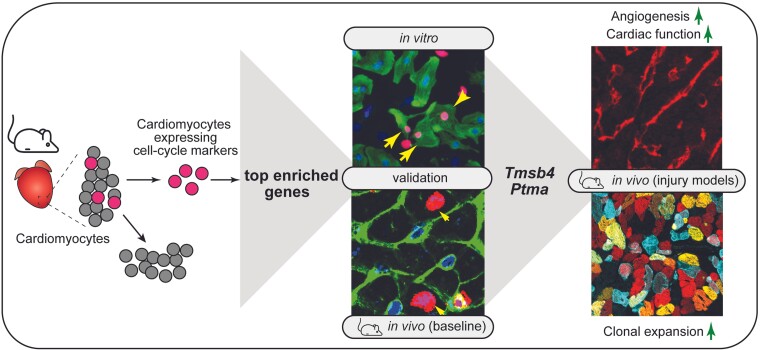
Abstract
Aims
The adult mammalian heart is a post-mitotic organ. Even in response to necrotic injuries, where regeneration would be essential to reinstate cardiac structure and function, only a minor percentage of cardiomyocytes undergo cytokinesis. The gene programme that promotes cell division within this population of cardiomyocytes is not fully understood. In this study, we aimed to determine the gene expression profile of proliferating adult cardiomyocytes in the mammalian heart after myocardial ischaemia, to identify factors to can promote cardiac regeneration.
Methods and results
Here, we demonstrate increased 5-ethynyl-2’deoxyuridine incorporation in cardiomyocytes 3 days post-myocardial infarction in mice. By applying multi-colour lineage tracing, we show that this is paralleled by clonal expansion of cardiomyocytes in the borderzone of the infarcted tissue. Bioinformatic analysis of single-cell RNA sequencing data from cardiomyocytes at 3 days post ischaemic injury revealed a distinct transcriptional profile in cardiomyocytes expressing cell cycle markers. Combinatorial overexpression of the enriched genes within this population in neonatal rat cardiomyocytes and mice at postnatal day 12 (P12) unveiled key genes that promoted increased cardiomyocyte proliferation. Therapeutic delivery of these gene cocktails into the myocardial wall after ischaemic injury demonstrated that a combination of thymosin beta 4 (TMSB4) and prothymosin alpha (PTMA) provide a permissive environment for cardiomyocyte proliferation and thereby attenuated cardiac dysfunction.
Conclusion
This study reveals the transcriptional profile of proliferating cardiomyocytes in the ischaemic heart and shows that overexpression of the two identified factors, TMSB4 and PTMA, can promote cardiac regeneration. This work indicates that in addition to activating cardiomyocyte proliferation, a supportive environment is a key for regeneration to occur.
Keywords: Cardiomyocyte, Proliferation, Cardiac ischaemia, Regeneration, Gene therapy
Graphical Abstract
Graphical abstract.
1. Introduction
During mammalian cardiac maturation, cardiomyocytes lose their proliferative capacity1,2 and retain a low basal turnover rate (less than 1% annually).3–5 While it is known that the mitogenic activity of cardiomyocytes increases in response to ischaemic injuries, this does not result in a sufficient amount of cell division for adequate regeneration, but rather polyploidization and multinucleation.6,7 In contrast to the adult heart, mammalian neonates and invertebrates can regenerate the myocardium by de-differentiation and proliferation of existing cardiomyocytes.8,9 Thus, efforts to try to promote cardiomyocyte cell-cycle re-entry in adult mammalian hearts have become a key focus within the field.10 To date, several studies have focused on characterizing the transcriptomic signature of proliferating cardiomyocytes in young neonates and invertebrates to provide insight into potential mechanisms to stimulate adult cardiomyocyte regeneration.8,11–13 However, less is known about the gene profile of adult proliferating cardiomyocytes and whether this can be utilized to uncover specific genes relevant for promoting cytokinesis in these highly organized cells. Here, we used multicolour lineage tracing to demonstrate cardiomyocyte proliferation in the adult mouse heart after myocardial infarction (MI). To further capture and characterize the small percentage of proliferating cardiomyocytes, we analyzed single-cell RNA sequencing (scRNA-seq) datasets from healthy and injured mouse hearts.14 We bioinformatically selected cardiomyocytes that expressed cell-cycle genes and characterized their gene expression signature compared to non-proliferating cardiomyocytes. Using a combinatorial screen based on the top enriched genes within the proliferating cardiomyocytes, we identified Tβ4 (encoded by Tmsb4) and Tα1 (encoded by Ptma) as factors that provide a permissive environment for cardiomyocytes to proliferate and regenerate the heart in vivo. Taken together, this work demonstrates that in addition to the necessary triggers to induce cell cycle activity, successful regeneration requires a suitable microenvironment.
2. Methods
2.1. Mice
Animal studies were performed according to the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes. Animal experiments were approved by the institutional policies and regulations of the Animal Welfare Committee of the Royal Netherlands Academy of Arts and Sciences (HI 13.2304, AVD8011002015250 16.2305/IVD366) and following the guidelines for the care and use of laboratory animals. Mice were housed with 12:12 hour light: dark cycle in a temperature-controlled room with access to food and water ad libitum. For all animal experiments, we used 8–9 weeks-old male and female mice as indicated. The number of mice used represents the minimum required to achieve statistical significance based on previous experience and power calculations. Mice were randomly allocated into experimental groups and the investigator was blinded to the experimental group, where possible.
2.2. MI
For cardiac surgeries, mice were injected subcutaneously with buprenorphine (0.05–0.1 mg/kg) as an analgesic at least 30 min prior to surgery to alleviate pain or distress. When multiple surgeries take place on the same day, all animals received buprenorphine at the same time in the morning. After 30 min (or longer) mice were anaesthetized with a mix of fentanyl (0.05 mg/kg), midazolam (5 mg/kg) and dexmedetomidine (0.125 mg/kg) via intraperitoneal injection and supplemented with 1–2% isoflurane to maintain a surgical plane of anaesthesia. Immediately after the surgery, anaesthesia was reversed using atipamezole. Mice received a second subcutaneous injection of buprenorphine (0.05–0.1 mg/kg) 8–12 h after the first dose to provide additional pain relief. The third dose of buprenorphine (0.05–0.1 mg/kg) was subcutaneously administered approximately 12 h later (the next morning after surgery). After the anaesthesia, mice were intubated, and the tracheal tube was connected to a ventilator (Uno Microventilator UMV-03). Hair was removed from the thorax and neck, and the surgical site was cleaned with iodine and 70% ethanol. The skin was incised left of the midline to allow access to the third intercostal space. Pectoral muscles were retracted, and the intercostal muscles cut caudal to the third rib. Wound hooks were placed to allow access to the heart.
For MI surgeries, the left anterior descending artery was identified and carefully isolated. For permanent occlusion of the left anterior descending artery (LAD), a 7.0 silk suture was tied around the LAD. For ischaemia-reperfusion infarctions, a 7.0 silk suture was tied around the LAD and a 2–3 mm PE 10 tubing. After 1 hour of ischaemia, the tubing and the tied suture were removed. In a subset of experiments, mice received intra-cardiac injections of AAV9 into the free wall of the left ventricle by two 15μL injections at a dose of 3 × 1010 viral genomes (in PBS) per animal immediately following reperfusion using a Hamilton syringe. After cardiac surgeries were completed, the muscle and rib cage were closed with 5.0 silk suture, and the skin was closed with a wound clip.
2.3. AAV9 injections in juvenile mice
Where applicable, mice were injected intraperitoneally (i.p) with the indicated adenoassoicated virus serotype 9 (AAV9) viruses at a dose 1 × 1011 viral genomes per animal in sterile phosphate buffered saline (PBS).
2.4. Edu injections in mice
Were applicable, mice were injected with the nucleoside analogue 5-ethynyl-2’deoxyuridine (Edu) (50μg/g; either Thermo Fisher Scientific, Bleiswijk, the Netherlands, or Santa Cruz Biotechnology, Inc. Heidelberg, Germany) dissolved in sterile PBS.
2.5. Echocardiography
Cardiac function was determined by two-dimensional transthoracic echocardiography on sedated mice (2–2.5% isoflurane) on a Visual Sonic Ultrasound system with a 30 MHz transducer. Echocardiographic M-mode measurements were recorded from the parasternal short-axis view at the level of the papillary muscles. Analysis of heart rate, wall thickness, ejection fraction (EF) end-diastolic/systolic dimensions were analyzed using LV Trace in the Visual Sonics analysis package.
2.6. Organ collection for histological analysis
At the relevant endpoint, mice were sedated with isoflurane (3–4%) followed by cervical dislocation. Adult hearts and livers were excised from animals, washed in ice-cold PBS and fixed with 4% formalin at room temperature (RT) for 48 h, embedded in paraffin and sectioned at 4μm. Paraffin sections were stained with haematoxylin and eosin (H&E) for routine histological analysis and Sirius Red (SR) for the detection of collagen according to standard procedures. Slides were visualized using a Zeiss Axioskop 2Plus with an AxioCamHRc and DM4000.
2.7. Immunohistochemistry
Paraffin-embedded sections were dewaxed and rehydrated through ethanol to water gradient. Antigen retrieval was performed in boiling 10 mmol/L Tris-EDTA, pH 9.0 for 20 min and allowed to cool to RT for an additional 30 min. Non-specific binding was blocked with 1% w/v bovine serum albumin (BSA), 0.1% v/v Tween-20 in PBS 1 hour at RT. Sections were incubated overnight with antibodies against mouse sarcomeric alpha-actinin (ACTN2, 5μg/mL, A7732, Sigma Aldrich), rabbit anti-phospho Histone 3 (Ser10) (#06–570, 2μg/ml, Merck Millipore, Darmstadt, Germany), mouse anti-cardiac troponin T (#MA5–12960, 1:200 Thermo Fisher Scientic) or goat anti-PECAM1 (#abAF3628, 1:50, R&D Systems). Sections were washed 3 × in PBS to remove excess unbound antibodies prior to incubation with secondary antibodies (1:400, Invitrogen Alexa-Fluor® dyes) for 1 hour at RT. The sections were then washed 3 × 10 min in PBS. To detect EdU incorporation, we utilized the Click-iT Plus Edu Imaging Kit (C10640, Thermo Fisher Scientific) according to manufacturers’ instructions. Cell membranes were stained by incubating with FITC-labelled wheat-germ-agglutinin (WGA) (1:200, Sigma Aldrich, The Netherlands) for 30 min. To visualize nuclei, sections were incubated with 4′,6-diamidino-2-phenylindole (DAPI; 1:5000, D3571 Life technologies, The Netherlands) for 15 min. Sections were rinsed 3 × 10 min in PBS prior to mounting in with ProLongTM antifade (Thermo Fisher Scientific).
2.8. Quantification of cardiomyocyte cell size and proliferation status
For cardiomyocyte cell size and proliferation status, 4μm paraffin embedded hearts were analyzed in the four-chamber view of the heart. Cardiomyocyte cell size was quantified using ImageJ from paraffin sections stained with WGA. Cross-sectional area (μm2) was used as a measurement of cell size and 10 cardiomyocytes were randomly counted per area (infarct region, border zone, and remote) in four mice per group. The percentage of proliferating cardiomyocytes in Supplementary material online, Figure S2 was quantified in paraffin sections stained with either WGA, α-actinin, DAPI, and EdU or pH3 by manual counts in ImageJ. A total of nine images were analyzed per heart: three images in the border zone, infarct and remote regions. Only DAPI positive cardiomyocytes were analyzed.
2.9. Generation of cardiomyocyte-specific confetti mice
To generate cardiomyocyte-specific inducible Confetti mice for clonal analysis, transgenic αMHC-MerCreMer mice were crossed with R26R-Confetti mice (a gift from Professor Hans Clevers at the Hubrecht Institute; also available from Jackson Laboratories; strain: Gt(ROSA)26Sortm1(CAG-Brainbow2.1) Cle/J). Adult (8-week-old) male and female mice were used for studies shown in Supplementary material online, Figure S4 and only males were used for Figure 4. Upon Cre mediated recombination, 1 of the 4 colours encoded by the locus, nuclear green fluorescent protein (GFP), membrane-bound cyan (CFP), yellow fluorescent protein (YFP) or red flurescent protein is expressed. αMHC-MerCreMe-Confetti mice were given a single intraperitoneal (i.p) injection of tamoxifen (T5648; Sigma-Aldrich, Zwijndrecht, The Netherlands) at a dose of 2 mg/30 g mouse) as optimized in Supplementary material online, Figure S3C. While a higher degree of recombination was observed with a higher dose of tamoxifen, this also resulted in significant fluid accumulation in the pericardial region. We therefore selected the lower dose for all experiments. Tamoxifen was prepared fresh daily by dissolving in 5% w/v ethanol in sunflower oil by gentle agitation at 37°C until completely in solution. Mice were given a wash-out period of 7 days prior to surgery to reduce residual circulating tamoxifen.
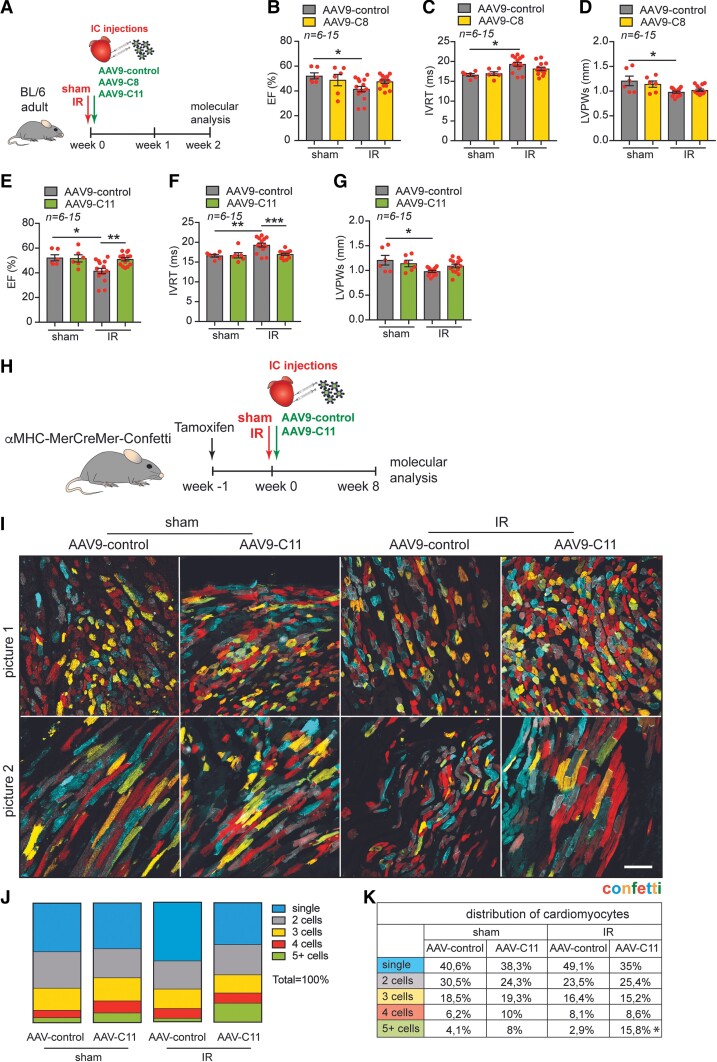
Figure 4.
Therapeutic AAV9 delivery of combination 11 improves cardiac function and promotes clonal expansion of cardiomyocytes post-ischaemic injury. (A) Experimental design. (B–D) Quantification of (B) EF and (C) IVRT and (D) LVPWs from AAV9-control and AAV9-C8 injected mice post-surgery (n = 6 sham, n = 15 IR). (E–G) Quantification of (E) EF (F) IVRT and (G) LVPWs from AAV9-control and AAV9-C11 injected mice post-surgery. (H) Experimental design. (I) Representative images of cardiac tissue (border zone) from aMHC-MerCreMer-Confetti mice injected with combination 11 (C11) post-injury (n = 6). (J–K) Cardiomyocyte clonal analysis based on quantification of adjacent cardiomyocytes with the same colour considered to arise from the proliferation of single-labeled cells (mononucleated cardiomyocytes (monochromatic cardiomyocytes)). Data are shown as mean ± SEM. *P< 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 using one-way ANOVA followed by Sidak’s multiple comparisons test (for B–G) and Dunnett’s multiple comparisons test (for J–K). Scale bar represents 100 mm. IC = intracardiac injection, IR = ischaemia/reperfusion.
2.10. Histological processing of tissue from αMHC-merCreMer-confetti mice
For general histological analysis, whole hearts were fixed in 4% v/v paraformaldehyde (PFA)/PBS for 48 h at RT under gentle agitation and dehydrated in an increasing ethanol gradient, embedded in paraffin and cut into 4μm sections. For analysis of endogenous fluorescence in the αMHC-MerCreMer-Confetti mice, hearts were briefly fixed in 4% v/v PFA solution for 4 h at 4°C under gentle agitation. Hearts were rinsed in PBS and immersed in 30% w/v sucrose solution (prepared in PBS). Once hearts had sunk to the bottom of the tube (∼24 h), hearts were embedded in optimal cutting temperature (OCT) freezing medium (Leica Biosystems, The Netherlands) and flash frozen.
2.11. Confocal imaging
Confocal fluorescent images were obtained with either a Leica SP5 (confetti hearts) or SP8 microscope (standard immunohistochemistry) and processed using ImageJ (Fiji, The Netherlands).
2.12. Cardiomyocyte clonal analysis
2D images of 10μm frozen heart sections were imaged for manual quantification of cardiomyocyte clones. Cells adjacent to each other with the same fluorescent protein were considered a clone. Single-coloured cardiomyocytes were considered mononucleated, whereas cells expressing more than one fluorescent protein were considered binucleated. The ImageJ counter plugin was used for analysis. All images were analyzed blindly by three to four investigators.
2.13. Plasmid and AAV production
Human or mouse cDNA was used to amplify the protein-coding regions for each gene for cloning into a AAV-multiple cloning site vector (pAAV-MCS) and expression in recombinant AAV vectors. AAVs were generated by the AAV Vector Unit at ICGEB Trieste (http://www.icgeb.org/avu-core-facility.html) as described previously.15 Briefly, HEK293T cells were transfected with a triple-plasmid for packaging of AAV of serotype 6 or 9, for in vitro and in vivo analysis, respectively. AAV viral stocks were obtained by CsCl2 gradient centrifugation and titration of AAV viral particles was determined by qPCR for quantification of viral genomes, as described previously.16 Viral titres ranged from 1.3 × 1011 to 3.8 × 1013vg/mL.
2.14. Viral infection
In our in vitro and in vivo assessments, we performed a screen of 15 different combinations of factors each consisting of 2–14 genes. To be consistent with the amount of virus infected in vitro and in vivo between the different combinations, we maintained the same number of viral genomes, irrespective of the number of genes. Therefore, the amount of viral genomes encoding a gene was dependent on the number of factors in each combination.
2.15. RNA isolation and quantitative PCR
Total RNA was extracted from the left ventricular free wall with TRIzol (Fisher Scientific) according to manufacturers’ instructions. For gene expression analysis, 1 μg of RNA was reverse transcribed to synthesize cDNA using iScript Reverse transcriptase kit (Bio-Rad, Veenendaal, The Netherlands). To quantify changes in gene expression, quantitative poylmerase chain reaction (qPCR) was performed using iQ SYBR Green supermix (Bio-Rad) in a CFX96 PCR system. All values were normalized to Hprt1 or Gapdh. See Supplementary material online, Table S2 for primer sequences.
2.16. Single cell sequencing transcriptomic analysis
To identify the transcriptomic signature of cardiomyocytes that are potentially proliferating, we used a previously generated single-cell sequencing dataset containing cells from a healthy and ischaemic hearts (3 days after ischaemia/reperfusion injury).14 This dataset is accessible in the GEO database which accession number GSE146285. For this analysis we used the 14 days post-sham surgery (Sham 14d) and the 3 days post-IR (IR 3d) samples. After analysis and clustering with the RaceID2 algorithm as described in Gladka et al.,14 we selected all cells with a cardiomyocyte transcriptomic profile. In this group of cells, we identified cardiomyocytes expressing either Pcna, Ccnd1, Cdk6, Cdk4, or Mki67 by a read count of at least 1 after normalization by the RaceID2 algorithm. Next, we determined the differential expression of these select cells in comparison with the remaining cardiomyocyte population by DESeq2 using pooled dispersion.16 These data sets were generated using the SORT-seq protocol,17 which utilizes flow cytometry to exclude cell doublets and sort single cells into 384 well plates, prior to barcoding, pooling and sequencing. A uniform distribution of read counts and unique molecular identifiers was observed across all cardiomyocytes, including the selected cells expressing proliferative genes (Supplementary material online, Figure S14A-B). This indicates exclusion of cell doublets within the analysis.
2.17. Isolation of ventricular cardiomyocytes from neonatal rats
Cardiomyocytes were isolated by enzymatic dissociation of 1–2-day-old neonatal rat hearts. Briefly, pups were placed on ice for 5–10 min to induce anaesthesia, followed by decapitation directly into liquid nitrogen. After decapitation, hearts were collected, ventricles were separated from the atria and cut into small pieces in a balanced salt solution prior to enzymatic digestion using trypsin (0.1% #15400054, Thermo Fisher Scientific) under constant stirring at 37°C using a stirring flask. The supernatant containing intact cardiomyocytes was collected, centrifuged at 300xg for 4 min and resuspended in Ham F10 medium (Thermo Fisher Scientific, no. 11550043) supplemented with 5% FBS, 10% L-glutamine and 10% Pen-Strep. Collected cells were seeded onto uncoated 100 mm plastic dishes for 1.5 h at 37°C in 5% CO2 humidified atmosphere. The supernatant, which consists mainly of cardiomyocytes, was collected, and cells were counted and plated on gelatinized six-well plates at a concentration of 1 × 106 cells per well. After 24 h, the medium was changed to Ham F10 supplemented with Insulin-Transferrin-Sodium Selenite Supplement (Roche), 10% L-glutamine and 10% Pen-Strep. Cells were infected with AAV serotype 6 encoding the indicated factors or an empty vector control at a total of 5 × 103 viral genomes per cell. Cells were analysed 48 h after viral transduction. EdU was added at a final concentration of 10μM in the final 24 h. Cells were washed twice in PBS and fixed in 4% PFA:PBS (v/v) for 15 min followed by two additional PBS rinses. EdU incorporation was assessed using the Click-iT Plus Edu Imaging Kit, according to manufacturers’ instructions. Cardiomyocytes were identified using either goat anti-cardiac troponin I (1:200 Hytest/Biotrend #4T21/2) or ACTN2 (5μg/mL, A7732, Sigma Aldrich).
2.18. Statistics
The number of samples (n) used in each experiment is indicated in the legends or shown in the figures and indicates biological replicates. Results are presented as the mean ± standard error of the mean (SEM). Statistical analyses were performed using Prism (GraphPad Software Inc. version 6). Two groups were statistically compared using the Student’s t-test. Multiple groups were statistically compared with ordinary one-way analysis of variance (ANOVA) or two-way ANOVA. Outliers were defined by Grubbs’ test (alpha = 0.05). Data are represented as mean ± SEM. Differences were considered statistically significant at P < 0.05. In the figures, asterisks indicate statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001), which is also denoted within the figure legends. All representative images of hearts or cells were selected from at least three independent experiments with similar results, unless indicated differently in the figure legend.
2.19. Study approval
All experiments were performed in accordance with the guidelines of the Animal Welfare Committee of the Royal Netherlands Academy of Arts and Sciences and conducted in accordance with protocols approved by the ethics committee of the Hubrecht Institute in Utrecht.
3. Results
3.1. Temporal and spatial analysis of cardiac proliferation after ischaemia
In an attempt to better define the active proliferative phase in the heart following ischaemic injury, we performed a temporal and spatial analysis of proliferation after MI. Mice were given an MI by permanent occlusion of the LAD after which their hearts were analyzed at various timepoints. We isolated the infarcted myocardial wall together with the border zone and the remote area to the ischaemic zone and performed a qPCR-array of established cell-cycle regulators. Corresponding regions in mice exposed to sham surgeries were used as controls (see Supplementary material online, Figure S1A-E). This identified a cell-cycle gene expression profile post-MI, with the most prominent induction observed at 3 and 14 days after cardiac ischaemia. In response to injury, several cardiac cell types, especially fibroblasts, undergo dynamic changes, including activation of the cell cycle to mount the regenerative response.18 To assess the contribution of cycling cardiomyocytes at the timepoints where the mRNA expression was the most profound (3 and 14 days post-MI), mice were given an MI and injected with EdU 24 h prior to heart isolation (see Supplementary material online, Figure S2A). The percentage of EdU positive cardiomyocytes was increased at 3 days post-MI in the infarct and border zone (see Supplementary material online, Figure S2B, D). However, at 14 days post-MI, there was no increase in EdU incorporation in comparison to the sham controls (see Supplementary material online, Figure S2C-D). To validate our findings, we stained for the mitosis marker, phospho-histone H3 (pH3), which corroborated our EdU analysis at 3 days post-MI (see Supplementary material online, Figure S2E-F). Additionally, we observed a significant increase in pH3 positive cardiomyocytes in the infarct region compared with sham hearts at 14 days post-MI (see Supplementary material online, Figure S2E, G).
3.2. Clonal analysis of cardiomyocyte proliferation after MI
While our data provide support of cell cycle activity in the adult heart, it does not distinguish between cell division and binucleation. It is also known that in response to injury, cardiomyocytes undergo hypertrophy, which may also activate cell cycle genes in the absence of cytokinesis.7 Both polyploidization and binucleation result in EdU incorporation and pH3 expression.7 To further characterize de novo cardiomyocyte cell division and whether they undergo clonal expansion (a single cell giving rise to more than one cell), we performed lineage tracing in cardiomyocytes. Specifically, we utilized the stochastic four-colour reporter allele, R26R-LoxSTOPLox-Confetti, crossed with the αMHC-MerCreMer mouse to generate inducible cardiomyocyte-specific confetti mice (αMHC-MerCreMer-Confetti; Supplementary material online, Figure S3A). Upon tamoxifen-induced recombination, cells express either one (a mononucleated cell) or two (binucleated cell) fluorescent colours (see Supplementary material online, Figure S3B). Because the percentage of proliferating cardiomyocytes is low, we opted to pre-label all cardiomyocytes using a high dose of tamoxifen (see Supplementary material online, Figure S3C). This is in contrast to a recent study, which analyzed the clonality of cardiomyocyte proliferation using a low dose of tamoxifen to label only a small number of cardiomyocytes.19 Given that cardiomyocyte proliferation is a rare event, we reasoned that fluorescently tagging only a few cardiomyocytes may result in undetected clonal clusters. Using our approach, we expected to see an increase in cardiomyocyte clonal expansion post-ischaemic injury but not under baseline conditions (see Supplementary material online, Figure S3D). When tested in vivo (see Supplementary material online, Figure S4A), we observed a significant increase in the percentage of mononucleated cells in larger clusters (5 or more cells) in the border zone of MI hearts compared with sham (20.4% vs. 4.4%; Supplementary material online, Figure S4B-D and Supplementary material online, Figure S5A-H). In addition, we observed a decrease in the percentage of mononucleated single cells in the remote region of the heart (see Supplementary material online, Figure S4B-D and Supplementary material online, Figure S5A-H). Taken together, these results suggest that mononucleated cardiomyocytes can undergo clonal expansion in response to cardiac ischaemia.
3.3. Single-cell analysis to identify the transcriptomic signature of proliferating cardiomyocytes
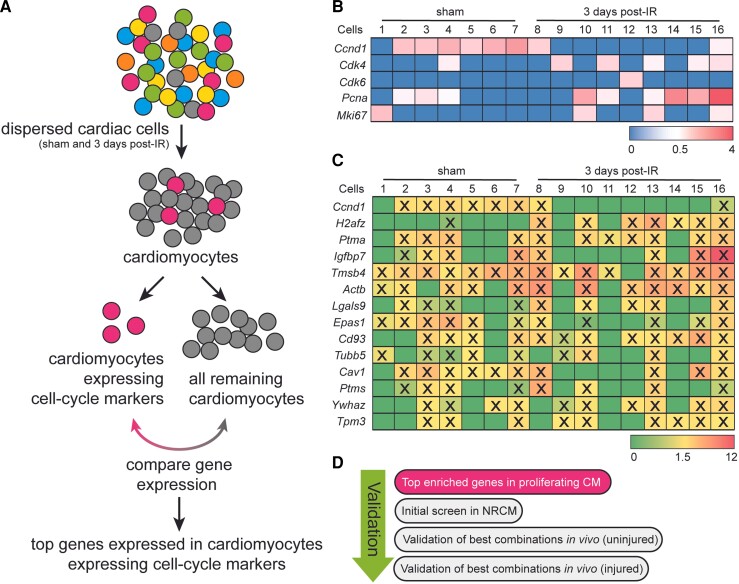
Next, we sought to identify genes that can promote endogenous cardiomyocyte cell proliferation. Given that the heart contains a heterogeneous population of cardiomyocytes and that proliferation events are rare, we utilized our recent scRNA-seq dataset14 to identify cardiomyocytes expressing cell-cycle genes (Figure 1A). We focused specifically on single cells from mice that had received cardiac ischaemic injury or sham surgery 3 days prior, as this was the timepoint where we observed an increase in cardiomyocyte proliferation (see Supplementary material online, Figure S2). To specifically characterize gene expression signatures within cardiomyocytes, we first bioinformatically selected cells enriched for cardiomyocyte marker genes. While flow cytometry sorting of cells aims to exclude cell doublets by forward and side-scattering of events, we additionally verified doublet exclusion by assessing the total read counts and unique molecular identifiers of all cardiomyocytes [as previously described14 and see Methods for details]. All cardiomyocytes were then re-analyzed using the RaceID2 algorithm20 and by K-medoids clustering of 1-Pearson correlation coefficients. RaceID2 is an algorithm that was developed to detect rare cell populations from scRNA-seq data.20 Using this approach, we were unable to detect a unique cluster of cardiomyocytes expressing cell cycle genes. To overcome this, we bioinformatically screened for cardiomyocytes expressing one or more of the following cell cycle genes; Ccnd1, Cdk4, Cdk6, Pcna or Mki67 with a minimum of 1 read count per cell after normalization. In doing so we selected 16 cardiomyocytes from the total population of both sham and ischaemia-reperfused (IR) hearts for follow up analysis (Figure 1B). We then performed differential gene expression analysis of these 16 cells (in bulk) in comparison with the rest of the cardiomyocytes to profile their molecular signature and identify unique factors that may contribute to cardiomyocyte proliferation. A total of 854 genes were >2 fold enriched with a P value >0.01 (see Supplementary material online, Dataset S1). We selected 14 genes that were enriched within the 16 identified cardiomyocytes. Based on the individual gene expression within each cell, we defined 15 different gene combinations to overexpress in cardiomyocytes for further analysis (Figure 1C) as outlined in Figure 1D.
Figure 1.
Gene profile of cardiomyocytes expressing cell-cycle markers. (A) Experimental workflow to identify proliferating cardiomyocytes. (B) Expression level of the selected proliferation markers in the 16 identified cardiomyocytes. (C) Top 14 enriched genes expressed in sixteen identified cardiomyocytes, compared to all other cardiomyocytes. (D) Validation strategies. X = indicates the expression of a selected factor in the individual cell.
3.4. Combinatorial gene expression promotes cardiomyocyte proliferation in neonatal rat cardiomyocytes
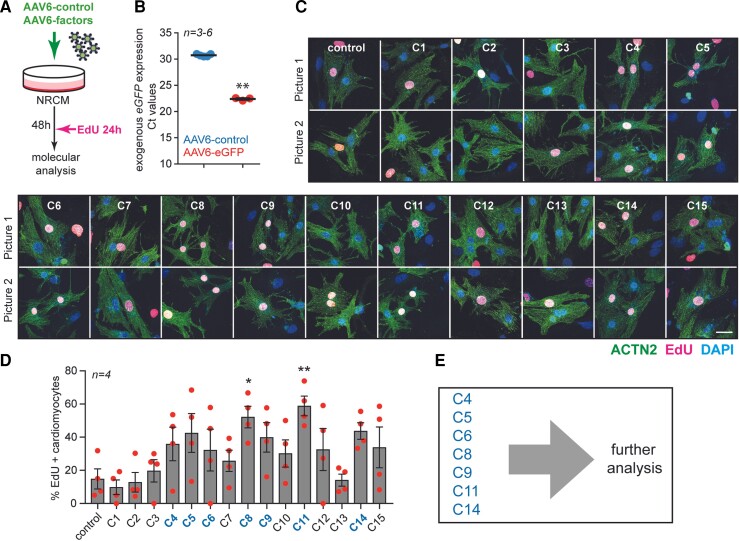
To test the proliferative capacity of the identified factors we cloned the top 14 genes into a CMV-driven plasmid using the human coding sequence (which shares >85% sequence homology with the mouse, apart from Lgals9, Cd93, and Cav1: see Supplementary material online, Figure S6A for homology across the mouse, human and rat) and delivered the genes using an adeno-associated virus serotype type 6 (AAV6) vector to neonatal rat cardiomyocytes (NRCMs; Figure 2A and Supplementary material online, Figure S6B). We first infected NRCMs with an AAV6 encoding enhanced green fluorescent protein (eGFP) to ascertain its infectious capacity. qPCR analysis for eGFP revealed a substantial overexpression compared with AAV6-control-treated cells (Figure 2B). Next, we generated 15 gene cocktails based on the expression patterns of the top 14-upregulated genes within each cell (see Supplementary material online, Figure S6C; hereinafter referred to as combination 1–15) and infected NRCMs with a total 5 × 103 viral genomes per cell. The total viral particles used per condition were kept consistent, irrespective of the number of encoded factors within the cocktail combination (see methods for details; viral infection). To investigate the proliferative potential of the proposed gene combinations, we incubated cells with EdU 24 h prior to analysis (Figure 2A). Several of the gene cocktails increased the percentage of EdU positive cardiomyocytes, but only combination 8 (Ccnd1, H2afz, Ptma, Igfbp7, Tmsb4, Actb, Lgals9, Epas1, Cd93, Cav1 and Ptms) and combination 11 (Ptma, Tmsb4) significantly increased EdU incorporation in cardiomyocytes (Figure 2C-D). Based on this first screen, we selected the 7 most promising combinations for follow-up experiments (Figure 2E).
Figure 2.
Proliferative potential of different combinations of factors delivered to NRCMs. (A) Experimental design. (B) mRNA expression level of eGFP (Ct values) to assess the infection efficiency in cultured NRCM. (C) Representative images (two pictures per condition) of NRCM infected with the indicated combinations (n = 4, ±100 cells per condition). Cells were stained with ACTN2, EdU and DAPI. (D) Quantification of the percentage of EdU-positive cardiomyocytes. (E) List of the top combinations selected for follow-up analysis. Scale bar represents 50 mm. Data are shown as the mean ± SEM. *P < 0.05 and **P < 0.01 compared to control using unpaired, two-tailed Student’s t-test or one-way ANOVA followed by Dunnett’s multiple comparisons test.
3.5. Combinations 8 and 11 induce cardiomyocyte proliferation in vivo
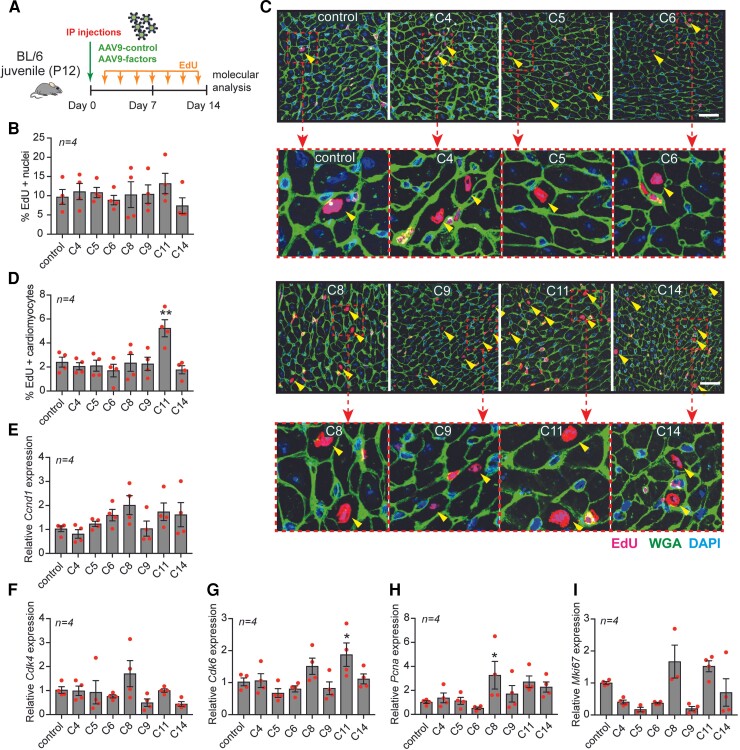
We next sought to identify whether our selected gene combinations could promote cardiomyocyte proliferation in mouse hearts at postnatal day 12 (P12) when most cardiomyocytes have withdrawn from the cell cycle.21 We incorporated the genes from the selected top 7 combinations into the cardiotropic AAV9 vector (see Supplementary material online, Figure S6B) and injected mice at P12 with a viral cocktail by intraperitoneal (i.p.) injection and analyzed hearts two weeks later (Figure 3A and Supplementary material online, Figure S7A). We have previously shown that a cardiomyocyte can be infected by more than one virus in vivo.22 Mice received EdU injections every other day in order to identify all cells that either had proliferated or were actively proliferating (Figure 3A). We isolated hearts and confirmed by qPCR that the delivered factors for each combination were overexpressed compared to the levels detected in hearts from mice treated with a control virus (see Supplementary material online, Figure S7B-P). While we did not detect an effect on heart weight to body weight ratio (see Supplementary material online, Figure S8A), we did observe a decrease in cardiomyocyte size in response to most of the gene combinations (see Supplementary material online, Figure S8B). There were no detectable changes in cardiac morphology or fibrosis in response to any of the combinations tested (see Supplementary material online, Figure S8C-D). Although AAV9 has a high tropism for cardiomyocytes, it may also display tropism for other tissues, including the liver.23 We therefore also assessed EdU incorporation in the liver in response to systemic gene delivery. While no increase in proliferation was observed with any of the combinations, we detected a decrease in proliferation in response to combinations 11 and 14 (see Supplementary material online, Figure S8E-F). In histological heart sections, total EdU incorporation (all cell types) was unchanged across all conditions (Figure 3B). However, injection with condition 11 resulted in an increase in the percentage of EdU positive cardiomyocytes (Figure 3C-D). mRNA analysis of cell cycle markers in ventricular homogenates showed an increased expression of Cdk6 with combination 11 and an increased expression of Pcna with combination 8 (Figure 3E-I). Although there was a trend towards increased gene expression of Ccnd1, Cdk4 and Mki67 for combinations 8 and 11, this did not reach statistical significance (Figure 3E-I). In addition, we observed a significant increase in PECAM1 (endothelial cell marker) positive areas in hearts treated with combinations 6, 8, 9, and 11 (C6, C8, C9 and C11), suggesting an enhanced angiogenic response upon treatment with these conditions (see Supplementary material online, Figure S8G-H).
Figure 3.
AAV9-mediated gene delivery of combinations 8 and 11 induces cardiomyocyte proliferation in vivo. (A) Experimental design. (B) Quantification of total proliferation in the heart based on the percentage of EdU positive nuclei from mice treated with the indicated combinations (n = 4, 750–1000 cardiomyocytes from 5 sections per mouse). (C) Immunofluorescent staining for EdU, WGA, ACTN2 and DAPI of hearts treated with the indicated combinations and zoomed-in magnifications of EdU positive cardiomyocytes. (D) Quantification of the percentage of EdU-positive cardiomyocytes. (E-I) mRNA expression levels of (E) Ccnd1, (F) Cdk4, (G) Cdk6, (H) Pcna, and (I) Mki67 on heart tissue from mice treated with the indicated combinations. Scale bar represents 50 mm. Data are shown as the mean ± SEM. *P < 0.05 and **P < 0.01 compared to control using one-way ANOVA followed by Dunnett’s multiple comparisons test.
3.6. Therapeutic delivery of combination 8 and combination 11 improves cardiac function post-ischaemic injury
Our in vitro and in vivo proliferation screens with our gene combinations highlighted combinations 8 and 11 to contain potential regenerative factors. To assess whether our selected factors could promote cardiomyocyte regeneration in response to injury, we performed an IR injury. Combinations 8 and 11 were directly injected into the myocardial wall during reperfusion (Figure 4A). We have previously shown that this method of delivery induces robust gene expression in the heart.24 As expected, echocardiographic analysis revealed a decrease in EF (Figure 4B, E and Supplementary material online, Table S1) and an increase in the isovolumic relaxation time (IVRT) in response to ischaemic injury, indicating poor cardiac contractility and myocardial relaxation (Figure 4C, F and Supplementary material online, Table S1). In addition, the left ventricular posterior weight in systole (LVPWs) was significantly reduced, indicating cardiac dilation (Figure 4D, G). While combination 8 did not significantly improve the decline in cardiac function in response to ischaemia as assessed by EF, IVRT and LVPWs (Figure 4B-D), combination 11 restored cardiac function after the injury as indicated by EF and IVRT (Figure 4E-F). However, we did not detect any significant effect on the LVPWs in response to either combination 8 or 11 (Figure 4D, G). While mRNA analysis confirmed the overexpression of all genes delivered in both conditions (see Supplementary material online, Figure S9A-D), we did not observe any differences in mRNA expression of proliferation genes in the infarcted region (see Supplementary material online, Figure S10A-E) but only in the remote region of the hearts treated with combination 11 (see Supplementary material online, Figure S10F-J). PECAM1 expression was also increased by combination 11 in both sham and post-IR hearts, suggesting increased angiogenesis (see Supplementary material online, Figure S10K-L). Together these data show that cardiac delivery of TMSB4 and PTMA (combination 11) improves cardiac function and enhances angiogenesis in response to ischaemic injury.
3.7. Combination 11 increases the clonal expansion of cardiomyocytes post-ischaemic injury
In our initial analysis, we validated the expression of our individual factors by mRNA analysis. To confirm the overexpression of TMSB4 and PTMA in combination 11, we performed immunohistochemistry in mice that were injected at P12 and in sham and IR treated mice (see Supplementary material online, Figure S11). In our AAV9-C11 treated mice (both juvenile, sham and IR groups), we observed an increased nuclear expression of PTMA in cardiomyocytes (see Supplementary material online, Figure S11A, C, E and G). Of note, we also observed increased nuclear expression in non-cardiomyocytes in mice treated with AAV9-C11, which may be due to a paracrine effect of secreted TMSB4 and PTMA overexpression in cardiomyocytes (see Supplementary material online, Figure S11A, C, E and G). Increased TMSB4 was observed in the interstitial space of the hearts of mice treated with AAV9-C11, which is consistent with our previous findings that TMSB4 is secreted from cardiomyocytes24 (see Supplementary material online, Figure S11B, D, F and H).
To assess whether the observed cardioprotective effects of TMSB4 and PTMA were due to an improvement in cardiac regeneration in these mice, we next performed cardiomyocyte clonal analysis in αMHC-MerCreMer-Confetti mice treated with combination 11 after cardiac ischaemia, during reperfusion (Figure 4H). To induce expression of the confetti locus in cardiomyocytes, adult mice were injected with a single dose of tamoxifen. After a one-week wash-out period, mice were given either a sham or an IR surgery and injected with either AAV9-control or combination 11 (AAV9-TMSB4 and AAV9-PTMA) directly into the myocardial wall after reperfusion. Eight weeks later, hearts were isolated and analyzed for cardiomyocyte proliferation by assessing clonal expansion (Figure 4I-K). We observed a significant increase in the percentage of large clusters of mononucleated cells (5 or more cells) in IR hearts treated with AAV9-combination 11 compared to IR hearts treated with AAV9-control (15.8% vs. 2.9%; Figure 4J-K and see Supplementary material online, Figure S12).
3.8. TMSB4 and PTMA independently modulate cardiomyocyte proliferation
We initially hypothesized that a combinatorial approach would be necessary to induce cell proliferation. Our data provided evidence that the selective combined overexpression of TMSB4 and PTMA could promote cardiomyocyte proliferation. To test whether the selected genes could stimulate proliferation independently, we infected NRCMs with AAV6 encoding each factor or in combination. EdU incorporation analysis in cardiomyocytes indicated that both TMSB4 and PTMA can independently induce cardiomyocyte proliferation in vitro (see Supplementary material online, Figure S13).
4. Discussion
The inability of the adult mammalian heart to regenerate is a significant impediment to the successful recovery and survival of patients with myocardial ischaemia. Here, we determined that cardiomyocytes proliferate clonally in response to cardiac ischaemia (see Supplementary material online, Figure S1–2). We utilized scRNA-seq data of the adult mouse ischaemic heart to identify rare cardiomyocytes expressing cell cycle genes and their gene expression signature (Figure 1).14 Overexpression of the top enriched genes in vitro and in vivo identified TMSB4 and PTMA as factors that promote cardiomyocyte proliferation. Specifically, the overexpression of TMSB4 and PTMA increased EdU incorporation in vitro (Figure 2) and in vivo in juvenile mice under baseline conditions (Figure 3). Injection of AAV9 encoding TMSB4 and PTMA during reperfusion after cardiac ischaemia increased the mRNA expression of cell cycling genes two-weeks post-infarction especially in the remote region of the heart (see Supplementary material online, Figure S10). This was found to induce a therapeutic benefit as assessed by echocardiographic parameters (Figure 4) and increased angiogenesis (see Supplementary material online, Figure S10). Finally, to determine cell division, we performed clonal analysis and found that TMSB4 and PTMA promoted cardiomyocyte regeneration post-MI (Figure 4). Further in vitro analysis indicated that both factors could independently stimulate cardiomyocyte proliferation (see Supplementary material online, Figure S13).
While TMSB4 and PTMA have not been shown to directly regulate the cell cycle, these proteins may act as cardiokines to promote a cell and tissue microenvironment that is more permissible to regeneration. TMSB4 encodes thymosin β4 (Tβ4), a ubiquitously expressed protein that is known for its regulation of filamentous actin polymerization by scavenging globular actin.25 However, it exerts pleiotropic roles that are independent of actin-binding, several of which have demonstrated a functional role in the ischaemic heart.26,27 In cardiomyocytes, Tβ4 was shown to promote cardiomyocyte cell survival by phosphorylation of Akt and subsequently reduced infarct size, cardiac fibrosis, cardiomyocyte apoptosis and promoted neoangiogenesis, resulting in preservation of EF.26,27 Administration of recombinant thymosin α1 (Tα1), encoded by PTMA, has previously also been shown to reduce infarct size by promoting cardiomyocyte cell survival via Akt.28 Tα1 is the active ingredient in ZADAXIN®, which is used Worldwide as an immunomodulatory drug to treat viral hepatitis, HIV/AIDS and certain cancers.29 Most recently, Tα1 was shown to reduce mortality in patients with severe coronavirus disease 2019 (COVID-19) by increasing circulating T cells.30 Tα1 exerts immune dampening effects by promoting an increase in regulatory T cells (Treg), inhibiting cytokine production.29 Treg-cells have been shown to have a beneficial effect after MI by modulating macrophage polarization.31 Furthermore, conditioned medium from Treg cells was shown to stimulate cardiomyocyte, but not fibroblast proliferation in vitro.32 Intra-myocardial injection of Treg cells after coronary artery ligation reduced infarct size and improved cardiac function and increased the number of EdU-positive cardiomyocytes in the peri-infarct region.32 Moreover, in neonatal heart regeneration, Tregs are essential for cardiomyocyte proliferation and subsequent regeneration.33
While further studies are required to elucidate the underlying mechanisms by which the combinatorial gene delivery of TMSB4 and PTMA promote cardiomyocyte clonal expansion and preserved EF, we postulate that Tβ4 and Tα1 stimulate a regenerative environment by promoting cardiomyocyte survival and neoangiogenesis. It is already known that a permissive environment, including enhanced angiogenesis, immune cell activation and the composition of extracellular matrix9,34 is necessary for cardiac regeneration. Indeed, increased expression of PECAM1, which marks endothelial cells, was observed in both juvenile and adult sham and infarcted mice that received Tβ4 and Tα1 (see Supplementary material online, Figures S8 and S10). Recently, we have coincidently shown that the transcription factor ZEB2 in cardiomyocytes promotes the secretion of Tβ4 and Tα1 and subsequent angiogenesis and improvement in cardiac function.24 Given that Tα1 is known to exert immunomodulatory effects by increasing Treg,29 which has been demonstrated to promote cardiomyocyte proliferation,32,33 we postulate that this axis may contribute to the increase in cardiomyocyte proliferation observed in this study. However, Tβ4 and Tα1 also induced proliferation of NRCMs (which consists predominantly of cardiomyocytes and some fibroblasts; Figure 2), alone and in combination, suggesting that alternative mechanisms may also contribute to the observed increase in cardiomyocyte proliferation. For example, regulation of the actin cytoskeleton by Tβ425 may promote cytokinesis of cardiomyocytes.35 Furthermore, several combinations tested in this study included TMSB4 and PTMA in their gene cocktail (combination 2–4, 7–9, 10 and 13; see Figure 1C and Supplementary material online, Figure S6C), yet not all these combinations increased cardiomyocyte proliferation in vitro or in vivo. This could be due to either cell cycle inhibition by the combined expression of select genes or a specific factor within the cocktail. Alternatively, it may highlight that a certain threshold of gene expression is required to achieve therapeutic effects (as the same total viral genomes were injected per mouse, irrespective of the number of factors injected). Additionally, the pleiotropic nature of Tβ4 leading to a diverse range of effects within different cells and tissues, as also observed in this study (increased EdU incorporation in the heart (Figure 3C-E) compared with decreased EdU incorporation in the liver (see Supplementary material online, Figure S8E-F)) requires further elucidation.
We observed an increase in clonal expansion of mononucleated cardiomyocytes in response to cardiac ischaemia and combination 11 (see Supplementary material online, Figure S4 and Figure S7). Current evidence suggests that mononucleated cardiomyocytes have a higher propensity towards proliferation than binucleated cells, the latter of which is the predominant cell type in the adult murine heart.36 Our data is in contrast to a recent clonal analysis of cardiomyocyte cell division, which only detected very few rare two-cell clones after cardiac ischaemia.19 However, this study utilized sparse labelling of cardiomyocytes, which may therefore capture significantly fewer proliferation events. In contrast, labelling the majority of cardiomyocytes with a maximal dose of tamoxifen, as represented here, may also over-estimate cell division due to chance labelling of adjacent cardiomyocytes with the same fluorescent protein. However, we did not utilize this technology to provide a direct quantification of cell division, but rather to compare between different conditions (sham vs. ischaemia and AA9-control vs. AAV9-combination 11).
Here, we utilized single cell data analysis to identify the transcriptomic profile of cells that expressed candidate cell cycle genes. This approach has limitations as it does not prove that these cells are either cycling or proliferating. Furthermore, it is currently not possible to identify whether these cells are multinucleated or polyploid cells. Future studies that can enrich for proliferating mononucleated cardiomyocytes may provide further insight into genes that can promote cell-cycle re-entry. In addition, utilizing a combinatorial approach to determine cardiomyocyte proliferation together with the confetti model, would provide a more quantitative analysis of the therapeutic effects observed in response to Tβ4 and Tα1.
While it will be necessary to determine the mechanisms by which Tβ4 and Tα1 potentiate cardiac regeneration, our study highlights the beneficial effects of using two clinically approved factors that could be repurposed to manage acute and chronic cardiac ischaemia to promote muscularization, vascularization, and improve cardiac function.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ Contributions
M.M.G., A.K.Z.J. and E.v.R. designed experiments. M.M.G., A.K.Z.J., S.J.v.K., M.C.P., D.V. and L.K. performed all experiments. M.M.G., A.K.Z.J., S.J.v.K., M.C.P. and B.M. analysed data. M.G. and L.Z. provided models and materials. M.M.G., A.K.Z.J. and E.v.R. wrote the manuscript.
Supplementary Material
Acknowledgements
We gratefully acknowledge Hesther de Ruiter, Jeroen Korving, and Mathias Baumann from the Hubrecht Institute for technical support and Dr. Jose Gomez-Arroyo from the University of Cincinnati for graphics.
Contributor Information
Monika M Gladka, Hubrecht Institute, Royal Netherlands Academy of Arts and Sciences (KNAW) and University Medical Center Utrecht, The Netherlands.
Anne Katrine Z Johansen, Hubrecht Institute, Royal Netherlands Academy of Arts and Sciences (KNAW) and University Medical Center Utrecht, The Netherlands.
Sebastiaan J van Kampen, Hubrecht Institute, Royal Netherlands Academy of Arts and Sciences (KNAW) and University Medical Center Utrecht, The Netherlands.
Marijn M C Peters, Hubrecht Institute, Royal Netherlands Academy of Arts and Sciences (KNAW) and University Medical Center Utrecht, The Netherlands; Department of Cardiology, Regenerative Medicine Center Utrecht, University Medical Center Utrecht, The Netherlands.
Bas Molenaar, Hubrecht Institute, Royal Netherlands Academy of Arts and Sciences (KNAW) and University Medical Center Utrecht, The Netherlands.
Danielle Versteeg, Hubrecht Institute, Royal Netherlands Academy of Arts and Sciences (KNAW) and University Medical Center Utrecht, The Netherlands.
Lieneke Kooijman, Hubrecht Institute, Royal Netherlands Academy of Arts and Sciences (KNAW) and University Medical Center Utrecht, The Netherlands.
Lorena Zentilin, AAV Vector Unit, International Centre for Genetic Engineering and Biotechnology (ICGEB), Trieste, Italy.
Mauro Giacca, School of Cardiovascular Medicine and Science, King’s College London, London, United Kingdom.
Eva van Rooij, Hubrecht Institute, Royal Netherlands Academy of Arts and Sciences (KNAW) and University Medical Center Utrecht, The Netherlands; Department of Cardiology, Regenerative Medicine Center Utrecht, University Medical Center Utrecht, The Netherlands.
Funding
This work was supported by Horizon 2020 research and innovation programme REANIMA (E.v.R.), an NWO-TOP grant (E.v.R.), and an ERA-CVD grant (E.v.R). M.M.G. was funded by a Dr. Dekker postdoctoral fellowship from the Dutch Heart Foundation (NHS2016T009).
Data availability
The authors declare that the main data supporting the findings of this study are available within the article and its Supplementary Information file. All sequencing data that support the findings of this study are available in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are accessible through the GEO Series accession number GSE146285 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE146285] (for SCS data).
Translational perspective.
Ischaemic heart disease represents a leading cause of morbidity and mortality worldwide. Clinical management includes pharmacotherapy, surgery, and lifestyle changes. While current therapeutic strategies improve cardiac function, they do not address the loss of viable myocardium that results from ischaemic damage. The inherently low regenerative capacity of cardiomyocytes in the adult myocardium represents an obstacle for successful regeneration of tissue following ischaemia. Here, we identified two factors, thymosin β4 and prothymosin α, that promote cardiomyocyte regeneration and improve cardiac function of the ischaemic heart.
References
- 1. Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol 1996;28:1737–1746. [DOI] [PubMed] [Google Scholar]
- 2. Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol 1996;271:H2183–H2189. [DOI] [PubMed] [Google Scholar]
- 3. Alkass K, Panula J, Westman M, Wu TD, Guerquin-Kern JL, Bergmann O. No evidence for cardiomyocyte number expansion in preadolescent mice. Cell 2015;163:1026–1036. [DOI] [PubMed] [Google Scholar]
- 4. Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisen J. Evidence for cardiomyocyte renewal in humans. Science 2009;324:98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, Lee RT. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 2013;493:433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Herget GW, Neuburger M, Plagwitz R, Adler CP. DNA Content, ploidy level and number of nuclei in the human heart after myocardial infarction. Cardiovasc Res 1997;36:45–51. [DOI] [PubMed] [Google Scholar]
- 7. Leone M, Engel FB. Advances in heart regeneration based on cardiomyocyte proliferation and regenerative potential of binucleated cardiomyocytes and polyploidization. Clin Sci (Lond) 2019;133:1229–1253. [DOI] [PubMed] [Google Scholar]
- 8. Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science 2011;331:1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Uygur A, Lee RT. Mechanisms of cardiac regeneration. Dev Cell 2016;36:362–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sadek H, Olson EN. Toward the goal of human heart regeneration. Cell Stem Cell 2020;26:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cui M, Wang Z, Chen K, Shah AM, Tan W, Duan L, Sanchez-Ortiz E, Li H, Xu L, Liu N, Bassel-Duby R, Olson EN. Dynamic transcriptional responses to injury of regenerative and non-regenerative cardiomyocytes revealed by single-nucleus RNA sequencing. Dev Cell 2020;55:665–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Honkoop H, de Bakker DE, Aharonov A, Kruse F, Shakked A, Nguyen PD, de Heus C, Garric L, Muraro MJ, Shoffner A, Tessadori F, Peterson JC, Noort W, Bertozzi A, Weidinger G, Posthuma G, Grun D, van der Laarse WJ, Klumperman J, Jaspers RT, Poss KD, van Oudenaarden A, Tzahor E, Bakkers J. Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart. Elife 2019;8:e50163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Z, Cui M, Shah AM, Ye W, Tan W, Min YL, Botten GA, Shelton JM, Liu N, Bassel-Duby R, Olson EN. Mechanistic basis of neonatal heart regeneration revealed by transcriptome and histone modification profiling. Proc Natl Acad Sci U S A 2019;116:18455–18465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gladka MM, Molenaar B, de Ruiter H, van der Elst S, Tsui H, Versteeg D, Lacraz GPA, Huibers MMH, van Oudenaarden A, van Rooij E. Single-cell sequencing of the healthy and diseased heart reveals cytoskeleton-associated protein 4 as a new modulator of fibroblasts activation. Circulation 2018;138:166–180. [DOI] [PubMed] [Google Scholar]
- 15. Arsic N, Zentilin L, Zacchigna S, Santoro D, Stanta G, Salvi A, Sinagra G, Giacca M. Induction of functional neovascularization by combined VEGF and angiopoietin-1 gene transfer using AAV vectors. Mol Ther 2003;7:450–459. [DOI] [PubMed] [Google Scholar]
- 16. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muraro MJ, Dharmadhikari G, Grun D, Groen N, Dielen T, Jansen E, van Gurp L, Engelse MA, Carlotti F, de Koning EJ, van Oudenaarden A. A single-cell transcriptome atlas of the human pancreas. Cell Syst 2016;3:385–394. e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fu X, Khalil H, Kanisicak O, Boyer JG, Vagnozzi RJ, Maliken BD, Sargent MA, Prasad V, Valiente-Alandi I, Blaxall BC, Molkentin JD. Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J Clin Invest 2018;128:2127–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sereti KI, Nguyen NB, Kamran P, Zhao P, Ranjbarvaziri S, Park S, Sabri S, Engel JL, Sung K, Kulkarni RP, Ding Y, Hsiai TK, Plath K, Ernst J, Sahoo D, Mikkola HKA, Iruela-Arispe ML, Ardehali R. Analysis of cardiomyocyte clonal expansion during mouse heart development and injury. Nat Commun 2018;9:754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grun D, Muraro MJ, Boisset JC, Wiebrands K, Lyubimova A, Dharmadhikari G, van den Born M, van Es J, Jansen E, Clevers H, de Koning EJP, van Oudenaarden A. De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell 2016;19:266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alvarez R J, Wang BJ, Quijada PJ, Avitabile D, Ho T, Shaitrit M, Chavarria M, Firouzi F, Ebeid D, Monsanto MM, Navarrete N, Moshref M, Siddiqi S, Broughton KM, Bailey BA, Gude NA, Sussman MA. Cardiomyocyte cell cycle dynamics and proliferation revealed through cardiac-specific transgenesis of fluorescent ubiquitinated cell cycle indicator (FUCCI). J Mol Cell Cardiol 2019;127:154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johansen AK, Molenaar B, Versteeg D, Leitoguinho AR, Demkes C, Spanjaard B, de Ruiter H, Akbari Moqadam F, Kooijman L, Zentilin L, Giacca M, van Rooij E. Postnatal cardiac gene editing using CRISPR/Cas9 with AAV9-mediated delivery of short guide RNAs results in mosaic gene disruption. Circ Res 2017;121:1168–1181. [DOI] [PubMed] [Google Scholar]
- 23. Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther 2008;16:1073–1080. [DOI] [PubMed] [Google Scholar]
- 24. Gladka MM, Kohela A, Molenaar B, Versteeg D, Kooijman L, Monshouwer-Kloots J, Kremer V, Vos HR, Huibers MMH, Haigh JJ, Huylebroeck D, Boon RA, Giacca M, van Rooij E. Cardiomyocytes stimulate angiogenesis after ischemic injury in a ZEB2-dependent manner. Nat Commun 2021;12:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Skruber K, Read TA, Vitriol EA. Reconsidering an active role for G-actin in cytoskeletal regulation. J Cell Sci 2018;131:jcs203760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou B, Honor LB, Ma Q, Oh JH, Lin RZ, Melero-Martin JM, von Gise A, Zhou P, Hu T, He L, Wu KH, Zhang H, Zhang Y, Pu WT. Thymosin beta 4 treatment after myocardial infarction does not reprogram epicardial cells into cardiomyocytes. J Mol Cell Cardiol 2012;52:43–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bock-Marquette I, Saxena A, White MD, Dimaio JM, Srivastava D. Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature 2004;432:466–472. [DOI] [PubMed] [Google Scholar]
- 28. Cannavo A, Rengo G, Liccardo D, Pironti G, Scimia MC, Scudiero L, De Lucia C, Ferrone M, Leosco D, Zambrano N, Koch WJ, Trimarco B, Esposito G. Prothymosin alpha protects cardiomyocytes against ischemia-induced apoptosis via preservation of akt activation. Apoptosis 2013;18:1252–1261. [DOI] [PubMed] [Google Scholar]
- 29. Cynthia W, Tuthill RSK. Thymosin apha 1–A peptide immune modulator with a broad range of clinical applications. J Clin Exp Pharmacol 2013;3:1000133. [Google Scholar]
- 30. Liu Y, Pan Y, Hu Z, Wu M, Wang C, Feng Z, Mao C, Tan Y, Liu Y, Chen L, Li M, Wang G, Yuan Z, Diao B, Wu Y, Chen Y. Thymosin alpha 1 reduces the mortality of severe coronavirus disease 2019 by restoration of lymphocytopenia and reversion of exhausted T cells. Clin Infect Dis 2020;71:2150–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weirather J, Hofmann UD, Beyersdorf N, Ramos GC, Vogel B, Frey A, Ertl G, Kerkau T, Frantz S. Foxp3+ CD4+ T cells improve healing after myocardial infarction by modulating monocyte/macrophage differentiation. Circ Res 2014;115:55–67. [DOI] [PubMed] [Google Scholar]
- 32. Zacchigna S, Martinelli V, Moimas S, Colliva A, Anzini M, Nordio A, Costa A, Rehman M, Vodret S, Pierro C, Colussi G, Zentilin L, Gutierrez MI, Dirkx E, Long C, Sinagra G, Klatzmann D, Giacca M. Paracrine effect of regulatory T cells promotes cardiomyocyte proliferation during pregnancy and after myocardial infarction. Nat Commun 2018;9:2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li J, Yang KY, Tam RCY, Chan VW, Lan HY, Hori S, Zhou B, Lui KO. Regulatory T-cells regulate neonatal heart regeneration by potentiating cardiomyocyte proliferation in a paracrine manner. Theranostics 2019;9:4324–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zlatanova I, Pinto C, Silvestre JS. Immune modulation of cardiac repair and regeneration: the art of mending broken hearts. Front Cardiovasc Med 2016;3:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ali H, Braga L, Giacca M. Cardiac regeneration and remodelling of the cardiomyocyte cytoarchitecture. FEBS J 2020;287:417–438. [DOI] [PubMed] [Google Scholar]
- 36. Patterson M, Barske L, Van Handel B, Rau CD, Gan P, Sharma A, Parikh S, Denholtz M, Huang Y, Yamaguchi Y, Shen H, Allayee H, Crump JG, Force TI, Lien CL, Makita T, Lusis AJ, Kumar SR, Sucov HM. Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat Genet 2017;49:1346–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the main data supporting the findings of this study are available within the article and its Supplementary Information file. All sequencing data that support the findings of this study are available in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are accessible through the GEO Series accession number GSE146285 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE146285] (for SCS data).