Key Words: competing endogenous RNA, inflammatory pathway, low-level laser therapy, macrophage, miR-330-5p, neurological function, photobiomodulation, RNA-seq, spinal cord injury, STAT3
Abstract
As a classic noninvasive physiotherapy, photobiomodulation, also known as low-level laser therapy, is widely used for the treatment of many diseases and has anti-inflammatory and tissue repair effects. Photobiomodulation has been shown to promote spinal cord injury repair. In our previous study, we found that 810 nm low-level laser therapy reduced the M1 polarization of macrophages and promoted motor function recovery. However, the mechanism underlying this inhibitory effect is not clear. In recent years, transcriptome sequencing analysis has played a critical role in elucidating the progression of diseases. Therefore, in this study, we performed M1 polarization on induced mouse bone marrow macrophages and applied low-level laser therapy. Our sequencing results showed the differential gene expression profile of photobiomodulation regulating macrophage polarization. We analyzed these genes using gene ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses. Networks of protein-protein interactions and competing RNA endogenous networks were constructed. We found that photobiomodulation inhibited STAT3 expression through increasing the expression of miR-330-5p, and that miR-330-5p binding to STAT3 inhibited STAT3 expression. Inducible nitric oxide synthase showed trends in changes similar to the changes in STAT3 expression. Finally, we treated a mouse model of spinal cord injury using photobiomodulation and confirmed that photobiomodulation reduced inducible nitric oxide synthase and STAT3 expression and promoted motor function recovery in spinal cord injury mice. These findings suggest that STAT3 may be a potential target of photobiomodulation, and the miR-330-5p/STAT3 pathway is a possible mechanism by which photobiomodulation has its biological effects.
Introduction
Spinal cord injury (SCI), as a neurological disease, can be caused by external factors, such as trauma, car accidents, or falling from heights, and often leads to the loss of motor, sensory, and autonomic nervous function (Tashiro et al., 2021; Assunção Silva et al., 2022; Griffey and Yamamoto, 2022). Bone marrow-derived macrophages (BMDMs) are the most important inflammatory cells in secondary SCI and are recruited to the injury area after injury. M1 polarization of BMDMs aggravates the inflammatory response, promotes neuronal apoptosis, and worsens the progression of SCI. Therefore, it is important to understand how to effectively control M1 polarization following SCI (Ren and Young, 2013).
Photobiomodulation (PBM), also known as low-level laser therapy, is a widely used noninvasive physiotherapy with minimal side effects that reduces inflammation, repairs scars, and promotes bone repair (Sarvestani et al., 2017; Song et al., 2017). PBM has been reported to have a good repair function for SCI (Song et al., 2017; Yang et al., 2021). We previously demonstrated the safety of 810 nm low-level laser irradiation in pigs, showed that 810 nm laser therapy regulated BMDM polarization in mice, and preliminarily explored the possible mechanism of PBM (Zhang et al., 2020; Zheng et al., 2021; Zuo et al., 2022). For example, PBM stimulated the release of various neurotrophic factors by activating the protein kinase A/cAMP response element binding pathway in macrophages, and inhibition of the Notch1-hypoxia-inducible factor-1α/nuclear factor-κB signaling pathway attenuated macrophage M1 polarization (Zhang et al., 2020; Ma et al., 2022). However, the specific mechanism of PBM regulation of BMDM polarization needs to be further studied.
In many diseases, transcriptome sequencing can be used to analyze the differential genes and signaling pathways in disease occurrence and progression, which is of great value for elucidating the disease pathogenesis and clarifying target genes causing a disease (Cao et al., 2021; Pan et al., 2021). However, no previous studies have performed transcriptome sequencing to analyze the potential targets and possible mechanism of PBM regulation of M1 polarization to promote SCI repair. Therefore, in this study, we used transcriptome sequencing to investigate the differential gene expression profile and possible signaling pathways by which PBM regulates M1 macrophage polarization to promote SCI repair, which will help explain the possible mechanism of PBM.
Methods
Animals
Because the incidence of SCI is much higher in men than in women (Wu et al., 2022), and female mice had a lower inflammatory response after SCI than male mice (Farooque et al., 2006), we used male mice for this study. We obtained male C57BL/6 mice (specific-pathogen-free level, 6–8 weeks) from the Animal Center of Air Force Medical University, Xi’an, China (license No. SYXK (Shaan) 2019-001). The animal experimental protocol was approved by the Animal Protection and Utilization Committee of Air Force Military Medical University (approval No. IACUC-20210358; date: March 1, 2021). The mice were raised under standard conditions (temperature of 22–25°C, relative humidity of 45–65%, 12-hour light/dark cycle, and food and water available at all times), with three mice in each cage. A total of 80 mice were included in the experiment, and 69 survived for experimental study. Mice were randomly divided into a sham group (n = 15), SCI groups (n = 33), and PBM treatment groups (n = 15), and mice in the SCI groups were randomly divided into 3, 7, 14, and 28 days subgroups. Grouping details are described in Additional Figure 1 (935.5KB, tif) .
SCI model
The SCI model was generated as described previously (Jiang et al., 2017). Mice were anesthetized by injecting 0.6% sodium pentobarbital (10 mL/kg) intraperitoneally. A longitudinal incision was made with T9 as the center, the skin and subcutaneous tissue were cut successively, T8–T10 spinous processes and lamina were exposed, and the T9 lamina was removed to expose the spinal cord. The forceps were completely inserted into both sides of the spinal cord in the vertical direction, and the spinal cord is clamped with the forceps maximally closed for 30 seconds at the T9 level to cause SCI, which was followed by hemostasis and suturing. After the operation, micturition was performed by manually squeezing the bladder every day. Only laminectomy was performed in the sham group.
Cell extraction, culture, and transfection
The male C57BL/6 mice were killed by an overdose of 0.6% sodium pentobarbital (10 mL/kg) and were placed in 70% ethanol for 15 minutes. The intact hindlimb femurs and tibias were removed using aseptic techniques. The medullary cavity was repeatedly washed with precooled phosphate-buffered saline to obtain a mixed suspension, which was then collected in a 15 mL centrifuge tube. The red blood cell lysate was added for 10 minutes. The mixture was centrifuged at 300 × g for 5 minutes and the supernatant was discarded. The cells were gently resuspended in Dulbecco’s modified Eagle’s medium (Cytiva-HyClone Laboratories Inc., South Logan, UT, USA) containing 10% fetal bovine serum (Biological Industries, Kibbutz Beit-Haemek, Israel) and 10 ng/mL macrophage colony-stimulating factor (Sino Biological, Beijing, China). BMDMs were cultured at 37°C with 5% CO2 for 7 days. M1 polarization of BMDMs was induced by lipopolysaccharide (100 ng/mL, Sigma-Aldrich, St. Louis, MO, USA) + interferon-γ (20 ng/mL, PeproTech, Cranbury, NJ, USA) (Zhang et al., 2020). miR-330-5p mimics, miR-330-5p inhibitors and negative control (NC) were obtained from General Biology Co., Ltd. (Chuzhou, Anhui Province, China).
PBM treatment in in vivo and in vitro models
A previous percutaneous irradiation model was modified for subcutaneous irradiation in mice (Ma et al., 2022). SCI mice were anesthetized and fixed. The low-level laser fiber was placed subcutaneously in the injured area and mice were irradiated (808 ± 5 nm) once a day for 1 hour each time for 28 days. In the in vitro experiment, cells were placed on an ultraclean worktable and subjected to 808 ± 5 nm low-level laser irradiation every 12 hours for 48 hours. The parameters for PBM are described in detail in Additional Table 1.
Additional Table 1.
Specific parameters of photobiomodulation
| Parameter | In vivo | In vitro |
|---|---|---|
| Center wavelength (nm) | 808 | 808 |
| Spectral bandwidth (nm) | <5 | <5 |
| Operating mode | Continuous wave | Continuous wave |
| Frequency (Hz) | 50 kHz | 50 kHz |
| Beam spot size at target (cm²) | 0.2 | 4.5 |
| Irradiance at garget (mW/cm²) | 50 | 6 |
| Exposure duration (min) | 50 | 7 |
| Radiant exposure (J/cm²) | 150 | 2.52 |
| Radiant energy (J) | 30 | 11.34 |
| Number of points irradiated | Once a day | Twice a day |
RNA sequencing
BMDMs were harvested after treatment, and total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). mRNA enrichment, fragmentation, reverse transcription, library construction, sequencing, and data analysis were performed at Genergy Biotechnology Co. Ltd. (Shanghai, China). The thresholds for determining differentially expressed transcripts were P < 0.05 and fold change ≥ 1. Then, differentially expressed transcripts were evaluated by functional and pathway enrichment analyses using the Gene Ontology (GO; http://geneontology.org/) (Ashburner et al., 2000; Gene Ontology Consortium, 2021) and Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.kegg.jp/) databases. Significantly enriched pathways were determined when P < 0.05 and at least two genes were involved. Protein-protein interaction (PPI) networks were constructed using the STRING database (https://cn.string-db.org/) (Szklarczyk et al., 2021). The miRanda database (http://www.miranda.org/) was used to build a competing endogenous RNA (ceRNA) network with long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs).
Western blot assay
BMDMs were washed with phosphate-buffered saline after treatment, and incubated with radioimmunoprecipitation assay buffer containing phosphatase inhibitor. In the in vivo experiment, mice were deeply anesthetized and sacrificed at 3, 7, 14, and 28 days post-operation, and a 0.5-cm segment of spinal cord centered on the injury site was collected. Radioimmunoprecipitation assay buffer containing phosphatase inhibitor was added for grinding. All proteins were harvested after being digested at 4°C for 20 minutes. The samples were transferred to a 1.5 mL centrifuge tube and centrifuged for 20 minutes at 4°C and 12,000 × g. The precipitate was discarded. Bicinchoninic acid protein analysis kits (Pierce, Rockford, IL, USA) were used to detect protein concentrations. The total protein extracts were separated by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat milk. Next, the membranes were incubated with the corresponding antibodies overnight at 4°C. The specific primary antibodies are as follows: rabbit anti-signal transducer and activator of transcription 3 (STAT3, 1:1000, Cell Signaling Technology, Danvers, MA, USA, Cat# 12640, RRID: AB_2629499), rabbit anti-inducible nitric oxide synthase (iNOS, 1:1000, Cell Signaling Technology, Cat# 13120, RRID: AB_2687529), mouse anti-CD86 (1:500, Santa Cruz Biotechnology, Dallas, TX, USA, Cat# sc-28347, RRID: AB_627200) and mouse anti-β-actin (1:1000, Proteintech, Wuhan, Hubei Province, China, Cat# 66009-1-Ig, RRID: AB-2687938). The membrane was washed with phosphate-buffered saline/Tween at room temperature (25°C). Subsequently, the secondary antibodies were added and incubated for 1 hour at room temperature (goat anti-rabbit horseradish peroxidase antibody, 1:3000, InCellGene, Cimarron Path San Antonio, TX, USA, Cat# SA-10011; goat anti-mouse horseradish peroxidase antibody, 1:3000, InCellGene, Cat# SA-10010). After samples were washed with phosphate-buffered saline/Tween, the chemiluminescence image was obtained with an Amersham Imager 600 (GE Healthcare, Stockholm, Sweden). The experiment was repeated three times and ImageJ software (v1.4.67; National Institutes of Health, Bethesda, MD, USA) (Schneider et al., 2012) was used to calculate the optical density of proteins.
Quantitative polymerase chain reaction
As described in a previous study (Ju et al., 2018), total RNA was extracted from BMDMs using the TRIzol reagent (Invitrogen). RT mix was used to obtain complementary DNA. SYBR Select Master Mix was used for quantitative polymerase chain reaction (PCR). CFX (Invitrogen) and the CFX Connect Real-time PCR System (Bio-Rad, Hercules, CA, USA) settings were 95°C for 15 seconds, followed by 40 cycles of 95°C for 5 seconds and 60°C for 34 seconds. The data were analyzed with the 2–ΔΔCt method (Livak and Schmittgen, 2001). MicroRNA (miRNA) primers and the internal reference U6 were synthesized by General Biology Co., Ltd. Sequences of all primers were shown in Additional Table 2.
Additional Table 2.
The primer sequences used in this study
| Gene | Sequence (5’-3’) |
|---|---|
| mmu-miR-330-5p | Forward: CCC TCT CTG GGC CTG TGT CTT AG Reverse: ATC CAG TGC AGG GTC CGA GG |
| U6 | Forward: GGA ACG ATA CAG AGA AGA TTA GC |
| Reverse: TGG AAC GCT TCA CGA ATT TGC G |
Luciferase assay
PmirGLO-mutant (Mut) STAT3 3′-untranslated regions (3′-UTRs, General Biology Co., Ltd.) or PmirGLO-wild type (WT) STAT3 3′-UTRs (General Biology Co., Ltd.) and miR-330-5p mimics or NC mimics (General Biology Co., Ltd.) were cotransfected into 293T cells (National Collection of Authenticated Cell Culture, Shanghai, China; Cat# GNHu17). The Luciferase Reporter Assay System (E1910, Promega, Madison, WI, USA) was used to detect luciferase activity.
Luminex assay
After 7 days, spinal cord tissue from the sham, SCI, and SCI + PBM groups were collected. Before the experiment, the lysate (protease inhibitor) was added at 150–250 µL per 20 mg of tissue then ground for 60 seconds, and the sample was diluted twice. The Mouse Premixed Multi-Analyte Kit (R&D, Minneapolis, MN, USA; Cat# LXSAMSM-10) was maintained at room temperature for approximately 30 minutes before use. Data were read using the Luminex200 instrument (Luminex Corp., Austin, TX, USA).
Functional assessment
The Basso Mouse Scale (BMS) was used to evaluate the motor function of the mice hindlimbs on days 1, 3, 7, 14, and 28 after injury and was scored from 0 to 9 according to the previously described scoring system (posterior ankle mobility, coordination, paw posture, trunk stability, and tail posture, higher scores representing better functional recovery) (Basso et al., 2006).
The Louisville swimming scale (LSS) is an 18-point scale (0–17) divided into 0–5, 6–11, and 12–17. It is usually used to assess the recovery of hindlimb motor function in mice, with higher scores representing better functional recovery. The LSS assesses swimming performance based on three main components: forelimb dependence, hindlimb activity, and body posture (Smith et al., 2006). Two experienced researchers who were not involved in the experiment conducted the evaluation.
Statistical analysis
Statistics were analyzed using GraphPad Prism (version 9.0.0 for Windows, GraphPad Software, San Diego, CA, USA, www.graphpad.com). Two groups were compared using Student’s t-test, and multiple groups were compared using one-way analysis of variance with least significant difference test. The measured data were expressed as mean ± standard deviation. P < 0.05 was considered statistically significant.
Results
Identification of differentially expressed mRNAs, lncRNAs, and circRNAs
We extracted and cultured BMDMs in vitro and sequenced cells after M1 induction and irradiation. Screening of the sequencing results identified 18,176 differentially expressed mRNAs, 1712 differentially expressed lncRNAs, and 71 differentially expressed circRNAs between M0 and M1 macrophages (Figure 1A). We also identified 3612 differentially expressed mRNAs, 218 differentially expressed lncRNAs, and 25 differentially expressed circRNAs between M1 and M1 + PBM macrophages (Figure 1B). In summary, we detected extensive expression differences between M0, M1, and M1 + PBM macrophages. Additionally, we obtained the intersection of the differentially expressed mRNAs, lncRNAs, and circRNAs in the M0 vs. M1 and M1 vs. M1 + PBM groups, as visualized in a Venn diagram in Figure 2.
Figure 1.
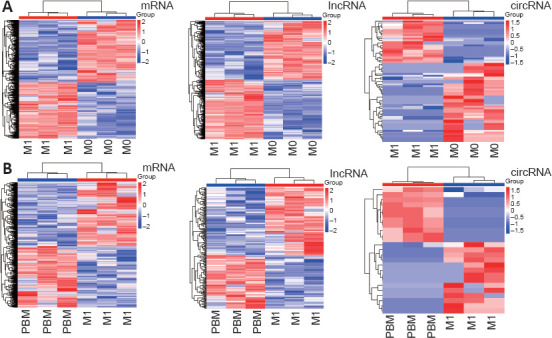
Identification of differentially expressed mRNAs, lncRNAs, and circRNAs in M0 vs. M1 and M1 vs. M1 + PBM macrophages.
(A) Differentially expressed mRNAs, lncRNAs, and circRNAs between M0 and M1 macrophages identified by RNA sequencing (three samples per group). (B) Differentially expressed mRNAs, lncRNAs, and circRNAs between M1 and M1 + PBM macrophages. Each line represents a gene; red indicates upregulation and blue indicates downregulation. circRNAs: Circular RNAs; lncRNAs: long noncoding RNAs; PBM: photobiomodulation.
Figure 2.
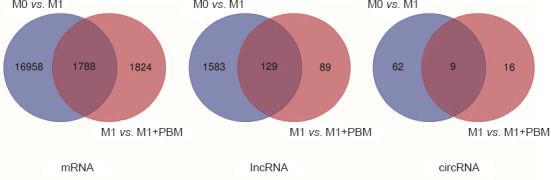
Venn diagram of differentially expressed genes in the M0 vs. M1 and M1 vs. M1 + PBM groups.
The intersection of differentially expressed genes in the M0 vs. M1 comparison and M1 vs. M1+PBM comparison is presented. circRNA: Circular RNA; lncRNA: long noncoding RNA; PBM: photobiomodulation.
GO enrichment analysis results of differentially expressed genes
We performed a GO enrichment analysis of the overlapping differentially expressed genes in the M0 vs. M1 and M1 vs. M1 + PBM groups (Figure 3A–C). In the biological process category, the differentially expressed mRNAs were mainly enriched for cellular metabolic processes. In the cellular component category, the differentially expressed mRNAs were mainly expressed in intracellular, membrane-bounded organelles. In the molecular function category, the differentially expressed mRNAs were mostly related to protein binding. The lncRNAs were mainly enriched in the GO terms antigen processing and presentation, major histocompatibility complex class II protein complex, and major histocompatibility complex class II protein complex binding. The circRNAs were mainly enriched in the intracellular distribution of mitochondria, cyclin K-cyclin-dependent kinase 13 complex, and phosphatidylinositol 3-kinase regulator activity. These results suggest potential biological processes by which differentially expressed genes induced by PBM inhibit M1 macrophage polarization.
Figure 3.
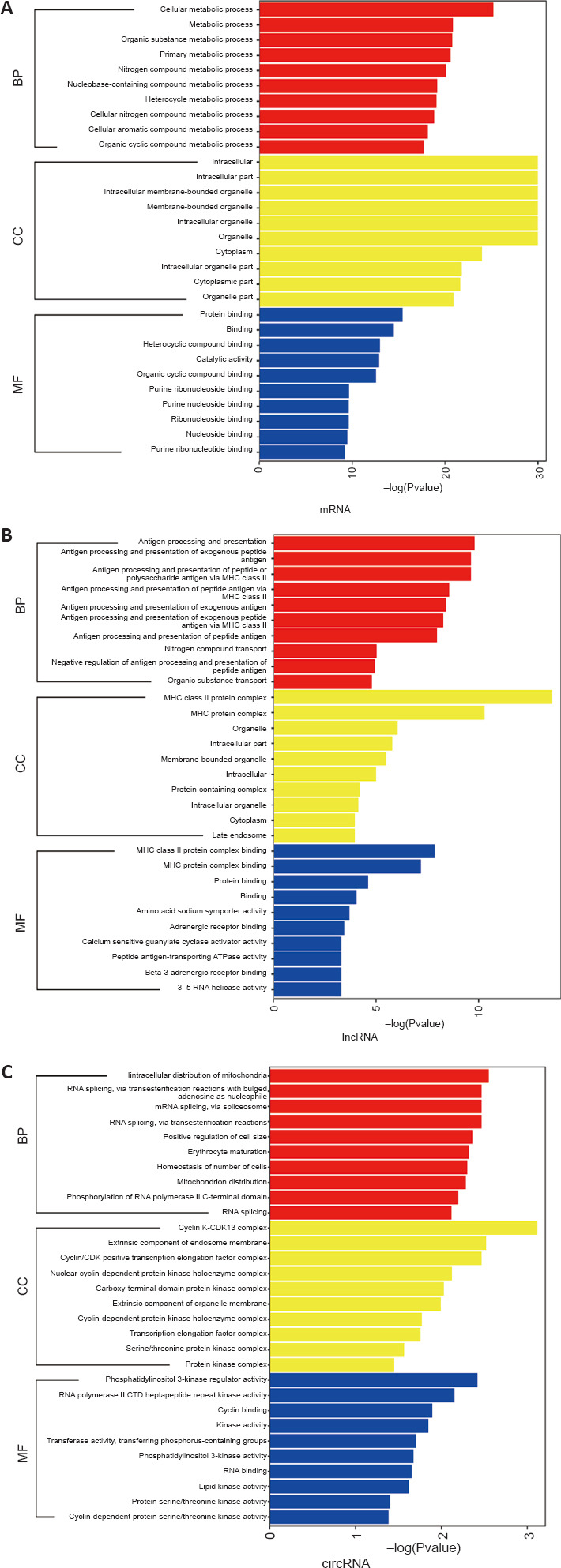
GO enrichment analysis of differentially expressed genes between M0 vs. M1 and M1 vs. M1 + PBM groups.
(A–C) GO enrichment analysis of differentially expressed genes in mRNA (A), lncRNA (B) and circRNA (C), according to three categories: biological process, cellular component, and molecular function. circRNA: Circular RNA; GO: Gene Ontology; lncRNA: long noncoding RNA; PBM: photobiomodulation.
KEGG pathway enrichment analysis results of differentially expressed genes
Additionally, we conducted a KEGG enrichment analysis of all differentially expressed genes (Figure 4A–C). The differentially expressed mRNAs, lncRNAs, and circRNAs were mainly involved in inflammation-related pathways, such as the nuclear factor-κB signaling pathway, tumor necrosis factor signaling pathway, phosphatidylinositol 3-kinase/protein kinase B (AKT) signaling pathway, and Janus kinase/STAT signaling pathway. These signaling pathways may play essential roles in M1 macrophages.
Figure 4.
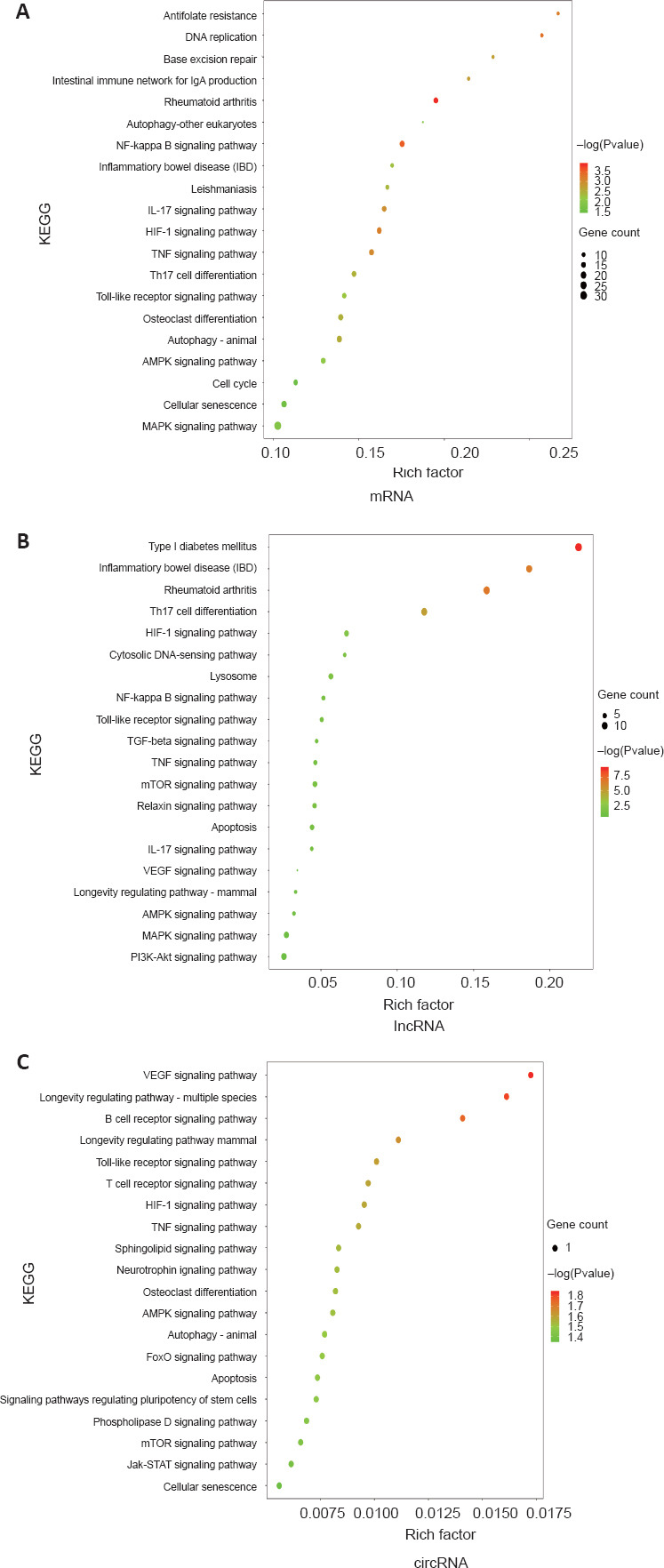
KEGG pathway enrichment analysis of differentially expressed genes between M0 vs. M1 and M1 vs. M1 + PBM groups.
(A–C) KEGG pathway enrichment analysis of differentially expressed genes in mRNA (A), lncRNA (B) and circRNA (C), which suggested important roles of inflammation-related pathways. The size of the circle represents the number of genes. Darker red indicates a higher P-value. circRNA: Circular RNA; KEGG: Kyoto Encyclopedia of Genes and Genomes; lncRNA: long noncoding RNA; PBM: photobiomodulation.
PPI network analysis results
With the differentially expressed mRNAs summarized in Figure 2, we constructed a PPI network using the STRING database. The network indicated potential interactions among the genes. The PPI network based on the top 50 differentially expressed genes was presented in Figure 5.
Figure 5.
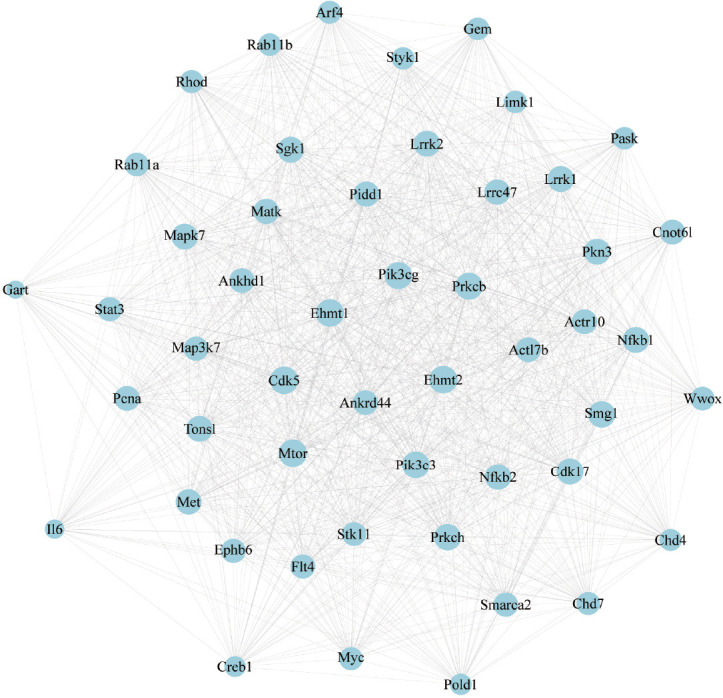
PPI network analysis of differentially expressed genes between M0 vs. M1 and M1 vs. M1 + PBM groups.
PPI network of the interactions among the top 50 genes. Interactions between genes are indicated by a straight line. PBM: Photobiomodulation; PPI: protein-protein interaction.
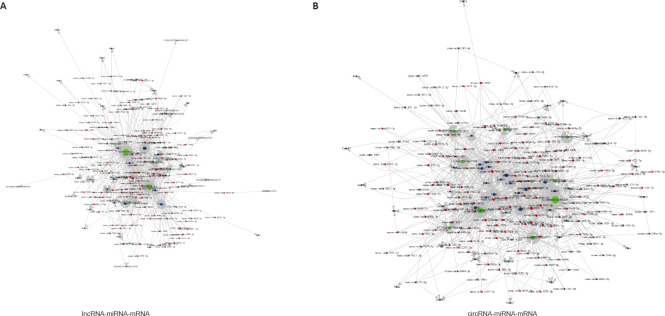
Construction of lncRNA-miRNA-mRNA and circRNA-miRNA-mRNA networks
In the PPI network, multiple genes were associated with inflammation, such as IL-6, STAT3, and SGK1. We identified key genes by building ceRNA networks with lncRNAs and circRNAs using the top 50 genes in the PPI network (Figure 6A and B). We found that the miR-330-5p/STAT3 axis was included in both ceRNA networks. STAT3 and miR-330-5p are involved in the inflammatory response. STAT3 promoted inflammation, whereas miR-330-5p inhibited inflammation in a lung injury model (Yu et al., 2021, 2022). In addition, in the RNA sequencing analysis, STAT3 was highly expressed in M1 macrophages and was decreased after PBM. Therefore, miR-330-5p/STAT3 may mediate the effects of PBM and was evaluated in subsequent experiments.
Figure 6.
Construction of lncRNA-miRNA-mRNA (A) and circRNA-miRNA-mRNA (B) networks.
Red represents miRNA, green represents lncRNA, and blue represents mRNA. Connections are represented by a straight line. circRNA: Circular RNA; lncRNA: long noncoding RNA; miRNA: microRNA.
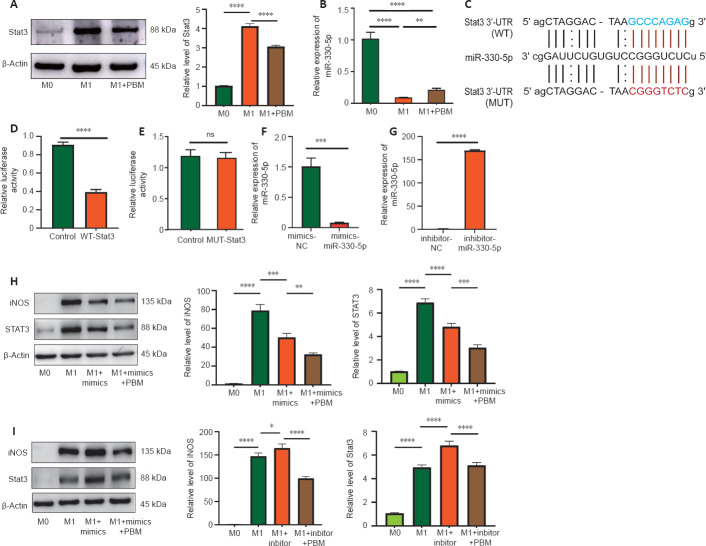
Regulatory effects of the miR-330-5p/STAT3 axis on M1 polarization of BMDMs
We detected the expression of miR-330-5p and STAT3 in macrophages. According to western blotting results, STAT3 expression in M1 macrophages was higher than that in M0 macrophages, and was decreased after PBM treatment (Figure 7A). Quantitative PCR results showed that miR-330-5p expression in M1 macrophages was lower than that in M0 macrophages, and was increased after treatment with PBM (Figure 7B). We then predicted the binding sites of miR-330-5p and STAT3 using StarBase (Figure 7C). The luciferase reporter assay showed that the activity of WT-STAT3 in miR-330-5p mimics was reduced, but the activity of Mut-STAT3 was not changed. These results showed that miR-330-5p binds to STAT3 directly (Figure 7D and E). Furthermore, we transfected miR-330-5p mimics and inhibitors into macrophages. Quantitative PCR showed that, compared with that after the transfection with NC, the expression of miR-330-5p was higher after the transfection of miR-330-5p mimics and was lower after the transfection of the miR-330-5p inhibitor, indicating that transfection was successful (Figure 7F and G). Western blot assay showed that the transfection of miR-330-5p mimics into M1 macrophages inhibited STAT3 and iNOS expression, with further reductions in expression after the combined use of miR-330-5p mimics and PBM (Figure 7H). In contrast, transfection of miR-330-5p inhibitors increased STAT3 and iNOS expression in M1 macrophages, and PBM attenuated the miR-330-5p inhibitor-induced increases in STAT3 and iNOS expression (Figure 7I). These findings showed that miR-330-5p, a potential target of PBM, reduced STAT3 expression and macrophage polarization.
Figure 7.
Regulatory effects of the miR-330-5p/STAT3 axis on M1 polarization of BMDMs.
(A) STAT3 protein expression, as evaluated by western blotting, in M0, M1, and M1 + PBM macrophages. Data were normalized by M0. (B) qPCR detection of miR-330-5p expression in M0, M1, and M1 + PBM macrophages. (C) Bioinformatics analysis identified a potential miR-330-5p binding site of STAT3. (D, E) Luciferase reporter assay demonstrating the luciferase activity of cells cotransfected with miR-335-5p mimics and WT-STAT3 or Mut-STAT3. (F, G) qPCR results for the expression of miR-330-5p mimics and inhibitors. (H, I) Western blotting analysis of iNOS and STAT3 expression. Data were normalized by M0. The data are expressed as mean ± SD (n = 6 mice per group). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (one-way analysis of variance followed by least significant difference test [A, B, H, I] or Student’s t-test [D–G]). 3′-UTR: 3′-Untranslated region; BMDM: bone marrow-derived macrophage; iNOS: inducible nitric oxide synthase; Mut: mutant; ns: not significant; PBM: photobiomodulation; qPCR: quantitative polymerase chain reaction; STAT3: signal transducers and activators of transcription 3; WT: wild type.
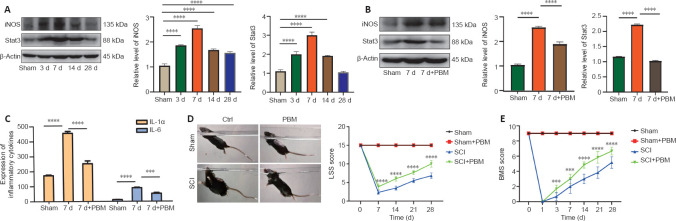
PBM reduces STAT3 and iNOS expression and promotes SCI repair in vivo
We collected spinal cord tissue for western blotting at 3, 7, 14, and 28 days after SCI. The expression levels of iNOS and STAT3 were highest on day 7 after injury and decreased on day 14 after injury compared with sham groups (Figure 8A). Furthermore, after 7 days of treatment with PBM, iNOS and STAT3 expression levels were decreased compared with SCI groups (Figure 8B). We also detected the expression of CD86, which was consistent with iNOS expression (Additional Figure 2A (1.7MB, tif) and B (1.7MB, tif) ). In addition, a Luminex assay showed that the levels of inflammatory factors IL-1α and IL-6 also were decreased after 7 days of PBM treatment compared with SCI groups. These results showed that PBM inhibited STAT3 and iNOS expression in mice after SCI, which was consistent with the results of the in vitro assays (Figure 8C). Furthermore, we evaluated the motor functional recovery of SCI mice after PBM treatment. After 28 days of PBM treatment, the BMS and LSS scores of mice were increased compared with SCI groups, indicating that PBM promoted the recovery of motor function after SCI (Figure 8D and E). Overall, our data showed that PBM had a significant therapeutic effect on SCI model mice.
Figure 8.
PBM reduces STAT3 and iNOS expression in the injured spinal cord and promotes SCI repair in vivo.
(A) Western blotting of iNOS and STAT3 expression (n = 6 mice per group). (B) Western blotting of iNOS and STAT3 expression after 7 days of SCI and PBM treatment (n = 6 mice per group). (C) Luminex assay was used to detect the expression of inflammatory cytokines (n = 3 mice per group). (D, E) BMS and LSS scores were used to evaluate motor function recovery in mice after PBM treatment (n = 6 mice per group). Higher LSS scores indicate better recovery of motor function. Motor function of the sham group and sham + PBM group were the same. Motor function in the PBM group was better than that in SCI group. The data are expressed as mean ± SD. ***P < 0.001, ****P < 0.0001, vs. SCI group (one-way analysis of variance followed by least significant difference test [A, B, H, I] or Student’s t-test [D–F]). BMS: Basso Mouse Scale; IL: interleukin; iNOS: inducible nitric oxide synthase; LSS: Louisville swimming scale; PBM: photobiomodulation; SCI: spinal cord injury; STAT3: signal transducers and activators of transcription 3.
Discussion
The secondary inflammatory response is critical for functional recovery after SCI, and BMDMs are critical for the regulation of secondary inflammation (Gensel and Zhang, 2015). Therefore, strategies to effectively change the polarization direction of BMDMs and regulate the inflammatory response may be valuable for SCI repair. Genome sequencing can identify genes associated with disease occurrence and development (Wu et al., 2021; Young et al., 2022). In this study, we performed transcriptome sequencing for the first time at the cellular level for PBM regulation of macrophage polarization, which included lncRNAs, mRNAs, and circRNAs. These results indicate potential target genes that warrant further investigation.
To identify the biological features and signaling pathways associated with these differential genes, GO and KEGG analyses were performed. GO enrichment analysis is performed from three aspects: biological process, cellular component and molecular function. The GO analysis indicated that the differentially expressed genes were enriched in immune and metabolic responses, which is consistent with the biological function of macrophages (Greene et al., 2022; Pan et al., 2022). This provides reliable information to help further our understanding of macrophage regulation of PBM. The KEGG enrichment analysis identified that multiple common signaling pathways were altered, such as the nuclear factor-κB signaling pathway, hypoxia-inducible factor-1 signaling pathway, tumor necrosis factor signaling pathway, phosphatidylinositol 3-kinase/AKT signaling pathway, and Janus kinase/STAT signaling pathway. These signaling pathways have been reported to be involved in nerve injury, oxidative stress, osteogenic differentiation and inflammatory response (Gao et al., 2022; Geng et al., 2022; Lin et al., 2022; Qiu et al., 2022; Wang et al., 2022b, c). In SCI, selective inhibition of nuclear factor-κB in astrocytes improved SCI recovery (Brambilla et al., 2005). Autophagy induced by activation of the hypoxia-inducible factor-1α/BNIP3 signaling pathway promoted nerve survival and axonal regeneration (Li et al., 2019). Elimination of tumor necrosis factor in macrophages and neutrophils led to a decrease in lesion volume after SCI (Ellman et al., 2020). Activation of the phosphatidylinositol 3-kinase/AKT pathway reduced endothelial cell apoptosis and microvascular injury induced by SCI (Li et al., 2020). This finding provides a direction for further study of the signaling pathways that may be involved in PBM. Furthermore, we conducted a PPI analysis of the differential genes to assess molecular interactions. Our results showed interactions among the top 50 differentially expressed genes, of which many have been reported to be involved in inflammatory response. For example, inhibition of STAT3 reduced renal ischemia/reperfusion injury, and miR-221-3p inhibited inflammatory response in osteoarthritis by targeting Janus kinase 3/STAT3 (Quero et al., 2019; Wang et al., 2022a). SGK1 plays an important role in pulmonary fibrosis, diabetic renal fibrosis and liver cirrhosis by upregulating the nuclear factor-κB pathway and then stimulating the expression of many inflammatory mediators (Lu et al., 2022). Nfkb2 increases the proinflammatory gene response driven by RelA in intestinal epithelial cells (Chawla et al., 2021). The signaling pathways associated with these top genes were consistent with the KEGG analysis.
In addition, we constructed the ceRNA network using the top 50 genes (lncRNA-miRNA-mRNA, and circRNA-miRNA-mRNA). lncRNA is involved in the occurrence of various diseases and performs biological functions through molecular scaffolds, RNA-binding proteins, or ceRNAs (Ju et al., 2018, 2019; Liu et al., 2021). circRNAs are noncoding covalently closed circular RNAs produced by alternative splicing. The 3′ and 5′ ends of circRNAs are covalently linked to a covalent closed-loop structure without a free end. These RNAs are evolutionarily conserved and form a ceRNA interaction with miRNAs (Li et al., 2018; Patop et al., 2019; Chen, 2020; Xiao et al., 2020). The importance of the interaction between miRNAs and mRNAs, which degrades mRNAs and inhibits their function, is now well-established (Borchert et al., 2006; Krol et al., 2010). These analyses are helpful for us to elucidate the potential mechanism of PBM regulating macrophage polarization from the perspective of noncoding RNA.
Finally, our study verified the interaction between miR-330-5p and STAT3. In previous studies, enhancing miR-330-5p expression inhibited the progression of bladder cancer and pancreatic cancer (Chen et al., 2019, 2020). It was also reported that miR-330-5p inhibited intervertebral disc degeneration by targeting cartilage intermediate layer protein (Li et al., 2021). Furthermore, upregulation of miR-330-5p expression inhibited oxidative stress and macrophage inflammation (Liu et al., 2019), and miR-330-5p targeting TIM3 inhibited myocardial ischemia-reperfusion injury induced by NLRP3 activation (Zuo et al., 2021). We found that miR-330-5p reduced STAT3 expression and inhibited M1 macrophage polarization. The results in vivo were consistent with those at the cellular level. iNOS and STAT3 expression were increased after SCI and decreased after PBM treatment. The PBM treatment improved motor function, which indicates that our PBM protocol has a therapeutic effect on mice after SCI at both the cellular and animal levels. This finding provides theoretical support for the clinical application of PBM.
There were some limitations in this study to address. First, the bioinformatics analysis in this experiment was performed only at the cellular level, which may differ from the tissue level. Therefore, in a future study, we intend to perform tissue-level transcriptome sequencing and combined analysis with cellular-level transcriptome sequencing, which will be important to better clarify the therapeutic value of PBM. Second, at the animal level, we only made a preliminary observation of the phenotype, and the mechanism has not been explained. This will need to be studied in depth in future studies.
In summary, through transcriptome sequencing and bioinformatics analysis, we identified the gene expression profile of PBM regulation of macrophage polarization and analyzed the functional enrichment of differential genes. These findings are important for understanding the potential key pathways and genes involved in PBM regulation of macrophage polarization, and are helpful for clarifying the potential mechanism of PBM in the treatment of SCI.
Additional files:
Additional Table 1: Specific parameters of photobiomodulation.
Additional Table 2: The primer sequences used in this study.
Additional Figure 1 (935.5KB, tif) : Animal grouping.
Animal grouping.
BMS: Basso Mouse Scale; LSS: Louisville swimming scale; PBM: photobiomodulation; SCI: spinal cord injury.
Additional Figure 2 (1.7MB, tif) : PBM reduces CD86 expression in SCI mice.
PBM reduces CD86 expression in SCI mice.
(A) Expression of CD86 in control group and at 3, 7, 14, and 28 days after SCI (n = 6 mice per group). (B) Expression of CD86 after 7 days of SCI and PBM treatment. The data are expressed as mean ± SD (n = 6 mice per group). **P < 0.01, ***P < 0.001, ****P < 0.0001 (one-way analysis of variance followed by least significant difference test). PBM: Photobiomodulation; SCI: spinal cord injury.
Footnotes
Funding: This study was supported by the National Natural Science Foundation of China, Nos. 81070996 (to ZW), 81572151 (to XYH); and Shaanxi Provincial Key R&D Program, China, Nos. 2020ZDLSF02-05 (to ZW), 2021ZDLSF02-10 (to XYH).
Conflicts of interest: The authors declare that they have no competing interests.
Availability of data and materials: All raw sequencing data has been uploaded to NCBI database (accession number PRJNA780778). In addition, the datasets used in the project are available from the corresponding author.
C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: McCollum L, Yu J, Song LP; T-Editor: Jia Y
References
- 1.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology:tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. (2000);25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assunção Silva RC, Pinto L, Salgado AJ. Cell transplantation and secretome based approaches in spinal cord injury regenerative medicine. Med Res Rev. (2022);42:850–896. doi: 10.1002/med.21865. [DOI] [PubMed] [Google Scholar]
- 3.Basso DM, Fisher LC, Anderson AJ, Jakeman LB, McTigue DM, Popovich PG. Basso Mouse Scale for locomotion detects differences in recovery after spinal cord injury in five common mouse strains. J Neurotrauma. (2006);23:635–659. doi: 10.1089/neu.2006.23.635. [DOI] [PubMed] [Google Scholar]
- 4.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. (2006);13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 5.Brambilla R, Bracchi-Ricard V, Hu WH, Frydel B, Bramwell A, Karmally S, Green EJ, Bethea JR. Inhibition of astroglial nuclear factor kappaB reduces inflammation and improves functional recovery after spinal cord injury. J Exp Med. (2005);202:145–156. doi: 10.1084/jem.20041918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao QQ, Li S, Lu Y, Wu D, Feng W, Shi Y, Zhang LP. Transcriptome analysis of molecular mechanisms underlying facial nerve injury repair in rats. Neural Regen Res. (2021);16:2316–2323. doi: 10.4103/1673-5374.310700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chawla M, Mukherjee T, Deka A, Chatterjee B, Sarkar UA, Singh AK, Kedia S, Lum J, Dhillon MK, Banoth B, Biswas SK, Ahuja V, Basak S. An epithelial Nfkb2 pathway exacerbates intestinal inflammation by supplementing latent RelA dimers to the canonical NF-κB module. Proc Natl Acad Sci U S A. (2021);118:e2024828118. doi: 10.1073/pnas.2024828118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Yang X, Zhao J, Xiong M, Almaraihah R, Chen Z, Hou T. Circ_0008532 promotes bladder cancer progression by regulation of the miR-155-5p/miR-330-5p/MTGR1 axis. J Exp Clin Cancer Res. (2020);39:94. doi: 10.1186/s13046-020-01592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. (2020);21:475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 10.Chen S, Chen JZ, Zhang JQ, Chen HX, Qiu FN, Yan ML, Tian YF, Peng CH, Shen BY, Chen YL, Wang YD. Silencing of long noncoding RNA LINC00958 prevents tumor initiation of pancreatic cancer by acting as a sponge of microRNA-330-5p to down-regulate PAX8. Cancer letters. (2019);446:49–61. doi: 10.1016/j.canlet.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 11.Ellman DG, Lund MC, Nissen M, Nielsen PS, Sørensen C, Lester EB, Thougaard E, Jørgensen LH, Nedospasov SA, Andersen DC, Stubbe J, Brambilla R, Degn M, Lambertsen KL. Conditional ablation of myeloid TNF improves functional outcome and decreases lesion size after spinal cord injury in mice. Cells. (2020);9:2407. doi: 10.3390/cells9112407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farooque M, Suo Z, Arnold PM, Wulser MJ, Chou CT, Vancura RW, Fowler S, Festoff BW. Gender-related differences in recovery of locomotor function after spinal cord injury in mice. Spinal Cord. (2006);44:182–187. doi: 10.1038/sj.sc.3101816. [DOI] [PubMed] [Google Scholar]
- 13.Gao Y, Hu M, Niu X, Li M, Xu L, Xiao Y, Zhang J, Wang H, Li L, Chu B, Lv P. Dl-3-n-butylphthalide improves neuroinflammation in mice with repeated cerebral ischemia-reperfusion injury through the Nrf2-mediated antioxidant response and TLR4/MyD88/NF-κB signaling pathway. Oxid Med Cell Longev. (2022);2022:8652741. doi: 10.1155/2022/8652741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gene Ontology Consortium (2021) The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng B, Wang X, Park KH, Lee KE, Kim J, Chen P, Zhou X, Tan T, Yang C, Zou X, Janssen PM, Cao L, Ye L, Wang X, Cai C, Zhu H. UCHL1 protects against ischemic heart injury via activating HIF-1αsignal pathway. Redox biology. (2022);52:102295. doi: 10.1016/j.redox.2022.102295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gensel JC, Zhang B. Macrophage activation and its role in repair and pathology after spinal cord injury. Brain Res. (2015);1619:1–11. doi: 10.1016/j.brainres.2014.12.045. [DOI] [PubMed] [Google Scholar]
- 17.Greene CJ, Nguyen JA, Cheung SM, Arnold CR, Balce DR, Wang YT, Soderholm A, McKenna N, Aggarwal D, Campden RI, Ewanchuk BW, Virgin HW, Yates RM. Macrophages disseminate pathogen associated molecular patterns through the direct extracellular release of the soluble content of their phagolysosomes. Nat Commun. (2022);13:3072. doi: 10.1038/s41467-022-30654-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffey CJ, Yamamoto A. Macroautophagy in CNS health and disease. Nat Rev Neurosci. (2022);23:411–427. doi: 10.1038/s41583-022-00588-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang W, Li M, He F, Zhou S, Zhu L. Targeting the NLRP3 inflammasome to attenuate spinal cord injury in mice. J Neuroinflammation. (2017);14:207. doi: 10.1186/s12974-017-0980-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ju C, Zhou R, Sun J, Zhang F, Tang X, Chen KK, Zhao J, Lan X, Lin S, Zhang Z, Lv XB. LncRNA SNHG5 promotes the progression of osteosarcoma by sponging the miR-212-3p/SGK3 axis. Cancer Cell Int. (2018);18:141. doi: 10.1186/s12935-018-0641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ju C, Liu R, Zhang YW, Zhang Y, Zhou R, Sun J, Lv XB, Zhang Z. Mesenchymal stem cell-associated lncRNA in osteogenic differentiation. Biomed Pharmacother. (2019);115:108912. doi: 10.1016/j.biopha.2019.108912. [DOI] [PubMed] [Google Scholar]
- 22.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. (2010);11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 23.Li H, Kong R, Wan B, Yang L, Zhang S, Cao X, Chen H. Initiation of PI3K/AKT pathway by IGF-1 decreases spinal cord injury-induced endothelial apoptosis and microvascular damage. Life Sci. (2020);263:118572. doi: 10.1016/j.lfs.2020.118572. [DOI] [PubMed] [Google Scholar]
- 24.Li S, Liu J, Chen L. MiR-330-5p inhibits intervertebral disk degeneration via targeting CILP. J Orthop Surg Res. (2021);16:440. doi: 10.1186/s13018-021-02582-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. (2018);71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Han W, Wu Y, Zhou K, Zheng Z, Wang H, Xie L, Li R, Xu K, Liu Y, Wang X, Xiao J. Stabilization of hypoxia inducible factor-1αby dimethyloxalylglycine promotes recovery from acute spinal cord injury by inhibiting neural apoptosis and enhancing axon regeneration. J Neurotrauma. (2019);36:3394–3409. doi: 10.1089/neu.2018.6364. [DOI] [PubMed] [Google Scholar]
- 27.Lin MJ, Lin P, Wen KC, Chiang HM, Lu MC. Jelly Fig (Ficus awkeotsang Makino) Exhibits antioxidative and anti-inflammatory activities by regulating reactive oxygen species production via NFκB signaling pathway. Antioxidants (Basel) (2022);11:981. doi: 10.3390/antiox11050981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Huang GQ, Ke ZP. Silence of long intergenic noncoding RNA HOTAIR ameliorates oxidative stress and inflammation response in ox-LDL-treated human macrophages by upregulating miR-330-5p. J Cell Physiol. (2019);234:5134–5142. doi: 10.1002/jcp.27317. [DOI] [PubMed] [Google Scholar]
- 29.Liu SJ, Dang HX, Lim DA, Feng FY, Maher CA. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer. (2021);21:446–460. doi: 10.1038/s41568-021-00353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. (2001);25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Lu RQ, Zhang YY, Zhao HQ, Guo RQ, Jiang ZX, Guo R. SGK1, a critical regulator of immune modulation and fibrosis and a potential therapeutic target in chronic graft-versus-host disease. Front Immunol. (2022);13:822303. doi: 10.3389/fimmu.2022.822303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Y, Li P, Ju C, Zuo X, Li X, Ding T, Liang Z, Zhang J, Li K, Wang X, Zhu Z, Zhang Z, Song Z, Quan H, Hu X, Wang Z. Photobiomodulation attenuates neurotoxic polarization of macrophages by inhibiting the Notch1-HIF-1α/NF-κB signalling pathway in mice with spinal cord injury. Front Immunol. (2022);13:816952. doi: 10.3389/fimmu.2022.816952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan X, Zhu Q, Pan LL, Sun J. Macrophage immunometabolism in inflammatory bowel diseases: From pathogenesis to therapy. Pharmacol Ther. (2022);238:108176. doi: 10.1016/j.pharmthera.2022.108176. [DOI] [PubMed] [Google Scholar]
- 34.Pan Y, Abdureyim M, Yao Q, Li X. Analysis of differentially expressed genes in endothelial cells following tumor cell adhesion, and the role of PRKAA2 and miR-124-3p. Front Cell Dev Biol. (2021);9:604038. doi: 10.3389/fcell.2021.604038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J. (2019);38:e100836. doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu Y, Yang J, Ma L, Song M, Liu G. Limonin isolated from pomelo seed antagonizes Aβ25-35-mediated neuron injury via PI3K/AKT signaling pathway by regulating cell apoptosis. Front Nutr. (2022);9:879028. doi: 10.3389/fnut.2022.879028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quero L, Tiaden AN, Hanser E, Roux J, Laski A, Hall J, Kyburz D. miR-221-3p drives the shift of M2-macrophages to a pro-inflammatory function by suppressing JAK3/STAT3 activation. Front Immunol. (2019);10:3087. doi: 10.3389/fimmu.2019.03087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren Y, Young W. Managing inflammation after spinal cord injury through manipulation of macrophage function. Neural Plast. (2013);2013:945034. doi: 10.1155/2013/945034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarvestani FK, Dehno NS, Nazhvani SD, Bagheri MH, Abbasi S, Khademolhosseini Y, Gorji E. Effect of low-level laser therapy on fracture healing in rabbits. Laser Ther. (2017);26:189–193. doi: 10.5978/islsm.17-OR-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ:25 years of image analysis. Nat Methods. (2012);9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith RR, Burke DA, Baldini AD, Shum-Siu A, Baltzley R, Bunger M, Magnuson DS. The Louisville Swim Scale:a novel assessment of hindlimb function following spinal cord injury in adult rats. J Neurotrauma. (2006);23:1654–1670. doi: 10.1089/neu.2006.23.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song JW, Li K, Liang ZW, Dai C, Shen XF, Gong YZ, Wang S, Hu XY, Wang Z. Low-level laser facilitates alternatively activated macrophage/microglia polarization and promotes functional recovery after crush spinal cord injury in rats. Sci Rep. (2017);7:620. doi: 10.1038/s41598-017-00553-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. The STRING database in 2021:customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. (2021);49:D605–612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tashiro S, Tsuji O, Shinozaki M, Shibata T, Yoshida T, Tomioka Y, Unai K, Kondo T, Itakura G, Kobayashi Y, Yasuda A, Nori S, Fujiyoshi K, Nagoshi N, Kawakami M, Uemura O, Yamada S, Tsuji T, Okano H, Nakamura M. Current progress of rehabilitative strategies in stem cell therapy for spinal cord injury:a review. NPJ Regen Med. (2021);6:81. doi: 10.1038/s41536-021-00191-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Xiong M, Fan Y, Liu C, Wang Q, Yang D, Yuan Y, Huang Y, Wang S, Zhang Y, Niu S, Yue J, Su H, Zhang C, Chen H, Zheng L, Huang K. Mecp2 protects kidney from ischemia-reperfusion injury through transcriptional repressing IL-6/STAT3 signaling. Theranostics. (2022a);12:3896–3910. doi: 10.7150/thno.72515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang SJ, Qiu ZZ, Chen FW, Mao AL, Bai JC, Hong YJ, Zhang ZP, Zhu WA, Zhang ZW, Zhou H. Bone marrow mesenchymal stem cell-derived extracellular vesicles containing miR-181d protect rats against renal fibrosis by inhibiting KLF6 and the NF-κB signaling pathway. Cell Death Dis. (2022b);13:535. doi: 10.1038/s41419-022-04875-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Cao W, Sun C, Wang Y, Wang M, Wu J. Development of pH-sensitive dextran-based methotrexate nanodrug for rheumatoid arthritis therapy through inhibition of JAK-STAT pathways. Int J Pharm. (2022c);622:121874. doi: 10.1016/j.ijpharm.2022.121874. [DOI] [PubMed] [Google Scholar]
- 48.Wu J, Fang Z, Liu T, Hu W, Wu Y, Li S. Maximizing the utility of transcriptomics data in inflammatory skin diseases. Front Immunol. (2021);12:761890. doi: 10.3389/fimmu.2021.761890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Y, Zhang Z, Wang F, Wang W. Current status of traumatic spinal cord injury caused by traffic accident in Northern China. Sci Rep. (2022);12:13892. doi: 10.1038/s41598-022-16930-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao MS, Ai Y, Wilusz JE. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. (2020);30:226–240. doi: 10.1016/j.tcb.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang M, Yang Z, Wang P, Sun Z. Current application and future directions of photobiomodulation in central nervous diseases. Neural Regen Res. (2021);16:1177–1185. doi: 10.4103/1673-5374.300486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young DA, Barter MJ, Soul J. Osteoarthritis year in review:genetics, genomics, epigenetics. Osteoarthritis Cartilage. (2022);30:216–225. doi: 10.1016/j.joca.2021.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu W, Haque I, Venkatraman A, Menden HL, Mabry SM, Roy BC, Xia S, Prokop JW, Umar S, Geurts AM, Sampath V. SIGIRR mutation in human necrotizing enterocolitis (NEC) disrupts STAT3-dependent microRNA expression in neonatal gut. Cell Mol Gastroenterol Hepatol. (2022);13:425–440. doi: 10.1016/j.jcmgh.2021.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu Y, Sun H, Zhu L, Ji L, Liu H. Downregulating lncRNA PRNCR1 ameliorates LPS-induced pulmonary vascular endothelial cell injury by modulating miR-330-5p/TLR4 axis. J Biochem Mol Toxicol. (2021);35:e22644. doi: 10.1002/jbt.22644. [DOI] [PubMed] [Google Scholar]
- 55.Zhang J, Sun J, Zheng Q, Hu X, Wang Z, Liang Z, Li K, Song J, Ding T, Shen X, Zhang J, Qiao L. Low-level laser therapy 810-nm up-regulates macrophage secretion of neurotrophic factors via PKA-CREB and promotes neuronal axon regeneration in vitro. J Cell Mol Med. (2020);24:476–487. doi: 10.1111/jcmm.14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng Q, Zhang J, Zuo X, Sun J, Liang Z, Hu X, Wang Z, Li K, Song J, Ding T, Shen X, Ma Y, Li P. Photobiomodulation promotes neuronal axon regeneration after oxidative stress and induces a change in polarization from M1 to M2 in macrophages via stimulation of CCL2 in neurons:relevance to spinal cord injury. J Mol Neurosci. (2021);71:1290–1300. doi: 10.1007/s12031-020-01756-9. [DOI] [PubMed] [Google Scholar]
- 57.Zuo W, Tian R, Chen Q, Wang L, Gu Q, Zhao H, Huang C, Liu Y, Li J, Yang X, Xu L, Zhang B, Liu Z. miR-330-5p inhibits NLRP3 inflammasome-mediated myocardial ischaemia-reperfusion injury by targeting TIM3. Cardiovasc Drugs Ther. (2021);35:691–705. doi: 10.1007/s10557-020-07104-8. [DOI] [PubMed] [Google Scholar]
- 58.Zuo X, Liang Z, Zhang J, Wang S, Zheng Q, Ma Y, Li P, Ding T, Hu X, Wang Z. Photobiomodulation and diffusing optical fiber on spinal cord's impact on nerve cells from normal spinal cord tissue in piglets. Lasers Med Sci. (2022);37:259–267. doi: 10.1007/s10103-020-03231-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Animal grouping.
BMS: Basso Mouse Scale; LSS: Louisville swimming scale; PBM: photobiomodulation; SCI: spinal cord injury.
PBM reduces CD86 expression in SCI mice.
(A) Expression of CD86 in control group and at 3, 7, 14, and 28 days after SCI (n = 6 mice per group). (B) Expression of CD86 after 7 days of SCI and PBM treatment. The data are expressed as mean ± SD (n = 6 mice per group). **P < 0.01, ***P < 0.001, ****P < 0.0001 (one-way analysis of variance followed by least significant difference test). PBM: Photobiomodulation; SCI: spinal cord injury.