] [Google Scholar]
- 75.Wang P, et al. Shifts in microbial communities in soil, rhizosphere and roots of two major crop systems under elevated CO2 and O3. Sci. Rep. 2017;7:15019. doi: 10.1038/s41598-017-14936-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Santos-Medellín C, et al. Prolonged drought imparts lasting compositional changes to the rice root microbiome. Nat. Plants. 2021;7:1065–1077. doi: 10.1038/s41477-021-00967-1. [DOI] [PubMed] [Google Scholar]
- 77.Bazany KE, Wang JT, Delgado-Baquerizo M, Singh BK, Trivedi P. Water deficit affects inter-kingdom microbial connections in plant rhizosphere. Environ. Microbiol. 2022 doi: 10.1111/1462-2920.16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu H, Brettell LE, Qiu Z, Singh BK. Microbiome-mediated stress resistance in plants. Trends Plant Sci. 2020;25:733–743. doi: 10.1016/j.tplants.2020.03.014. [DOI] [PubMed] [Google Scholar]
- 79.Berendsen RL, et al. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018;12:1496–1507. doi: 10.1038/s41396-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carrion VJ, et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science. 2019;366:606–612. doi: 10.1126/science.aaw9285. [DOI] [PubMed] [Google Scholar]
- 81.Rolfe SA, Griffiths J, Ton J. Crying out for help with root exudates: adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr. Opin. Microbiol. 2019;49:73–82. doi: 10.1016/j.mib.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 82.Gao M, et al. Disease-induced changes in plant microbiome assembly and functional adaptation. Microbiome. 2021;9:187. doi: 10.1186/s40168-021-01138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yuan J, et al. Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome. 2018;6:156. doi: 10.1186/s40168-018-0537-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim JY, et al. Cellular export of sugars and amino acids: role in feeding other cells and organisms. Plant. Physiol. 2021;187:1893–1915. doi: 10.1093/plphys/kiab228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schulz-Bohm K, et al. Calling from distance: attraction of soil bacteria by plant root volatiles. ISME J. 2018;12:1252–1262. doi: 10.1038/s41396-017-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Trivedi P, Schenk PM, Wallenstein MD, Singh BK. Tiny microbes, big yields: enhancing food crop production with biological solutions. Microb. Biotechnol. 2017;10:999–1003. doi: 10.1111/1751-7915.12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mendes R, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 88.Leach JE, Triplett LR, Argueso CT, Trivedi P. Communication in the phytobiome. Cell. 2017;169:587–596. doi: 10.1016/j.cell.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 89.Wu J, Yu S. Effect of root exudates of Eucalyptus urophylla and Acacia mearnsii on soil microbes under simulated warming climate conditions. BMC Microbiol. 2019;19:224. doi: 10.1186/s12866-019-1604-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Q, et al. The effects of warming on root exudation and associated soil N transformation depend on soil nutrient availability. Rhizosphere. 2021;17:100263. doi: 10.1016/j.rhisph.2020.100263. [DOI] [Google Scholar]
- 91.Ulrich DEM, et al. Root exudate composition reflects drought severity gradient in blue grama (Bouteloua gracilis) Sci. Rep. 2022;12:12581. doi: 10.1038/s41598-022-16408-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Snelders, N. C., Petti, G. C., van den Berg, G. C. M., Seidl, M. F. & Thomma, B. P. H. J. An ancient antimicrobial protein co-opted by a fungal plant pathogen for in planta mycobiome manipulation. Proc. Natl Acad. Sci. USA10.1073/pnas.2110968118 (2021). [DOI] [PMC free article] [PubMed]
- 93.Seybold H, et al. A fungal pathogen induces systemic susceptibility and systemic shifts in wheat metabolome and microbiome composition. Nat. Commun. 2020;11:1910. doi: 10.1038/s41467-020-15633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Agler MT, et al. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016;14:e1002352. doi: 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Venturi V, da Silva DP. Incoming pathogens team up with harmless ‘resident’ bacteria. Trends Microbiol. 2012;20:160–164. doi: 10.1016/j.tim.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 96.Trivedi P, Trivedi C, Grinyer J, Anderson IC, Singh BK. Harnessing host-vector microbiome for sustainable plant disease management of phloem-limited bacteria. Front. Plant Sci. 2016;7:1423. doi: 10.3389/fpls.2016.01423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hosni T, et al. Sharing of quorum-sensing signals and role of interspecies communities in a bacterial plant disease. ISME J. 2011;5:1857–1870. doi: 10.1038/ismej.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Trivedi P, et al. Huanglongbing alters the structure and functional diversity of microbial communities associated with citrus rhizosphere. ISME J. 2012;6:363–383. doi: 10.1038/ismej.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hamonts K, et al. Field study reveals core plant microbiota and relative importance of their drivers. Environ. Microbiol. 2018;20:124–140. doi: 10.1111/1462-2920.14031. [DOI] [PubMed] [Google Scholar]
- 100.Lebreton L, et al. Temporal dynamics of bacterial and fungal communities during the infection of Brassica rapa roots by the protist Plasmodiophora brassicae. PLoS ONE. 2019 doi: 10.1371/journal.pone.0204195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blaustein RA, Lorca GL, Meyer JL, Gonzalez CF, Teplitski M. Defining the core citrus leaf- and root-associated microbiota: factors associated with community structure and implications for managing huanglongbing (citrus greening) disease. Appl. Environ. Microbiol. 2017 doi: 10.1128/aem.00210-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saijo Y, Loo EP. Plant immunity in signal integration between biotic and abiotic stress responses. N. Phytol. 2020;225:87–104. doi: 10.1111/nph.15989. [DOI] [PubMed] [Google Scholar]
- 103.Lebeis SL, et al. Plant microbiome. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science. 2015;349:860–864. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- 104.Janda M, et al. Temporary heat stress suppresses PAMP-triggered immunity and resistance to bacteria in Arabidopsis thaliana. Mol. Plant. Pathol. 2019;20:1005–1012. doi: 10.1111/mpp.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hacquard S, Spaepen S, Garrido-Oter R, Schulze-Lefert P. Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 2017;55:565–589. doi: 10.1146/annurev-phyto-080516-035623. [DOI] [PubMed] [Google Scholar]
- 106.Chen QL, et al. Host identity determines plant associated resistomes. Environ. Pollut. 2020;258:113709. doi: 10.1016/j.envpol.2019.113709. [DOI] [PubMed] [Google Scholar]
- 107.Gonzalez R, et al. Plant virus evolution under strong drought conditions results in a transition from parasitism to mutualism. Proc. Natl Acad. Sci. USA. 2021 doi: 10.1073/pnas.2020990118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Suryanarayanan TS, Shaanker RU. Can fungal endophytes fast-track plant adaptations to climate change? Fungal Ecol. 2021 doi: 10.1016/j.funeco.2021.101039. [DOI] [Google Scholar]
- 109.Stringlis IA, et al. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl Acad. Sci. USA. 2018;115:E5213–E5222. doi: 10.1073/pnas.1722335115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Doll S, et al. Accumulation of the coumarin scopolin under abiotic stress conditions is mediated by the Arabidopsis thaliana THO/TREX complex. Plant J. 2018;93:431–444. doi: 10.1111/tpj.13797. [DOI] [PubMed] [Google Scholar]
- 111.Lekberg Y, et al. Nitrogen and phosphorus fertilization consistently favor pathogenic over mutualistic fungi in grassland soils. Nat. Commun. 2021;12:3484. doi: 10.1038/s41467-021-23605-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Singh BK, Trivedi P. Microbiome and the future for food and nutrient security. Microb. Biotechnol. 2017;10:50–53. doi: 10.1111/1751-7915.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Arie T. Fusarium diseases of cultivated plants, control, diagnosis, and molecular and genetic studies. J. Pestic. Sci. 2019;44:275–281. doi: 10.1584/jpestics.J19-03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yan Z, Xiong C, Liu H, Singh BK. Sustainable agricultural practices contribute significantly to One Health. J. Sustain. Agric. Env. 2022 doi: 10.1002/sae2.12019. [DOI] [Google Scholar]
- 115.Edlinger A, et al. Agricultural management and pesticide use reduce the functioning of beneficial plant symbionts. Nat. Ecol. Evol. 2022;6:1145–1154. doi: 10.1038/s41559-022-01799-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Haas SE, Hooten MB, Rizzo DM, Meentemeyer RK. Forest species diversity reduces disease risk in a generalist plant pathogen invasion. Ecol. Lett. 2011;14:1108–1116. doi: 10.1111/j.1461-0248.2011.01679.x. [DOI] [PubMed] [Google Scholar]
- 117.Kirkby KA, Lonergan PA, Allen SJ. Three decades of cotton disease surveys NSW, Australia. Crop. Sci. Pasture Sci. 2013;64:774–779. doi: 10.1071/CP13143. [DOI] [Google Scholar]
- 118.Araki M, Ishii T. Towards social acceptance of plant breeding by genome editing. Trends Plant Sci. 2015;20:145–149. doi: 10.1016/j.tplants.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 119.Hickey LT, et al. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019;37:744–754. doi: 10.1038/s41587-019-0152-9. [DOI] [PubMed] [Google Scholar]
- 120.Qiu Z, Egidi E, Liu H, Kaur S, Singh BK. New frontiers in agriculture productivity: optimised microbial inoculants and in situ microbiome engineering. Biotechnol. Adv. 2019;37:107371. doi: 10.1016/j.biotechadv.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 121.Jeger M, et al. Global challenges facing plant pathology: multidisciplinary approaches to meet the food security and environmental challenges in the mid-twenty-first century. CABI Agric. Bio. 2021;2:20. doi: 10.1186/s43170-021-00042-x. [DOI] [Google Scholar]
- 122.APS. Phytobiomes: A Roadmap for Research and Translation (American Phytopathological Society, 2016).
- 123.Pozo MJ, Zabalgogeazcoa I, Vazquez de Aldana BR, Martinez-Medina A. Untapping the potential of plant mycobiomes for applications in agriculture. Curr. Opin. Plant Biol. 2021;60:102034. doi: 10.1016/j.pbi.2021.102034. [DOI] [PubMed] [Google Scholar]
- 124.Trivedi P, Mattupalli C, Eversole K, Leach JE. Enabling sustainable agriculture through understanding and enhancement of microbiomes. N. Phytol. 2021;230:2129–2147. doi: 10.1111/nph.17319. [DOI] [PubMed] [Google Scholar]
- 125.Lamichhane JR, Venturi V. Synergisms between microbial pathogens in plant disease complexes: a growing trend. Front. Plant Sci. 2015;6:00385. doi: 10.3389/fpls.2015.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Garrett KA, et al. Climate change effects on pathogen emergence: artificial intelligence to translate big data for mitigation. Annu. Rev. Phytopathol. 2022 doi: 10.1146/annurev-phyto-021021-042636. [DOI] [PubMed] [Google Scholar]
- 127.Delgado-Baquerizo M. Simplifying the complexity of the soil microbiome to guide the development of next-generation SynComs. J. Sustain. Agric. Environ. 2022;1:9–15. doi: 10.1002/sae2.12012. [DOI] [Google Scholar]
- 128.Liu H, et al. Effective colonisation by a bacterial synthetic community promotes plant growth and alters soil microbial community. J. Sustain. Agric. Environ. 2022;1:30–42. doi: 10.1002/sae2.12008. [DOI] [Google Scholar]
- 129.Liu H, et al. Inner plant values: diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 2017;8:2552. doi: 10.3389/fmicb.2017.02552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Singh BK, Trivedi P, Egidi E, Macdonald CA, Delgado-Baquerizo M. Crop microbiome and sustainable agriculture. Nat. Rev. Microbiol. 2020;18:601–602. doi: 10.1038/s41579-020-00446-y. [DOI] [PubMed] [Google Scholar]
- 131.Li J, Wang J, Liu H, Macdonald CA, Singh BK. Application of microbial inoculants significantly enhances crop productivity: a meta-analysis of studies from 2010 to 2020. J. Sustain. Agric. Environ. 2022;1:216–225. doi: 10.1002/sae2.12028. [DOI] [Google Scholar]
- 132.Matsumoto H, et al. Bacterial seed endophyte shapes disease resistance in rice. Nat. Plants. 2021;7:60–72. doi: 10.1038/s41477-020-00826-5. [DOI] [PubMed] [Google Scholar]
- 133.Berg G, Raaijmakers JM. Saving seed microbiomes. ISME J. 2018;12:1167–1170. doi: 10.1038/s41396-017-0028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mendes LW, Raaijmakers JM, de Hollander M, Mendes R, Tsai SM. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 2018;12:212–224. doi: 10.1038/ismej.2017.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.de Vries FT, Griffiths RI, Knight CG, Nicolitch O, Williams A. Harnessing rhizosphere microbiomes for drought-resilient crop production. Science. 2020;368:270–274. doi: 10.1126/science.aaz5192. [DOI] [PubMed] [Google Scholar]
- 136.Rizzo DM, Lichtveld M, Mazet JAK, Togami E, Miller SA. Plant health and its effects on food safety and security in a One Health framework: four case studies. One Health Outlook. 2021;3:6. doi: 10.1186/s42522-021-00038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Porfirio LL, Newth D, Finnigan JJ, Cai Y. Economic shifts in agricultural production and trade due to climate change. Palgrave Commun. 2018;4:111. doi: 10.1057/s41599-018-0164-y. [DOI] [Google Scholar]
- 138.Carvajal-Yepes M, et al. A global surveillance system for crop diseases. Science. 2019;364:1237–1239. doi: 10.1126/science.aaw1572. [DOI] [PubMed] [Google Scholar]
- 139.Blumenthal DM. Interactions between resource availability and enemy release in plant invasion. Ecol. Lett. 2006;9:887–895. doi: 10.1111/j.1461-0248.2006.00934.x. [DOI] [PubMed] [Google Scholar]
- 140.Mallon CA, Elsas JDV, Salles JF. Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol. 2015;23:719–729. doi: 10.1016/j.tim.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 141.Feng Y, et al. Temperature thresholds drive the global distribution of soil fungal decomposers. Glob. Change Biol. 2022;28:2779–2789. doi: 10.1111/gcb.16096. [DOI] [PubMed] [Google Scholar]
- 142.Hurtt GC, et al. Harmonization of land-use scenarios for the period 1500–2100: 600 years of global gridded annual land-use transitions, wood harvest, and resulting secondary lands. Clim. Change. 2011;109:117–161. doi: 10.1007/s10584-011-0153-2. [DOI] [Google Scholar]
- 143.Lawrence DM, et al. The Land Use Model Intercomparison Project (LUMIP) contribution to CMIP6: rationale and experimental design. Geosci. Model. Dev. 2016;9:2973–2998. doi: 10.5194/gmd-9-2973-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dufresne JL, et al. Climate change projections using the IPSL-CM5 Earth System Model: from CMIP3 to CMIP5. Clim. Dyn. 2013;40:2123–2165. doi: 10.1007/s00382-012-1636-1. [DOI] [Google Scholar]
- 145.Hempel S, Frieler K, Warszawski L, Schewe J, Piontek F. A trend-preserving bias correction—the ISI-MIP approach. Earth Syst. Dyn. 2013;4:219–236. doi: 10.5194/esd-4-219-2013. [DOI] [Google Scholar]
- 146.Kinnunen M, et al. A conceptual framework for invasion in microbial communities. ISME J. 2016;10:2773–2775. doi: 10.1038/ismej.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bewick S, Staniczenko PPA, Li B, Karig DK, Fagan WF. Invasion speeds in microbial systems with toxin production and quorum sensing. J. Theor. Biol. 2017;420:290–303. doi: 10.1016/j.jtbi.2017.01.034. [DOI] [PubMed] [Google Scholar]
- 148.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jayathilake PG, et al. A mechanistic individual-based model of microbial communities. PLoS ONE. 2017;12:e0181965. doi: 10.1371/journal.pone.0181965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Daly AJ, et al. Individual-based modelling of invasion in bioaugmented sand filter communities. Processes. 2018 doi: 10.3390/pr6010002. [DOI] [Google Scholar]
- 151.Liu X, Wang M, Nie Y, Wu X-L. Successful microbial colonization of space in a more dispersed manner. ISME Commun. 2021;1:68. doi: 10.1038/s43705-021-00063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Deutsch CA, et al. Increase in crop losses to insect pests in a warming climate. Science. 2018;361:916–919. doi: 10.1126/science.aat3466. [DOI] [PubMed] [Google Scholar]
- 153.Zhang X, et al. Climate change increases risk of Fusarium ear blight on wheat in central China. Ann. Appl. Biol. 2014;164:384–395. doi: 10.1111/aab.12107. [DOI] [Google Scholar]
- 154.Duku C, Sparks AH, Zwart SJ. Spatial modelling of rice yield losses in Tanzania due to bacterial leaf blight and leaf blast in a changing climate. Clim. Change. 2016;135:569–583. doi: 10.1007/s10584-015-1580-2. [DOI] [Google Scholar]
- 155.Cunniffe NJ, Cobb RC, Meentemeyer RK, Rizzo DM, Gilligan CA. Modeling when, where, and how to manage a forest epidemic, motivated by sudden oak death in California. Proc. Natl Acad. Sci. USA. 2016;113:5640–5645. doi: 10.1073/pnas.1602153113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hyatt-Twynam SR, et al. Risk-based management of invading plant disease. N. Phytol. 2017;214:1317–1329. doi: 10.1111/nph.14488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Thompson RN, Gilligan CA, Cunniffe NJ. Control fast or control smart: when should invading pathogens be controlled? PLoS Comput. Biol. 2018;14:e1006014. doi: 10.1371/journal.pcbi.1006014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mastin AJ, Gottwald TR, van den Bosch F, Cunniffe NJ, Parnell S. Optimising risk-based surveillance for early detection of invasive plant pathogens. PLoS Biol. 2020;18:e3000863. doi: 10.1371/journal.pbio.3000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bebber DP, et al. Many unreported crop pests and pathogens are probably already present. Glob. Chang. Biol. 2019;25:2703–2713. doi: 10.1111/gcb.14698. [DOI] [PubMed] [Google Scholar]
- 160.Diffenbaugh NS. Verification of extreme event attribution: using out-of-sample observations to assess changes in probabilities of unprecedented events. Sci. Adv. 2020;6:eaay2368. doi: 10.1126/sciadv.aay2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Garrett KA, Dendy SP, Frank EE, Rouse MN, Travers SE. Climate change effects on plant disease: genomes to ecosystems. Annu. Rev. Phytopathol. 2006;44:489–509. doi: 10.1146/annurev.phyto.44.070505.143420. [DOI] [PubMed] [Google Scholar]
- 162.Botero D, et al. Genome-scale metabolic model of Xanthomonas phaseoli pv. manihotis: an approach to elucidate pathogenicity at the metabolic level. Front. Genet. 2020;11:837. doi: 10.3389/fgene.2020.00837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gerlin L, et al. Genome-scale investigation of the metabolic determinants generating bacterial fastidious growth. mSystems. 2020 doi: 10.1128/mSystems.00698-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Xu N, Yang Q, Yang X, Wang M, Guo M. Reconstruction and analysis of a genome-scale metabolic model for Agrobacterium tumefaciens. Mol. Plant Pathol. 2021;22:348–360. doi: 10.1111/mpp.13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Heinken A, Basile A, Hertel J, Thinnes C, Thiele I. Genome-scale metabolic modeling of the human microbiome in the era of personalized medicine. Annu. Rev. Microbiol. 2021;75:199–222. doi: 10.1146/annurev-micro-060221-012134. [DOI] [PubMed] [Google Scholar]
- 166.Kim MS, Zhang H, Shim WB. Application of game theory to explore the dynamics of host−pathogen association in phytobiomes. Phytobiomes J. 2018 doi: 10.1094/PBIOMES-04-18-0019-P. [DOI] [Google Scholar]
- 167.Trivedi P, et al. Keystone microbial taxa regulate the invasion of a fungal pathogen in agro-ecosystems. Soil. Biol. Biochem. 2017;111:10–14. doi: 10.1016/j.soilbio.2017.03.013. [DOI] [Google Scholar]
- 168.Delgado-Baquerizo M, et al. Multiple elements of soil biodiversity drive ecosystem functions across biomes. Nat. Ecol. Evol. 2020;4:210–220. doi: 10.1038/s41559-019-1084-y. [DOI] [PubMed] [Google Scholar]
- 169.Liu Q, et al. Rhizosphere fungal dynamics in sugarcane during different growth growth stages. Int. J. Mol. Sci. 2023;24:5701. doi: 10.3390/ijms24065701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Garrett KA, et al. Network analysis: a systems framework to address grand challenges in plant pathology. Annu. Rev. Phytopathol. 2018;56:559–580. doi: 10.1146/annurev-phyto-080516-035326. [DOI] [PubMed] [Google Scholar]
- 171.Skelsey P, Cooke DE, Lynott JS, Lees AK. Crop connectivity under climate change: future environmental and geographic risks of potato late blight in Scotland. Glob. Chang. Biol. 2016;22:3724–3738. doi: 10.1111/gcb.13368. [DOI] [PubMed] [Google Scholar]
- 172.Newlands NK. Model-based forecasting of agricultural crop disease risk at the regional scale, integrating airborne inoculum, environmental, and satellite-based monitoring data. Front. Environ. Sci. 2018 doi: 10.3389/fenvs.2018.00063. [DOI] [Google Scholar]
- 173.Picault S, et al. EMULSION: transparent and flexible multiscale stochastic models in human, animal and plant epidemiology. PLoS Comput. Biol. 2019;15:e1007342. doi: 10.1371/journal.pcbi.1007342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zou Y, et al. Nucleic acid purification from plants, animals and microbes in under 30 seconds. PLoS Biol. 2017;15:e2003916. doi: 10.1371/journal.pbio.2003916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Paul R, Ostermann E, Gu Z, Ristaino JB, Wei Q. DNA extraction from plant leaves using a microneedle patch. Curr. Protoc. Plant Biol. 2020;5:e20104. doi: 10.1002/cppb.20104. [DOI] [PubMed] [Google Scholar]
- 176.Xu H, et al. An ultraportable and versatile point-of-care DNA testing platform. Sci. Adv. 2020;6:eaaz7445. doi: 10.1126/sciadv.aaz7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Almogy G, et al. Cost-efficient whole genome-sequencing using novel mostly natural sequencing-by-synthesis chemistry and open fluidics platform. bioRxiv. 2022 doi: 10.1101/2022.05.29.493900. [DOI] [Google Scholar]
- 178.Zarco-Tejada PJ, et al. Divergent abiotic spectral pathways unravel pathogen stress signals across species. Nat. Commun. 2021;12:6088. doi: 10.1038/s41467-021-26335-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li Z, et al. Non-invasive plant disease diagnostics enabled by smartphone-based fingerprinting of leaf volatiles. Nat. Plants. 2019;5:856–866. doi: 10.1038/s41477-019-0476-y. [DOI] [PubMed] [Google Scholar]
- 180.McNish IG, Smith KP. Oat crown rust disease severity estimated at many time points using multispectral aerial photos. Phytopathology. 2022;112:682–690. doi: 10.1094/PHYTO-09-20-0442-R. [DOI] [PubMed] [Google Scholar]
- 181.Tanner F, et al. Sensor-based phenotyping of above-ground plant–pathogen interactions. Plant Methods. 2022;18:35. doi: 10.1186/s13007-022-00853-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schmale DG, III, Ross SD. Highways in the sky: scales of atmospheric transport of plant pathogens. Annu. Rev. Phytopathol. 2015;53:591–611. doi: 10.1146/annurev-phyto-080614-115942. [DOI] [PubMed] [Google Scholar]
- 183.Vélez-Rodríguez Z, Torres-Pratts H, Maldonado-Ramírez SL. Use of drones to recover fungal spores and pollen from the lower atmosphere. Caribb. J. Sci. 2020;50:159–170. doi: 10.18475/cjos.v50i1.a16. [DOI] [Google Scholar]
- 184.O’Shea J. Digital disease detection: a systematic review of event-based internet biosurveillance systems. Int. J. Med. Inf. 2017;101:15–22. doi: 10.1016/j.ijmedinf.2017.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Guerra CA, et al. Tracking, targeting, and conserving soil biodiversity. Science. 2021;371:239–241. doi: 10.1126/science.abd7926. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.