Abstract
OBJECTIVE
Environmental exposures may have greater predictive power for type 2 diabetes than polygenic scores (PGS). Studies examining environmental risk factors, however, have included only individuals with European ancestry, limiting the applicability of results. We conducted an exposome-wide association study in the multiancestry Personalized Environment and Genes Study to assess the effects of environmental factors on type 2 diabetes.
RESEARCH DESIGN AND METHODS
Using logistic regression for single-exposure analysis, we identified exposures associated with type 2 diabetes, adjusting for age, BMI, household income, and self-reported sex and race. To compare cumulative genetic and environmental effects, we computed an overall clinical score (OCS) as a weighted sum of BMI and prediabetes, hypertension, and high cholesterol status and a polyexposure score (PXS) as a weighted sum of 13 environmental variables. Using UK Biobank data, we developed a multiancestry PGS and calculated it for participants.
RESULTS
We found 76 significant associations with type 2 diabetes, including novel associations of asbestos and coal dust exposure. OCS, PXS, and PGS were significantly associated with type 2 diabetes. PXS had moderate power to determine associations, with larger effect size and greater power and reclassification improvement than PGS. For all scores, the results differed by race.
CONCLUSIONS
Our findings in a multiancestry cohort elucidate how type 2 diabetes odds can be attributed to clinical, genetic, and environmental factors and emphasize the need for exposome data in disease-risk association studies. Race-based differences in predictive scores highlight the need for genetic and exposome-wide studies in diverse populations.
Graphical Abstract
Introduction
The complex etiology of type 2 diabetes includes genetic, lifestyle, and environmental risk factors. Genome-wide association studies (GWAS) have revealed numerous risk loci for type 2 diabetes, and polygenic scores are well-established as predictive of disease risk (1–3). The impact of environmental exposures, or the exposome, is less well understood, and most studies have examined individual exposures and not considered mixtures and polyexposure effects.
Genetic risk and the effects of environmental exposures differ among racial and ethnic groups (4–7), but work on cumulative effects of environmental exposures using exposome-wide association studies (ExWAS) and risk score modeling, while promising, is limited to populations with European ancestry and has not examined sex differences (6–8). Therefore, studying diverse populations to ensure translational, inclusive results is imperative (7).
We conducted an ExWAS, similar to a GWAS, for type 2 diabetes in the Personalized Environment and Genes Study (PEGS), a diverse, North Carolina-based cohort with extensive health and internal and external exposure data. We detected novel exposure/disease associations and used machine-learning techniques to consider multiple exposures. To understand the relationship between disease odds and environmental exposures, genetics, and clinical factors, we built on the ExWAS results and compared the predictive performance of a polyexposure score (PXS), genome-wide polygenic score (PGS), and overall clinical score (OCS) built using established clinical factors. We further evaluated their performance with self-reported race- and sex-stratified analyses.
Research Design and Methods
Study Participants and Data
PEGS collects extensive survey data and whole-genome sequencing (WGS) data from individuals of varying age, race, education level, and socioeconomic status. We used data for 9,414 PEGS participants on genetic, environmental, and health outcomes for multiple phenotypes (Freeze 1.1) (9), which are outlined in Table 1A and the Supplementary Material.
Table 1A.
Data components from the PEGS cohort used in the study
| Data component | Description | Participants (n) |
|---|---|---|
| Health and Exposure Survey | Demographics, health, family history of disease, environmental exposures, socioeconomic status, and lifestyle factors | 9,414 |
| External Exposome Survey | Residential and occupational environmental exposures | 3,519 |
| Internal Exposome Survey | Medication use, physical activity, stress, sleep, diet, genetics, and reproductive history | 2,962 |
| WGS data | WGS data for PEGS participants | 4,737 |
Exposome Data
PEGS participants provide health and exposure information through three surveys. Beginning in 2013, participants were administered the Health and Exposure Survey, which asks for general health information, individual and family medical histories, and lifestyle and occupational exposures (n = 9,414). The External Exposome Survey covers exogenous exposures that include chemical and environmental exposures at home and work (n = 3,519). The Internal Exposome Survey covers endogenous exposures, such as medications and lifestyle factors such as sleep habits, stress, physical activity, and diet (n = 2,962) (Table 1A). The surveys were created using established scales and forms, as detailed in the Supplementary Material.
Genetic Data
The Broad Institute performed WGS for 4,737 PEGS participants with the most complete survey data. Samples were alig-ned to the hg38 human reference assembly, and joint genotyping was performed using the WGS germline single nucleotide polymorphism (SNP) and Indel workflow based on the Genome Analysis Tool Kit (GATK) (10). Quality control was performed using FastQC (11), the Picard Tools suite (12), chrXY (12) for consistency checks of self-reported and genotype-inferred sex, and fastStructure (13) for ancestry consistency. As detailed in the Supplementary Material, the raw data were independently analyzed using DeepVariant (14), and final consensus genotypes were those with identical values in DeepVariant and GATK output as well as the highest-quality variants identified by GATK alone.
Definition of Type 2 Diabetes and Covariates
The Health and Exposure Survey asks whether participants have been diagnosed with diabetes, their age at diagnosis, and current and past treatments but not diabetes type. To minimize phenotype misspecification, we used a cutoff of 20 years at diagnosis and excluded participants diagnosed with gestational diabetes. Supple-mentary Fig. 1 provides details.
As detailed in the Supplementary Material, we used the following covariates: age at survey completion, BMI at survey completion, household income, sex, and self-reported race (White, Black, and other).
Statistical Analysis
ExWAS
We conducted ExWAS to test the association of single variables with type 2 diabetes. After processing exposome and covariate data, we excluded variables with <10 observations per response category. We used the resulting 662 binary and ordered factor exposome variables in the following logistic regression model, with T2D indicating type 2 diabetes:
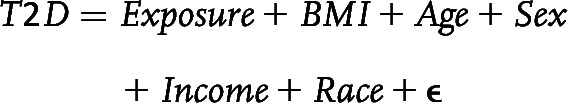 |
We used a Benjamini-Hochberg false discovery rate (FDR) of q < 0.10.
Deletion/Substitution/Addition Algorithm for Multiexposure Models
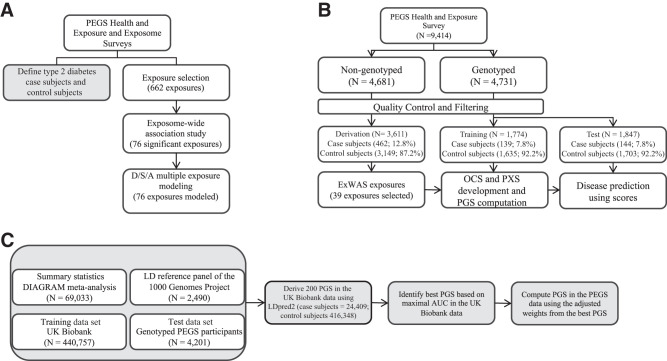
To examine concurrent exposures, we input ExWAS significant exposures to the deletion/substitution/addition (DSA) algorithm (15), which uses a series of deletions, substitutions, and additions to select an optimal multiexposure regression model of input variables for multiple regression analysis. Figure 1A summarizes the ExWAS workflow. As a sensitivity analysis, we stratified the population by self-reported race and sex and added smoking as a covariate because of the multiple smoking factors (i.e., smoking >100 cigarettes in a lifetime, smoking indoors, and smoking at home) significant in the ExWAS results to ensure the selection process was not driven by collinearity among these variables.
Figure 1.
A: ExWAS design workflow. We classified type 2 diabetes from Health and Exposure Survey data. We selected exposures for ExWAS from PEGS questionnaire responses and individually modeled these exposures for associations with type 2 diabetes. We used DSA modeling to select the most parsimonious model for exposures associated with type 2 diabetes (FDR < 0.10). B: Risk score design and workflow. We divided participants into three data sets based on genotyping status. The derivation data set comprised nongenotyped participants and was used to derive risk score features with an ExWAS. We split genotyped participants into training and test data sets. We developed OCS and PXS in the training data set using lasso regression. We computed PXS using 13 exposure variables and OCS using four clinical variables. We developed a PGS using 122,359 SNPs in UK Biobank data. We tested risk scores for association with type 2 diabetes in the training data set. We then used the held-out test data set to evaluate the predictive accuracy of the computed risk scores for type 2 diabetes. C: PGS computation workflow. We used four data sets for PGS development. We used summary statistics from the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) consortium meta-analysis, the linkage disequilibrium (LD) reference panel of the 1000 Genomes Project, and UK Biobank data as the training data set and the genotyped PEGS participants as the test data set. We used LDpred2 to derive 200 polygenic risk scores in UK Biobank data from which we selected the best PGS based on the AUC. We used the adjusted weights of the best PGS to compute PGS for the genotyped PEGS participants.
Classification Scores
To assess the effects of multiple factors on the odds of disease and determine whether PXS provides additional information, we computed OCS, PGS, and PXS for participants. For optimal use of the population, we divided PEGS participants into 1) a derivation data set of participants without WGS (n = 3,611) and 2) training (n = 1,774) and 3) test (n = 1,847) data sets created by randomly splitting participants with WGS into two approximately equal-sized data sets. Figure 1B outlines the design of the score analyses.
Computation of Scores.
We developed a multiancestry PGS using UK Biobank data (16) based on summary statistics from a meta-analysis of GWAS data from the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium (17) (see Supplementary Table 1). We used LDpred2 (18) to develop a grid of 200 candidate PGS using 122,359 SNPs in UK Biobank data. We selected the best PGS based on the area under the curve (AUC) and, using the adjusted weights from the best score, computed PGS for the genotyped PEGS participants. Figure 1C outlines the PGS computation workflow, and the Supplementary Material provides additional details.
We computed OCS and PXS similarly to PGS, using the ExWAS results in the derivation data set analogous to GWAS summary statistics and the coefficients from the least absolute shrinkage and selection operator (lasso) model in the training data set analogous to adjusted SNP weights. Separately for OCS and PXS, we used lasso with 10-fold cross-validation to select nonredundant (i.e., coefficients not shrunken to 0) yet predictive (at the minimum cross-validated error in the score) variables in the training data set while controlling for age, sex, and the first 10 genetic principal components (R function cv.glmnet, package glmnet version 4.1-1) (19). We computed OCS and PXS as weighted sums of the selected variables, using lasso coefficients as weights. For OCS, we used BMI and prediabetes, hypertension, and high cholesterol status as these are known predictors for type 2 diabetes and were previously used to develop a risk score in a cohort of exclusively European ancestry (8,20). For PXS, 13 of the 39 exposures identified from ExWAS in the derivation data set were retained by lasso in the training data set (see Supplementary Table 2). We standardized OCS, PXS, and PGS separately so that each had a mean of 0 and an SD of 1 across all participants.
Classification Score Analyses.
We analyzed the classification scores in a data set comprising participants with WGS data and complete data on the disease phenotype, age, sex, the selected exposure variables, and several clinical variables. Figure 1B describes the score modeling design.
After quality control and filtering, the analyses included 3,611, 1,774, and 1,847 participants with 462, 139, and 144 cases in the derivation, training, and test data sets, respectively. Table 1B outlines the demographics of each data set. We used logistic regression to fit type 2 diabetes in the training data set for each score, controlling for age, sex, and the first 10 principal components that describe genetic ancestry (R function glm, package stats version 4.0.1) (19). We used the training data set model fits to predict the odds of assignment to the case group for the test data set and assessed the predictive performance of each score based on the AUC. We estimated the proportion of variance explained by each score with the Nagelkerke pseudo-R2 metric (21), which is the R2 for the full model (control variables and the predictive score) minus the R2 for control variables only. We modeled associations between the scores and disease status. We then compared the continuous net reclassification index (NRI) for each model to the NRI of the OCS model to determine whether the addition of these terms improved the model (R function nribin, package nricens version 1.6) (19). We further stratified our models by self-reported race (Black and White) and self-reported sex (male and female) separately and controlled for age, sex, and the first 10 principal components for each subgroup, except for the sex-stratified models, which did not include sex as a covariate.
Table 1B.
Demographics of participants in the derivation, training, and test data sets used in the predictive score analyses
| Demographic variable | Derivation data setn = 3,611 | Training data setn = 1,774 | Test data setn = 1,847 |
|---|---|---|---|
| Type 2 diabetes phenotype, n (%) | |||
| Case subjects | 462 (12.8) | 139 (7.8) | 144 (7.8) |
| Control subjects | 3,149 (87.2) | 1,635 (92.2) | 1,703 (92.2) |
| Sex, n (%) | |||
| Female | 2,348 (65.0) | 1,206 (68.0) | 1,254 (67.9) |
| Male | 1,263 (35.0) | 568 (32.0) | 593 (32.1) |
| Race, n (%) | |||
| White | 2,439 (67.5) | 1,468 (82.8) | 1,490 (80.7) |
| Black | 1,020 (28.3) | 231 (13.0) | 253 (13.7) |
| Other | 152 (4.2) | 75 (4.2) | 104 (5.6) |
| Ethnicity, n (%) | |||
| Non-Hispanic/non-Latino | 3,506 (97.1) | 1,708 (96.8) | 1,807 (97.8) |
| Hispanic/Latino | 105 (2.9) | 52 (2.9) | 32 (1.7) |
| Age, mean (SD), years | 30.61 (16.4) | 49.4 (14.6) | 49.5 (14.5) |
| BMI, mean (SD), kg/m2 | 29.14 (7.1) | 27.9 (6.5) | 28 (6.6) |
As an additional test of whether environmental exposures in PEGS data provide further information over PGS or OCS, we separately modeled the association of PGS or OCS along with each lasso-selected exposure variable for disease status in the test data set, using logistic regression and controlling for age, sex, and the 10 principal components. To account for multiple testing, we used q < 0.10. For a sensitivity analysis comparing the associations of OCS, PXS, and PGS with type 2 diabetes, we computed OCS without prediabetes. The Supplementary Material provides details. We conducted all analyses in R version 4.0.1 software (19).
Results
As expected, participants with type 2 diabetes had 17% higher BMI and were 40% older than subjects not diagnosed. Of the participants with type 2 diabetes, 60% were White compared with 71% in the overall cohort, while 32% of participants with the disease were Black compared with 22% in the overall cohort. There was also income disparity, with average income of $40,000–$49,000 for individuals with the disease and $50,000–$59,000 for the overall cohort. Supplementary Table 3 provides demographic information for the PEGS participants included in the ExWAS.
ExWAS Results
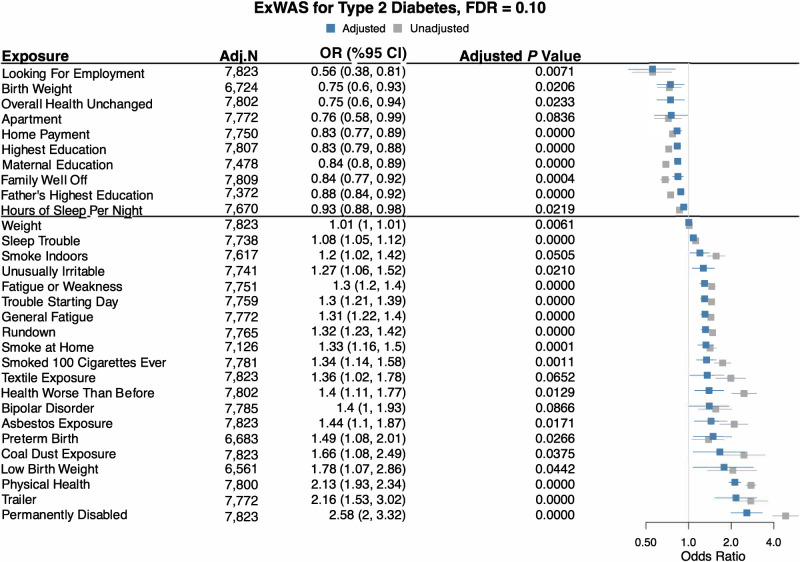
We conducted an ExWAS with 78 exposures from the Health and Exposure Survey, 289 from the Internal Exposome Survey, and 295 from the External Exposome Survey. After adjusting for age, BMI, income, sex, and self-reported race, 28, 0, and 48 exposures were significant, respectively. Figure 2 shows the odds ratios (ORs) and CIs for all statistically significant exposures from the Health and Exposure Survey at q < 0.10. A sensitivity analysis with a more stringent FDR of q < 0.05 showed the results are highly consistent (shown in Supplementary Fig. 2). Supplementary Figs. 3–14 show the sensitivity analysis results for the Internal and External Exposome Surveys overall and stratified by self-reported race and sex with q < 0.10 and q < 0.05.
Figure 2.
ExWAS results for the PEGS Health and Exposure Survey. This survey requests information on lifestyle, environmental, and occupational exposures. We regressed all exposures on type 2 diabetes with the covariates of age, BMI, income, and sex. Adj. N value is the number of participants with complete case data in each analysis. We used exponentiated ORs and 95% CIs to assess association direction and an FDR cutoff of 10% to select exposures. Blue denotes adjusted ORs and CIs, and gray denotes unadjusted ORs and CIs.
As shown in Fig. 2, several biological and chemical exposures, namely, asbestos (OR 1.44 [95% CI 1.10, 1.87], adjusted P = 0.02) and coal dust exposure (1.66 [1.08, 2.49], P = 0.04), smoking >100 cigarettes in a lifetime (1.34 [1.14, 1.58], P = 0.001), smoking indoors (1.20 [1.02, 1.42], P = 0.05), and smoking at home (1.33 [1.16, 1.50], P < 0.00001), were significantly associated with increased odds of type 2 diabetes.
For socioeconomic factors and mental health variables, which we considered as exposures, living in a trailer was associated with increased disease odds (OR 2.16 [95% CI 1.53, 3.02], P < 0.00001), while having a mortgage was associated with decreased odds (0.83 [0.77, 0.89], P < 0.00001). Family income (0.84 [0.77, 0.92], P < 0.00001) and maternal (0.84 [0.80, 0.89], P < 0.00001) and paternal (0.88 [0.84, 0.92], P < 0.00001) education levels were inversely associated with odds. Unusual irritability (1.27 [1.06, 1.52], P = 0.02) and trouble sleeping (1.08 [1.05, 1.12], P < 0.00001) were associated with increased odds.
Medication and lifestyle variables associated with increased disease odds include use of any medication (OR 1.60 [95% CI 1.43, 1.79], P < 0.00001) and acid reflux medication (1.99 [1.31, 2.99], P = 0.009). Diet variables associated with increased odds include frequently eating at a buffet (1.69 [1.3, 2.18], P = 0.001), frequent cream cheese consumption (1.36 [1.11, 1.65], P = 0.02), frequent fast-food consumption (1.31 [1.09, 1.58], P = 0.02), and consumption of fish oil (1.72 [1.09, 2.47], P = 0.02), low-calorie soda (1.21 [1.09, 2.47], P = 0.003), and Splenda (1.19 [1.07, 1.31], P = 0.008). Skim milk consumption was associated with decreased odds (0.74 [0.59, 0.89], P = 0.02). Diagnosis with a sleep disorder (2.41 [1.64, 3.54], P < 0.00001) was also associated with increased odds.
For a further investigation of the effects of sex, self-reported race, and smoking, we separately conducted several sensitivity analyses that are detailed in the Supplementary Material and Supplementary Figs. 5–8.
DSA Results
We used DSA to evaluate multivariable models and follow-up ExWAS-identified univariate associations. The most parsimonious model for Health and Exposure Survey data included maternal and paternal education level, smoking at home, smoking indoors, smoking >100 cigarettes in a lifetime, having a mortgage, trouble sleeping, home type, asbestos exposure, coal dust exposure, and education level. The most parsimonious model for Exposome Survey data included use of medication for high cholesterol, high blood pressure, and stroke; dietary variables of frequently eating at a buffet, frequent fast-food consumption, consumption of large quantities of nuts, and consumption of Splenda; sleep-related variables of hours of sleep/week, hours awake during the day, and self-reported high temperature of sleeping area; and high heart rate.
Classification Score Results
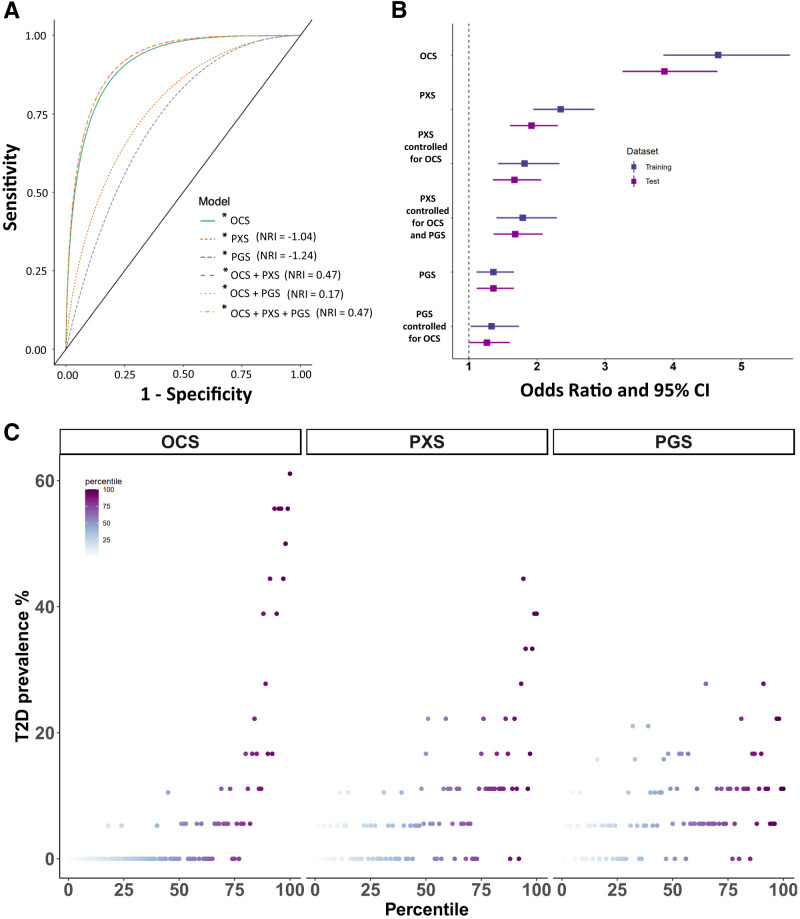
All classification scores were significantly associated with type 2 diabetes (P < 0.05) in the training and test data sets (Supplementary Table 4). Based on the AUC, OCS had the highest discriminative power, followed by PXS and PGS. The odds of having type 2 diabetes relative to not having it increased with all scores, with the ORs for OCS>PXS>PGS. Notably, PXS had a much higher OR (1.921 [95% CI 1.604, 2.304]) and AUC (0.775 [95% CI 0.737, 0.813]) than PGS (OR 1.360 [95% CI 1.115, 1.662]; AUC 0.731 [95% CI 0.692. 0.770]) (Fig. 3A and B).
Figure 3.
A: Sensitivity and specificity of OCS, PXS, PGS, and their combinations for association with type 2 diabetes odds. A higher AUC indicates better predictive accuracy. A positive NRI indicates improved reclassification. B: ORs and their 95% CIs for OCS, PXS, PGS, and their combinations for association with type 2 diabetes odds. For the combined risk score models, the displayed ORs are the values after controlling for other scores as noted. Dark blue denotes values from the training data set, and purple denotes values from the test data set. C: Predictive score percentile plots. Participants in the test data set were binned into 100 groups based on the percentile for each score separately. Type 2 diabetes (T2D) prevalence is plotted against the score percentile for each score.
For a relative comparison of the scores, we classified individuals with scores in the lower quartile (bottom 25 percentiles) and upper quartile (top 25 percentiles) as low and high risk. High-risk individuals categorized by OCS, PXS, and PGS were 50.7-, 4.9-, and 2.4-fold, respectively, more likely to have type 2 diabetes compared with low-risk individuals (see Supplementary Table 5). The proportion of variance explained, estimated using the Nagelkerke pseudo-R2, was the highest for OCS (0.331), followed by PXS (0.06) and PGS (0.011). Based on overall continuous NRI, the classification accuracy of the OCS model improved by 44.6% with the addition of PXS alone and 17.4% with the addition of PGS alone. As expected, adding both PXS and PGS yielded the highest overall continuous NRI of 46.9%, although this is a small improvement over the addition of PXS alone (Supplementary Table 4).
Also as expected, OCS showed the sharpest increase in disease odds from lowest to highest percentile, followed by PXS and PGS (Fig. 3C). The self-reported race- and sex-stratified analyses produced overall association and classification accuracy results in concordance with those of the multiancestry analyses, with differences in predictive scores across self-reported race and sex. In the test data set, the mean of each score was significantly higher for Black compared with White participants (Supplementary Fig. 15). For sex, PXS was significantly higher for women (Supplementary Fig. 16). The Supplementary Material provides details.
In the test data set, OCS computed without prediabetes had an OR of 2.802 (95% CI 2.316, 3.419) and AUC of 0.833 (95% CI 0.801, 0.866) (Supplementary Table 6). Compared with OCS computed with all selected clinical factors, this represents a decrease in OR by 1.067 and AUC by 0.086. The general trend of association with disease odds remained the same, with the ORs for OCS>PXS>PGS. Using low-/high-risk categories for predictive scores in the association models, OCS computed without prediabetes had an OR of 10.765 (95% CI 5.221, 25.269) compared with 50.741 (95% CI 15.61, 311.848) for OCS computed with all selected clinical factors (Supplementary Tables 5 and 7). Similarly, the addition of PXS, PGS, and PXS and PGS together resulted in a continuous NRI of 46.4%, 25.0%, and 58.7%, respectively (Supplementary Table 6). Adding PXS and PGS together to OCS computed without prediabetes resulted in a much higher NRI (58.7%) than adding them to OCS computed with prediabetes (46.9%) (Supplementary Tables 4 and 6).
To examine whether environmental exposures provide additional information on disease odds compared with OCS and PGS, we individually added each exposure variable selected by lasso to the OCS and PGS models, and nine and three of the 13 selected exposures, respectively, remained significant at q < 0.10 (Supplementary Tables 8 and 9). Notably, among the significant results in the PGS plus exposure models, the OR for smoking was 1.864 after controlling for PGS. These results indicate that individual exposures provide additional information over PGS and OCS for disease odds.
Conclusions
By performing an ExWAS, we discovered emerging risk factors for diabetes such as asbestos exposure. Further, by implementing robust modeling approaches, we provide support for adding environmental risk factors into decision support frameworks for public health interventions and, eventually, personalized medicine. The results of both the ExWAS and predictive score analysis confirm prior findings of the association of individual environmental factors (22,23) and the cumulative effects of multiple environmental factors with type 2 diabetes odds using PXS (8,9). Our results from modeling combinations of OCS, PXS, and PGS as well as OCS or PGS with individual environmental exposures provide insights into how disease odds can be attributed to clinical, genetic, and nongenetic factors.
ExWAS
The ExWAS results revealed associations of smoking, asbestos exposure, and coal dust exposure with type 2 diabetes. The associations of asbestos and coal dust exposure are novel and highlight the usefulness of our method in discovering previously unknown associations between environmental exposures and diseases, with reverse causality highly unlikely. While studies have loosely linked impaired lung function to type 2 diabetes, this understudied aspect could have implications for disease treatment and prevention (24,25). Our finding of an association of sleep disorder with the disease is another direction for future work. While disrupted sleep patterns may be artifacts of BMI-related diseases such as sleep apnea, this relationship warrants further investigation, particularly due to findings of increased insulin resistance in laboratory studies of interrupted sleep (26–28).
Classification Scores
Despite the smaller sample size in our data (N = 9,414 in PEGS vs. N = 502,536 in He et al.), our classification score analyses replicated several results reported in He et al. (8), who used a polyexposure risk score to predict type 2 diabetes. Their score included alcohol use, diet, early life factors, household and income information, sleep, and smoking, while our PXS included household-, income- and sleep-related variables and smoking but excluded diet-related variables due to low sample sizes. Our results show similar predictive power of the classification scores as determined by AUC or C statistic, namely OCS>PXS>PGS. Additionally, the NRI and AUC values for our score models are in alignment with He et al. (8) (see Fig. 3A), with the AUC for PXS higher than for PGS and the NRI for OCS+PXS higher than for OCS+PGS.
Determined by AUC, OCS had the highest predictive accuracy (0.919) when calculated with prediabetes, which is a strong predictor of type 2 diabetes (Supplementary Figs. 17 and 18). While PXS alone had moderate predictive accuracy, its addition to OCS improved disease classification accuracy by 44.6%. In the self-reported race- and sex-stratified analyses, the addition of PXS to OCS improved classification accuracy by 26% in the Black subgroup, 44.6% in the White subgroup, 51.4% in women, and 35% in men (Supplementary Tables 10–15). Because people with prediabetes are often unaware of their status (29–31), we conducted a sensitivity analysis comparing the classification scores with OCS computed without prediabetes (Supplementary Tables 6 and 7).
Utility of PXS
Several studies have shown the usefulness of PGS in predicting risk for complex diseases and improving clinical decision making in precision medicine (1,32–34). However, studies assessing the predictive utility of PXS are limited. Our results show PXS has moderate power to determine associations, with larger effect size and greater power and improvement in reclassification than PGS. Accordingly, PXS may be a stronger indicator of type 2 diabetes odds than PGS. By pseudo-R2, PXS explains a much larger proportion of variance in disease odds than PGS (PXS, 0.060; PGS, 0.011). By OR, PXS has a much larger effect size than PGS (PXS, 1.921; PGS, 1.360). PXS also had a much stronger association with disease odds than PGS and resulted in a much greater improvement in reclassification as determined by NRI (PXS, 44.6%; PGS, 17.4%). In addition, data for PXS can be obtained through questionnaires and are thus less expensive and easier to obtain than the genotyping or WGS data required for PGS. Importantly, unlike PGS, PXS is potentially modifiable.
Race Differences
While racial and ethnic differences have been found in associations of genetic and nongenetic factors with disease risk (4–7), there are few studies in multiancestry, non-European, and mixed populations (8,22,35,36). PGS built using data from European populations are several-fold less accurate in predicting disease risk/odds in non-European populations (5,7,23). We used data from the racially diverse PEGS cohort to compute multiancestry classification scores, which have higher predictive accuracy compared with race-stratified scores (18,37). Our results show clear differences for the scores between Black and White subgroups (Supplementary Fig. 8 and Supplementary Tables 10–12).
The importance of the sociocultural context of the varying results for Black and White subgroups cannot be understated. Race is a social construct correlated with health, economic, and exposure disparities. While socioeconomic status and other aspects of health disparities are important risk factors for many diseases, they are both a component and a confounder of our scores and may also be drivers of exposure disparities (23,38,39). Instead of stratifying scores by self-reported race, we developed multiancestry scores while controlling for genetic ancestry using the eigenvectors from the principal components computed from WGS data. Our results show environmental exposures have high predictive accuracy for type 2 diabetes, underlining the critical nature of the role of often modifiable exposures in health disparities. Further, as the first study to evaluate PXS in a multiancestry cohort, we take an important first step in contextualizing PXS, just as PGS are calculated and contextualized in ancestry-based subgroups, to understand health disparities.
Limitations
Despite its contributions, this study has limitations. First, because exposure data are self-reported, there may have been recall bias.
Second, some exposure variables were not included in the analyses due to insufficient sample sizes.
Third, although, by definition, we identify associations rather than causation, reverse causality between some PXS-identified exposures is possible (e.g., sleep and income). Owing to the observational nature of PEGS data, we cannot ascertain the direction of causality, potentially confounding variables, or comorbidities. For example, while socioeconomic status plays a substantial role in environmental exposures and is both a component and confounder of many diseases, it may play a role in exposure disparities.
Fourth, with any high-dimensional modeling, the FDR rate should be considered when interpreting the results.
Fifth, the small sample sizes of some of the stratified analyses may have affected statistical power, so the results should be interpreted with appropriate caution.
Sixth, we recognize that PXS is not as easily translatable as PGS. Unlike PGS, which can be computed once and used throughout an individual’s lifetime to assess disease risk, PXS is based on temporal factors and hence may need to be recomputed because of factors’ variability.
Finally, bias may exist because of differences between PEGS participants with genotyping data and those with exposure data only. Accordingly, further validation and replication in independent data sets are required.
Implications
Our results support the strong potential utility of PXS in precision medicine to prioritize patients for further screening or lifestyle modification and its higher accuracy than PGS. Understanding how individual and multiple exposures influence disease risk, coupled with knowledge of the proportion of risk due to genetics, can inform interventions to limit specific environmental exposures. Further, it is much less expensive and easier to collect the questionnaire-based environmental exposure data required for PXS than the genetic data required for PGS. This highlights the need for collecting individual exposure information and considering it in disease-risk association and prediction studies.
Within the field of human genetics, methods development for polygenic scores is rapidly advancing. Overall, our results support the expansion of methods development to PXS, both alone and in combination with other risk scores, to increase understanding of the complex interactions among genetic and environmental factors in disease etiology. Another area of focus in future work should be the collection of data on currently unmeasured exposures to increase the accuracy of the proportion of risk explained by predictive scores. These scores can substantially underestimate the proportion of risk explained as they are based on available exposure data. Including additional data on exposures will enable the building of risk/odds models with interactive effects to uncover and understand interactions.
Article Information
Acknowledgments. The authors would like to thank the PEGS participants for their contributions to this work. The authors would also like to thank the staff at DLH Corporation, namely Samantha Shuptrine, Rebecca Ritter, Nathaniel MacNell, Jamie Glover, Jennifer Emerson, and Nicole Edwards, for their support in creating and maintaining the PEGS cohort and data. Additionally, the authors would like to thank Hannah Collins, National Institute of Environmental Health Sciences, for help with manuscript preparation.
Funding. Financial support was received from intramural funds from the National Institutes of Health, National Institute of Environmental Health Sciences.
The opinions expressed in this article are those of the authors and do not necessarily reflect the views of the National Institutes of Health.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. F.S.A. wrote the manuscript, analyzed data, and contributed to discussion. D.L. wrote the manuscript, analyzed data, and contributed to discussion. A.B. reviewed and edited the manuscript, analyzed data, and contributed to discussion. X.T. reviewed and edited the manuscript, analyzed data, and contributed to discussion. J.S.H. reviewed and edited the manuscript, analyzed data, and contributed to discussion. E.Y.L. analyzed data and contributed to discussion. J.B. reviewed and edited the manuscript and contributed to discussion. S.H.S. collected data. D.C.F. reviewed and edited the manuscript and contributed to discussion. C.P.S. reviewed and edited the manuscript and contributed to discussion. J.H. developed research concepts, contributed to the manuscript, and edited the manuscript. A.A.M.-R. developed research concepts and contributed to, wrote, and revised the manuscript. A.A.M.-R. takes responsibility for the contents of the article. A.A.M.-R. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.21399177.
F.S.A. and D.L. contributed equally.
J.H. and A.A.M.-R. contributed equally.
References
- 1. Lambert SA, Abraham G, Inouye M. Towards clinical utility of polygenic risk scores. Hum Mol Genet 2019;28:R133–R142 [DOI] [PubMed] [Google Scholar]
- 2. Mars N, Koskela JT, Ripatti P, et al.; FinnGen . Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat Med 2020;26:549–557 [DOI] [PubMed] [Google Scholar]
- 3. Padilla-Martínez F, Collin F, Kwasniewski M, Kretowski A. Systematic review of polygenic risk scores for type 1 and type 2 diabetes. Int J Mol Sci 2020;21:1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Khera AV, Emdin CA, Drake I, et al. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med 2016;375:2349–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belsky DW, Moffitt TE, Sugden K, et al. Devel-opment and evaluation of a genetic risk score for obesity. Biodemogr Soc Biol 2013;59:85–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ware EB, Schmitz LL, Faul J, et al. Heterogeneity in polygenic scores for common human traits. 5 February 2017. [preprint]. bioRxiv:106062 [Google Scholar]
- 7. Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet 2019;51:584–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He Y, Lakhani CM, Rasooly D, Manrai AK, Tzoulaki I, Patel CJ. Comparisons of polyexposure, polygenic, and clinical risk scores in risk prediction of type 2 diabetes. Diabetes Care 2021;44:935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee EY, Akhtari F, House JS, et al. Questionnaire-based exposome-wide association studies (ExWAS) reveal expected and novel risk factors associated with cardiovascular outcomes in the Personalized Environment and Genes Study. Environ Res 2022;212:113463. [DOI] [PubMed] [Google Scholar]
- 10. Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11.0.1–11.0.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Andrews S. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics; 2010. Accessed 29 September 2022. Available from https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 12. Broad Institute . Picard tools. 2021. Accessed 29 July 2022. Available from https://broadinstitute.github.io/picard/
- 13. Raj A, Stephens M, Pritchard JK. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 2014;197:573–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poplin R, Chang PC, Alexander D, et al. A universal SNP and small-indel variant caller using deep neural networks. Nat Biotechnol 2018;36:983–987 [DOI] [PubMed] [Google Scholar]
- 15. Sinisi SE, van der Laan MJ. Deletion/substitution/addition algorithm in learning with applications in genomics. Stat Appl Genet Mol Biol 2004;3:Article18. [DOI] [PubMed] [Google Scholar]
- 16. Sudlow C, Gallacher J, Allen N, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scott RA, Scott LJ, Mägi R, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes 2017;66:2888–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Privé F, Arbel J, Vilhjálmsson BJ. LDpred2: better, faster, stronger. Bioinformatics 2020;36:5424–5431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. R Core Team . R: A language and environment for statistical computing. Vienna, Austria, R Foun-dation for Statistical Computing, 2020 [Google Scholar]
- 20. Wilson PW, Meigs JB, Sullivan L, Fox CS, Nathan DM, D’Agostino RB Sr. Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring Study. Arch Intern Med 2007;167:1068–1074 [DOI] [PubMed] [Google Scholar]
- 21. Nagelkerke NJ. A note on a general definition of the coefficient of determination. Biometrika 1991;78:691–692 [Google Scholar]
- 22. Morales J, Welter D, Bowler EH, et al. A standardized framework for representation of ancestry data in genomics studies, with application to the NHGRI-EBI GWAS Catalog. Genome Biol 2018;19:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vilhjálmsson BJ, Yang J, Finucane HK, et al.; Schizophrenia Working Group of the Psychiatric Genomics Consortium, Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) study . Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet 2015;97:576–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peng Y, Zhong GC, Wang L, et al. Chronic obstructive pulmonary disease, lung function and risk of type 2 diabetes: a systematic review and meta-analysis of cohort studies. BMC Pulm Med 2020;20:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heianza Y, Arase Y, Tsuji H, et al. Low lung function and risk of type 2 diabetes in Japanese men: the Toranomon Hospital Health Management Center Study 9 (TOPICS 9). Mayo Clin Proc 2012;87:853–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Punjabi NM, Shahar E, Redline S, Gottlieb DJ, Givelber R; Sleep Heart Health Study Investigators . Sleep-disordered breathing, glucose intolerance, and insulin resistance: the Sleep Heart Health Study. Am J Epidemiol 2004;160:521–530 [DOI] [PubMed] [Google Scholar]
- 27. Kent BD, Grote L, Bonsignore MR, et al.; European Sleep Apnoea Database collaborators . Sleep apnoea severity independently predicts glycaemic health in nondiabetic subjects: the ESADA study. Eur Respir J 2014;44:130–139 [DOI] [PubMed] [Google Scholar]
- 28. Reynolds AC, Banks S. Total sleep deprivation, chronic sleep restriction and sleep disruption. Prog Brain Res 2010;185:91–103 [DOI] [PubMed] [Google Scholar]
- 29. Abraham TM, Fox CS. Implications of rising prediabetes prevalence. Diabetes Care 2013;36:2139–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Campbell MD, Sathish T, Zimmet PZ, et al. Benefit of lifestyle-based T2DM prevention is influenced by prediabetes phenotype. Nat Rev Endocrinol 2020;16:395–400 [DOI] [PubMed] [Google Scholar]
- 31. Centers for Disease Control and Prevention . About Prediabetes & Type 2 Diabetes. 2021. Accessed 11 February 2022. Available from https://www.cdc.gov/diabetes/prevention/about-prediabetes.html
- 32. Khera AV, Chaffin M, Aragam KG, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018;50:1219–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med 2020;12:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet 2018;19:581–590 [DOI] [PubMed] [Google Scholar]
- 35. Hall MA, Dudek SM, Goodloe R, et al. Environment-wide association study (EWAS) for type 2 diabetes in the Marshfield Personalized Medicine Research Project Biobank. In: Pacific Symposium on Biocomputing. Altman RB, Dunker AK, Hunter L, Murray TA, Klein TE, Ritchie MD, Eds. Singapore, World Scientific, 2014, pp. 200–211 [PMC free article] [PubMed] [Google Scholar]
- 36. Need AC, Goldstein DB. Next generation disparities in human genomics: concerns and remedies. Trends Genet 2009;25:489–494 [DOI] [PubMed] [Google Scholar]
- 37. Weissbrod O, Kanai M, Shi H, et al. Leveraging fine-mapping and non-European training data to improve trans-ethnic polygenic risk scores. Nat Genet 2022;54:450–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gee GC, Payne-Sturges DC. Environmental health disparities: a framework integrating psychosocial and environmental concepts. Environ Health Perspect 2004;112:1645–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bagby SP, Martin D, Chung ST, Rajapakse N. From the outside in: biological mechanisms linking social and environmental exposures to chronic disease and to health disparities. Am J Public Health 2019;109(Suppl. 1):S56–S63 [DOI] [PMC free article] [PubMed] [Google Scholar]