Abstract
Glucuronoyl esterases (GEs) are microbial enzymes able to cleave covalent linkages between lignin and carbohydrates in the plant cell wall. GEs are serine hydrolases found in carbohydrate esterase family 15 (CE15), which belongs to the large α/β hydrolase superfamily. GEs have been shown to reduce plant cell wall recalcitrance by hydrolysing the ester bonds found between glucuronic acid moieties on xylan polysaccharides and lignin. In recent years, the exploration of CE15 has broadened significantly and focused more on bacterial enzymes, which are more diverse in terms of sequence and structure to their fungal counterparts. Similar to fungal GEs, the bacterial enzymes are able to improve overall biomass deconstruction but also appear to have less strict substrate preferences for the uronic acid moiety. The structures of bacterial GEs reveal that they often have large inserts close to the active site, with implications for more extensive substrate interactions than the fungal GEs which have more open active sites. In this review, we highlight the recent work on GEs which has predominantly regarded bacterial enzymes, and discuss similarities and differences between bacterial and fungal enzymes in terms of the biochemical properties, diversity in sequence and modularity, and structural variations that have been discovered thus far in CE15.
Keywords: carbohydrate esterase, enzymology, lignin-carbohydrate complex, lignocellulose, protein structure
Introduction
Lignin–carbohydrate complexes and plant cell wall recalcitrance
The plant cell wall is a complex network mainly consisting of polysaccharides. Cellulose is typically the most abundant polysaccharide and it coalesces via strong hydrogen bonding into crystalline fibres that in turn are coated and cross-linked by different heteropolysaccharides [1]. The cell wall can be further reinforced by lignin, where lignin monomers polymerize in radical coupling reactions. In order to use the plant cell wall polymers as a nutrient source, microorganisms need to produce an arsenal of different hydrolytic and oxidative enzymes to tackle this recalcitrant matrix [2]. Likewise, full utilization of renewable biomass by enzymatic processing in biorefineries requires a variety of enzymatic activities, usually formulated in so-called enzyme cocktails. A feature that is often overlooked is that during lignification, covalent bonds are formed not only between the lignin monomers but also between lignin and the exposed carbohydrates of the cell wall, particularly xylan, forming so-called lignin–carbohydrate complexes (LCCs) [1,3]. It has been estimated that virtually all lignin in softwood, and a major fraction of lignin also in hardwood, can be found covalently bound to carbohydrates [4]. Correspondingly, a significant proportion of the plant cell wall carbohydrates can be bound to lignin, as in beechwood, where around a third of the glucuronic acid (GlcA) moieties are estimated to participate in LCCs [5]. LCCs greatly add to the recalcitrance of the cell wall, and while these structures are difficult to study, various lignin–carbohydrate bonds have been proposed, including benzyl ethers, benzyl esters, γ-esters, phenyl glycosides and ferulate esters (Figure 1) [3]. While ferulate-mediated cross-links to lignin can indirectly be cleaved by feruloyl esterases that cleave bonds between feruloyl and arabinofuranosyl moieties, when it comes to direct enzymatic cleavage of LCCs, the only known enzymes with this ability to date are the glucuronoyl esterases (GEs) which hydrolyse the ester bonds between GlcA moieties in glucuronoxylan and lignin. The importance of GlcA moieties for cell wall recalcitrance has recently been highlighted [6] and indeed points to these residues as being key in forming the LCC. Here, we summarize the current literature on GEs, with a focus on their activities, modularity and structure.
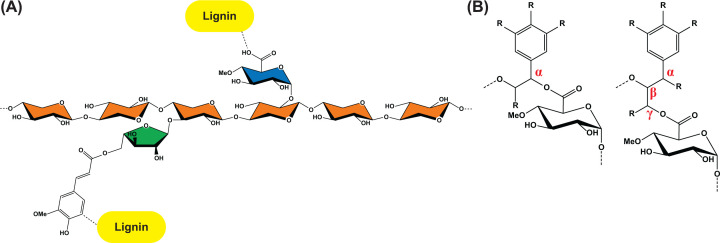
Figure 1. Lignin–carbohydrate complexes involving xylan.
(A) Lignin can bind covalently to xylan in direct ester linkages to glucuronic acid (GlcA) moieties (often 4-O-methylated) or via feruloylated side chains. The xylose units are coloured orange, GlcA blue, and arabinofuranose green. (B) The ester bonds between (4-O-Me)GlcA and lignin has been identified in two configurations, either α-linked (benzyl) or γ-linked.
Discovery of glucuronoyl esterases and characterization of fungal enzymes
The history of the discovery of GEs has been covered in past reviews [7,8] and will only be briefly introduced here. The first GE was discovered in 2006 in a serendipitous study of the fungus Schizophyllum commune and was found to be able to cleave the methyl ester of 4-O-methyl-d-GlcA [9]. This pointed toward a biological role in cleaving LCCs, and together with follow-up characterization of the GE Cip2 from Trichoderma reesei (anamorph of Hypocrea jecorina, [10]) led to the establishment of the new carbohydrate esterase family 15 (CE15) in the Carbohydrate-Active Enzymes database (www.cazy.org; [11]). Characterization of several fungal CE15 enzymes followed and began shedding light on the substrate preferences and structures of fungal GEs.
Fungal GEs have been shown to be specific for glucuronate esters and show no activity on galacturonate esters [12,13] or other esterase substrates such as feruloyl- or acetyl esters [9,12,14,15]. The standard substrates used have been various GlcA-based esters, with or without 4-O-methylation and with variation of the alcohol substituent that represents the ‘lignin side’ of LCCs (Figure 1). As a general observation, GEs tend to prefer bulkier substrates that more closely resemble proposed LCC structures that include aromatic moieties, that is, with activity on benzyl glucuronate or larger substrates clearly preferred to smaller methyl or allyl esters [16–18]. 4-O-methylation has been shown to be important for activity in several cases, with faster hydrolysis rates of substrates containing this moiety compared with ones with unsubstituted hydroxyl groups at the 4 position [14,19]. On more complex substrates that may better mimic the LCCs found in the cell wall, GE activity has been demonstrated on chemically methyl esterified 4-O-methylglucuronoxylan [20], as well as LCC-rich substrates. These include the first demonstration of activity by the GE from Sodiomyces alcalophilus (Acremonium alcalophilum) on LCCs extracted from spruce- and birchwood by observing increased amounts of carboxylic acids using phosphorus NMR [18]. GEs have also been shown to improve sugar release from corn fibre by enzyme cocktails [17], release xylooligosaccharides from LCC extractions from birch wood and act synergistically with a xylanase [21,22]. Expression of fungal GEs in plants have interestingly shown clear phenotypes, including thinner cell walls with reduced cross-linking, changes in cell wall composition, and improved saccharification, which further support the role of GEs cleaving LCCs in muro [23,24]. As such, GEs are potential important components of enzyme cocktails in biorefineries, and indeed have been included in several patents related to utilization of renewable lignocellulose [25,26].
Bacterial glucuronoyl esterases
Until recently, bacterial GEs had not been studied apart from two cases. The first was the C-terminal domain from the multidomain CesA enzyme from Ruminococcus flavefaciens [20], a cellulosomal protein also containing an acetyl xylan esterase and a dockerin domain, and the second was a GE from a marine metagenome [27]. Broader and more in-depth studies followed in 2018, with the detailed characterization of ten GEs from the three bacterial species Opitutus terrae, Solibacter usitatus (Candidatus Solibacter usitatus Ellin6076) and Spirosoma linguale [28], each having been found in soil environments. Similar to fungal GEs, several of the bacterial enzymes displayed a strong preference for bulkier substrates rather than methyl derivatives, but interestingly some showed little or no discrimination toward the alcohol moiety. Of note, the previously held fact that CE15 enzymes were strictly limited to glucuronate esters was here revealed to not be the case, as several of the bacterial enzymes had comparable kinetic parameters for galacturonate esters, such as OtCE15A-C, SuCE15A and SlCE15C [28]. Additionally, significantly increased sugar release from corn cob biomass was shown when GE-lacking enzyme cocktails were supplemented with bacterial GEs, echoing the former results using fungal enzymes and showing that also bacterial enzymes can greatly boost plant cell wall degradation, presumably by loosening up the cell wall matrix. A correlation between boosting ability and activity on model substrates could be observed, indicating that the model substrates may serve as relevant screening candidates to identify industrially relevant enzymes [28]. Expression analysis of the S. linguale CE15 genes revealed different responses of all three genes, with corncob biomass eliciting the strongest up-regulation compared with when the bacteria were grown on glucose, which correlates with studies of fungi, such as Phanerochaete carnosa or Malbranchea cinnamomea whose GEs are mainly up-regulated during growth on complex biomass substrates compared with glucose or yeast extract and peptides [29,30].
The bacterial enzymes do not appear to have a strong need for 4-O-methylation on the substrate uronic acid, as they rapidly hydrolyse unmethylated substrates. One of the fastest GEs on these comes from Teredinibacter turnerae, a bacterium that has been isolated from the gills of a wood-boring shipworm. The TtCE15A enzyme clearly preferred bulkier substrates, and was also shown to be competitively inhibited by both xylooligosaccharides and aromatic compounds, indicating that it interacts intimately with both sides of the substrate [31]. In a follow-up study, the enzyme was shown to have the capacity to liberate sugars from extracted birchwood LCCs, without being hindered by the main chain xylan polysaccharide [32]. These results indicate that GEs are likely enzymes acting early during plant cell wall degradation, as they can clearly act on GlcA esters even when these are attached to xylan, rather than being used in later degradation stages to remove residual smaller lignin-bound sugars. TtCE15A was also found to be less prone than other proteins to permanently adsorb to biomass and LCCs [32], which suggests that the enzyme and perhaps GEs in general have the ability to avoid unproductive binding to or denaturation by lignin.
Another recently characterized bacterial GE was CkGE15A from the hyperthermophilic bacterium Caldicellulosiruptor kristjanssonii [33], a species with a growth optimum of 78°C [34]. As expected, the enzyme showed a high thermal stability, with a melting temperature (Tm) of 72°C [33]. That the Tm was clearly below the optimal growth temperature of the bacterium could be attributed to either missing glycosylation, which has been shown in related species [35], or that the catalytic domain was studied in isolation (see section on multimodularity below). While most fungal GEs characterized to date are unstable above ∼50°C, CuGE from Cerrena unicolor was shown to have a Tm of 70°C [16], which is very close to that of CkGE15A. The activity of CkGE15A on model substrates was comparatively low [33], though its full potential has not been evaluated as synthetic substrates auto-hydrolyse rapidly above ∼40°C. While this enzyme might show much higher activities at a temperature closer to its Tm, other enzymes only have very weak activity even when assayed close to their expected physiological temperature, such as OtCE15B [28] and the enzyme BeCE15A from the gut symbiont Bacteroides eggerthii [36]. The latter enzyme would be expected to act optimally at 37°C, though at this temperature no substrate saturation could be achieved, and only kcat/KM values were determined, which could point toward yet undiscovered LCC structures as true substrates.
GE diversity
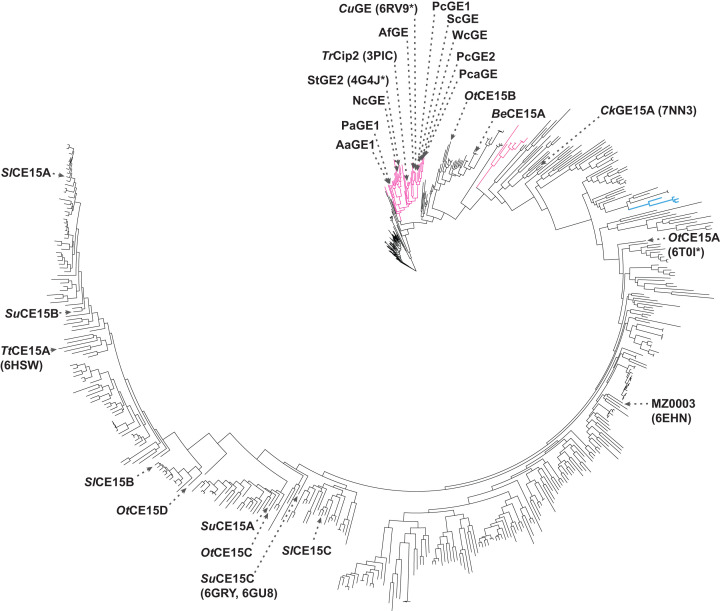
The wider substrate range of bacterial enzymes is also reflected in their protein sequences. Several phylogenetic trees of CE15 have been constructed to visualize this diversity [28], sometimes including also enzymes not incorporated in CAZy [8], and also peptide pattern recognition has been used to map the family [37]. In Figure 2, the phylogenetic tree of CE15 and its current members is shown, to showcase the wider sequence diversity among bacterial enzymes compared with fungal counterparts within CAZy. Interestingly, for most species that encode more than one CE15 member, these are found in different parts of the tree which suggests that they could have different activities. The biochemical characterization that has been performed further supports this notion, as different enzymes from the same species tend to have significantly different specificity profiles toward model and natural substrates [28].
Figure 2. Phylogenetic tree of CE15.
Fungal members are shown in magenta, archaeal in blue, and bacterial in black. Biochemically characterized enzymes are indicated with names, including PDB codes of those with determined structures in parentheses (asterisks indicate that more structures are found in the Protein Data Bank). The tree was constructed by aligning the catalytic domains of all CE15 members (n = 577, after removal of fragment and duplicate sequences) using ClustalOmega, tree building using IQ-TREE with automated identification of substitution model (WAG+F+I+G4), and visualisation by iTOL.
Also in terms of multimodularity, there is quite a lot of diversity within the family. While most GEs are found as single catalytic domains, fusion to other protein modules is not uncommon. Many fungal GEs have been shown to have appended modules from carbohydrate-binding module family 1 (CBM1) [8], and CBM-mediated binding to cellulose has been demonstrated for TrCip2 and CuGE [10,38]. For bacterial enzymes, the most common CBM partners come from CBM2, a family including cellulose-, chitin-, and xylan-binding proteins. No binding data are currently available for these however, and it is an open question what part(s) of the cell wall these CBM2 modules target. One studied bacterial GE that is coupled to several CBMs and that has been thoroughly investigated is CkGE15A [33,39]. The full-length CkXyn10C-GE15A enzyme does not comprise only the GE, but consists of two N-terminal CBM22 modules, linked to a glycoside hydrolase family 10 (GH10) xylanase, three additional CBM9 modules before the GE domain, which is followed by cadherin and surface-layer homology domains thought to anchor the large enzyme to the outer Gram-positive cell wall. Interestingly, the CBMs display various binding properties, and collectively enable the protein to bind virtually all major plant cell wall glycans [33,39]. Likely, also other CBM-appended GEs benefit from CBMs to help guide them to their complex LCC target substrate, or close proximity thereof.
The combination of more than one catalytic domain, as seen in the C. kristjanssonii enzyme, is fairly uncommon but not a unique case within CE15. Such multicatalytic architectures suggest a common substrate, and not surprisingly the majority of multicatalytic GEs consist of fusions to putative xylan-active enzymes such as xylanases, as in CkXyn10C-GE15A. The first studied bacterial GE, CesA from R. flavefaciens is linked to a CE3 acetyl xylan esterase [20,40], and likely both domains benefit from each other’s action as well as other enzymes within a larger multi-enzyme cellulosome. Also BeCE15A is found linked to a glycoside hydrolase domain, from GH8. GH8 is a polyspecific family, and the C-terminal domain of the full-length BeCE15A-Rex8A was found to be a reducing-end-specific xylose-releasing exo-oligoxylanase (Rex) [36], which as the name implies hydrolyses xylooligosaccharides but not xylan. While a xylanase-GE fusion appears logical to enable a tandem attack of the cell wall, the fusion of an oligosaccharide-targeting domain to a GE is more puzzling. The weak activity of BeCE15A on GE substrates could however indicate that the natural substrate for this two-domain enzyme is yet to be discovered, especially as several more such CE15-GH8 fusions exist in the family [11]. Also the characterised OtCE15C is fused to another catalytic domain (GH106), though the latter has not been investigated either as a single enzyme or in synergy with the OtCE15C GE domain. It is possible that deeper investigation of such fusions of different catalytic domains will be a useful strategy both to find new activities and LCC targets in the future.
Structural comparisons
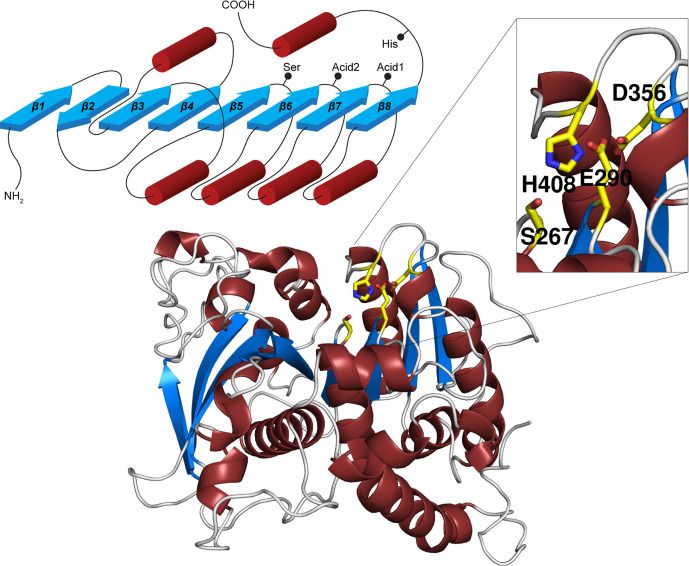
Among CE families in the CAZy database, CE15 along with CE1, CE5, CE7 and CE19 belong to the α/β hydrolase (ABH) superfamily (http://www.ebi.ac.uk/interpro/entry/InterPro/IPR029058/) while CE2, CE3, CE6, CE12, CE16, CE17 and CE20 belong to the SGNH-hydrolase superfamily (http://www.ebi.ac.uk/interpro/entry/InterPro/IPR036514/), and remaining CE families have other folds (or have been deleted from CAZy) [11]. Despite some structural similarities (a β-sheet sandwiched by helical elements) and catalytic triads consisting of a Ser nucleophile, a His, and an acidic residue, the two superfamilies are clearly distinct, though sometimes mixed up in the literature. A key difference – aside from differences in arrangement and number of secondary structure elements – is that in SGNH-hydrolases, the His and acidic residues reside on the same loop, while in ABHs all catalytic residues reside on distinct loops (Figure 3) [41]. In canonical ABHs (according to secondary structure numbering in Figure 3), the Ser nucleophile is positioned at the end of strand β5, the acid at the end of strand β7 (Acid1 in figure) and the catalytic His in a long loop following strand β8. In contrast, the fungal GEs which were structurally determined initially have the catalytic Glu at the end of strand β6 (Acid2 in figure), though other fungal sequences and most bacterial sequences have an Asp at the canonical ABH position (Acid1). A subdivision of the family has been recently suggested based on the position of the acid, leading to the CE15-A (Acid1) and CE15-B (Acid2) groups of sequences [38]. Some bacterial enzymes, exemplified in Figure 3 by OtCE15A, have acids in both positions and might be evolutionary intermediates as mutagenesis shows some functional redundancy, though both residues are important for maximum catalytic turnover [42].
Figure 3. The overall fold of GEs.
Top left, the ABH topology, with red cylinders representing helices and blue arrows β-strands. The locations of the catalytic residues are indicated. Bottom, cartoon representation of OtCE15A, and top right, close-up of the active site which shows the dual catalytic acids. Catalytic residues are shown as yellow sticks.
As already shown by initial crystallographic ligand binding studies (Figure 4) [43], the GlcA moiety of the esters cleaved by GEs binds in a pocket located above the main parallel β-sheet of the ABH fold (Figure 3), which is located on a fairly open surface in the structurally characterized fungal members of CE15 [38,43,44]. In structurally characterized bacterial enzymes, sequence insertions create high ridges on one or two sides of the substrate binding surface (Figure 3), indicating that the enzymes might interact more intimately with their complex substrate than their fungal counterparts [28,31,39,42,45,46].
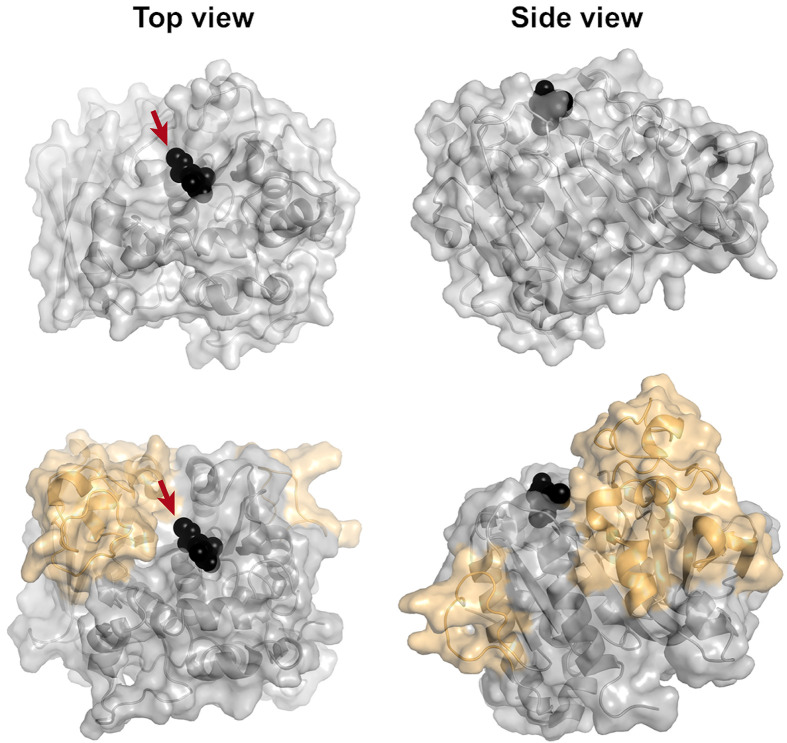
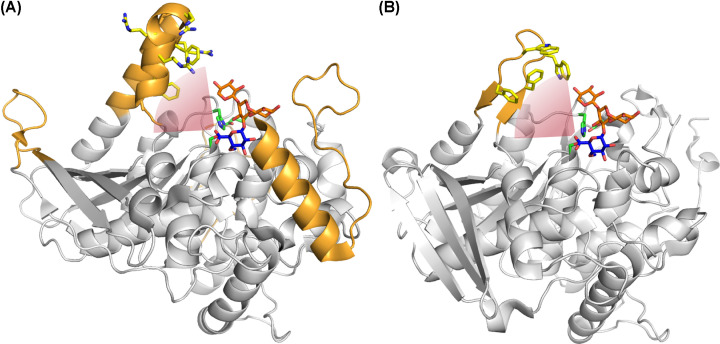
Figure 4. Comparison of fungal and bacterial GE structures.
Top, surface view of the fungal StGE2 (PDB code: 4G4G), and bottom, surface view of OtCE15A (PDB code: 6GRW), illustrating the large inserts relative to the fungal structure, in orange. Me-4-O-MeGlcA bound to StGE2 (and superposed onto the OtCE15A structure) is shown as black spheres. The attachment point for lignin is indicated with arrows.
The initial structure of StGE2 in complex with a small substrate highlighted extensive hydrogen bonding interactions (Figure 5A) involving all GlcA hydroxyls [43]. More recently, interactions with glucuronoxylooligosaccharides have been crystallographically characterized with OtCE15A and later CuGE (Figure 5B,C) [38,42], showing that GEs directly interact also with the xylan chain through van der Waals interactions – including through a conserved Trp residue – and thus, in contrast to some other CEs in CAZy, are true carbohydrate-active enzymes. However, hydrogen bond interactions are scarce, which perhaps allows the enzymes to accommodate diverse substrates as would be expected in LCCs. The interaction with the lignin portion of LCC substrates has been more difficult to assess, and thus far no structures of Michaelis complexes of GEs have been solved. However, the large inserts close to the active site in bacterial enzymes have been proposed to interact with lignin as they in several cases have a surface rich in aromatic residues facing the presumed ‘lignin side’ of the substrate (Figure 6) [28,31,39]. Possibly, these regions could be involved in LCC interactions, both productively to access the complex lignin–carbohydrate substrate, or to help avoid unproductive binding to lignin as described for TtCE15A, above [32]. Structure-guided inhibition studies to probe the interactions between TtCE15A and small aromatic molecules or glucuronoxylooligosaccharides have also highlighted residues in the active site presumed to interact with lignin, as well as the conserved Trp that interacts with the GlcA moiety and the xylan chain [31].
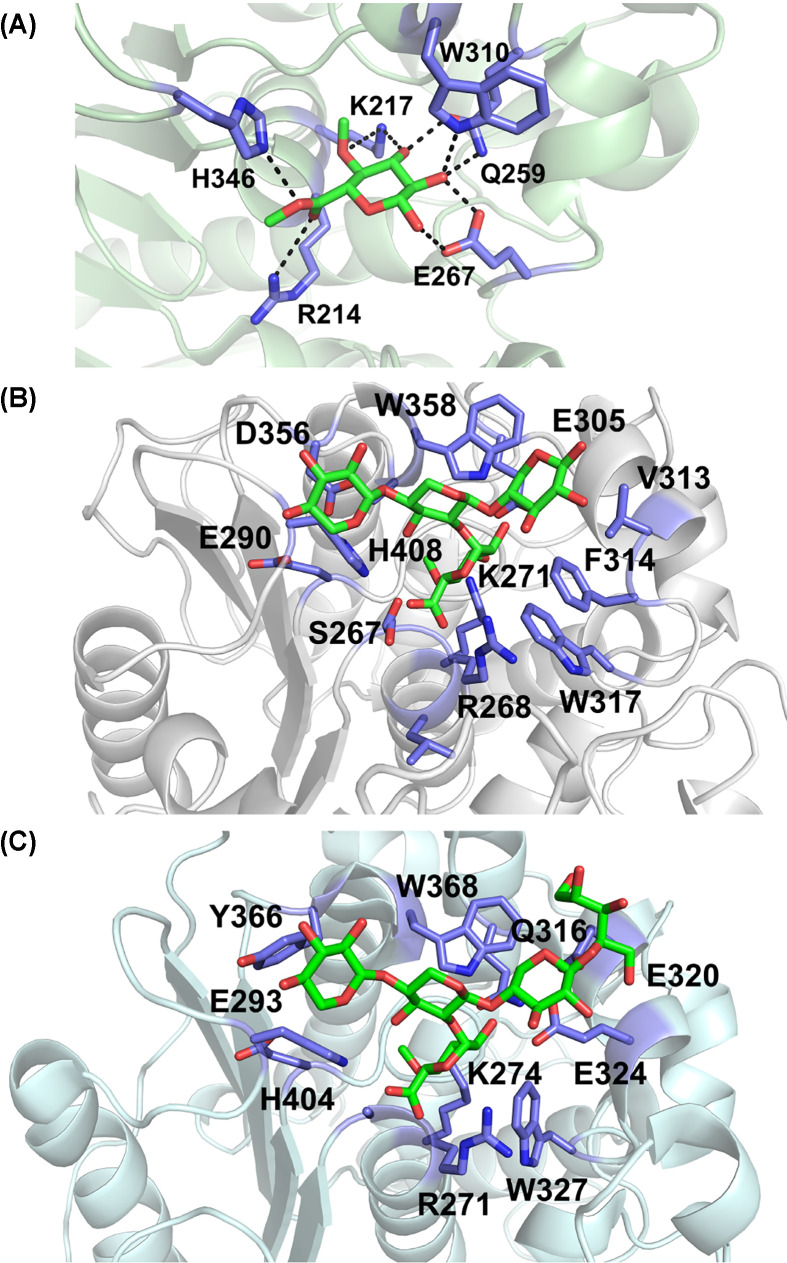
Figure 5. Close up on active sites structures of GEs with bound ligands in green sticks and binding/active site residues in purple sticks.
(A) Structure of StGE2 nucleophile mutant in complex with Me-4-O-MeGlcA (PDB code: 4G4J). Hydrogen bonds are highlighted. (B) Structure of OtCE15A in complex with XUX (22-(4-O-methyl-α-d-glucuronyl)-xylotriose) (PDB code: 6T0I) (C) Structure of CuGE in complex with 4-O-methyl-d-glucuronyl-alpha-d-xylotetraitol (PDB code: 6RV9). Panels (B and C) are shown in same orientation.
Figure 6. Residues in bacterial insert regions putatively interacting with lignin.
(A) Structure of TtCE15A (PDB code: 6HSW) with XUX (22-(4-O-methyl-α-d-glucuronyl)-xylotriose) superposed from OtCE15A (PDB code: 6T0I). (B) Structure of CkGE15A (PDB code: 7NN3) with XUX superposed as in (A). Inserts relative to fungal structures are shown in orange, catalytic residues in green sticks, XUX in orange and blue sticks, and aromatic and positive residues in the putative lignin-interacting region in yellow sticks. The presumed attachment site and positioning of lignin is indicated with a red arc.
In addition to the aforementioned product complexes, covalent intermediates have been trapped in crystals with catalytically impaired versions of the same enzymes, where either the catalytic His or a conserved active site Arg had been mutated to Ala for OtCE15A [42,46], or using the wild-type CuGE [38]. The highly conserved Arg in the catalytic cleft is important for stabilization of the oxyanion hole during catalysis, a function which in many ABHs is fulfilled by main chain nitrogen atoms. Puzzlingly, some natural GEs are also devoid of this conserved residue (OtCE15B and BeCE15A) and as a result display low activity on model substrates [28,36]. Other ‘inactive’ GEs have been reported (with different sequence features), which may suggest yet undiscovered natural substrates in the family [47,48]. The accumulated structural information has been the basis for the first computational investigations of the catalytic mechanism of the model GE OtCE15A using QM/MM [46]. The study confirmed that for this enzyme, the deacylation step is rate-limiting, though only by a small margin. Experimentally, this is reflected by very little dependence of activity on the nature of the leaving group, which is characteristic of OtCE15A, while as stated above, many GEs strongly prefer bulky, lignin-like leaving groups. For these GEs, the rate-limiting step is likely to be acylation. Furthermore, the same study investigated in depth the effect of substituting the two catalytic acids and the conserved Arg, the latter being shown to be particularly important, as deacylation could not occur when this residue was mutated in silico and the covalent intermediate could be experimentally observed [46]. Another QM/MM study on the Thermothelomyces thermophila glucuronoyl esterase (TtGE) later confirmed the importance of the conserved Arg for stabilization of the oxyanion hole but found that for this specific GE the acylation step is rate-limiting [49]. Finally, MD simulations with OtCE15A highlighted the importance of considering dissociation from substrate when investigating overall performance on real biomass, as for a complex substrate this step had a similar energy barrier as catalysis itself [46].
As described in the ‘Diversity’ section above, some GEs are part of modular enzymes, the most extreme case characterized so far being CkXyn10C-GE15A [33,39]. Attempts to structurally investigate the full-length enzyme (not including C-terminal cell wall-binding domains) have failed due to production issues. However, the GE domain and one of the associated CBM9 modules could be crystallized, and small-angle X-ray scattering (SAXS) carried out on the smaller N-terminal CBM22-CBM22-Xyn10C construct [39]. The CE15 module of CkXyn10C-GE15A lacks two of the inserts commonly found in bacterial sequences but has a more protruding and aromatic rich middle insert (Figure 6), which was speculated to be involved in lignin interaction. Intriguingly, the SAXS study shows that the CBM22-CBM22-Xyn10C is not in an extended conformation at room temperature, but the biological significance is difficult to extrapolate to the full-length construct at elevated temperatures.
Outlook
GEs are intriguing enzymes that simultaneously need to interact with both carbohydrates and lignin, and by cleaving lignin–carbohydrate bonds, they can greatly reduce plant cell wall recalcitrance. Their ability to boost the action of other plant cell wall-degrading enzymes has been demonstrated for both fungal and bacterial enzymes, but more studies are needed to systematically compare the action of different GEs on different types of biomass. Furthermore, as lignin and polysaccharides are coupled through different bonds in the LCC, additional enzyme activities, yet to be discovered, might be needed to fully assess the action of GEs. The large inserts in bacterial GEs relative to fungal GEs may have roles in steering their interactions with lignin, which could also be clarified in future works. Lastly, the association of GEs with other catalytic domains and CBMs, as well as their co-localization in genomes with enzymes targeting other polysaccharides than xylan may point toward new activities remaining to be unravelled within CE15.
Summary
Glucuronoyl esterases (GEs) are enzymes belonging to carbohydrate esterase family 15 (CE15) that are able to cleave bonds between lignin and carbohydrates in the plant cell wall.
GEs interact both with the carbohydrate and lignin portions of their substrates, though the molecular basis for the interaction with the ‘lignin side’ remains elusive.
Bacterial GEs appear to be more diverse than fungal counterparts, both in terms of sequence and structure, where bacterial enzymes often have large inserts relative to the fungal enzymes.
Better defined substrates to mimic the lignin–carbohydrate complex (LCC) structures are needed to further characterize GEs, as several known CE15 enzymes have little activity on model substrates.
Though the importance of GEs for LCC breakdown is amply demonstrated, additional enzyme activities are probably needed to fully decouple lignin and polysaccharides in future biorefineries.
Acknowledgements
The authors would like to acknowledge Novo Nordisk Foundation, project numbers: NNF17OC0027698 and NNF21OC0071611.
Abbreviations
- ABH
α/β hydrolase
- CAZy
carbohydrate-active enzymes database
- CBM
carbohydrate-binding module
- CE15
carbohydrate esterase family 15
- GE
glucuronoyl esterase
- GH
glycoside hydrolase
- GlcA
glucuronic acid
- LCC
lignin–carbohydrate complex
- SAXS
small-angle X-ray scattering
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Terrett O.M. and Dupree P. (2019) Covalent interactions between lignin and hemicelluloses in plant secondary cell walls. Curr. Opin. Biotechnol. 56, 97–104 10.1016/j.copbio.2018.10.010 [DOI] [PubMed] [Google Scholar]
- 2.Rosso M.-N., Berrin J.-G. and Lomascolo A. (2022) Plant wastes and sustainable refineries: What can we learn from fungi? Cur. Opin. Green Sustain. Chem. 34, 100602 10.1016/j.cogsc.2022.100602 [DOI] [Google Scholar]
- 3.Giummarella N., Pu Y., Ragauskas A.J. and Lawoko M. (2019) A critical review on the analysis of lignin carbohydrate bonds. Green Chem. 21, 1573–1595 10.1039/C8GC03606C [DOI] [Google Scholar]
- 4.Tarasov D., Leitch M. and Fatehi P. (2018) Lignin–carbohydrate complexes: properties, applications, analyses, and methods of extraction: a review. Biotechnol. Biofuels 11, 269 10.1186/s13068-018-1262-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi N. and Koshijima T. (1988) Ester linkages between lignin and glucuronoxylan in a lignin-carbohydrate complex from beech (Fagus crenata) wood. Wood Sci. Technol. 22, 231–241 10.1007/BF00386018 [DOI] [Google Scholar]
- 6.Lyczakowski J.J., Wicher K.B., Terrett O.M., Faria-Blanc N., Yu X., Brown D.et al. (2017) Removal of glucuronic acid from xylan is a strategy to improve the conversion of plant biomass to sugars for bioenergy. Biotechnol. Biofuels 10, 224 10.1186/s13068-017-0902-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biely P. (2016) Microbial glucuronoyl esterases: 10 years after discovery. Appl. Environ. Microbiol. 82, 7014–7018 10.1128/AEM.02396-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monrad R.N., Eklöf J., Krogh K. and Biely P. (2018) Glucuronoyl esterases: diversity, properties and biotechnological potential. A review. Crit. Rev. Biotechnol. 38, 1121–1136 10.1080/07388551.2018.1468316 [DOI] [PubMed] [Google Scholar]
- 9.Spániková S. and Biely P. (2006) Glucuronoyl esterase – novel carbohydrate esterase produced by Schizophyllum commune. FEBS Lett. 580, 4597–4601 10.1016/j.febslet.2006.07.033 [DOI] [PubMed] [Google Scholar]
- 10.Li X.L., Spániková S., de Vries R.P. and Biely P. (2007) Identification of genes encoding microbial glucuronoyl esterases. FEBS Lett. 581, 4029–4035 10.1016/j.febslet.2007.07.041 [DOI] [PubMed] [Google Scholar]
- 11.Drula E., Garron M.-L., Dogan S., Lombard V., Henrissat B. and Terrapon N. (2021) The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 50, D571–D577 10.1093/nar/gkab1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duranová M., Hirsch J., Kolenová K. and Biely P. (2009) Fungal glucuronoyl esterases and substrate uronic acid recognition. Biosci. Biotechnol. Biochem. 73, 2483–2487 10.1271/bbb.90486 [DOI] [PubMed] [Google Scholar]
- 13.Wong D.W., Chan V.J., McCormack A.A., Hirsch J. and Biely P. (2012) Functional cloning and expression of the Schizophyllum commune glucuronoyl esterase gene and characterization of the recombinant enzyme. Biotechnol. Res. Int. 2012, 951267 10.1155/2012/951267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spániková S., Poláková M., Joniak D., Hirsch J. and Biely P. (2007) Synthetic esters recognized by glucuronoyl esterase from Schizophyllum commune. Arch. Microbiol. 188, 185–189 10.1007/s00203-007-0241-x [DOI] [PubMed] [Google Scholar]
- 15.Vafiadi C., Topakas E., Biely P. and Christakopoulos P. (2009) Purification, characterization and mass spectrometric sequencing of a thermophilic glucuronoyl esterase from Sporotrichum thermophile. FEMS Microbiol. Lett. 296, 178–184 10.1111/j.1574-6968.2009.01631.x [DOI] [PubMed] [Google Scholar]
- 16.d'Errico C., Jørgensen J.O., Krogh K.B., Spodsberg N., Madsen R. and Monrad R.N. (2015) Enzymatic degradation of lignin-carbohydrate complexes (LCCs): model studies using a fungal glucuronoyl esterase from Cerrena unicolor. Biotechnol. Bioeng. 112, 914–922 10.1002/bit.25508 [DOI] [PubMed] [Google Scholar]
- 17.d'Errico C., Börjesson J., Ding H., Krogh K.B., Spodsberg N., Madsen R.et al. (2016) Improved biomass degradation using fungal glucuronoyl-esterases-hydrolysis of natural corn fiber substrate. J. Biotechnol. 219, 117–123 10.1016/j.jbiotec.2015.12.024 [DOI] [PubMed] [Google Scholar]
- 18.Arnling Bååth J., Giummarella N., Klaubauf S., Lawoko M. and Olsson L. (2016) A glucuronoyl esterase from Acremonium alcalophilum cleaves native lignin-carbohydrate ester bonds. FEBS Lett. 590, 2611–2618 10.1002/1873-3468.12290 [DOI] [PubMed] [Google Scholar]
- 19.Huynh H.H., Ishii N., Matsuo I. and Arioka M. (2018) A novel glucuronoyl esterase from Aspergillus fumigatus-the role of conserved Lys residue in the preference for 4-O-methyl glucuronoyl esters. Appl. Microbiol. Biotechnol. 102, 2191–2201 10.1007/s00253-018-8739-5 [DOI] [PubMed] [Google Scholar]
- 20.Biely P., Malovíková A., Uhliariková I., Li X.L. and Wong D.W. (2015) Glucuronoyl esterases are active on the polymeric substrate methyl esterified glucuronoxylan. FEBS Lett. 589, 2334–2339 10.1016/j.febslet.2015.07.019 [DOI] [PubMed] [Google Scholar]
- 21.Mosbech C., Holck J., Meyer A.S. and Agger J.W. (2018) The natural catalytic function of CuGE glucuronoyl esterase in hydrolysis of genuine lignin-carbohydrate complexes from birch. Biotechnol. Biofuels 11, 71 10.1186/s13068-018-1075-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosbech C., Holck J., Meyer A. and Agger J.W. (2019) Enzyme kinetics of fungal glucuronoyl esterases on natural lignin-carbohydrate complexes. Appl. Microbiol. Biotechnol. 103, 4065–4075 10.1007/s00253-019-09797-w [DOI] [PubMed] [Google Scholar]
- 23.Tsai A.Y., Canam T., Gorzsás A., Mellerowicz E.J., Campbell M.M. and Master E.R. (2012) Constitutive expression of a fungal glucuronoyl esterase in Arabidopsis reveals altered cell wall composition and structure. Plant Biotechnol. J. 10, 1077–1087 10.1111/j.1467-7652.2012.00735.x [DOI] [PubMed] [Google Scholar]
- 24.Latha Gandla M., Derba-Maceluch M., Liu X., Gerber L., Master E.R., Mellerowicz E.J.et al. (2015) Expression of a fungal glucuronoyl esterase in Populus: effects on wood properties and saccharification efficiency. Phytochemistry 112, 210–220 10.1016/j.phytochem.2014.06.002 [DOI] [PubMed] [Google Scholar]
- 25.Börjesson J., Viksoe-Nielsen A., Spodsberg N. and Krogh K. (2018) Polypeptides having glucuronyl esterase activity and polynucleotides encoding same. U.S. Pat. US10731140B2 [Google Scholar]
- 26.Abramson M., Shani Z. and Shoseyov O. (2012) Transgenic plants with improved saccharification yields and methods of generating same. Int. Pat. WO2012059922 [Google Scholar]
- 27.De Santi C., Willassen N.P. and Williamson A. (2016) Biochemical characterization of a family 15 carbohydrate esterase from a bacterial marine arctic metagenome. PloS ONE 11, e0159345 10.1371/journal.pone.0159345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnling Bååth J., Mazurkewich S., Knudsen R.M., Poulsen J.N., Olsson L., Lo Leggio L.et al. (2018) Biochemical and structural features of diverse bacterial glucuronoyl esterases facilitating recalcitrant biomass conversion. Biotechnol. Biofuels 11, 213 10.1186/s13068-018-1213-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacDonald J., Doering M., Canam T., Gong Y., Guttman D.S., Campbell M.M.et al. (2011) Transcriptomic responses of the softwood-degrading white-rot fungus Phanerochaete carnosa during growth on coniferous and deciduous wood. Appl. Environ. Microbiol. 77, 3211–3218 10.1128/AEM.02490-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hüttner S., Nguyen T.T., Granchi Z., Chin-A-Woeng T., Ahrén D., Larsbrink J.et al. (2017) Combined genome and transcriptome sequencing to investigate the plant cell wall degrading enzyme system in the thermophilic fungus Malbranchea cinnamomea. Biotechnol. Biofuels 10, 265 10.1186/s13068-017-0956-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnling Bååth J., Mazurkewich S., Poulsen J.N., Olsson L., Lo Leggio L. and Larsbrink J. (2019) Structure-function analyses reveal that a glucuronoyl esterase from Teredinibacter turnerae interacts with carbohydrates and aromatic compounds. J. Biol. Chem. 294, 6635–6644 10.1074/jbc.RA119.007831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raji O., Arnling Bååth J., Vuong T.V., Larsbrink J., Olsson L. and Master E.R. (2021) The coordinated action of glucuronoyl esterase and α-glucuronidase promotes the disassembly of lignin-carbohydrate complexes. FEBS Lett. 595, 351–359 10.1002/1873-3468.14019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krska D. and Larsbrink J. (2020) Investigation of a thermostable multi-domain xylanase-glucuronoyl esterase enzyme from Caldicellulosiruptor kristjanssonii incorporating multiple carbohydrate-binding modules. Biotechnol. Biofuels 13, 68 10.1186/s13068-020-01709-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bredholt S., Sonne-Hansen J., Nielsen P., Mathrani I.M. and Ahring B.K. (1999) Caldicellulosiruptor kristjanssonii sp. nov., a cellulolytic, extremely thermophilic, anaerobic bacterium. Int. J. Syst. Bacteriol. 49, 991–996 10.1099/00207713-49-3-991 [DOI] [PubMed] [Google Scholar]
- 35.Chung D., Young J., Bomble Y.J., Vander Wall T.A., Groom J., Himmel M.E.et al. (2015) Homologous expression of the Caldicellulosiruptor bescii CelA reveals that the extracellular protein is glycosylated. PloS ONE 10, e0119508 10.1371/journal.pone.0119508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kmezik C., Krska D., Mazurkewich S. and Larsbrink J. (2021) Characterization of a novel multidomain CE15-GH8 enzyme encoded by a polysaccharide utilization locus in the human gut bacterium Bacteroides eggerthii. Sci. Rep. 11, 17662 10.1038/s41598-021-96659-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agger J.W., Busk P.K., Pilgaard B., Meyer A.S. and Lange L. (2017) A new functional classification of glucuronoyl esterases by peptide pattern recognition. Front Microbiol. 8, 309 10.3389/fmicb.2017.00309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ernst H.A., Mosbech C., Langkilde A.E., Westh P., Meyer A.S., Agger J.W.et al. (2020) The structural basis of fungal glucuronoyl esterase activity on natural substrates. Nat. Commun. 11, 1026 10.1038/s41467-020-14833-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krska D., Mazurkewich S., Brown H.A., Theibich Y., Poulsen J.N., Morris A.L.et al. (2021) Structural and functional analysis of a multimodular hyperthermostable xylanase-glucuronoyl esterase from Caldicellulosiruptor kristjansonii. Biochemistry 60, 2206–2220 10.1021/acs.biochem.1c00305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aurilia V., Martin J.C., McCrae S.I., Scott K.P., Rincon M.T. and Flint H.J. (2000) Three multidomain esterases from the cellulolytic rumen anaerobe Ruminococcus flavefaciens 17 that carry divergent dockerin sequences. Microbiology (Reading) 146, 1391–1397 10.1099/00221287-146-6-1391 [DOI] [PubMed] [Google Scholar]
- 41.Mølgaard A., Kauppinen S. and Larsen S. (2000) Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Structure 8, 373–383 10.1016/S0969-2126(00)00118-0 [DOI] [PubMed] [Google Scholar]
- 42.Mazurkewich S., Poulsen J.N., Lo Leggio L. and Larsbrink J. (2019) Structural and biochemical studies of the glucuronoyl esterase OtCE15A illuminate its interaction with lignocellulosic components. J. Biol. Chem. 294, 19978–19987 10.1074/jbc.RA119.011435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Charavgi M.D., Dimarogona M., Topakas E., Christakopoulos P. and Chrysina E.D. (2013) The structure of a novel glucuronoyl esterase from Myceliophthora thermophila gives new insights into its role as a potential biocatalyst. Acta Crystallogr. D. Biol. Crystallogr. 69, 63–73 10.1107/S0907444912042400 [DOI] [PubMed] [Google Scholar]
- 44.Pokkuluri P.R., Duke N.E., Wood S.J., Cotta M.A., Li X.L., Biely P.et al. (2011) Structure of the catalytic domain of glucuronoyl esterase Cip2 from Hypocrea jecorina. Proteins 79, 2588–2592 10.1002/prot.23088 [DOI] [PubMed] [Google Scholar]
- 45.De Santi C., Gani O.A., Helland R. and Williamson A. (2017) Structural insight into a CE15 esterase from the marine bacterial metagenome. Sci. Rep. 7, 17278 10.1038/s41598-017-17677-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zong Z., Mazurkewich S., Pereira C.S., Fu H., Cai W., Shao X.et al. (2022) Mechanism and biomass association of glucuronoyl esterase: an α/β hydrolase with potential in biomass conversion. Nat. Commun. 13, 1449 10.1038/s41467-022-28938-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hüttner S., Klaubauf S., de Vries R.P. and Olsson L. (2017) Characterisation of three fungal glucuronoyl esterases on glucuronic acid ester model compounds. Appl. Microbiol. Biotechnol. 101, 5301–5311 10.1007/s00253-017-8266-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dilokpimol A., Mäkelä M.R., Cerullo G., Zhou M., Varriale S., Gidijala L.et al. (2018) Fungal glucuronoyl esterases: Genome mining based enzyme discovery and biochemical characterization. N. Biotechnol. 40, 282–287 10.1016/j.nbt.2017.10.003 [DOI] [PubMed] [Google Scholar]
- 49.Viegas M.F., Neves R.P.P., Ramos M.J. and Fernandes P.A. (2022) QM/MM study of the reaction mechanism of thermophilic glucuronoyl esterase for biomass treatment. ChemPhysChem 23, e202200269 10.1002/cphc.202200269 [DOI] [PubMed] [Google Scholar]