Abstract
Diamond-Blackfan anemia (DBA) is a ribosomopathy that is characterized by macrocytic anemia, congenital malformations, and early onset during childhood. Genetic studies have demonstrated that most patients carry mutations in one of the 20 related genes, most of which encode ribosomal proteins (RP). Treatment of DBA includes corticosteroid therapy, chronic red blood cell transfusion, and other forms of immunosuppression. Currently, hematopoietic stem cell transplantation is the only cure for DBA. Interestingly, spontaneous remissions occur in 10-20% of transfusion-dependent DBA patients. However, there is no consistent association between specific mutations and clinical manifestations. In the past decades, researchers have made significant progress in understanding the pathogenesis of DBA, but it remains unclear how the ubiquitous RP haploinsufficiency causes the erythroid-specific defect in hematopoiesis in DBA patients, and why there is a difference in penetrance and spontaneous remission among individuals who carry identical mutations. In this paper, we provide a comprehensive review of the development of DBA animal models and discuss the future research directions for these important experimental systems.
Introduction
Diamond-Blackfan anemia (DBA) is a rare congenital hypoplastic anemia that manifests as moderate or severe macrocytic anemia associated with short stature, physical anomalies involving bone development, and a predisposition to malignancies.1-5 More than 90% of patients are diagnosed during their first year of life (median age 12 weeks).6 Elevated erythrocyte adenosine deaminase activity is found in more than 75% of DBA patients. Many DBA patients are dependent on corticosteroids or red blood cell transfusion. However, chronic life-long exposure to these treatments often leads to intolerance.1,7,8 Currently, hematopoietic stem cell transplantation is the only curative therapy.6,9
Erythroid failure in DBA patients is characterized by a significant reduction of erythroid precursors/progenitors in bone marrow, specifically, a blockade between the burst-forming unit-erythroid (BFU-E) and colony-forming unit-erythroid (CFU-E) stages or between the erythropoietin-independent and erythropoietin-dependent stages of erythroid development.10,11 The mutant genes that encode ribosomal proteins (RP) are responsible for the defect in ribosomal biogenesis in most DBA patients, directly affecting the synthesis of hemoglobin and the process of erythropoiesis. As a consequence, erythrocyte maturation is arrested leading to a toxic elevation of heme.7 In 2019, Ulirsch et al.12 described the genetic landscape of DBA. Heterozygous mutations in one of the 16 RP genes or three non-RP genes are found in approximately 70-80% of DBA patients.1,7,12-14 Mutations in RPS19, RPL5, RPS26, RPL11, RPL35a, RSL10, RPS24, and RPS17 have been identified in 70% of DBA patients. Mutations in GATA1 and TSR2 (encoding a chaperone protein of eS26/RPS26) have also been detected in DBA patients.15-17 HEATR3 was reported to be associated with uL5 (RPL11) and uL18 (RPL5).18 Recently, O'Donohue et al.13 reported homozygous missense and splice site variants on HEATR3 in six DBA patients from four distinct families, in which all parents are heterozygous for the variants found in their symptomatic children. A complete list of DBA-associated mutant genes that have been reported in patients is provided in Online Supplementary Table S1.
Although DBA patients share the unifying macrocytic anemia, they show a high degree of clinical heterogeneity, with disease severity and responsiveness to therapies varying among patients. Notably, spontaneous remissions occur in 10-20% of transfusion-dependent DBA patients by the age of 25 years, even in those who have never been independent of transfusions.7,19-21 In addition, asymptomatic family members of DBA patients with RPS7 or RPL15 mutations have been reported, and an identical mutant RP gene was detected in blood cells from a patient with DBA both prior to and after remission.22-24 To date, there is no obvious link to specific genetic mutations, gender, and treatment for the patients who attain remission or those who remain symptomatic with asymptomatic family members.25,26 Major unresolved questions in DBA remain how a ubiquitous RP deficiency is responsible for the erythroid-specific defect in hematopoiesis and why there is different penetrance among individuals or family members who carry the identical genetic mutation.
The molecular mechanism underlying the association between ribosome insufficiency and erythroid failure in DBA patients is not fully understood to date. It is well accepted that activation of p53 and dysregulated GATA1 contribute to the pathogenesis in DBA.15,16,27-30 Dysregulation of cell cycle progression, apoptosis, and heme have also been linked to DBA.31-33 Nemo-like kinase (NLK), a serine-threonine protein kinase, was reported to be hyperactive in erythroid progenitors from RPS19 and RPL11 knockdown human hematopoietic stem and progenitor cells.34 Recently, Wilkes et al.35 reported that upregulated microRNA (miRNA), miR-34 and miR-30, are associated with the downregulation of SATB1, a global chromatin organizer and transcriptional factor, in a preclinical model of DBA, suggesting that epigenetics may play an important role in the pathogenesis of DBA.
DBA provides a unique disease model for studying how RP deficiency has an impact on ribosome biogenesis and subsequent protein translation. However, because of the essential role of these proteins in fundamental cellular processes, it has been historically challenging to develop animal models that faithfully recapitulate the complete pathogenesis of DBA. Despite these challenges, since the first reviews on murine and zebrafish models of DBA, which were published in 2011 by McGowan and Taylor,36,37 research has advanced rapidly and is unraveling the pathogenesis of DBA. In this review, we focus on the murine models of DBA that recapitulate the hematologic features of DBA patients. We provide a comprehensive picture of recent progress and challenges in the development of animal models of DBA.
Zebrafish models
Several valuable zebrafish mutant lines have been developed, with data from these models providing many important insights into the role of RP defects in the regulation of erythropoiesis in DBA patients. Specifically, knockdown of rps19 in zebrafish recapitulates the hematopoietic and developmental phenotypes of DBA, including failure of erythropoiesis with severe anemia, cell cycle arrest, and increased apoptosis, as well as upregulation of p53.28,38,39 Bibikova et al.40 reported that gata1 expression was downregulated in rps19-deficient zebrafish, and that a tumor necrosis factor-a inhibitor, etanercept, could rescue the erythroid and developmental defects. Knockdown of rpl11 in zebrafish leads to morphological defects in the brain, head, and eyes, and pericardial edema.29 Danilova et al.41 characterized the molecular and cellular impact of uL5 (rpl11) deficiency. The rpl11 mutant zebrafish had anemia, decreased hematopoietic stem cells, and activation of the Tp53 pathway with altered expression of genes involved in cell cycle arrest (cdkn1a and ccng1) and apoptosis (bax and puma). Taylor et al.42 reported that the hematopoietic defects caused by mutant rps29 depend on p53 activation. Moreover, they observed abnormal regulation of metabolic pathways with a shift from glycolysis to aerobic respiration, including upregulation of genes involved in gluconeogenesis, decreased biosynthesis, and increased catabolism. Nucleotide metabolism was also affected in the rps29-knockdown model because of upregulation of adenosine deaminase (ADA) and xanthine dehydrogenase/oxidase.38,43 Although no RPL18 mutations have been reported in DBA patients, mutant rpl18 causes maturation arrest of red blood cells in zebrafish, due to increased p53 activation and JAK2STAT3 activation.44 Thus, in zebrafish models, researchers have successfully recapitulated maturation arrest of red blood cells, growth retardation, decreased globin synthesis, increased adenosine deaminase and apoptosis, and p53 activation, which are observed in DBA patients.28 The advantage of zebrafish models is that they are relatively easy to establish and can be used for studying developmental hematopoiesis, which is difficult in mouse models. Zebrafish models provide a useful tool for in vivo drug screening to identify possible DBA therapeutics. Oyarbide et al.45 published an excellent review covering these aspects in 2019.
Mouse models harboring the mutations found in patients with Diamond-Blackfan anemia
Rps19 (eS19)
RPS19 (eS19) is the most frequently mutated gene in DBA patients, being found in 25% of them. This gene has been intensively studied in both human cells and mouse models, allowing valuable insights into all aspects of DBA caused by the RPS19 mutation.
RNA interference (RNAi) exploits an endogenous mechanism of gene regulation that can be adapted to target and suppress specific genes in vivo.46 Short hairpin (sh)RNA containing a specific sequence homologous to a target RNA transcript is embedded within a natural miRNA backbone; upon delivery to the cell, shRNA is processed to form short interfering (si)RNA which can bind to a target RNA and functionally inactivate it.47 shRNA can silence transcripts without modifying the genomic locus, thus they can be reversible. Using transgenic RNAi, Jaako et al.48 engineered a mouse model containing shRNA, which could be regulated by doxycycline, targeting the Rps19 transcript, thereby creating an inducible regulation of Rps19 expression in adult mice. They demonstrated that Rps19-deficient mice developed macrocytic anemia along with leukocytopenia and viable platelet counts. The severity of the disease phenotype was correlated with loss of Rps19 expression and could be rescued by overexpression of Rps19 or inactivation of Trp53. They also demonstrated that chronic eS19 deficiency caused irreversible exhaustion of hematopoietic stem cells and impaired proliferation and apoptosis in hematopoietic progenitors, resulting in bone marrow failure.
Matsson et al.49 described mice in which they engineered constitutive disruption of the Rps19 gene by deletion of the 5’ untranslated region and exons 1 to 4. Whereas zygotes homozygous for the knockout (Rps19-/-) could not develop to normal blastocysts leading to embryonic lethality, heterozygous mice with a single copy of wildtype Rps19 (Rps19+/-) maintained normal ribosomal/extra-ribosomal functions. Interestingly, no erythrocyte abnormality was detected in the Rps19+/- mice. Kubik-Zahorodna et al.50 reported that homozygous mice with an Arginine 67 deletion (Arg67del) in eS19 had growth retardation, macroscopic skeletal deformation, hydrocephalus, and behavioral defects without obvious hematologic abnormalities, whereas heterozygous mice had no phenotype. Devlin et al.51 described transgenic mouse models with either a constitutive or Prion-Cre-mediated conditional missense mutation at the highly conserved amino acid 62 of eS19 (RPS19R62W). They demonstrated that the constitutive mutation was embryonic lethal, but the conditional RPS19R62W mutation exerted a dominant negative effect which manifested with many, but not all, of the pathological features of human DBA, including growth retardation, mild anemia with reduced numbers of erythroid progenitors, and significant inhibition of terminal erythroid maturation.
Xenotransplantation of CD34+ cells from DBA patients with RPS19 haploinsufficiency into sublethally irradiated immunocompromised non-obese diabetic/severe combined immunodeficient-b2 microglobulin null mice provided conclusive evidence that RPS19 mutation is the cause of the disease phenotype in DBA patients.43 Additionally, Fly-
gare et al. and Zivny et al.52,53 reported that overexpression of RPS19 in DBA CD34+ cells with RPL19 mutation improved engraftment and erythropoiesis.
Rpl11 (uL5)
Heterozygous loss-of-function mutations in the RPL11 (uL5) gene are found in 5-20% of DBA patients.1 MorgadoPalacin et al.54 described transgenic mouse models with a Cre-mediated conditional knockout of exons 3 and 4 of Rpl11. When using a ubiquitous Cre recombinase (Tg.pCAGCre) which was constitutively expressed at early stages of development, they could not produce any Rpl11+/D pups, suggesting that a single copy of Rpl11 is not sufficient to support embryonic development. When they induced Rpl11 deletion in adult mice with Rpl11loxP/loxP and a ubiquitous tamoxifen-inducible Cre recombinase (Tx-Cre), they found that complete deletion of Rpl11 (Rpl11D/D) in adult mice (1.5 months old) was lethal (survival time <8 weeks after deletion). Furthermore, when compared to their wildtype littermates, adult mice with Rpl11 haploinsufficiency (Rpl11+/D) had lower red blood cell counts and macrocytosis because of a significant decrease in erythroblasts in bone marrow. It was shown that partial loss of Rpl11 impairs erythroid maturation, reduces p53 responses, and increases cMYC levels. The data also recapitulated the Cdkn1a and Bax molecular defects observed in some DBA patients. The authors concluded that diploid Rpl11 is required for embryonic development, and that partial loss of Rpl11 in adult mice causes non-lethal DBA-like anemia and increases susceptibility to cancer.54
Using a mouse model with Rpl11+/loxP Tx-Cre, Doty et al.55 recently collected data from single-cell analysis of erythropoiesis in adult mice with Rpl11+/D. They found that the transcriptional pathways regulated by GATA1, GATA1-target genes, and genes involved in mitotic spindle formation were significantly downregulated in adult mice with Rpl11+/D, as observed in DBA patients.15 They also reported that Rpl11 haploinsufficiency uniquely caused upregulation of several mitochondrial genes, p53 and CDKN1 pathway genes, as well as DNA damage checkpoint genes.55
Rpl5 (uL18) and Rps24 (eS24)
RPL5 (uL18) is one of the most commonly mutated RP genes found in DBA patients (7%).1 In 2016, Kazerounian et al.56 reported two mouse models with conditional knockout of Rpl5 or Rps24 (eS24). A pGK-gb2 Loxp/FRT-flanked neomycin cassette was inserted into either Rpl5 or Rps24, resulting in deletion of exons 1-8 for Rpl5 and exons 2-3 for Rps24. They demonstrated that homozygous deletion of Rpl5 or Rps24 was embryonic lethal. Heterozygous Rpl5 and Rps24 mice were born normal and did not develop any sign of disease, including anemia. Real-time quantitative polymerase chain reaction (qPCR) and immunoblot analysis demonstrated no significant reduction of Rpl5 and Rps24 expression, suggesting that one copy of wildtype Rpl5 and Rps24 is sufficient to prevent the development of anemia. However, some older mice with heterozygous deletion of Rpl5 or Rps24 developed soft tissue sarcoma after the age of 17 months. Although the incidence was low, the authors concluded that prolonged RP deficiency may increase the risk of cancer, as observed in some DBA patients. Later, Kazerounian et al.57 described a mouse model in which doxycycline could be used to regulate Rpl5-targeting shRNA to induce downregulation of Rpl5 expression. They suggested that mice with inducible Rpl5targeting shRNA recapitulate the major features of DBA, including mild anemia, reticulocytopenia, and bone marrow erythroblastopenia.
Recently, Yu et al.58 reported a mouse model with an intronic ENU-induced mutation in the Rpl5 gene (Rpl5Skax23Jus/+), which alters the sixth nucleotide in intron 1 (c.3+6C>T). Rpl5Skax23-Jus/+ mice had a profound delay in erythroid maturation and increased mortality at embryonic day (E) 12.5, which improved by E14.5. Macrocytic anemia and cardiac defects were observed in newborn mice with Rpl5+/-. Penetrance of a kinky tail phenotype was 100%. Most of the diseased mice died within 3 weeks after birth due to severe pancytopenia or ventricular septal defect. However, the anemia observed at birth in Rpl5+/- mice was, surprisingly, resolved completely in adult mice that survived to 7 weeks,58 suggesting that epigenetic regulation may play an important role in spontaneous remission observed in DBA patients.
Rps7 (eS7)
Watkins-Chow et al.59 reported a mouse model with ENUinduced mutations in the Rps7 (eS7) gene (Rps7Mtu and Rps7Zma). Although the mutant mice did not show any defects in hematopoiesis, Rps7 haploinsufficiency caused decreased mouse size, abnormal skeletal morphology, mid-ventral white spotting, and eye malformations, phenotypes that also occur in mice with haploinsufficiency for other RP subunits. In addition, significant apoptosis occurred in the central nervous system along with subtle behavioral phenotypes, suggesting that eS7 is particularly important for central nervous system development. The data reported by Sato et al.60 also suggest that RPS7 deficiency contributes to the pathogenesis of DBA indirectly through other mechanisms, such as protein-protein interactions in ribosome assembly.
Rps20 (uS10)
Recently, a missense mutation in RPS20 (uS10) was reported in patients with DBA.14 However, mice with heterozygous Rps20 mutation showed only dark skin.61
Gata1
GATA1 is an essential hematopoietic transcription factor in
early hematopoietic stem and progenitor cells for the specification of erythropoiesis, as well as in megakaryocytes and eosinophils.62 Although GATA1 mutations are found in <1% of DBA patients, it has been reported that a reduction of GATA1 expression is observed at early hematopoietic stem and progenitor cell stages in uncultured bone marrow cells from DBA patients with diverse RP gene mutations,15,16,40,63 suggesting that GATA1 deficiency may be a second hit in some patients with RP gene mutations. In humans and mice, GATA1 and Gata1 are located on the X chromosome, and RP haploinsufficiency impairs the translation of GATA1 mRNA.15,16 Fujiwara et al.64 reported that mouse embryos lacking Gata1 died at approximately E11.5 because of maturation arrest of erythroid progenitors. Female heterozygotes (Gata1+/-) were born pale due to random X chromosome inactivation. Interestingly, those mice recovered during the neonatal period, presumably as a result of in vivo selection for progenitors able to express Gata1. Later Gutiérrez et al.65 described mouse models with inducible Gata1 deletion using conditional Gata1 knockout mediated by either interferon-Cre recombinase (Mx-Cre) or Tx-Cre. They found that Mx-Cre-mediated Gata1 deletion, although not producing anemia, caused maturation arrest of erythroid cells at the proerythroblast stage, thrombocytopenia, and excessive proliferation of megakaryocytes in spleens from adult Gata1-null mice. TxCre-mediated Gata1 deletion caused depletion of the erythroid compartment in both bone marrow and spleen, resulting in severe anemia in adult mice. Furthermore, formation of both early and late erythroid progenitors was significantly reduced in adult bone marrow in the absence of Gata1.65
Mouse models with molecular defects not found in patients with Diamond-Blackfan anemia but relevant to its pathogenesis
Despite extensive efforts to sequence all genes coding RP in affected individuals, molecular lesions remain elusive in approximately 20-25% of cases of DBA, and the correlation between genotype and phenotype is not clear in DBA.1 Therefore, in efforts to develop mouse models mimicking the clinical features or pathogenesis of DBA in humans, researchers have explored the impact of other proteins with known roles in the maturation of erythrocytes, accumulation of heme, and bone development.
Rps14 (uS11)
The Rps14 (uS11) mutation, like the 5q deletion, has never been reported in DBA patients. In an attempt to mimic 5q deletion in mice, Barlow et al.66 investigated mice engineered with LIM domain only 2 (Lmo2)-Cre and Cd74+/loxNid67+/lox (containing the Rps14 gene for uS11, Cd74+/D Nid67+/D) and reported that haploinsufficiency of the CD74Nid67 interval caused a DBA-like phenotype, including macrocytic anemia, thrombocytopenia, and hypocellularity in bone marrow, which was associated with increased p53 expression and apoptosis, as observed in DBA samples.27 They also demonstrated that deletion of Trp53 completely restored the populations of hematopoietic stem cells, common myeloid progenitors, megakaryocyte-erythrocyte progenitors and granulocyte/macrophage progenitors, and reversed the dysplasia in bone marrow of mice carrying Cd74+/D Nid67+/D.
Flvcr
Feline leukemia virus subgroup C receptor (FLVCR) is a heme export protein, and delayed globin synthesis leads to excess heme in DBA patients.67 Although a DBA-causative mutation in the FLVCR1 gene has never been identified in DBA families,68 Keel et al.69 reported mouse models with constitutive Flvcr+/- and inducible Flvcr mutations (Flvcr+/flox;Mx-cre). They demonstrated that Flvcr-null mice lack definitive erythropoiesis and have craniofacial and limb deformities, while deletion of Flvcr in the neonatal period causes severe macrocytic anemia with proerythroblast maturation arrest, resembling the clinical features of DBA patients. Later, the same group reported the data from single-cell analyses of mice with Flvcr deletion.70 They demonstrated that the heme-GATA1 feedback loop regulates red cell differentiation and that deletion of Flvcr1 causes high levels of intracellular heme, which decreases GATA1 and expression of GATA1-target genes, as well as mitotic spindle genes in late stage erythroid cells (CD71+Ter119lo-hi). Their data suggest that excessive heme may be responsible for the progression of RP imbalance and prematurely lower GATA1, and may impede mitosis in patients in the late stages of DBA.
Rps6 (eS6)
Mouse embryos with haploinsufficiency of Rps6 (Rps6+/D) are small and die at gastrulation; genetic inactivation of p53 bypasses this checkpoint, prolonging the embryonic development until E12.5, at which point the embryos likely die from anemia.71 Two groups reported their observations in mice with inducible Rps6 deletion using conditional Rps6 knockout mediated by Mx-Cre.72,73 Regardless of the timing of deletion, induced in either the neonatal period (5-7 days after birth) or in adulthood (7-9 weeks), mice with Rps6+/D exhibited robust macrocytic anemia, granulocytopenia, lymphopenia, and progressive thrombocytosis. Elevated levels of erythroid ADA (eADA) were also observed in these mice, a finding present in >80% of DBA patients. McGowan et al.72 demonstrated that deletion of Trp53 rescued the red blood cell counts and burst-forming unit-erythroid or mature erythroid precursors in Rps6deficient mice, a finding that was also reported by Tiu et al.,30 collectively confirming that p53 plays an important role in the pathogenesis of DBA. In particular, Tiu et al.30 characterized limb development defects upon Rps6 deletion during the development of the limb bud mesenchyme; these defects are similar to the congenital birth defects found in DBA patients, including radial hypoplasia and defects in digit formation. They also observed a decrease in translation, specifically of transcripts controlling limb development, upon Rps6 deletion. This decrease could be rescued by knockdown of either p53 and/or the repressor of cap binding protein, 4E-BP1. 4E-BP1 represses the essential eukaryotic initiation factor eIF4E. Strikingly, it was also observed in cells with Rps19- and Rps14knockdown that the selective translation phenomena was driven by upregulation of p53 and 4E-BP1, suggesting a common pathway via the p53-4EBP1-eIF4E axis by which selective changes in translation occur upon ribosome perturbation.30
Other mutations
Mouse models with mutations in Rpl24 (eL24)74 and Rpl27a (eL27) have been described.75 These animals had non-hematologic phenotypes (Table 1).
Conclusions and future directions
Most molecular defects in DBA patients, are caused by haploinsufficiency in one of the RP genes.1,12 In 2005, DBA was recognized as the first ribosomopathy in humans.76 Ribosomopathies broadly comprise two categories: disorders caused by single-copy mutations in specific RP, and disorders associated with defects in ribosome biogenesis factors. The phenotypic patterns among different ribosomopathies are divergent but often share some overlapping features, including effects on bone marrow-derived cell lineages and skeletal tissues. It is unknown how the phenotypic diversity among the ribosomopathies is regulated. The common tissue specificities of the different ribosomopathies are very challenging to reconcile with the ubiquitous requirement for ribosomes in all cells.77
Due to the limited numbers of bone marrow cells in DBA patients, studies on the critical roles of ribosomes in protein biosynthesis have been very challenging. Researchers have successfully recapitulated the macrocytic anemia and growth retardation of DBA patients in zebrafish and mouse models with engineered mutations of Rps19, Rpl11, Rpl5, Rps24, Rps7, Rps14, and Gata1 (Table 1). Other animal models with mutant genes not found in DBA patients, such as Rps6 and Flvcr, have advanced our understanding of how ribosome dysfunction affects erythropoiesis and contributes to the pathogenesis of DBA. Haploinsufficiency of Rpl11 and Rpl5 in mice also produces manifestations resembling the clinical features of DBA. While in some models the RP composition of ribosomes varies,78 Khajuria et al.63 recently reported that in hematopoietic cells, RP haploinsufficiency did not demonstrate an altered ribosome composition. Rather, the impaired lineage commitment, a characteristic of hematopoiesis in DBA patients, arises from reduced cellular levels of ribosomes, suggesting that ribosome levels are rate-limiting and selectively regulate translation and lineage commitment in human hematopoiesis.
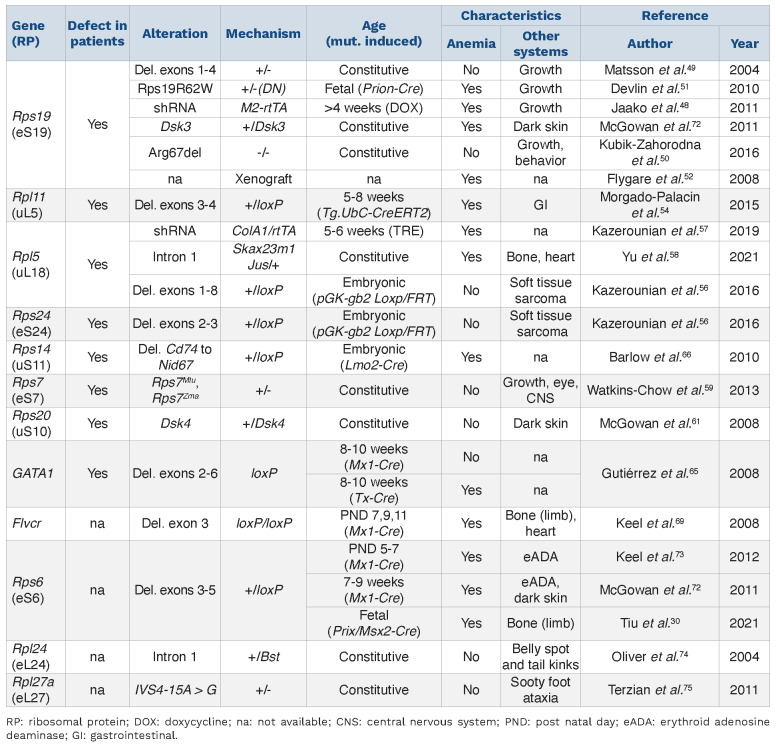
Table 1.
Mouse models with Diamond-Blackfan anemia-associated mutations.
DBA patients with RP gene mutations were identified as being exclusively heterozygous. From mouse models, we learned that homozygous RP gene mutations are embryonic lethal, and fetuses with heterozygous deletions occurring in early development cannot survive to birth, such as homozygous Rps19 fetuses and heterozygous Rpl11 fetuses.49,54 This suggests that gene dosage and the timing of mutation hits play critical roles in recapitulating the clinical manifestations of DBA patients. Although each of the reported mouse models has various limitations in recapitulating the phenotype observed in DBA patients, such as the presence of a combination of anemia and physical anomalies, the models suggest that the complexity of haploinsufficiency of various RP depends on tissue specificity, resulting in varying disease severity in children with DBA. In the past decades, we have learnt much about the genetic changes in DBA,12 but we know very little about the role of epigenetics in the pathogenesis of DBA. It is striking that most DBA patients are diagnosed during their first year of life, when epigenetic regulation is required for developmental hematopoiesis, as the neonate adapts to normoxia and rapid growth. However, this age specificity and the dynamics of developmental hematopoiesis have been underappreciated in most of the reported DBA mouse models.
Spontaneous remissions occur in 10-20% of transfusiondependent DBA patients, and some DBA patients experience periods of anemia remission and relapse as a result of unknown mechanisms.24 Additionally, asymptomatic family members of DBA patients with RPS7 or RPL15 mutation have been reported, and the mutation persisted in the blood cells from a DBA patient after remission.22-24 Furthermore, female mice with Gata1+/- were born pale due to the random inactivation of the X chromosome bearing the normal allele, but recovered during the neonatal period, presumably as a result of in vivo selection for progenitors able to express Gata1.64 Recently, it was reported that the severe anemia and cardiac defects in young mice with Rpl5+/- could resolve spontaneously in surviving adults.58 Collectively, these data suggest that epigenetic regulation may contribute to the pathogenesis of DBA.
Fetal hematopoietic stem cells may follow various tracks to migrate into adult hematopoiesis based on the status of the hematopoiesis switch at which the RP mutations occur. In addition, the effects of mutant RP on the assembled ribosome and subsequent translation likely dictate the spectrum and severity of the disease. Previous reports suggest that ribosome levels are rate-limiting and selectively regulate translation and lineage commitment in human hematopoiesis.63 Therefore, insufficient ribosomes most likely alter the dynamics of hematopoiesis during development in early childhood. Elevated fetal hemoglobin levels are often detected in DBA patients.6 It has been reported that Pten deficiency in mice sustains fetallike hematopoiesis at an age at which the fetal-to-adulthematopoiesis switch should be completed,79 and the timing of Pten deficiency determines the disease severity in mice with juvenile leukemia.80 Recently, Strahm et al.81 reported favorable outcomes of hematopoietic stem cell transplantation in children and adolescents with DBA, suggesting the benefit of correcting the molecular defects at an early stage of development. Emerging data support the idea that developmental hematopoiesis plays critical roles in pediatric hematopoietic disorders. Therefore, understanding the molecular mechanisms of fetal-toadult hematopoiesis will be key for future efforts to mimic DBA in mouse models. In the future, investigation of the timing of mutations in different developmental stages may help to uncover how developmental hematopoiesis contributes to the pathogenesis of DBA, particularly in pediatric patients.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Anupama Narla for her helpful suggestions and comments while planning the manuscript. We also thank Dr. Rhonda Perriman for her critical editing of the final version of the manuscript.
Funding Statement
Funding: KMS is supported by the NIH/R01 DK107286, Department of Defense Bone Marrow Failure Program (BM180024), DBA Foundation, SPARK program in Translational Research, Stanford Maternal Child Health Research Institute, and California Institute of Regenerative Medicine (CIRM 12475). MB is a New York Stem Cell Foundation Robertson Investigator and is supported by the New York Stem Cell Foundation and National Institutes of Health grant R01HD086634. MCW and AS are supported by the NIDDK T32 Training in Pediatric Nonmalignant Hematology and Stem Cell Biology (DK098132-06A1). MCW is also supported by K01 (DK123140), a Maternal Child Health Research Institute fellowship (1111239-442-JHACT), and Instructor K Award (1258512-700WAGZG).
References
- 1.Da Costa LM, Marie I, Leblanc TM. Diamond-Blackfan anemia. Hematology Am Soc Hematol Educ Program. 2021;2021(1):353-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter BP, Giri N, Savage SA, et al. Cancer in the National Cancer Institute inherited bone marrow failure syndrome cohort after fifteen years of follow-up. Haematologica. 2018;103(1):30-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipton JM, Alter BP. Heritable cancer: rounding up the not so usual suspects. Pediatr Blood Cancer. 2017;64(2):219-220. [DOI] [PubMed] [Google Scholar]
- 4.Lipton JM, Federman N, Khabbaze Y, et al. Osteogenic sarcoma associated with Diamond-Blackfan anemia: a report from the Diamond-Blackfan Anemia Registry. J Pediatr Hematol Oncol. 2001;23(1):39-44. [DOI] [PubMed] [Google Scholar]
- 5.Vlachos A, Rosenberg PS, Atsidaftos E, et al. Incidence of neoplasia in Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry. Blood. 2012;119(16):3815-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Da Costa L, Leblanc T, Mohandas N. Diamond-Blackfan anemia. Blood. 2020;136(11):1262-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Da Costa L, Narla A, Mohandas N. An update on the pathogenesis and diagnosis of Diamond-Blackfan anemia. F1000Res. 2018;7:F1000 Faculty Rev-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipton JM, Atsidaftos E, Zyskind I, et al. Improving clinical care and elucidating the pathophysiology of Diamond Blackfan anemia: an update from the Diamond Blackfan Anemia Registry. Pediatr Blood Cancer. 2006;46(5):558-564. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Lodish HF, Sieff CA. Critical issues in Diamond-Blackfan anemia and prospects for novel treatment. Hematol Oncol Clin North Am. 2018;32(4):701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohene-Abuakwa Y, Orfali KA, Marius C, et al. Two-phase culture in Diamond Blackfan anemia: localization of erythroid defect. Blood. 2005;105(2):838-846. [DOI] [PubMed] [Google Scholar]
- 11.Iskander D, Psaila B, Gerrard G, et al. Elucidation of the EP defect in Diamond-Blackfan anemia by characterization and prospective isolation of human EPs. Blood. 2015;125(16):25532557. [DOI] [PubMed] [Google Scholar]
- 12.Ulirsch JC, Verboon JM, Kazerounian S, et al. The genetic landscape of Diamond-Blackfan anemia. Am J Hum Genet. 2018;103(6):930-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Donohue MF, Da Costa L, Lezzerini M, et al. HEATR3 variants impair nuclear import of uL18 (RPL5) and drive DiamondBlackfan anemia. Blood. 2022;139(21):3111-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhar S, Zhou F, Reineke LC, et al. Expansion of germline RPS20 mutation phenotype to include Diamond-Blackfan anemia. Hum Mutat. 2020;41(11):1918-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludwig LS, Gazda HT, Eng JC, et al. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat Med. 2014;20(7):748-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sankaran VG, Ghazvinian R, Do R, et al. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest. 2012;122(7):2439-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gripp KW, Curry C, Olney AH, et al. Diamond-Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28. Am J Med Genet A. 2014;164A(9):2240-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calvino FR, Kharde S, Ori A, et al. Symportin 1 chaperones 5S RNP assembly during ribosome biogenesis by occupying an essential rRNA-binding site. Nat Commun. 2015;6:6510-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ball SE, McGuckin CP, Jenkins G, et al. Diamond-Blackfan anaemia in the U.K.: analysis of 80 cases from a 20-year birth cohort. Br J Haematol. 1996;94(4):645-653. [DOI] [PubMed] [Google Scholar]
- 20.Willig TN, Niemeyer CM, Leblanc T, et al. Identification of new prognosis factors from the clinical and epidemiologic analysis of a registry of 229 Diamond-Blackfan anemia patients. DBA group of Societe d'Hematologie et d'Immunologie Pediatrique (SHIP), Gesellshaft fur Padiatrische Onkologie und Hamatologie (GPOH), and the European Society for Pediatric Hematology and Immunology (ESPHI). Pediatr Res. 1999;46(5):553-561. [DOI] [PubMed] [Google Scholar]
- 21.Janov AJ, Leong T, Nathan DG, et al. Diamond-Blackfan anemia. Natural history and sequelae of treatment. Medicine (Baltimore). 1996;75(2):77-78. [DOI] [PubMed] [Google Scholar]
- 22.Akram T, Fatima A, Klar J, et al. Aberrant splicing due to a novel RPS7 variant causes Diamond-Blackfan anemia associated with spontaneous remission and meningocele. Int J Hematol. 2020;112(6):894-899. [DOI] [PubMed] [Google Scholar]
- 23.Smetanina NS, Mersiyanova IV, Kurnikova MA, et al. Clinical and genomic heterogeneity of Diamond Blackfan anemia in the Russian Federation. Pediatr Blood Cancer. 2015;62(9):1597-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wlodarski MW, Da Costa L, O'Donohue MF, et al. Recurring mutations in RPL15 are linked to hydrops fetalis and treatment independence in Diamond-Blackfan anemia. Haematologica. 2018;103(6):949-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narla A, Vlachos A, Nathan DG. Diamond Blackfan anemia treatment: past, present, and future. Semin Hematol. 2011;48(2):117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubickova A, Maceckova Z, Vojta P, et al. Missense mutation in RPS7 causes Diamond-Blackfan anemia via alteration of erythrocyte metabolism, protein translation and induction of ribosomal stress. Blood Cells Mol Dis. 2022;97:102690. [DOI] [PubMed] [Google Scholar]
- 27.Dutt S, Narla A, Lin K, et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117(9):2567-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112(13):5228-5237. [DOI] [PubMed] [Google Scholar]
- 29.Chakraborty A, Uechi T, Higa S, et al. Loss of ribosomal protein L11 affects zebrafish embryonic development through a p53dependent apoptotic response. PLoS One. 2009;4(1):e4152-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiu GC, Kerr CH, Forester CM, et al. A p53-dependent translational program directs tissue-selective phenotypes in a model of ribosomopathies. Dev Cell. 2021;56(14):2089-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuda T, Kato T, Kiyotani K, et al. p53-independent p21 induction by MELK inhibition. Oncotarget. 2017;8(35):57938-57947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doty RT, Phelps SR, Shadle C, et al. Coordinate expression of heme and globin is essential for effective erythropoiesis. J Clin Invest. 2015;125(12):4681-4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang B, Wang C, Wan Y, et al. Decoding the pathogenesis of Diamond-Blackfan anemia using single-cell RNA-seq. Cell Discov. 2022;8(1):41-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkes MC, Siva K, Chen J, et al. Diamond Blackfan anemia is mediated by hyperactive Nemo-like kinase. Nat Commun. 2020;11(1):3344-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkes MC, Scanlon V, Shibuya A, et al. Downregulation of SATB1 by miRNAs reduces megakaryocyte/erythroid progenitor expansion in preclinical models of Diamond-Blackfan anemia. Exp Hematol. 2022;111:66-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGowan KA, Mason PJ. Animal models of Diamond Blackfan anemia. Semin Hematol. 2011;48(2):106-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor AM, Zon LI. Modeling Diamond Blackfan anemia in the zebrafish. Semin Hematol. 2011;48(2):81-88. [DOI] [PubMed] [Google Scholar]
- 38.Danilova N, Bibikova E, Covey TM, et al. The role of the DNA damage response in zebrafish and cellular models of Diamond Blackfan anemia. Dis Model Mech. 2014;7(7):895-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uechi T, Nakajima Y, Chakraborty A, et al. Deficiency of ribosomal protein S19 during early embryogenesis leads to reduction of erythrocytes in a zebrafish model of Diamond-Blackfan anemia. Hum Mol Genet. 2008;17(20):3204-3211. [DOI] [PubMed] [Google Scholar]
- 40.Bibikova E, Youn MY, Danilova N, et al. TNF-mediated inflammation represses GATA1 and activates p38 MAP kinase in RPS19-deficient hematopoietic progenitors. Blood. 2014;124(25):3791-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danilova N, Sakamoto KM, Lin S. Ribosomal protein L11 mutation in zebrafish leads to haematopoietic and metabolic defects. Br J Haematol. 2011;152(2):217-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor AM, Humphries JM, White RM, et al. Hematopoietic defects in rps29 mutant zebrafish depend upon p53 activation. Exp Hematol. 2012;40(3):228-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amsterdam A, Nissen RM, Sun Z, et al. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101(35):12792-12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C, Lu M, Lin S, et al. The nuclear gene rpl18 regulates erythroid maturation via JAK2-STAT3 signaling in zebrafish model of Diamond-Blackfan anemia. Cell Death Dis. 2020;11(2):135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oyarbide U, Topczewski J, Corey SJ. Peering through zebrafish to understand inherited bone marrow failure syndromes. Haematologica. 2019;104(1):13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemann MT, Fridman JS, Zilfou JT, et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33(3):396-400. [DOI] [PubMed] [Google Scholar]
- 47.Dickins RA, Hemann MT, Zilfou JT, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37(11):1289-1295. [DOI] [PubMed] [Google Scholar]
- 48.Jaako P, Flygare J, Olsson K, et al. Mice with ribosomal protein S19 deficiency develop bone marrow failure and symptoms like patients with Diamond-Blackfan anemia. Blood. 2011;118(23):6087-6896. [DOI] [PubMed] [Google Scholar]
- 49.Matsson H, Davey EJ, Draptchinskaia N, et al. Targeted disruption of the ribosomal protein S19 gene is lethal prior to implantation. Mol Cell Biol. 2004;24(9):4032-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kubik-Zahorodna A, Schuster B, Kanchev I, et al. Neurological deficits of an Rps19(Arg67del) model of Diamond-Blackfan anaemia. Folia Biol (Praha). 2016;62(4):139-147. [PubMed] [Google Scholar]
- 51.Devlin EE, Dacosta L, Mohandas N, et al. A transgenic mouse model demonstrates a dominant negative effect of a point mutation in the RPS19 gene associated with Diamond-Blackfan anemia. Blood. 2010;116(15):2826-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flygare J, Olsson K, Richter J, et al. Gene therapy of Diamond Blackfan anemia CD34(+) cells leads to improved erythroid development and engraftment following transplantation. Exp Hematol. 2008;36(11):1428-1435. [DOI] [PubMed] [Google Scholar]
- 53.Zivny J, Jelinek J, Pospisilova D, et al. Diamond Blackfan anemia stem cells fail to repopulate erythropoiesis in NOD/SCID mice. Blood Cells Mol Dis. 2003;31(1):93-97. [DOI] [PubMed] [Google Scholar]
- 54.Morgado-Palacin L, Varetti G, Llanos S, et al. Partial loss of Rpl11 in adult mice recapitulates Diamond-Blackfan anemia and promotes lymphomagenesis. Cell Rep. 2015;13(4):712-722. [DOI] [PubMed] [Google Scholar]
- 55.Doty RT, Yan X, Meng C, et al. Single-cell analysis of erythropoiesis in Rpl11 haploinsufficient mice reveals insight into the pathogenesis of Diamond-Blackfan anemia. Exp Hematol. 2021;97:66-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kazerounian S, Ciarlini PD, Yuan D, et al. Development of soft tissue sarcomas in ribosomal proteins L5 and S24 heterozygous mice. J Cancer. 2016;7(1):32-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kazerounian S, Yuan D, Alexander MS, et al. Rpl5-inducible mouse model for studying Diamond-Blackfan anemia. Discoveries (Craiova). 2019;7(3):e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu L, Lemay P, Ludlow A, et al. A new murine Rpl5 (uL18) mutation provides a unique model of variably penetrant Diamond-Blackfan anemia. Blood Adv. 2021;5(20):4167-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watkins-Chow DE, Cooke J, Pidsley R, et al. Mutation of the Diamond-Blackfan anemia gene Rps7 in mouse results in morphological and neuroanatomical phenotypes. PLoS Genet. 2013;9(1):e1003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sato Y, Yano S, Ewis AA, et al. SRY interacts with ribosomal proteins S7 and L13a in nuclear speckles. Cell Biol Int. 2011;35(5):449-452. [DOI] [PubMed] [Google Scholar]
- 61.McGowan KA, Li JZ, Park CY, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40(8):963-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferreira R, Ohneda K, Yamamoto M, et al. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol. 2005;25(4):1215-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khajuria RK, Munschauer M, Ulirsch JC, et al. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell. 2018;173(1):90-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fujiwara Y, Browne CP, Cunniff K, et al. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci U S A. 1996;93(22):12355-12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gutierrez L, Tsukamoto S, Suzuki M, et al. Ablation of Gata1 in adult mice results in aplastic crisis, revealing its essential role in steady-state and stress erythropoiesis. Blood. 2008;111(8):4375-4385. [DOI] [PubMed] [Google Scholar]
- 66.Barlow JL, Drynan LF, Hewett DR, et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nat Med. 2010;16(1):59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Z, Keel SB, Shimamura A, et al. Delayed globin synthesis leads to excess heme and the macrocytic anemia of Diamond Blackfan anemia and del(5q) myelodysplastic syndrome. Sci Transl Med. 2016;8(338):67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quigley JG, Gazda H, Yang Z, et al. Investigation of a putative role for FLVCR, a cytoplasmic heme exporter, in Diamond-Blackfan anemia. Blood Cells Mol Dis. 2005;35(2):189-192. [DOI] [PubMed] [Google Scholar]
- 69.Keel SB, Doty RT, Yang Z, et al. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science. 2008;319(5864):825-828. [DOI] [PubMed] [Google Scholar]
- 70.Doty RT, Yan X, Lausted C, et al. Single-cell analyses demonstrate that a heme-GATA1 feedback loop regulates red cell differentiation. Blood. 2019;133(5):457-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Panic L, Tamarut S, Sticker-Jantscheff M, et al. Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol Cell Biol. 2006;26(23):8880-8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McGowan KA, Pang WW, Bhardwaj R, et al. Reduced ribosomal protein gene dosage and p53 activation in low-risk myelodysplastic syndrome. Blood. 2011;118(13):3622-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keel SB, Phelps S, Sabo KM, et al. Establishing Rps6 hemizygous mice as a model for studying how ribosomal protein haploinsufficiency impairs erythropoiesis. Exp Hematol. 2012;40(4):290-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oliver ER, Saunders TL, Tarle SA, et al. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development. 2004;131(16):3907-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Terzian T, Dumble M, Arbab F, et al. Rpl27a mutation in the sooty foot ataxia mouse phenocopies high p53 mouse models. J Pathol. 2011;224(4):540-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leger-Silvestre I, Caffrey JM, Dawaliby R, et al. Specific role for yeast homologs of the Diamond Blackfan anemia-associated Rps19 protein in ribosome synthesis. J Biol Chem. 2005;280(46):38177-38185. [DOI] [PubMed] [Google Scholar]
- 77.Mills EW, Green R. Ribosomopathies: there's strength in numbers. Science. 2017;358(6363):eaan2755. [DOI] [PubMed] [Google Scholar]
- 78.Shi Z, Fujii K, Kovary KM, et al. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genomewide. Mol Cell. 2017;67(1):71-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vara N, Liu Y, Yan Y, et al. Sustained fetal hematopoiesis causes juvenile death from leukemia: evidence from a dual-age-specific mouse model. Blood Adv. 2020;4(15):3728-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu YL, Yan Y, Webster C, et al. Timing of the loss of PTEN protein determines disease severity in a mouse model of myeloid malignancy. Blood. 2016;127(15):1912-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Strahm B, Loewecke F, Niemeyer CM, et al. Favorable outcomes of hematopoietic stem cell transplantation in children and adolescents with Diamond-Blackfan anemia. Blood Adv. 2020;4(8):1760-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.