Abstract
The discovery of oxidative cleavage of glycosidic bonds by enzymes currently known as lytic polysaccharide monooxygenases (LPMOs) has profoundly changed our current understanding of enzymatic processes underlying the conversion of polysaccharides in the biosphere. LPMOs are truly unique enzymes, harboring a single copper atom in a solvent-exposed active site, allowing them to oxidize C-H bonds at the C1 and/or C4 carbon of glycosidic linkages found in recalcitrant, often crystalline polysaccharides such as cellulose and chitin. To catalyze this challenging reaction, LPMOs harness and control a powerful oxidative reaction that involves Fenton-like chemistry. In this essay, we first draw a brief portrait of the LPMO field, notably explaining the shift from the monooxygenase paradigm (i.e., using O2 as cosubstrate) to that of a peroxygenase (i.e., using H2O2). Then, we briefly review current understanding of how LPMOs generate and control a hydroxyl radical (HO•) generated through Cu(I)-catalyzed H2O2 homolysis, and how this radical is used to create the proposed Cu(II)-oxyl species, abstracting hydrogen atom of the C-H bond. We also point at the complexity of analyzing redox reactions involving reactive oxygen species and address potential deficiencies in the interpretation of existing LPMO data. Being the first copper enzymes shown to enable site-specific Fenton-like chemistry, and maybe not the only ones, LPMOs may serve as a blueprint for future research on monocopper peroxygenases.
Keywords: copper, Fenton, LPMO, peroxygenase, polysaccharides
A bit of history
In 1974, Eriksson et al. showed that cellulose degradation by a fungal secretome was more efficient in the presence of oxygen. At the time, this finding directed attention to an enzyme called cellobiose dehydrogenase (initially called cellobiose oxidase [1]). It was not until 2010 that it was shown that bacterial and fungal secretomes contain enzymes, today referred to as lytic polysaccharide monooxygenases (LPMOs), which oxidize polysaccharides (such as chitin and cellulose) in an oxygen-dependent manner and that, by doing so, increase the efficiency of classical hydrolases. The existence of these enzymes was first revealed by studying chitin degradation [2]. First glimpses of what was coming appeared in 2005, when Vaaje-Kolstad et al. showed that a protein, at the time thought to be a family 33 carbohydrate-binding module (CBM33), boosts the activity of chitinases [3,4], and in 2007/2008, when Merino & Cherry and Karkehabadi et al. showed that proteins, at the time thought to be a family 61 glycoside hydrolases (GH61), are structurally similar to CBM33s and boost the activity of cellulases [5,6].
In the past decade, LPMOs have gained a lot of attention, because of their intriguing catalytic ability and mechanism, their documented value as a key component of enzyme cocktails for industrial processing of lignocellulosic biomass [7–10], and the emerging notion that these taxonomically widespread and abundant enzymes may be involved in other biological processes [11], such as cellular development [12,13] and virulence [14–16]. Today, LPMOs are classified as auxiliary activities (AA) in the carbohydrate-active enzymes (CAZy) database and distributed in eight families, namely AA9 to AA11 and AA13 to AA17. LPMOs have the unique ability to act on solid–liquid interfaces, meaning that they can degrade polysaccharides that are embedded in an insoluble, sometimes even crystalline structure [17,18]. This is very different from classical hydrolytic enzymes, such as cellulases, which act on single ‘decrystallized’ polysaccharide chains [2]. LPMOs have evolved to overcome the exceedingly high energy barrier (approximately 95–104 kcal/mol; [19–21]) associated with C-H bond activation in the glycosidic bonds of crystalline polysaccharides. The chemistry afforded by combining and orienting the 20 naturally occurring amino acids does not suffice to overcome this barrier; hence, LPMOs have evolved an active site equipped for controlled metal-catalyzed generation of a highly oxidative oxygen species.
While the first crystallographic studies of proteins known today as LPMOs showed the presence of a single metal-binding site [3,6], the nature of this metal ion remained unclear [2,22], until it was established in 2011 [23,24] that LPMOs are monocopper enzymes. The copper is bound by a highly conserved arrangement of two histidine residues referred to as the histidine-brace [24–27]. Although the catalytic mechanism of LPMOs remains unknown in part (see below), it is generally accepted that LPMO action entails the formation of a powerful reactive oxygen species on the copper ion, possibly a copper(II)-oxyl ([CuO]+) [19,28–30]. In cellulose-active LPMOs, this oxygen species abstracts a hydrogen atom from the C1 or the C4 of the scissile glycosidic bond, followed by substrate hydroxylation through an oxygen-rebound mechanism. Hydroxylation destabilizes the glycosidic bond, leading to cleavage and formation of one normal and one oxidized new chain end [23,31].
Regardless of the fine mechanistic details underlying the formation of the copper(II)-oxyl species, oxygen atoms must be recruited at some point during catalysis. In the first study showing that LPMOs catalyze oxidative cleavage of polysaccharides, experiments with 18O2 showed incorporation of one 18O in the oxidized products [2]. Thus, when the nature of the bound metal ion was found to be copper [23,24,32], by analogy to enzymes such as particulate methane monooxygenase and ammonia monooxygenase, the logical conclusion was that LPMOs are monooxygenases. The monooxygenase reaction requires two externally delivered electrons and one O2 molecule to catalyze the following reaction: R-H + O2 + 2H+ +2e− → R-OH + H2O. Accordingly, typical set-ups for LPMO reactions entail mixing the enzyme with a substrate and an electron source, under aerobic conditions. While ascorbic acid is commonly used as electron source, LPMO reactions can be fueled by a wide range of reductants as well as by enzymes capable of delivering electrons, such as cellobiose dehydrogenase [33–35].
The LPMO copper site is remarkably solvent-exposed and its reactivity will be heavily affected by the presence of substrate, which will shield the copper from solvent, displace water molecules, and change the electronic environment of the metal ion. Such effects have indeed been revealed by both crystallographic studies [36,37], modeling [38], and EPR spectroscopy [38–40]. Importantly, when not bound to substrate, LPMOs may, upon reduction, engage in off-pathway reactions. First, reduced LPMOs may react with molecular oxygen to generate hydrogen peroxide [41] in what is referred to as an oxidase reaction. Reduced LPMOs may also react with hydrogen peroxide in the reaction solution and this may lead to oxidative damage to the LPMO active site (see below). The occurrence of such off-pathway reactions will depend on whether or not an appropriate substrate is present, and on the substrate concentration.
Intriguingly, in contrast with other copper monooxygenases, LPMOs contain only one copper ion and can thus store only one of the two electrons needed in a monooxygenase reaction. The path by which the second electron reaches the catalytic center is not immediately obvious since, during catalysis, the LPMO is secluded from the solvent by the polymeric, insoluble substrate (Figure 1). Several researchers have proposed the existence of electron-transport paths in LPMOs, but the enzymes do not show conserved structural features that might relate to such paths. This issue has sometimes been referred to as the ‘second electron conundrum’.
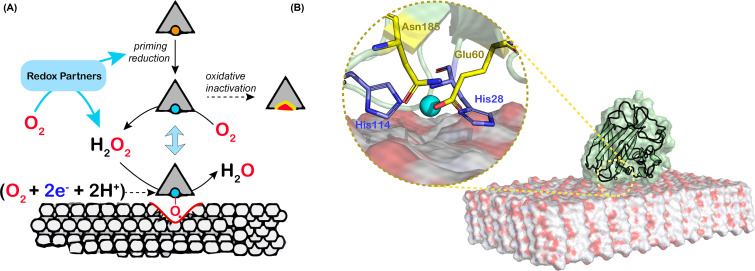
Figure 1. Schematic view of the peroxygenase action of LPMOs on a crystalline polysaccharide.
(A) The LPMO-Cu(II) form (copper atom shown as orange sphere) is reduced to the Cu(I) form (blue sphere) by a redox partner (small organic reductant, redox protein, photoactivated molecule). LPMO-Cu(I) molecules remaining in the unbound state (e.g. because of lack of a genuine substrate) can catalyze an oxidase reaction (production of H2O2) and are susceptible to oxidative inactivation. LPMO-Cu(I) molecules bound to the polysaccharide will catalyze a peroxygenase reaction leading to chain cleavage. The alternative, hypothetical monooxygenase reaction is shown in between parenthesis and entails the delivery of two electrons and two protons per catalytic cycle. (B) Model of an LPMO (the AA10 from Serratia marcescens) bound to β-chitin [38]. The inset shows the entrance to a tunnel that connects the active site to the bulk solvent, including the residues Glu60 and Asn185 (shown as yellow sticks) that limit tunnel accessibility to small molecules only (such as O2, H2O2, or H2O). The histidine brace is shown as blue sticks.
Although perhaps somewhat less visible in the literature, kinetic data obtained using standard ‘monooxygenase reaction conditions’ raise additional intriguing issues. First, reported progress curves for LPMO reactions tend to be nonlinear, which reflects that LPMO reactions are hard to control, with possible reductant depletion and/or enzyme inactivation. Second, the reported ‘monooxygenase’ rates tend to be excessively low, usually below 1 per min (see ref. [42], for an overview).
Intrigued by these observations and considerations, and inspired by a study on light-driven LPMO catalysis showing unprecedented high catalytic rates [43], we questioned established views on LPMO catalysis and proposed, in 2016, based on a large series of experiments [44,45], that LPMOs are in fact peroxygenases, i.e. using hydrogen peroxide rather than molecular oxygen as the cosubstrate (Figure 1). Originally, our findings and proposal were presented under the title ‘Fenton-type chemistry by a copper enzyme: molecular mechanism of polysaccharide oxidative cleavage’ [44]. This proposal was controversial [46,47] because it entailed the existence of a new type of peroxygenase catalysis not requiring any other cofactor than a single copper (rather than a heme). At the same time, this proposal was attractive because it was, and still is, compatible with existing data and provided reasonable explanations for outstanding issues:
Apparent monooxygenase activity may very well be peroxygenase activity. In all typical LPMO reaction set-ups, H2O2 will be formed in situ, through (i) oxidation of the reductant (either LPMO-catalyzed—i.e. via the oxidase activity—or via abiotic, free metal-catalyzed oxidation of the reductant; [48,49]) and/or (ii) the possible oxidase activity of an enzymatic redox partner such as cellobiose dehydrogenase [50,51]. This also applies to the experiment with 18O2 conducted in 2010, where H218O2 was likely formed in situ. Thus, in these typical ‘monooxygenase’ reaction set-ups, the peroxygenase reaction can take place.
The ‘2nd electron conundrum’. A peroxygenase reaction (R-H + H2O2 →R-OH + H2O) only requires a priming reduction of the LPMO and does not require delivery of electrons during catalysis (Figure 1).
The excessively low catalytic rate of LPMOs. Both our original work [44,45] and follow-up studies [52–60] have shown that the peroxygenase reaction is orders of magnitude faster than the (apparent) monooxygenase reaction and that under ‘peroxygenase conditions’ kinetic parameters for LPMOs resemble those of other peroxygenases [52,54]. Today, it is clear that under standard ‘monooxygenase’ conditions (i.e. presence of a reductant, such as ascorbic acid, under aerobic conditions, with no added H2O2), the LPMO reaction rate is limited by the usually very slow in-situ generation of H2O2.
The instability of LPMOs. Reactions with ‘too much’ H2O2 lead to oxidative damage of the LPMO active site, showing that the regularly observed inactivation of LPMOs under turnover conditions is due to an autocatalytic process [45]. It is now generally accepted that reduced LPMOs that meet H2O2 in the absence of substrate catalyze a slow peroxidase reaction that may lead to damage [61]. Such damage, either caused by too much added H2O2 or by too high levels of in-situ generated H2O2, explains why LPMO reactions tend to show nonlinear progress curves and signs of enzyme inactivation. This may also explain why Scott et al. observed that addition of catalase, which will help keeping H2O2 levels low, improved the performance of an LPMO-containing cellulase cocktail in saccharification of lignocellulosic biomass [62].
Today, it is clear that LPMOs are efficient peroxygenases and most researchers would believe that the activity and stability of these enzymes, in laboratory reactions and industrial bioprocessing plants alike, depend on access to H2O2.
LPMOs are truly special: controlled, site-specific Fenton-like chemistry
At the time when LPMOs were still considered to be monooxygenases, it had been anticipated, based on calculations and chemical intuition, that the reactive species capable of abstracting a hydrogen atom from the C1 or C4 in crystalline cellulose would need to be a Cu(II)-oxyl ([CuO]+) or perhaps its protonated form, Cu(III)-hydroxo ([CuOH]2+) [28,63–65] (see Figure 2). While it had been shown that reduced LPMOs react with molecular oxygen [66], the resulting Cu(II)-superoxo ([CuO2]+) species was not considered sufficiently powerful to abstract the hydrogen atom [19,28]. Interestingly, almost all later calculations, considering a monooxygenase reaction, a peroxygenase reaction, or both, predicted Cu(II)-oxyl as the most likely species catalyzing hydrogen atom abstraction (haa), the rate-limiting step during substrate oxidation [19,29,67,68].
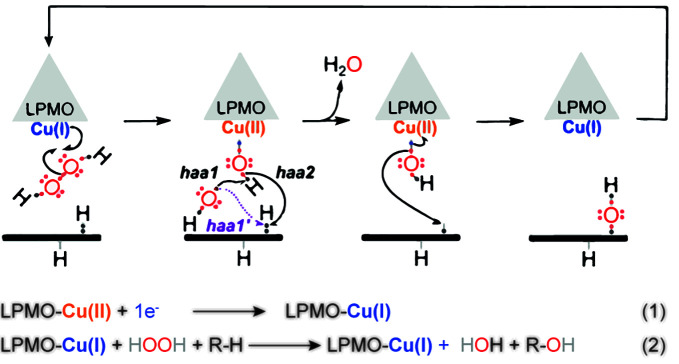
Figure 2. Reaction scheme proposed when the LPMO peroxygenase reaction was first described.
LPMO-Cu(II) is first reduced to LPMO-Cu(I) (‘priming reduction’), followed by H2O2 binding and homolytic bond cleavage. This cleavage leads to the Fenton-like generation of a hydroxyl radical, catalyzing haa either from the Cu(II)-hydroxide (haa1) or from the substrate (haa1’). The former scenario generates a Cu(II)-oxyl intermediate that can abstract a hydrogen atom from the substrate (haa2). Later studies have shown that haa1 (followed by haa2), rather than haa1’, is the most plausible scenario. The formation of the Cu(II)-hydroxide/substrate radical complex is followed by hydroxylation of the substrate via an oxygen-rebound mechanism and concomitant regeneration of the Cu(I) center. This picture and most of the legend were taken from Figure 2 in [44].
Several possible catalytic scenarios, all converging toward a common point, i.e. a Cu(II)-hydroxide/substrate radical complex (Figure 2, third state), were considered when the peroxygenase activity of LPMOs was first described (see the Supplementary Materials of [44]). These scenarios involve either heterolytic or homolytic cleavage of the H2O2 molecule and the latter scenario was considered most plausible [44] (Figure 2). Subsequent studies by multiple groups have concluded that, indeed, homolytic cleavage occurs, leading to the formation of a Cu(II)-hydroxide and a hydroxyl radical (HO•) [19,29,67,68]. Such homolytic cleavage has been experimentally evidenced by the detection of hydroxyl radicals in work by Bissaro et al. [68] and Jones et al. [55], using EPR spectroscopy.
The notion that LPMO activity involves Cu(I)-catalyzed formation of a hydroxyl radical from H2O2 supports the original suggestion that LPMOs enable controlled, site-specific exploitation of the power of Fenton chemistry [44]. The ability to generate hydroxyl radicals is not without risks as evidenced by the autocatalytic inactivation of LPMOs through reactions with H2O2 in the absence of substrate ([45]; Figure 3; more below). Notably, for LPMOs studied so far, this reaction seems rather slow (∼103 M−1 s−1) [30] compared with the productive reaction with substrate (∼106 M−1 s−1) [30,52] (note that these rates may vary between LPMOs). This rate difference shows that the confinement of the LPMO-substrate complex provides the appropriate molecular environment to generate the hydroxyl radical and then use it in a productive manner. In a later study, Wang et al. referred to this as a ‘caged radical’ [29]. Such tight enzymatic control is truly remarkable given that such radical is highly reactive, with a half-live of about 1 ns in biological systems, and underscores the importance of the substrate in protecting the LPMO from autocatalytic inactivation.
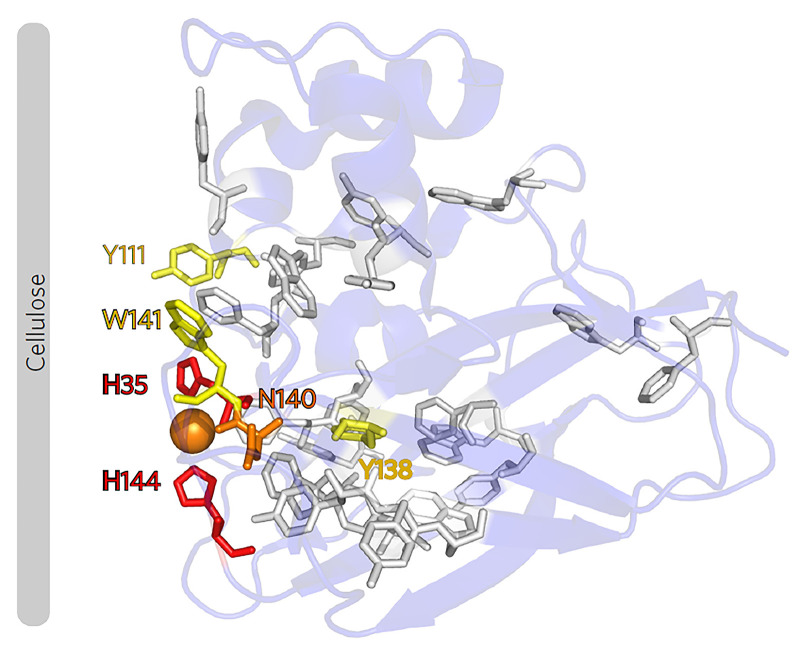
Figure 3. Oxidative damage in an AA10 LPMO.
Exposure of this LPMO to H2O2 in the absence of substrate showed that oxidative damage starts at the two catalytic histidines (shown as red sticks) and propagates to Tyr and Trp residues further away from the catalytic center [45]. The color code indicates the degree of oxidative damage, from high (red) to low (yellow). In the same study, it was also shown that productive binding to cellulose has a protective effect. Although the focus of this early work was not on protective mechanisms, these results clearly show that the radical generated in the catalytic center may propagate through the enzyme via aromatic residues. Figure taken from [45].
While it is clear that LPMOs are efficient peroxygenases and while it is now well established that H2O2 will be formed in situ in LPMO reactions, one key remaining question is whether LPMOs catalyze monooxygenase reactions at all. There is no doubt that reduced LPMOs can activate molecular oxygen [41,66]. Furthermore, there are computational indications that substrate-binding stabilizes the Cu(II)-superoxide species [40], which is otherwise prone to dissociation [66]. Modeling studies suggest that a monooxygenase reaction is feasible (via a Cu(II)-oxyl), and mechanisms for the timely delivery of the electrons and protons needed to reach the Cu(II)-oxyl state have been proposed [43,69]. Yet, such delivery mechanisms remain to be experimentally validated. Interestingly, based on quantum mechanics/molecular mechanics (QM/MM) metadynamics simulations, Wang et al. [29] have proposed a monooxygenase reaction involving the formation of H2O2 (from O2) within the active site cavity formed by the LPMO-substrate complex on the productive catalytic pathway. This proposal has been used by some to claim that the peroxygenase reaction is some sort of ‘shunt’ for what essentially would be a monooxygenase reaction. While this proposed catalytic pathway cannot be excluded, it fails to explain several experimental observations (see above), including the ‘second electron conundrum’ and the well-documented inhibition of LPMO activity by H2O2-consuming enzymes. Also, we believe that the use of the term ‘shunt’ is misleading, since in the field where this term has originally been used to describe oxygen-dependent redox catalysis (P450 cytochromes), the shunt pathway refers to a ‘forced’, rather slow and inefficient pathway that requires exceedingly high amounts of H2O2 (several mM) to bypass the electron/proton delivery steps of the standard O2 pathway. For LPMOs, the situation is actually nearly the opposite, since recent kinetic data clearly show that the peroxygenase reaction is orders of magnitude faster than the monooxygenase reaction [47,52,54–56] and that stable peroxygenase reactions with total turnover numbers of tenths of thousands can occur at the low steady-state H2O2 concentrations (µM range) found in microbial ecosystems [59].
Assessment of the LPMO literature in light of recent findings
Due to the many biotic and abiotic processes that may be going on in typical LPMO reactions, reliable kinetic characterization of LPMO catalysis is challenging, as discussed elsewhere [70,71]. Data on the catalytic action of LPMOs need to be analyzed with great care, for example due the possible occurrence of enzyme inactivation, and the occurrence of multiple H2O2-consuming and -producing (side-) reactions. It cannot be emphasized enough that many published LPMO reactions likely were limited by in-situ generation of H2O2 and not by, e.g. the concentration of active LPMO. Older LPMO studies with quantitative statements on LPMO activity may need some reassessment.
When reading the literature, one also has to realize that the use of H2O2 vs O2 as cosubstrate has been, and to some extent still is, controversial. The literature contains several claims that may have been colored by the ongoing debate and that perhaps need some critical reassessment:
It has been claimed by some that autocatalytic inactivation of LPMOs primarily happens in H2O2-driven reactions and that this shows that the peroxygenase reaction is not natural or optimal, in contrast with the apparent monooxygenase reaction. There are no data in the literature supporting such claims. It is actually well documented that for natural H2O2-dependent enzymes, such as peroxidases and peroxygenases, high amounts of H2O2 lead to high enzyme rates but also to enzyme damage [45,47]. Such damage may not always be seen under ‘monooxygenase conditions,’ but this is simply due to the steady-state concentration of H2O2 being lower, as is the reaction rate.
It has been claimed that the peroxygenase reaction is less specific [47]. However, we note that most published studies do not report differences between product profiles generated in apparent monooxygenase and peroxygenase reactions (e.g. [72]). Generally, one may see trace amounts of nonstandard products, e.g. in mass spectra, and the occurrence of such products varies between reactions. We would argue that oxidative damage to the catalytic center, which starts at the histidines [45], could lead to loss of specificity, for example due to a less well-controlled hydroxyl radical or minor changes in the enzyme-substrate interaction.
Concluding remarks
Metal-containing redox enzymes are difficult to study due to the many possible off-pathway reactions, the susceptibility of the enzyme to oxidative damage, and challenges related to obtaining reliable kinetic data. The LPMOs are no exception and research on these enzymes has pointed at one challenge in particular, namely conclusive identification of the true oxygen cosubstrate of what could be a monooxygenase or a peroxygenase reaction. The quest to understand the nature of LPMO catalysis has led to a much-debated paradigm change, with LPMOs going from being considered as monooxygenases to being recognized as efficient peroxygenases that may possibly act as slow monooxygenases. While the peroxygenase reaction of LPMOs provided plausible answers to outstanding questions in the field (as discussed above), and seems ‘logical’ in the context of how fungal secretomes work and have access to various oxygen species [42,59], it was met by considerable skepticism. One (understandable) reason for such skepticism likely was that LPMOs are truly unique peroxygenases, with an at the time unprecedented active site architecture. Indeed, the large majority of known peroxygenases are heme enzymes [73] and, to the best of our knowledge, LPMOs are the only monocopper enzymes for which peroxygenase activity has been convincingly demonstrated. Of note, multiple studies have shown that LPMOs and heme-peroxygenases have similar catalytic efficiencies [52,54,56,58,61].
Interestingly, other types of enzymes have also been suggested to make use of Fenton-type O–O homolysis of H2O2 to catalyze oxidative transformations, such as some heme-peroxidases, some families of cytochrome P450 and the nonheme HppE [74]. In these cases, the HO• species is either directly used as a substrate oxidant or, as in LPMO catalysis, used to generate the enzymatic active species (e.g. Compound I (Cpd I), Por+•Fe(IV)═O in P450s). HppE is a particularly interesting case since it was shown that this nonheme monoiron epoxidase involved in fosfomycin production, is not an oxidase (i.e. does not use O2) as previously thought, but a peroxidase, using H2O2 [75]. Clearly, identification of the oxygen cosubstrate is challenging and it is important to carefully consider the entire chain of reactive oxygen species when studying the enzymes discussed above. It cannot be excluded that additional ‘mistakes,’ similar to those made for LPMOs and HppE, will be discovered in the future. In this light, it is interesting to note that modeling studies suggest that a peroxygenase reaction is energetically feasible for particulate methane monooxygenase, which needs to overcome an even higher energy barrier than LPMOs [76].
Despite major advances achieved in the LPMO field over the past decade, several important questions remain. For example, the species likely abstracting the hydrogen from the substrate, the Cu(II)-oxyl, has not yet been observed experimentally. Turning to biology, questions remain as to the levels of O2 and H2O2 in biomass-degrading ecosystems and the actual levels of LPMO activity in such systems. Another interesting issue, of utmost importance for both understanding biological systems and industrial applications, concerns the stability of LPMOs under turnover conditions and the possible existence of protective mechanisms. The surfaces and cores of LPMOs contain aromatic amino acids that vary in number and location; it is quite conceivable that at least in some LPMOs, these residues may participate in so-called hole-hopping pathways [77] that help dissipating potentially damaging radicals from the catalytic center (Figure 3). Indeed, recent studies demonstrate the formation of aromatic radicals in LPMOs [55,56,78,79].
Perhaps, one of the most intriguing issues concerns the huge multiplicity of LPMOs in fungi [80] and the proven roles of LPMOs in microbial pathogenicity [11]. The true substrates of these many LPMOs remain unknown. While it is easy to envisage a need for powerful, Fenton-like chemistry for degradation of crystalline cellulose or complex copolymeric substructures in plant cell walls, it is not at all clear what type of recalcitrant substrate, if any, is being degraded/targeted by several LPMOs reported in the literature, such as CbpD, a virulence factor produced by Pseudomonas aeruginosa [16]. Further research is needed to reveal how Nature employs the power of site-specific, well-controlled Fenton-type chemistry to solve highly challenging catalytic tasks, some of which are well known, while others remain to be discovered.
Summary
LPMOs are abundant enzymes that play a key role in the enzymatic conversion of recalcitrant polysaccharides, such as cellulose and chitin, and that may have other functions related to cellular development and microbial virulence.
Since their discovery, LPMOs have progressed from being considered slow monooxygenases to being considered efficient peroxygenases.
These unique peroxygenases, with a single copper ion as the only cofactor, harness the power of Fenton-type chemistry, i.e. copper-catalyzed homolytic cleavage of hydrogen peroxide to generate a hydroxy radical, in a controlled, site-specific manner.
Topics for further studies include the detection of the hydrogen-abstracting oxygen species, predicted to be a Cu-oxyl rather than the hydroxyl radical, the substrates of LPMOs involved in cellular function rather than biomass conversion, and the existence of hole-hopping mechanisms that protect LPMOs from autocatalytic inactivation.
Acknowledgements
V.G.H.E. and B.B. acknowledge the contribution of former and present colleagues to LPMO research, and are thankful for inspiring discussions.
Abbreviations
- AA
auxiliary activities
- CBM33
a family 33 carbohydrate-binding module
- EPR
electron paramagnetic resonance
- GH61
a family 61 glycoside hydrolase
- haa
hydrogen atom abstraction
- LPMO
lytic polysaccharide monooxygenase
Contributor Information
Bastien Bissaro, Email: bastien.bissaro@inrae.fr.
Vincent G.H. Eijsink, Email: vincent.eijsink@nmbu.no.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
V.G.H.E. acknowledges support from the Research Council of Norway over many years, and recent support from the European Research Council, through the ERC‐Synergy program, for the project ‘CUBE – Unraveling the secrets of Cu‐based catalysts for C−H activation’ [grant number 856446]. B.B. thanks INRAE, the AgreenSkills (Marie-Curie COFUND) programme and the French National Research Agency (ANR) for support of the past and present research.
Author Contribution
B.B. and V.G.H.E. wrote the manuscript.
References
- 1.Ayers A.R., Ayers S.B. and Eriksson K.‐E. (1978) Cellobiose oxidase, purification and partial characterization of a hemoprotein from Sporotrichum pulverulentum. Eur. J. Biochem. 90, 171–181 10.1111/j.1432-1033.1978.tb12588.x [DOI] [PubMed] [Google Scholar]
- 2.Vaaje-Kolstad G., Westereng B., Horn S.J., Liu Z., Zhai H., Sørlie M.et al. (2010) An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222 10.1126/science.1192231 [DOI] [PubMed] [Google Scholar]
- 3.Vaaje-Kolstad G., Houston D.R., Riemen A.H.K., Eijsink V.G.H. and van Aalten D.M.F. (2005) Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J. Biol. Chem. 280, 11313–11319 10.1074/jbc.M407175200 [DOI] [PubMed] [Google Scholar]
- 4.Vaaje-Kolstad G., Horn S.J., van Aalten D.M.F., Synstad B. and Eijsink V.G.H. (2005) The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J. Biol. Chem. 280, 28492–28497 10.1074/jbc.M504468200 [DOI] [PubMed] [Google Scholar]
- 5.Merino S.T. and Cherry J. (2007) Progress and challenges in enzyme development for biomass utilization. Adv. Biochem. Eng. Biotechnol. 108, 95–120 10.1007/10_2007_066 [DOI] [PubMed] [Google Scholar]
- 6.Karkehabadi S., Hansson H., Kim S., Piens K., Mitchinson C. and Sandgren M. (2008) The first structure of a glycoside hydrolase family 61 member, Cel61B from Hypocrea jecorina, at 1.6 A resolution. J. Mol. Biol. 383, 144–154 10.1016/j.jmb.2008.08.016 [DOI] [PubMed] [Google Scholar]
- 7.Johansen K.S. (2016) Discovery and industrial applications of lytic polysaccharide mono-oxygenases. Biochem. Soc. Trans. 44, 143–149 10.1042/BST20150204 [DOI] [PubMed] [Google Scholar]
- 8.Müller G., Várnai A., Johansen K.S., Eijsink V.G.H. and Horn S.J. (2015) Harnessing the potential of LPMO-containing cellulase cocktails poses new demands on processing conditions. Biotechnol. Biofuels 8, 1–9 10.1186/s13068-015-0376-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J., Arantes V., Pribowo A., Gourlay K. and Saddler J.N. (2014) Substrate factors that influence the synergistic interaction of AA9 and cellulases during the enzymatic hydrolysis of biomass. Energy Environ. Sci. 7, 2308–2315 10.1039/C4EE00891J [DOI] [Google Scholar]
- 10.Cannella D. and Jørgensen H. (2014) Do new cellulolytic enzyme preparations affect the industrial strategies for high solids lignocellulosic ethanol production? Biotechnol. Bioeng. 111, 59–68 10.1002/bit.25098 [DOI] [PubMed] [Google Scholar]
- 11.Vandhana T.M., Reyre J.-L., Dangudubiyyam S., Berrin J.-G., Bissaro B. and Madhuprakash J. (2022) On the expansion of biological functions of Lytic Polysaccharides Monooxygenases. New Phytol. 233, 2380–2396 10.1111/nph.17921 [DOI] [PubMed] [Google Scholar]
- 12.Gonçalves A.P., Heller J., Span E.A., Rosenfield G., Do H.P., Palma-Guerrero J.et al. (2019) Allorecognition upon fungal cell-cell contact determines social cooperation and impacts the acquisition of multicellularity. Curr. Biol. 29, 3006.e3–3017.e3 10.1016/j.cub.2019.07.060 [DOI] [PubMed] [Google Scholar]
- 13.Sabbadin F., Hemsworth G.R., Ciano L., Henrissat B., Dupree P., Tryfona T.et al. (2018) An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion. Nat Commun. 9, 756 10.1038/s41467-018-03142-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong E., Vaaje-Kolstad G., Ghosh A., Hurtado-Guerrero R., Konarev P.V., Ibrahim A.F.et al. (2012) The Vibrio cholerae colonization factor GbpA possesses a modular structure that governs binding to different host surfaces. PLoS Pathog. 8, e1002373 10.1371/journal.ppat.1002373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabbadin F., Urresti S., Henrissat B., Avrova A.O., Welsh L.R.J., Lindley P.J.et al. (2021) Secreted pectin monooxygenases drive plant infection by pathogenic oomycetes. Science 373, 774–779 10.1126/science.abj1342 [DOI] [PubMed] [Google Scholar]
- 16.Askarian F., Uchiyama S., Masson H., Sørensen H.V., Golten O., Bunæs A.C.et al. (2021) The lytic polysaccharide monooxygenase CbpD promotes Pseudomonas aeruginosa virulence in systemic infection. Nat. Commun. 12, 1–19 10.1038/s41467-021-21473-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eibinger M., Ganner T., Bubner P., Rosker S., Kracher D., Haltrich D.et al. (2014) Cellulose surface degradation by a lytic polysaccharide monooxygenase and its effect on cellulase hydrolytic efficiency. J. Biol. Chem. 289, 35929–35938 10.1074/jbc.M114.602227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song B., Li B., Wang X., Shen W., Park S., Collings C.et al. (2018) Real-time imaging reveals that lytic polysaccharide monooxygenase promotes cellulase activity by increasing cellulose accessibility. Biotechnol. Biofuels 11, 41 10.1186/s13068-018-1023-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hedegård E.D. and Ryde U. (2017) Targeting the reactive intermediate in polysaccharide monooxygenases. J. Biol. Inorg. Chem. 22, 1029–1037 10.1007/s00775-017-1480-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo Y.R. (2002) Chapter 3. Tabulated BDEs of C–H bonds. Handbook of Bond Dissociation Energies in Organic Compounds CRC Press, Boca Raton: 10.1201/9781420039863 [DOI] [Google Scholar]
- 21.Gao J., Thomas D.A., Sohn C.H. and Beauchamp J.L. (2013) Biomimetic reagents for the selective free radical and acid-base chemistry of glycans: application to glycan structure determination by mass spectrometry. J. Am. Chem. Soc. 135, 10684–10692 10.1021/ja402810t [DOI] [PubMed] [Google Scholar]
- 22.Harris P.V., Welner D., McFarland K.C., Re E., Navarro Poulsen J.C., Brown K.et al. (2010) Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry 49, 3305–3316 10.1021/bi100009p [DOI] [PubMed] [Google Scholar]
- 23.Phillips C.M., Beeson W.T., Cate J.H. and Marletta M.A. (2011) Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 6, 1399–1406 10.1021/cb200351y [DOI] [PubMed] [Google Scholar]
- 24.Quinlan R.J., Sweeney M.D., Lo Leggio L., Otten H., Poulsen J.-C.N., Johansen K.S.et al. (2011) Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. U.S.A. 108, 15079–15084, 10.1073/pnas.1105776108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciano L., Davies G.J., Tolman W.B. and Walton P.H. (2018) Bracing copper for the catalytic oxidation of C-H bonds. Nat. Catal. 1, 571–577 10.1038/s41929-018-0110-9 [DOI] [Google Scholar]
- 26.Walton P.H. and Davies G.J. (2022) Structural perturbations of substrate binding and oxidation state changes in a lytic polysaccharide monooxygenase. J. Biol. Inorg. Chem. 27, 705–713 10.1007/s00775-022-01966-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tandrup T., Muderspach S.J., Banerjee S., Santoni G., Ipsen J., Hernández-Rollán C.et al. (2022) Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding. IUCrJ. 9, 666–681, Available from: http://scripts.iucr.org/cgi-bin/paper?mf5061 10.1107/S2052252522007175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S., Ståhlberg J., Sandgren M., Paton R.S. and Beckham G.T. (2014) Quantum mechanical calculations suggest that lytic polysaccharide monooxygenases use a copper-oxyl, oxygen-rebound mechanism. Proc. Natl. Acad. Sci. U.S.A. 111, 149–154 10.1073/pnas.1316609111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang B., Johnston E.M., Li P., Shaik S., Davies G.J., Walton P.H.et al. (2018) QM/MM studies into the H2O2-dependent activity of lytic polysaccharide monooxygenases: evidence for the formation of a caged hydroxyl radical intermediate. ACS Catal. 8, 1346–1351 10.1021/acscatal.7b03888 [DOI] [Google Scholar]
- 30.Bissaro B., Streit B., Isaksen I., Eijsink V.G.H., Beckham G.T., DuBois J.L.et al. (2020) Molecular mechanism of the chitinolytic peroxygenase reaction. Proc. Natl. Acad. Sci. U.S.A. 117, 1504–1513 10.1073/pnas.1904889117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chylenski P., Bissaro B., Sørlie M., Røhr Å.K., Várnai A., Horn S.J.et al. (2019) Lytic polysaccharide monooxygenases in enzymatic processing of lignocellulosic biomass. ACS Catal. 9, 4970–4991 10.1021/acscatal.9b00246 [DOI] [Google Scholar]
- 32.Aachmann F.L., Sørlie M., Skjåk-Bræk G., Eijsink V.G.H. and Vaaje-Kolstad G. (2012) NMR structure of a lytic polysaccharide monooxygenase provides insight into copper binding, protein dynamics, and substrate interactions. Proc. Natl. Acad. Sci. U.S.A. 109, 18779–18784 10.1073/pnas.1208822109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kracher D., Scheiblbrandner S., Felice A.K.G., Breslmayr E., Preims M., Haltrich D.et al. (2016) Extracellular electron transfer systems fuel cellulose oxidative degradation. Science 352, 1098–1101 10.1126/science.aaf3165 [DOI] [PubMed] [Google Scholar]
- 34.Garajova S., Mathieu Y., Beccia M.R., Bennati-Granier C., Biaso F., Fanuel M.et al. (2016) Single-domain flavoenzymes trigger lytic polysaccharide monooxygenases for oxidative degradation of cellulose. Sci. Rep. 6, 1–9 10.1038/srep28276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frommhagen M., Westphal A.H., van Berkel W.J.H. and Kabel M.A. (2018) Distinct substrate specificities and electron-donating systems of fungal lytic polysaccharide monooxygenases. Front. Microbiol. 9, 1–22 10.3389/fmicb.2018.01080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frandsen K.E.H., Simmons T.J., Dupree P., Poulsen J.-C.N., Hemsworth G.R., Ciano L.et al. (2016) The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases. Nat. Chem. Biol. 12, 298–303 10.1038/nchembio.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tandrup T., Frandsen K.E.H., Johansen K.S., Berrin J.G. and Leggio L. (2018) Recent insights into lytic polysaccharide monooxygenases (LPMOs). Biochem. Soc. Trans. 46, 1431–1447 10.1042/BST20170549 [DOI] [PubMed] [Google Scholar]
- 38.Bissaro B., Isaksen I., Vaaje-Kolstad G., Eijsink V.G.H. and Røhr Å.K. (2018) How a lytic polysaccharide monooxygenase binds crystalline chitin. Biochemistry 57, 1893–1906 10.1021/acs.biochem.8b00138 [DOI] [PubMed] [Google Scholar]
- 39.Borisova A.S., Isaksen T., Dimarogona M., Kognole A.A., Mathiesen G., Várnai A.et al. (2015) Structural and functional characterization of a lytic polysaccharide monooxygenase with broad substrate specificity. J. Biol. Chem. 290, 22955–22969 10.1074/jbc.M115.660183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Courtade G., Ciano L., Paradisi A., Lindley P.J., Forsberg Z., Sørlie M.et al. (2020) Mechanistic basis of substrate-O2 coupling within a chitin-active lytic polysaccharide monooxygenase: An integrated NMR/EPR study. Proc. Natl. Acad. Sci. U.S.A. 117, 19178–19189 10.1073/pnas.2004277117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kittl R., Kracher D., Burgstaller D., Haltrich D. and Ludwig R. (2012) Production of four Neurospora crassa lytic polysaccharide monooxygenases in Pichia pastoris monitored by a fluorimetric assay. Biotechnol. Biofuels 5, 79 10.1186/1754-6834-5-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bissaro B., Varnai A., Røhr Å.K. and Eijsink V.G.H. (2018) Oxidoreductases and reactive oxygen species in lignocellulose biomass conversion. Microbiol. Mol. Biol. Rev. 82, e00029–18 10.1128/MMBR.00029-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cannella D., Möllers K.B., Frigaard N.-U., Jensen P.E., Bjerrum M.J., Johansen K.S.et al. (2016) Light-driven oxidation of polysaccharides by photosynthetic pigments and a metalloenzyme. Nat. Commun. 7, 11134 10.1038/ncomms11134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bissaro B., Røhr Å.K., Skaugen M., Forsberg Z., Horn S.J., Vaaje-Kolstad G.et al. (2016) Fenton-type chemistry by a copper enzyme: molecular mechanism of polysaccharide oxidative cleavage. bioRxiv 10.1101/097022 [DOI] [Google Scholar]
- 45.Bissaro B., Røhr Å.K., Müller G., Chylenski P., Skaugen M., Forsberg Z.et al. (2017) Oxidative cleavage of polysaccharides by monocopper enzymes depends on H2O2. Nat. Chem. Biol. 13, 1123–1128 10.1038/nchembio.2470 [DOI] [PubMed] [Google Scholar]
- 46.Möllers K.B., Mikkelsen H., Simonsen T.I., Cannella D., Johansen K.S., Bjerrum M.J.et al. (2017) On the formation and role of reactive oxygen species in light-driven LPMO oxidation of phosphoric acid swollen cellulose. Carbohydr. Res. 448, 182–186 10.1016/j.carres.2017.03.013 [DOI] [PubMed] [Google Scholar]
- 47.Hangasky J.A., Iavarone A.T. and Marletta M.A. (2018) Reactivity of O2 versus H2O2 with polysaccharide monooxygenases. Proc. Natl. Acad. Sci. U.S.A. 115, 4915–4920 10.1073/pnas.1801153115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stepnov A.A., Forsberg Z., Sørlie M., Nguyen G.S., Wentzel A., Røhr Å.K.et al. (2021) Unraveling the roles of the reductant and free copper ions in LPMO kinetics. Biotechnol. Biofuels 14, 1–14 10.1186/s13068-021-01879-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stepnov A.A., Christensen I.A., Forsberg Z., Aachmann F.L., Courtade G. and Eijsink V.G.H. (2022) The impact of reductants on the catalytic efficiency of a lytic polysaccharide monooxygenase and the special role of dehydroascorbic acid. FEBS Lett. 596, 53–70 10.1002/1873-3468.14246 [DOI] [PubMed] [Google Scholar]
- 50.Kracher D., Forsberg Z., Bissaro B., Gangl S., Preims M., Sygmund C.et al. (2020) Polysaccharide oxidation by lytic polysaccharide monooxygenase is enhanced by engineered cellobiose dehydrogenase. FEBS J. 287, 897–908 10.1111/febs.15067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loose J.S.M., Forsberg Z., Kracher D., Scheiblbrandner S., Ludwig R., Eijsink V.G.H.et al. (2016) Activation of bacterial lytic polysaccharide monooxygenases with cellobiose dehydrogenase. Protein Sci. 25, 2175–2186 10.1002/pro.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuusk S., Bissaro B., Kuusk P., Forsberg Z., Eijsink V.G.H., Sørlie M.et al. (2018) Kinetics of H2O2-driven degradation of chitin by a bacterial lytic polysaccharide monooxygenase. J. Biol. Chem. 293, 523–531 10.1074/jbc.M117.817593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuusk S., Kont R., Kuusk P., Heering A., Sørlie M., Bissaro B.et al. (2019) Kinetic insights into the role of the reductant in H2O2-driven degradation of chitin by a bacterial lytic polysaccharide monooxygenase. J. Biol. Chem. 294, 1516–1528 10.1074/jbc.RA118.006196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kont R., Bissaro B., Eijsink V.G.H. and Väljamäe P. (2020) Kinetic insights into the peroxygenase activity of cellulose-active lytic polysaccharide monooxygenases (LPMOs). Nat. Commun. 11, 1–10 10.1038/s41467-020-19561-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones S.M., Transue W.J., Meier K.K., Kelemen B. and Solomon E.I. (2020) Kinetic analysis of amino acid radicals formed in H2O2-driven CuI LPMO reoxidation implicates dominant homolytic reactivity. Proc. Natl. Acad. Sci. U.S.A. 117, 11916–11922 10.1073/pnas.1922499117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hedison T.M., Breslmayr E., Shanmugam M., Karnpakdee K., Heyes D.J., Green A.P.et al. (2021) Insights into the H2O2-driven catalytic mechanism of fungal lytic polysaccharide monooxygenases. FEBS J. 288, 4115–4128 10.1111/febs.15704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bissaro B., Kommedal E., Røhr Å.K. and Eijsink V.G.H. (2020) Controlled depolymerization of cellulose by light-driven lytic polysaccharide oxygenases. Nat. Commun. 11, 890 10.1038/s41467-020-14744-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rieder L., Petrović D., Väljamäe P., Eijsink V.G.H. and Sørlie M. (2021) Kinetic characterization of a putatively chitin-active LPMO reveals a preference for soluble substrates and absence of monooxygenase activity. ACS Catal. 11, 11685–11695 10.1021/acscatal.1c03344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang H., Gacias Amengual N., Botz A., Schwaiger L., Kracher D., Scheiblbrandner S.et al. (2022) Investigating lytic polysaccharide monooxygenase-assisted wood cell wall degradation with microsensors. Nat. Commun. 13, 1–11 10.1038/s41467-022-33963-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brander S., Tokin R., Ipsen J., Jensen P.E., Hernández-Rollán C., Nørholm M.H.H.et al. (2021) Scission of glucosidic bonds by a lentinus similis lytic polysaccharide monooxygenases is strictly dependent on H2O2 while the oxidation of saccharide products depends on O2. ACS Catal. 11, 13848–13859 10.1021/acscatal.1c04248 [DOI] [Google Scholar]
- 61.Kuusk S. and Väljamäe P. (2021) Kinetics of H2O2-driven catalysis by a lytic polysaccharide monooxygenase from the fungus Trichoderma reesei. J. Biol. Chem. 297, 101256 10.1016/j.jbc.2021.101256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scott B.R., Huang H.Z., Frickman J., Halvorsen R. and Johansen K.S. (2016) Catalase improves saccharification of lignocellulose by reducing lytic polysaccharide monooxygenase-associated enzyme inactivation. Biotechnol. Lett 38, 425–434 10.1007/s10529-015-1989-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J.Y. and Karlin K.D. (2015) Elaboration of copper-oxygen mediated C-H activation chemistry in consideration of future fuel and feedstock generation. Curr. Opin. Chem. Biol. 25, 184–193 10.1016/j.cbpa.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dhar D. and Tolman W.B. (2015) Hydrogen atom abstraction from hydrocarbons by a copper(III)-hydroxide complex. J. Am. Chem. Soc. 137, 1322–1329 10.1021/ja512014z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walton P.H. and Davies G.J. (2016) On the catalytic mechanisms of lytic polysaccharide monooxygenases. Curr. Opin. Chem. Biol. 31, 1–13 10.1016/j.cbpa.2016.04.001 [DOI] [PubMed] [Google Scholar]
- 66.Kjaergaard C.H., Qayyum M.F., Wong S.D., Xu F., Hemsworth G.R., Walton D.J.et al. (2014) Spectroscopic and computational insight into the activation of O2 by the mononuclear Cu center in polysaccharide monooxygenases. Proc. Natl. Acad. Sci. U.S.A. 111, 8797–8802 10.1073/pnas.1408115111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bertini L., Breglia R., Lambrughi M., Fantucci P., De Gioia L., Borsari M.et al. (2018) Catalytic mechanism of fungal lytic polysaccharide monooxygenases investigated by first-principles calculations. Inorg. Chem. 57, 86–97 10.1021/acs.inorgchem.7b02005 [DOI] [PubMed] [Google Scholar]
- 68.Bissaro B., Streit B., Isaksen I., Beckham G.T., Eijsink V.G., DuBois J.et al. (2020) Molecular mechanism of the chitinolytic peroxygenase reaction. Proc. Natl. Acad. Sci. U.S.A. 117, 1504–1513 10.1073/pnas.1904889117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X., Beeson W.T., Phillips C.M., Marletta M.A. and Cate J.H.D. (2012) Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure 20, 1051–1061 10.1016/j.str.2012.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eijsink V.G.H., Petrovic D., Forsberg Z., Mekasha S., Røhr Å.K., Várnai A.et al. (2019) On the functional characterization of lytic polysaccharide monooxygenases (LPMOs). Biotechnol. Biofuels 12, 58 10.1186/s13068-019-1392-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stepnov A.A. and Eijsink V.G.H. (2022) Looking at LPMO reactions through the lens of the HRP/Amplex Red assay. Methods Enzymol. 679, 163–189 10.1016/bs.mie.2022.08.049 [DOI] [PubMed] [Google Scholar]
- 72.Petrović D.M., Várnai A., Dimarogona M., Mathiesen G., Sandgren M., Westereng B.et al. (2019) Comparison of three seemingly similar lytic polysaccharide monooxygenases from Neurospora crassa suggests different roles in plant biomass degradation. J. Biol. Chem. 294, 15068–15081 10.1074/jbc.RA119.008196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sigmund M.C. and Poelarends G.J. (2020) Current state and future perspectives of engineered and artificial peroxygenases for the oxyfunctionalization of organic molecules. Nat. Catal. 3, 690–702 10.1038/s41929-020-00507-8 [DOI] [Google Scholar]
- 74.Wang B., Zhang X., Fang W., Rovira C. and Shaik S. (2022) How do metalloproteins tame the Fenton reaction and utilize •OH radicals in constructive manners? Acc. Chem. Res. 55, 2280–2290 10.1021/acs.accounts.2c00304 [DOI] [PubMed] [Google Scholar]
- 75.Wang C., Chang W.C., Guo Y., Huang H., Peck S.C., Pandelia M.E.et al. (2013) Evidence that the fosfomycin-producing epoxidase, HppE, is a non-heme-iron peroxidase. Science 342, 991–995 10.1126/science.1240373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peng W., Qu X., Shaik S. and Wang B. (2021) Deciphering the oxygen activation mechanism at the CuC site of particulate methane monooxygenase. Nat. Catal. 4, 266–273 10.1038/s41929-021-00591-4 [DOI] [Google Scholar]
- 77.Gray H.B. and Winkler J.R. (2015) Hole hopping through tyrosine/tryptophan chains protects proteins from oxidative damage. Proc. Natl. Acad. Sci. U.S.A. 112, 10920–10925 10.1073/pnas.1512704112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paradisi A., Johnston E.M., Tovborg M., Nicoll C.R., Ciano L., Dowle A.et al. (2019) Formation of a copper(II)-tyrosyl complex at the active site of lytic polysaccharide monooxygenases following oxidation by H2O2. J. Am. Chem. Soc. 141, 18585–18599 10.1021/jacs.9b09833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singh R.K., Blossom B.M., Russo D.A., Singh R., Weihe H., Andersen N.H.et al. (2020) Detection and characterization of a novel copper-dependent intermediate in a lytic polysaccharide monooxygenase. Chem. - A Eur. J. 26, 454–463, , 10.1002/chem.201903562 [DOI] [PubMed] [Google Scholar]
- 80.Berrin J., Rosso M. and Abou Hachem M. (2017) Fungal secretomics to probe the biological functions of lytic polysaccharide monooxygenases. Carbohydr. Res. 448, 155–160, 10.1016/j.carres.2017.05.010 [DOI] [PubMed] [Google Scholar]