Abstract
We have demonstrated that possession of the gene for thermostable direct hemolysin-related hemolysin (trh) coincides with the presence of the urease gene among clinical Vibrio parahaemolyticus strains and that the location of the two genes are in close proximity on the chromosome. Here, we cloned and sequenced the 15,754-bp DNA region containing the trh gene and the gene cluster for urease production from the chromosome of clinical V. parahaemolyticus (TH3996). We found 16 open reading frames (ORFs) and a lower G+C content (41%) compared with the total genome of this bacterium (46 to 47%). The ure cluster consisted of eight genes, namely, ureDABCEFG and ureR. ureR was located 5.2 kb upstream of the other seven genes in the opposite direction. The genetic organization and sequences of the ure genes resembled those found in Proteus mirabilis. Between ureR and the other ure genes, there were five ORFs, which are homologous with the nickel transport operon (nik) of Escherichia coli. We disrupted each of the ureR, ureC, and nikD genes in TH3996 by homologous recombination and analyzed the phenotype of the mutants. In the presence of urea these mutant strains had dramatically less urease activity than the strain they were derived from. Disruption of ureR, nikD, or ureC, however, had no effect on TRH production. The DNA region containing the trh, nik, and ure genes was found in only trh-positive strains and not in Kanagawa phenomenon-positive and environmental V. parahaemolyticus strains. At the end of the region, an insertion sequence-like element existed. These results suggest that the DNA region was introduced into V. parahaemolyticus in the past through a mechanism mediated by insertion sequences. This is the first reported case that the genes for an ATP-binding cassette-type nickel transport system, which may play a role in nickel transport through bacterial cytoplasmic membrane, are located adjacent to the ure cluster on the genome of an organism.
Vibrio parahaemolyticus is a gram-negative, halophilic marine bacterium which causes gastroenteritis in humans who are infected through consumption of raw or inadequately cooked seafood (15). V. parahaemolyticus strains that are isolated from diarrheal patients produce either the thermostable direct hemolysin (TDH) or the TDH-related hemolysin (TRH), or both, while hardly any isolates from the environment have these properties (15, 41). TDH and TRH, encoded by tdh and trh, respectively, are each composed of 165-amino-acid residues and show approximately 67% identity in their amino acid sequences. TDH and TRH show several common biological activities, such as hemolytic activity, enterotoxicity, cytotoxicity, and cardiotoxicity, and are considered to be the major virulence factors in the pathogenesis of V. parahaemolyticus (15, 35, 41, 47).
Urease is an enzyme that catalyzes the hydrolysis of urea to ammonia and carbon dioxide. It is found in a number of bacteria, plants, fungi, and algae (29). Several bacterial urease genes have been isolated and characterized. The types of organization of gene clusters for urease in most bacteria are basically similar, comprising structural genes, accessory genes involved in the incorporation of the nickel ion into the apoenzyme and, in some cases, genes involved in a nickel uptake or regulation of urease expression (30). In common with other Vibrio species, generally only a small population (several percent) of clinical V. parahaemolyticus strains produce urease (7, 13, 16, 46). The relatively rare urease-positive phenotype of V. parahaemolyticus is always associated with the possession of the trh gene (26, 36, 37, 46), making urease production a reasonably good clinical diagnostic marker for virulent (trh-positive) V. parahaemolyticus (46). Our previous research has shown that the association is due to a genetic linkage between the structural gene for urease (ureC) and trh on the chromosome of virulent V. parahaemolyticus strains (17). In one such strain, using long and accurate PCR (LA-PCR), the distance between trh and ureC was shown to be less than 8.5 kb (18). We also recently reported that V. parahaemolyticus and other Vibrio species possess two chromosomes and that, when present, the trh and ureC genes are localized on the smaller replicon (52).
In this study, to precisely understand the nature of the genetic linkage between the trh and ure genes in V. parahaemolyticus, we cloned and sequenced a DNA region (ca. 16 kb) containing the genes from a clinical V. parahaemolyticus strain. Furthermore, we constructed mutant strains with disruption in the genes on this DNA region to examine the role of the genes to urease production and hemolytic activity of V. parahaemolyticus.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Table 1 shows the bacterial strains and plasmids used in this study. Clinical V. parahaemolyticus strains, including TH3996 (51), were isolated at the Osaka International Airport quarantine station from patients with traveller's diarrhea. The bacterium was cultured at 37°C with shaking in Luria-Bertani (LB) broth (tryptone, 1%; yeast extract, 0.5%) with 3% NaCl. The Escherichia coli JM109 (53) and SM10λpir (28) strains that we used for general manipulation of plasmids and mobilization of plasmid into V. parahaemolyticus were grown in LB broth or on LB agar. TCBS agar (Nissui, Tokyo, Japan) was used for the screening of mutant strains. Antibiotics were used at the following concentrations: ampicillin (100 μg/ml), kanamycin (50 μg/ml), and chloramphenicol (5 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and description | Source or reference |
|---|---|---|
| E. coli | ||
| JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB)/F′ (traD36 proAB+ lacIqlacZΔM15) | 53 |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpir R6K | 28 |
| V. parahaemolyticus | ||
| TH3996 | Clinical isolate, trh+ ure+ | 51 |
| TH3996(trh) | TH3996, trh disrupted | 51 |
| TH3996(nikD) | TH3996, nikD disrupted | This study |
| TH3996(ureR) | TH3996, ureR disrupted | This study |
| TH3996(ureC) | TH3996, ureC disrupted | This study |
| Plasmids | ||
| pUC119 | Cloning vector, Ampr | 40 |
| pT7-Blue | Multicopy (ColE1 ori) TA cloning vector, Ampr | Novagen, Inc. |
| pKY719 | R6K-ori suicide vector for gene replacement, Cmr | 33 |
| pKS1 | 11.8-kb XhoI fragment cloned into the SalI site on pUC119 | This study |
| pKS2 | 7.3-kb BglII fragment cloned into the BamHI site on pUC119 | This study |
| pKS3 | 4.0-kb SpeI fragment cloned into the XbaI site on pUC119 | This study |
DNA manipulation.
Chromosomal DNA from V. parahaemolyticus TH3996 was extracted from overnight culture of the organism in LB broth with 3% NaCl using a previously described method (50). DNA used for subcloning or nucleotide sequence analysis was extracted from E. coli by using the QIAPrep Spin Plasmid Kit (Qiagen) according to the manufacturer's recommendations. Cloning, restriction endonuclease procedures, DNA ligation, and transformation of E. coli by plasmids were carried out using previously described standard protocols (40). All of the restriction enzymes and DNA ligation kits were purchased from Takara Shuzo (Otsu, Japan).
Southern blot analysis.
Digested chromosomal DNA was resolved in 0.8% agarose gel in 1× TBE buffer (0.09 M Tris-borate, 2 mM EDTA [pH 8.0]) and transferred onto a nylon membrane (GeneScreen Plus; Dupont). DNA was fixed to the membrane by UV cross-linking using a GS Gene Linker UV chamber (Bio-Rad). We prepared probes using the PCR DIG Probe Synthesis Kit (Boehringer Mannheim) with specific primers for each gene. Hybridization was carried out at 40°C, and hybridized DNA was detected with an alkaline phosphatase-labeled anti-digoxigenin monoclonal antibody (Boehringer Mannheim).
DNA sequence analysis.
The nucleotide sequences were determined with a Licor model 4000L automated DNA sequencer (Lincoln, Nebr.) using universal M13 IRD41 infrared-dye-labeled primers. Sequence data were analyzed using the DNASIS program (Hitachi Software, Tokyo, Japan) and the GENETYX sequence analysis program (Software Development, Tokyo, Japan). The homology of the open reading frames (ORFs) was ascertained via the National Center for Biotechnology Information using the BLAST network service to search GenBank.
Oligonucleotide primers and PCR conditions.
The following oligonucleotide primers were used: for the cloning of pKS3, 5′-TCGCATAATTGGTGTTGAA-3′ and 5′-TTTTGGCAGCATACCTTTG-3′; for the Cm probe, 5′-TTGCCCGCCTGATGAATGCTC-3′ and 5′-CCCTGCCACTCATCGCAGTA-3′; for the nikD mutant, 5′-GTCAAATTGTCAACTCGT-3′ and 5′-CAATGGTTTCGCCATTAG-3′; for the ureR mutant, 5′-CACAAAGAAAATGGCATAT-3′ and 5′-CGAAATTGCAGTGGTGTT-3′; for the ureC mutant, 5′-ACTAATGCTACAACAGTCAC-3′ and 5′-ATGCTGGAATGATGTTAGGT-3′; and for LA-PCR, 5′-CATTTCCGCTCTCATATGC-3′ and 5′-ATGCTGGAATGATGTTAGGT-3′.
PCR conditions for the construction of mutant strains and probes were as follows: after 2 min of denaturation at 94°C, a cycle of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min was repeated 30 times. To examine the distance from trh to ureC, PCR was performed using LA-PCR Kit (version 2; Takara Shuzo). The PCR conditions were as follows: after initial denaturation at 94°C for 3 min, a cycle of 98°C for 20 s, 58°C for 20 s, and 68°C for 12 min was repeated 30 times, ending with one extension period of 10 min at 72°C. The PCR reaction was carried out using a Perkin-Elmer Type 9700 thermal cycler. Custom-synthesized oligonucleotides for the PCR were purchased from Nisshinbo (Tokyo, Japan).
Construction of mutant strains.
Mutant strains, derived from V. parahaemolyticus TH3996, were constructed as previously described (51), except that the homologous recombination was done by a single-crossover event. Briefly, to construct the mutants, using the chromosomal DNA of V. parahaemolyticus TH3996 as a template, we amplified partial sequences of ureR, nikD, and ureC by PCR. Each amplified fragment was cloned into a pT7-Blue vector and then digested with BamHI and PstI. These fragments were cloned into a R6K-ori suicide vector, pKY719 (33), digested with BamHI and PstI. These plasmids were introduced into E. coli SM10λpir and then mated with V. parahaemolyticus TH3996. Conjugation was performed on nitrocellulose membrane laid over LB agar plates with incubation for 4 h at 37°C. The membrane was suspended in LB broth and inoculated onto TCBS agar plates containing 5 μg of chloramphenicol per ml and incubated overnight at 37°C. Chloramphenicol-resistant colonies were isolated and screened by Southern blot analysis for single-crossover mutation of ureR, nikD, or ureC.
Urease assay.
Bacteria were grown at 37°C with shaking in 100 ml of LB broth (3% NaCl) supplemented with 0.1% sterile urea after autoclaving of the medium. The organisms were harvested by centrifugation at 10,000 × g for 10 min, washed three times with 20 mM sodium phosphate (pH 7.0)–5 mM dithiothreitol–1 mM EDTA, and disrupted by sonication. Subsequently, supernatant was obtained after centrifugation at 4°C (12,000 × g, 20 min). The supernatant was used both for the urease assay and for the determination of protein quantity using the Bio-Rad DC protein assay kit (Bio-Rad). Urease activity was determined using an ammonia test kit (Wako Pure Chemical Industries, Ltd., Osaka, Japan) to measure the release of ammonia. The amount of ammonia produced was calculated by referring to a standard curve made for known concentrations of ammonium chloride. Urease activity was expressed as micromoles of urea hydrolyzed per minute per milligram of protein.
Hemolytic activity assay.
Wild-type and mutant strains were cultured at 37°C for 16 h with shaking in SPP medium (14) (5 g of NaCl, 10 g of peptone, 2 g of glucose, and 5 g of dibasic sodium phosphate per liter in distilled water, pH 7.6). The supernatant was obtained by centrifugation (12,000 × g, 10 min) at 4°C. The hemolytic activity was determined by a modification of the previously described method (14). Briefly, sheep erythrocytes were washed in phosphate-buffered saline (PBS) three times and adjusted to a hematocrit of 4% with PBS. Equal volumes (100 μl each) of supernatant serially diluted with PBS and erythrocyte suspension were mixed and incubated at 37°C in a water bath for 1 h and then centrifuged at 2,000 × g for 2 min. Supernatant samples (160 μl) of the reaction mixtures were taken for spectrophotometric measurement of released hemoglobin at 540 nm on a 96-well plate using a Multiskan Mcc/340 (Labosystems, Tokyo, Japan).
Rabbit ileal loop test.
We assessed the enterotoxic activity of mutant strains and the wild-type strain by using a previously described method (51).
Nucleotide sequence accession number.
The nucleotide sequence data reported here will appear in the DDBJ, EMBL and the GenBank sequence databases under accession no. AB038238.
RESULTS
Cloning and sequencing of the region containing the trh and ure cluster.
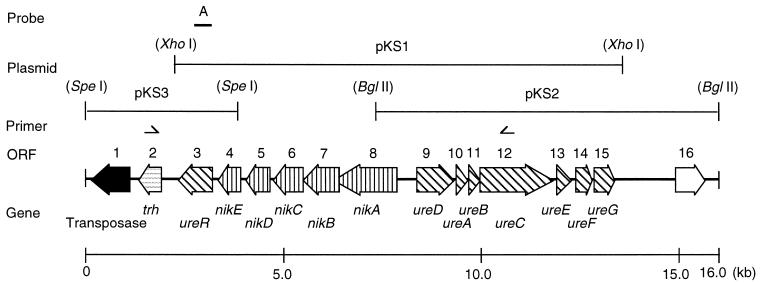
For the cloning of the region containing the trh and ure cluster, total chromosomal DNA was isolated from V. parahaemolyticus TH3996 and completely digested with several restriction enzymes. The DNA fragments were Southern blotted with a digoxigenin-labeled ure probe (17). The 11.8-kb XhoI fragment and the 7.3-kb BglII fragment which hybridized with the ure probe were cloned to the pUC119 vector at the SalI and BamHI sites (designated pKS1 and pKS2, respectively). The adjacent region of the 11.8-kb XhoI fragment, which as a 4.0-kb SpeI fragment hybridized with probe A (Fig. 1), was cloned into the XbaI site of the pUC19 vector (designated pKS3). In the end, the total region of approximately 16 kb that contains the trh and ure cluster was cloned (Fig. 1). After analysis of the restriction maps, each fragment was digested with appropriate restriction enzymes and subcloned into the corresponding sites of the pUC119 vector. The complete nucleotide sequence was determined in both directions.
FIG. 1.
Schematic representation of the region containing the trh and ure gene cluster and ORF assignment.
Nucleotide sequence analysis.
Nucleotide sequence analysis of the DNA region revealed that the region is 15,754 bp in size and possesses a total of 16 ORFs (Fig. 1 and Table 2). ORF1 as homologous with a transposase found in Vibrio anguillarum (88% identity; L40498) and was flanked by 18-bp inverted repeats (5′-GGCTTTGTTGCGTAATTC-3′) in both sides. ORF2 was the trh gene. ORF3 had some homology with two positive regulators for urease expression, P. mirabilis UreR (51% identity; A40644) and E. coli UreR (50% identity; P32326). In addition, the ORF contained a typical bacterial regulatory AraC family signature located at amino acid positions 215 to 257. Therefore, ORF3 was designated UreR.
TABLE 2.
ORFs identified in the region containing the trh and urease gene cluster
| ORF | Gene | Position
|
No. of aaa | Homologous ORFs in database
|
|||
|---|---|---|---|---|---|---|---|
| 5′ | 3′ | Accession no. | % Identity | Note (origin) | |||
| 1 | Transposase | 1042 | 125 | 306 | L40498 | 88 | Transposase (V. anguillarium) |
| 2 | trh | 1936 | 1370 | 189 | S67850 | 100 | TRH (V. parahaemolyticus) |
| 3 | ureR | 3220 | 2375 | 282 | A40644 | 53 | UreR (P. mirabilis) |
| 4 | nikE | 3990 | 3370 | 207 | AAC67221 | 41 | DppE (S. pyogenes) |
| 5 | nikD | 4682 | 3993 | 230 | AAC67220 | 32 | DppD (S. pyogenes) |
| 6 | nikC | 5485 | 4673 | 271 | AAB67065 | 31 | Putative transmembrane protein (S. typhimurium) |
| 7 | nikB | 6449 | 5490 | 320 | AAC67218 | 32 | DppB (S. pyogenes) |
| 8 | nikA | 7983 | 6346 | 546 | AAB67063 | 36 | Putative substrate-binding protein (S. typhimurium) |
| 9 | ureD | 8389 | 9228 | 280 | P17089 | 59 | UreD (P. mirabilis) |
| 10 | ureA | 9249 | 9548 | 100 | Q03282 | 90 | UreA (E. coli) |
| 11 | ureB | 9562 | 9882 | 107 | P17087 | 75 | UreB (P. mirabilis) |
| 12 | ureC | 9882 | 11582 | 567 | P17086 | 86 | UreC (P. mirabilis) |
| 13 | ureE | 11640 | 12116 | 159 | P17090 | 62 | UreE (P. mirabilis) |
| 14 | ureF | 12147 | 12809 | 221 | P17091 | 62 | UreF (P. mirabilis) |
| 15 | ureG | 12821 | 13456 | 212 | Q06206 | 88 | UreG (P. mirabilis) |
| 16 | 14836 | 15660 | 275 | ||||
aa, amino acid(s).
ORF4 and ORF8 showed striking amino acid sequence similarities to components of the peptide-binding-protein-dependent transport systems and ATP-binding cassette (ABC) transporters of several gram-positive and gram-negative bacteria, such as the dpp operon, membrane-associated complexes of five proteins belonging to the ABC transporter family of Streptococcus pyogenes (39), the Salmonella enterica serovar Typhimurium opp operon (12), the periplasmic binding-protein-dependent transport system, and the E. coli nik operon, which encode the specific transport system for nickel (34). To determine the possible contribution of these ORFs to urease production, ORF5 was disrupted by homologous recombination. The disruption of ORF5 resulted in the near abolishment of urease activity (see below) (Table 3). Thus, it seems likely that the five ORFs are components of an ABC transporter which mediates the transport of the nickel ion from the exterior to the cytosol and is designated nikABCDE.
TABLE 3.
Urease activities, hemolytic activities, and rabbit ileal loop test results of V. parahaemolyticus TH3996 and its isogenic mutant strains
| Strain | Mean urease activitya (μmol of NH3/min/mg of protein) ± SEM at:
|
Mean hemolytic activity (%)c ± SEM | Mean FAd ± SEM | |
|---|---|---|---|---|
| 5 hb | 16 hb | |||
| TH3996 | 2.500 ± 0.200 | 2.460 ± 0.200 | 100 | 0.35 ± 0.19 |
| TH3996 (nikD) | 0.020 ± 0.010 | 0.020 ± 0.010 | 110 ± 10 | 0.34 ± 0.23 |
| TH3996 (ureR) | 0.020 ± 0.010 | 0.020 ± 0.010 | 90 ± 10 | 0.27 ± 0.11 |
| TH3996 (ureC) | 0.010 ± 0.010 | 0.010 ± 0.010 | 90 ± 10 | 0.27 ± 0.09 |
| TH3996 (trh) | 2.490 ± 0.200 | 2.450 ± 0.020 | 0 | 0.12 ± 0.12 |
Values represent the means of three experiments, each conducted in duplicate.
Incubation time.
Hemolytic activity in culture supernatant after incubation of the strains in SPP medium at 37°C for 16 h with shaking. The percentage of hemolytic activity for the wild type was set at 100%. Values represent the means of six experiments.
FA, amount of accumulated fluid in milliliters per length (in centimeters) of ligated rabbit small intestine. Values represent the means of six experiments.
The nik operon was in the same direction of transcription as the ORF1 to ORF3 genes (Fig. 1). NikA, ORF8, comprised 546 amino acid residues with a predicted molecular mass of 61.0 kDa. Hydropathy plots of highly hydrophobic proteins, NikB (ORF7) and NikC (ORF6) could be organized in six potential transmembrane domains (data not shown). These features are consistent with the profile of integral cytoplasmic membrane proteins (21). NikD (ORF5) and NikE (ORF4) possessed a consensus ATP-binding motif consisting of two sites, A (GX2GXGKS) and B (DEX4LD), as described by Walker et al. (49). It seems likely that NikD and NikE are responsible for coupling energy to the nickel transport system.
ORF9 to ORF15 showed high homology with the ure clusters found in other bacterial species such as Proteus mirabilis and Klebsiella aerogenes. These seven contiguous ORFs were transcribed in the direction opposite to ORF1 to ORF8. ORF9 (UreD) showed homology with P. mirabilis UreD. ORF10 to ORF12 were highly homologous to UreA, UreB, and UreC, the three structural subunits of urease in P. mirabilis and K. aerogenes. Amino acid residues in the structural subunits, which have been shown to have functional significance in other bacterial ureases, were conserved in relative positions in the V. parahaemolyticus urease. Histidine residues at His-134, His-136, and His-246 of the α-subunit of the K. aerogenes urease were shown by site-directed mutagenesis to be involved in nickel binding (38). These histidine residues were present at the corresponding positions in the V. parahaemolyticus UreC as in the K. aerogenes UreC. Furthermore, the consensus sequence for the urease active sites (MVCHHLD) (27) was identified at amino acid positions 317 to 323 of the V. parahaemolyticus UreC. Additional amino acid residues with defined functional significance in the K. aerogenes urease include His-39 and His-41 in UreB and His-97 in UreA (38): these were also conserved in the appropriate positions in V. parahaemolyticus UreB and UreA. A high degree of homology of UreE, UreF, and UreG was also found with P. mirabilis UreE, UreF, and UreG, which encode the urease accessory proteins involved in the incorporation of nickel into the urease apoenzyme. UreE (ORF13) contained at its C terminus a polyhistidine tail, which suggests a role in Ni2+ binding and the insertion of the ion into apoenzyme. A similar histidine tail is present in the P. mirabilis and K. aerogenes UreE (23, 45). UreG (ORF15) contained, at amino acids 19 to 26, a signature for regions involved in ATP/GTP binding, which was similar to a motif found in the P. mirabilis UreG protein (44). This suggests involvement with nucleoside triphosphate hydrolysis during the assembly of urease or the insertion of Ni2+. We failed to find any significant homology of ORF16 protein with any sequences in GenBank.
The 16 ORFs appear to be grouped into two transcription directions. From ORF1 to ORF8 the genes are transcribed in the opposite direction to the remaining genes, ORFs 9 to 16. We found that the average G+C content of the DNA sequence, at 41.0%, was lower than the G+C content of the V. parahaemolyticus genome (46 to 47%) (1).
Construction of ureR, nikD, and ureC mutant strains.
To determine the possible contribution of a putative nickel transport gene operon and urease gene cluster to urease production and hemolytic activity, each of the genes (ureR, nikD, and ureC) was disrupted by homologous recombination in V. parahaemolyticus TH3996.
In the wild-type strain, hybridization of a BglII and XhoI digests of chromosomal DNA using probes specific for ureR or nikD showed a common 5.3-kb fragment. This fragment was replaced with 9.4-kb hybridizing fragments in the ureR and nikD mutants. The BglII digest of chromosomal DNA in the wild-type strain showed a single 7.3-kb fragment using a specific ureC probe. This fragment was also replaced with an 11.4-kb hybridizing fragment, a finding consistent with the size expected from insertion of the suicide vector into each gene, in the ureC mutant (data not shown). When the same digest of chromosomal DNA was examined using a Cm (a gene of chloramphenicol resistance) probe, each of the mutant strains (ureR, nikD, and ureC) was hybridized with the probe but no signal was detected in the wild-type strain. Mutant strains grew at a similar rate to that of the wild type (data not shown). We used the mutant strains to examine urease activity and hemolytic activity.
Urease activity of wild and mutant strains.
In P. mirabilis, which possesses the ureR gene, urease production is not regulated by the nitrogen regulatory system (ntr) or catabolite repression but is induced by urea and the induction is mediated by ureR (8). Because V. parahaemolyticus TH3996 has the ureR gene in common with P. mirabilis, we investigated whether or not the urease production of the TH3996 strain is affected by the presence of urea. In the LB broth with 3% NaCl without urea, there was little urease production. With 0.1% urea supplementing the medium, however, urease production was 100 times greater. These data suggest that urea is involved in the induction of V. parahaemolyticus urease. Therefore, the following experiments for examining urease production were conducted with media containing 0.1% urea. The urease activities of ureR, nikD, and ureC mutant strains were very low, even in the presence of 0.1% urea (Table 3). In contrast, under the same conditions the urease activity of the trh mutant strain (51) did not change compared with the wild-type strain. These results suggest that ureR, nikD, and ureC genes are essential for active urease production. From these results, we concluded that the ureR, nikD, and ureC genes are important for urease expression, whereas trh is not.
Hemolytic activity of mutant strains.
Next, we investigated the possible contribution of the ureR, nikD, and ureC genes to hemolysin (TRH) production by the TH3996 strain by comparing the hemolytic activity of wild-type and mutant strains. As shown in Table 3, the nikD, ureR, and ureC mutant strains retained the same hemolytic activity.
Enterotoxicity of mutants as shown by the rabbit ileal loop test.
To investigate the enterotoxicity of the mutant strains, we examined the enterotoxic activity of the wild-type and mutant strains in rabbit ileum. The wild-type or mutant strains (108 CFU) were injected into the ligated ileal loops of rabbits which were sacrificed after 16 h. After removal of the small intestine the fluid accumulation in the ligated ileal loops was measured (Table 3). Except for the trh mutant strain, no significant differences in fluid accumulation were observed in samples from wild-type and mutant strains. These results indicate that the presence or absence of urease production by the V. parahaemolyticus strain does not affect the enterotoxicity of the organism under these test conditions.
Distribution of ureR and nik operon in clinical V. parahaemolyticus strains.
In previous studies, we demonstrated that the possession of trh completely coincided with the presence of the ureC (a structural gene of urease) by clinical V. parahaemolyticus strains and that, in strain AQ4673, which possesses the two genes, the distance between trh and ureC is within several kilobases (17, 18). Consistent with these previous results, nucleotide sequence analysis of TH3996 showed that the distance between the two genes is 7,945 bp. To investigate the distance between the two genes of the other trh-positive clinical strains of V. parahaemolyticus, we performed LA-PCR using oligonucleotide primer pairs targeting trh and ureC (18). All of the strains for testing were amplified accurately to the same amplicon size as the AQ4673 strain (data not shown). Next, we used colony hybridization with specific probes for ureR and nikD to test for the presence or absence of the ureR and nik operon in clinical and environmental strains of V. parahaemolyticus. The ureR and nikD genes were present in all of the trh-positive strains (n = 8) but not in Kanagawa phenomenon (KP)-positive strains (n = 12) and the environmental strains (n = 6) (data not shown).
These data suggest that the DNA region containing the trh and urease gene cluster is present in trh-positive V. parahaemolyticus strains but absent in KP-positive and environmental V. parahaemolyticus.
DISCUSSION
Urease, which requires the nickel ion in the active site, catalyzes the hydrolysis of urea to ammonia and carbon dioxide, resulting in net increase in environmental pH (30). So far, urease gene clusters have been isolated in several bacteria, such as P. mirabilis (19, 20, 44), K. aerogenes (23, 31, 32), Helicobacter pylori (2, 4, 22), Bacillus sp. strain TB-90 (25), Streptococcus salivarius (42), and Yersinia enterocolitica (5, 43). The expression of bacterial urease genes is regulated by different mechanisms (3). For example, urease expression in K. aerogenes is activated only under nitrogen-limiting conditions (24). P. mirabilis urease is induced by urea and is mediated by the positive transcriptional regulator ureR (8). S. salivarius urease is regulated by pH (42).
We previously reported that the urease production by clinical V. parahaemolyticus strains correlates completely with the possession of the trh gene, and research by others has shown similar results (26, 36, 37). We have also found that the distance between the two genes is within several kilobases (17, 18), suggesting the genetic linkage between the two genes. In the present study, we demonstrated this genetic linkage. To investigate the coexistence and proximity of the trh and ure genes, we cloned and sequenced the region containing the trh and ure gene cluster on the chromosome of TH3996, a clinical V. parahaemolyticus strain that has the trh gene and produces urease.
Nucleotide sequencing showed that the ure gene cluster of V. parahaemolyticus is comprised of eight genes. The structural genes, ureA, ureB, and ureC, which encode subunits of the enzyme, are flanked immediately upstream by ureD, encoding a chaperone-like protein, and downstream by the ureE, ureF, and ureG accessory genes that encode the proteins required for incorporating the nickel ion into the metallocenter within the active site (30). In addition, ureR, the positive regulatory gene, lies 5.2 kb upstream of ureD and is oriented opposite the other seven genes. The predicted polypeptide sequence from this gene contains a putative helix-turn-helix DNA-binding motif, suggesting that this protein belongs to the AraC family of positive regulators. Members of the AraC family are primarily involved in the positive regulation of a number of genes (10).
As our data show, in the absence of urea, expression of V. parahaemolyticus urease is very low, but when 0.1% urea is present in the medium, urease expression is about 100 times greater. This increase was not seen in samples of the mutant strain which had undergone disruption of ureR. From this we conclude that V. parahaemolyticus urease is induced by urea and that this induction is mediated by ureR. The organization and sequences of the ure gene cluster of V. parahaemolyticus were similar to that of P. mirabilis. Furthermore, the regulation of the expression of urease is similar in the two species (Table 3). The major difference between the V. parahaemolyticus and P. mirabilis urease gene clusters was the presence or absence of a nickel transporter operon between ureR and ureD.
Between ureR and ureD of V. parahaemolyticus there were five overlapping genes that had significant homology to the nik operon of E. coli (34). These genes appear to encode a typical ABC transporter system, including ATP-binding polypeptides and integral cytoplasmic membrane proteins (9). NikB and NikC were highly hydrophobic proteins with six potential transmembrane domains, which may form the channel for the substrate transport. NikD and NikE contain Walker A and B motifs responsible for coupling energy to the nickel transport system (49). To investigate the role of the ABC transport system, we constructed a nikD disrupted mutant by homologous recombination. The mutant strain showed almost complete absence of urease activity compared with the wild-type strain. Consequently, it is plausible to suggest that the ABC transport system that we found contributes to nickel transport through the bacterial cytoplasmic membrane. ABC-type transporters of nickel ions from the extracellular milieu into bacterial cells, such as an ABC transporter in H. pylori (11), have also been found; however, similar genes have not been reported in other bacteria that possess the urease gene cluster. In H. pylori, the ABC transporter genes are not located close to the urease gene cluster on the genome (48). Interestingly, the nik genes of V. parahaemolyticus are located between the ureR and the ure genes. Thus, this is the first report of finding genes for the ABC-type nickel transport system adjacent to the urease gene cluster on the genome of an organism.
LA-PCR analysis showed the distance between the trh and ureC of all the trh-positive V. parahaemolyticus samples we tested to be precisely 7.9 kb. ureR and nik operon were consistently present in trh-positive strains (regardless of tdh possession) but not in KP-positive strains, which possess two copies of tdh, or in environmental strains. The sequencing results showed ORF1 to be a transposase. These findings suggest the possibility that the trh, nik operon, and ure gene cluster were transmitted into V. parahaemolyticus strains through a mechanism mediated by insertion sequences in the past.
Although the rabbit ileal loop test showed that the enterotoxicity of the ureR, ureC, and nikD mutant strains did not change compared with the TH3996 strain that they were derived from, we cannot completely rule out the possibility that the V. parahaemolyticus urease plays a certain role in the pathogenesis of this bacterium. In the case of Y. enterocolitica, urease appears to act as a virulence factor by enhancing the survival of bacteria during their passage through the stomach, presumably by neutralizing hydrogen ions which penetrate the bacterial cell wall (6). The contribution of urease to the pathogenesis of V. parahaemolyticus is currently being analyzed in our laboratory.
ACKNOWLEDGMENTS
This study was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan; a Grant for International Health Cooperation Research from the Ministry of Health and Welfare of Japan; and the “Research for the Future” Program of the Japan Society for the Promotion of Sciences (JSPS-RFTF 97L00704).
REFERENCES
- 1.Baumann P, Furniss A L, Lee J V. Facultatively anaerobic gram-negative rods, family Vibrionaceae, genus Vibrio. In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 518–538. [Google Scholar]
- 2.Clayton C L, Pallen M J, Kleanthous H, Wren B W, Tabaqchali S. Nucleotide sequence of two genes from Helicobacter pylori encoding for urease subunits. Nucleic Acids Res. 1990;18:362. doi: 10.1093/nar/18.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins C M, Orazio S E F D. Bacterial urease: structure, regulation of expression and role in pathogenesis. Mol Microbiol. 1993;9:907–913. doi: 10.1111/j.1365-2958.1993.tb01220.x. [DOI] [PubMed] [Google Scholar]
- 4.Cussae V, Ferrero R L, Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J Bacteriol. 1992;174:2466–2473. doi: 10.1128/jb.174.8.2466-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Koning-Ward T F, Ward A C, Robins-Browne R M. Characterization of the urease-encoding gene complex of Yersinia enterocolitica. Gene. 1994;145:25–32. doi: 10.1016/0378-1119(94)90318-2. [DOI] [PubMed] [Google Scholar]
- 6.De Koning-Ward T F, Robins-Browne R M. Contribution of urease to acid tolerance in Yersinia enterocolitica. Infect Immun. 1995;63:3790–3795. doi: 10.1128/iai.63.10.3790-3795.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eko F O. Urease production in Vibrio parahaemolyticus: a potential marker for virulence. Eur J Epidemiol. 1992;8:627–628. doi: 10.1007/BF00146387. [DOI] [PubMed] [Google Scholar]
- 8.Eric B N, Concaugh E A, Foxall P A, Island M D, Mobley H L T. Proteus mirabilis urease: transcriptional regulation by ureR. J Bacteriol. 1993;175:465–473. doi: 10.1128/jb.175.2.465-473.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fath M J, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallegos M T, Michan C, Gamos J L. The XylS/AraC family of regulators. Nucleic Acids Res. 1993;21:807–810. doi: 10.1093/nar/21.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendricks J K, Mobley H L. Helicobacter pylori ABC transporter: effect of allelic exchange mutagenesis on urease activity. J Bacteriol. 1997;179:5892–5902. doi: 10.1128/jb.179.18.5892-5902.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiles I D, Gallagher M P, Jamieson D J, Higgins C F. Molecular characterization of the oligopeptide permease of Salmonella typhimurium. J Mol Biol. 1987;195:125–142. doi: 10.1016/0022-2836(87)90332-9. [DOI] [PubMed] [Google Scholar]
- 13.Honda S, Matsumoto S, Miwatani T, Honda T. A survey of urease-positive Vibrio parahaemolyticus strains isolated from traveler's diarrhea, sea water and imported frozen sea foods. Eur J Epidemiol. 1992;8:861–864. doi: 10.1007/BF00145333. [DOI] [PubMed] [Google Scholar]
- 14.Honda T, Ni Y, Miwatani T. Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect Immun. 1988;56:961–965. doi: 10.1128/iai.56.4.961-965.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honda T, Iida T. The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct haemolysin and related haemolysin. Rev Med Microbiol. 1993;4:106–113. [Google Scholar]
- 16.Huq M I, Huber D, Kibryia G. Isolation of urease producing Vibrio parahaemolyticus strains from cases of gastroenteritis. Indian J Med Res. 1979;70:549–553. [PubMed] [Google Scholar]
- 17.Iida T, Suthienkul O, Park K-S, Tang G-Q, Yamamoto R K, Ishibashi M, Yamamoto K, Honda T. Evidence for genetic linkage the ure and trh genes in Vibrio parahaemolyticus. J Med Microbiol. 1997;46:639–645. doi: 10.1099/00222615-46-8-639. [DOI] [PubMed] [Google Scholar]
- 18.Iida T, Park K-S, Suthienkul O, Kozawa J, Yamaichi Y, Yamamoto K, Honda T. Close proximity of the tdh, trh and ure genes on the chromosome of Vibrio parahaemolyticus. Microbiology. 1998;144:2517–2523. doi: 10.1099/00221287-144-9-2517. [DOI] [PubMed] [Google Scholar]
- 19.Jones B D, Mobley H L T. Proteus mirabilis urease: nucleotide sequence determination and comparison with jack bean urease. J Bacteriol. 1989;171:6414–6422. doi: 10.1128/jb.171.12.6414-6422.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson D E, Russell R G, Lockatell C V, Zulty J C, Warren J W, Mobley H L T. Contribution of Proteus mirabilis urease to persistence, urolithiasis, and acute pyelonephritis in a mouse model of ascending urinary tract infection. Infect Immun. 1993;61:2748–2754. doi: 10.1128/iai.61.7.2748-2754.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 22.Labigue A, Cussac V, Courcoux P. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J Bacteriol. 1991;173:1920–1931. doi: 10.1128/jb.173.6.1920-1931.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee M H, Mulrooney S B, Renner M J, Markowitz Y, Hausinger R P. Klebsiella aerogenes urease gene cluster: sequence of ureD and demonstration that four accessory genes (ureD, ureE, ureF, and ureG) are involved in nickel metallocenter biosynthesis. Protein Sci. 1992;2:1042–1052. doi: 10.1128/jb.174.13.4324-4330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macaluso A, Best E A, Bender R A. Role of the nac gene product in the nitrogen regulation of some NTR-regulated operons of Klebsiella aerogenes. J Bacteriol. 1990;172:7249–7255. doi: 10.1128/jb.172.12.7249-7255.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeda M, Hidaka M, Nakamura A, Masaki H, Uozumi T. Cloning, sequencing, and expression of thermophilic Bacillus sp. strain TB-90 urease gene complex in Escherichia coli. J Bacteriol. 1994;176:432–442. doi: 10.1128/jb.176.2.432-442.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magalhaes M, Takeda Y, Magalhaes V, Tateno S. Brazilian urease-positive strains of Vibrio parahaemolyticus carry genetic potential to produce the TDH-related hemolysin. Mem Inst Oswaldo Cruz. 1992;87:167–168. doi: 10.1590/s0074-02761992000100027. [DOI] [PubMed] [Google Scholar]
- 27.Martin P R, Hausinger R P. Site-directed mutagenesis of the active cysteine in Klebsiella aerogenes urease. J Biol Chem. 1992;267:22024–22027. [PubMed] [Google Scholar]
- 28.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mobley H L T, Hausinger R P. Microbial urease: significance, regulation, and molecular characterization. Microbiol Rev. 1989;53:85–108. doi: 10.1128/mr.53.1.85-108.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mobley H L T, Island M D, Hausinger R P. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulrooney S B, Pankratz H S, Hausinger R P. Regulation of gene expression and cellular localization of cloned Klebsiella aerogenes urease. J Gen Microbiol. 1989;135:1769–1776. doi: 10.1099/00221287-135-6-1769. [DOI] [PubMed] [Google Scholar]
- 32.Mulrooney S B, Hausinger R P. Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J Bacteriol. 1990;172:5837–5843. doi: 10.1128/jb.172.10.5837-5843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagamune K, Yamamoto K, Honda T. Cloning and sequencing of a novel hemolysis gene of Vibrio cholerae. FEMS Microbiol Lett. 1995;128:265–269. doi: 10.1111/j.1574-6968.1995.tb07534.x. [DOI] [PubMed] [Google Scholar]
- 34.Navarro C, Wu L-F, Mandrand-Berthelot M-A. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol Microbiol. 1993;9:1181–1191. doi: 10.1111/j.1365-2958.1993.tb01247.x. [DOI] [PubMed] [Google Scholar]
- 35.Nishibuchi M, Fasano A, Russell R G, Kaper J B. Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect Immun. 1992;60:3539–3545. doi: 10.1128/iai.60.9.3539-3545.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okuda J, Ishibashi M, Abbott S L, Janda J M, Nishibuchi M. Analysis of the thermostable direct hemolysin (tdh) gene and the tdh-related hemolysin (trh) genes in urease-positive strains of Vibrio parahaemolyticus isolated on the West Coast of the United States. J Clin Microbiol. 1997;35:1965–1971. doi: 10.1128/jcm.35.8.1965-1971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osawa R, Okitsu T, Morozumi H, Yamai S. Occurrence of urease-positive Vibrio parahaemolyticus in Kanagawa, Japan, with specific reference to presence of thermostable direct hemolysin (TDH) and the TDH-related hemolysin genes. Appl Environ Microbil. 1996;62:725–727. doi: 10.1128/aem.62.2.725-727.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park I S, Hausinger R P. Site-directed mutagenesis of Klebsiella aerogenes urease-identification of histidine residues that appear to function in nickel ligation, substrate binding, and catalysis. Protein Sci. 1993;2:1034–1041. doi: 10.1002/pro.5560020616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Podbielski A, Leonard B A. The group A streptococcal dipeptide permease (Dpp) is involved in the uptake of essential amino acids and affects the expression of cysteine protease. Mol Microbiol. 1998;28:1323–1334. doi: 10.1046/j.1365-2958.1998.00898.x. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 41.Shirai H, Ito H, Hirayama T, Nakabayashi Y, Kumagai K, Takeda Y, Nishibuchi M. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect Immun. 1990;58:3568–3573. doi: 10.1128/iai.58.11.3568-3573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sissons C H, Perinpanayagam H E R, Hancock E M, Cutress T W. pH regulation of urease levels in Streptoccus salivarius. J Dent Res. 1990;69:1131–1137. doi: 10.1177/00220345900690050301. [DOI] [PubMed] [Google Scholar]
- 43.Skurnik M, Batsford S, Mertz A, Schlitz E, Toivanen P. The putative arthritogenic cationic 19-kilodalton antigen in Yersinia enterocolitica is a urease β-subunit. Infect Immun. 1993;61:2498–2504. doi: 10.1128/iai.61.6.2498-2504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sriwanthana B, Island M D, Mobley H L T. Sequence of the Proteus mirabilis urease accessory gene ureG. Gene. 1993;129:103–106. doi: 10.1016/0378-1119(93)90703-6. [DOI] [PubMed] [Google Scholar]
- 45.Sriwanthana B, Island M D, Maneval D, Mobley H L. Single-step purification of Proteus mirabilis urease accessory protein UreE, a protein with a naturally occurring histidine tail, by nickel chelate affinity chromatography. J Bacteriol. 1994;176:6836–6841. doi: 10.1128/jb.176.22.6836-6841.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suthienkul O, Ishibashi M, Iida T, Nettip N, Supavej S, Eampokalap B, Makino M, Honda T. Urease production correlates with possession of the trh gene in Vibrio parahaemolyticus strains isolated in Thailand. J Infect Dis. 1995;172:1405–1408. doi: 10.1093/infdis/172.5.1405. [DOI] [PubMed] [Google Scholar]
- 47.Takeda Y. Thermostable direct hemolysin of Vibrio parahaemolyticus. Pharmacol Ther. 1983;19:123–146. doi: 10.1016/0163-7258(82)90044-4. [DOI] [PubMed] [Google Scholar]
- 48.Tomb J F, White O, Kerlavage A R, Clayton R A, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 49.Walker J E, Saraste M, Runswich M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases, and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, et al., editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1987. pp. 2.4.1–2.4.2. [DOI] [PubMed] [Google Scholar]
- 51.Xu M, Yamamoto K, Honda T. Construction and characterization of an isogenic mutant of Vibrio parahaemolyticus having a deletion in the thermostable direct hemolysin-related hemolysin gene (trh) J Bacteriol. 1994;176:4757–4760. doi: 10.1128/jb.176.15.4757-4760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamaichi Y, Iida T, Park K S, Yamamoto K, Honda T. Physical and genetic map of the genome of Vibrio parahaemolyticus: presence of two chromosomes in Vibrio species. Mol Microbiol. 1999;31:1513–1521. doi: 10.1046/j.1365-2958.1999.01296.x. [DOI] [PubMed] [Google Scholar]
- 53.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]