Abstract
Periodontal disease affects a large percentage of the human population. Resorption of the alveolar bone of the jaw is a pivotal sequela of periodontal disease, because this bone is the attachment site for the periodontal ligaments that anchor the teeth. Using a murine model in which alveolar bone loss is induced by oral infection with Porphyromonas gingivalis, a gram-negative bacterium associated with human adult periodontal disease, we provide evidence suggesting that susceptibility to such bone loss is a genetically determined trait. AKR/J, DBA/2J, and BALB/cByJ or BALB/cJ mice were highly susceptible, while A/J, A/HeJ, 129/J, SJL/J, and C57BL/6J mice were much more resistant. When susceptible BALB/cJ and BALB/cByJ mice were crossed to resistant strains, two patterns were observed. (BALBc/ByJ × C57BL/6J)F1 offspring were susceptible, suggesting C57BL/6J has recessive resistance alleles, while (BALB/cJ × A/J)F1 mice were all resistant, suggesting that A/J mice have dominant resistance alleles. These results suggest a tractable genetic basis for P. gingivalis-induced alveolar bone loss and open the possibility of exploiting the mouse model to identify loci important for host susceptibility and resistance to periodontal disease.
Periodontal diseases are chronic inflammatory diseases which result in the breakdown of the supporting tissues of the teeth, including resorption of the alveolar bone of the jaw (32). Adult periodontitis is the most common form, but not all individuals are equally susceptible (7, 17). Attachment and bone levels decrease with age, but only 7 to 13% of individuals progress to advanced loss (15, 17, 21). Individuals living in similar environments develop disease at different rates, and 11% in a 15-year study showed no loss of attachment despite the presence of plaque and calculus and the lack of professional or self-care (22).
The gram-negative anaerobic bacterium Porphyromonas (Bacteroides) gingivalis is strongly associated with adult periodontitis (27, 30, 31). Again, not all individuals are equally susceptible to bone resorption when they are infected with this bacterium (17, 25), suggesting that host factors are important in the onset and progression of this disease.
Susceptibility to numerous infectious diseases appears to have a strong genetic component (23). Similarly, studies of adult-onset periodontal disease in humans suggest a genetic component (for reviews, see references 13 and 16). Studies involving monozygotic and dizygotic twins have suggested that about half of the population variance in disease could be accounted for by genetic factors (8, 24). Increased susceptibility to periodontal disease is found in conjunction with several inherited diseases (10, 28). Very recently, an interleukin-1 polymorphism has been reported to be a predictor of future severity in adult periodontal disease (19). With this possible exception, although many studies have pointed to genetic links with susceptibility to periodontal disease, no specific genes have been found (13).
Animal models, and murine models in particular, offer many advantages for analysis of genetic contributions to disease susceptibility. We have recently developed a murine model in which alveolar bone loss is induced after oral infection of specific-pathogen-free (SPF) mice with a human-derived strain of P. gingivalis (3, 4, 6). In this study, we have used this mouse model to determine whether inbred strains of mice and their F1 offspring differ in their susceptibility to bone loss.
MATERIALS AND METHODS
Animals.
SPF mice of strains A/J, A/HEJ, 129/J, SJL/J, C57BL/6J, AKR/J, DBA/2J, BALB/cByJ, and BALB/cJ were bred and raised at The Jackson Laboratory (Bar Harbor, Maine), as were F1 mice from two crosses: CAF1/J (F1 of BALB/cJ females × A/J males) and CByB6F1/J (F1 of BALB/cByJ females × C57BL/6J males). The animals were kept in the animal colony at Bates College, Lewiston, Maine, under conditions described previously (3). The animals within an experiment were age-matched females, 12 weeks old at the start of experiments. The experimental protocol was reviewed and approved by the Animal Care and Use Committee, Bates College.
Bacteria for bone loss induction.
P. gingivalis ATCC 53977 (A7A1-28) was maintained frozen in defibrinated sheep's blood at −70°C and by weekly transfer on supplemented blood agar (Trypticase soy agar base with 0.1% yeast extract, 5.0 μg of hemin per ml, 0.5 μg of menadione per ml, and 5% defibrinated sheep's blood). For experiments, bacteria were anaerobically grown under 5% CO2, 10% H2, and 85% N2 on supplemented blood agar at 37°C for 4 to 7 days.
Oral infection.
As described previously (3), mice were given sulfamethoxazole-trimethoprim (Sulfatrim; Goldline Laboratories, Ft. Lauderdale, Fla.) at 10 ml per pint in deionized water ad libitum for 10 days. This was followed by a 3-day antibiotic-free period. Mice were then infected: 109 CFU of live P. gingivalis suspended in 100 μl of phosphate-buffered saline with 2% carboxymethylcellulose was placed in a feeding needle. Three times at 2-day intervals, half of this volume was placed down the throat of each mouse, and half was placed directly into its oral cavity. Controls included sham-infected mice which received the antibiotic pretreatment and the carboxymethylcellulose without P. gingivalis. Forty-seven days after the first gavage, mice were euthanized by CO2.
Paper point sampling of the oral cavity.
A sterile medium-sized paper point (Johnson and Johnson, East Windsor, N.J.) was held against the gumline of the upper molars for 5 s and then vortexed in 1 ml of prereduced brain heart infusion broth supplemented with hemin and menadione. A 75-μl aliquot plated onto supplemented blood agar was incubated aerobically or anaerobically for 4 weeks. P. gingivalis colonies were identified by their black pigmentation and by gram stain (3). Other species were distinguished from one another by colony morphology and gram stain.
Alveolar bone loss.
Horizontal bone loss around the maxillary molars was assessed by a morphometric method (18). Horizontal bone loss is loss occurring in a horizontal plane, perpendicular to the alveolar bone crest, that results in a reduction of the crest height. Skulls were defleshed after 10 min of treatment in boiling water under a pressure of 15 lb/in2, immersed overnight in 3% hydrogen peroxide, pulsed for 1 min in bleach, and stained with 1% methylene blue. The distance from the cementoenamel junction (CEJ) to the alveolar bone crest (ABC) was measured at a total of 14 buccal sites per mouse (Fig. 1). Measurements were made under a dissecting microscope (×40) fitted with a video image marker measurement system (model VIA 170; Boeckeler Instruments, Inc., Tucson, Ariz.) standardized to give measurements in millimeters. Bone measurements were done a total of three times in a random and blinded protocol by two evaluators.
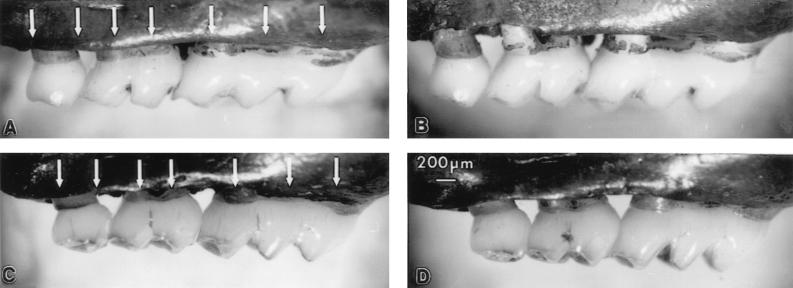
FIG. 1.
Alveolar bone loss caused by oral infection with P. gingivalis. Measurement of bone levels is made comparing the distance from the CEJ to the ABC at seven buccal sites on the three molars on the left side and seven sites on the right side of the maxilla (shown by the arrows in panels A and C). (A) Sham-infected BALB/cByJ mouse. (B) P. gingivalis-infected BALB/cByJ mouse. As alveolar bone resorbs in infected mice, the distance from the CEJ to the ABC increases, becoming greater than in sham mice. (C) Sham-infected C57BL/6J mouse. (D) P. gingivalis-infected C57BL/6J mouse. When there is no bone loss, the distances from the CEJ to the ABC are the same in sham-infected and infected mice.
The amount of change in the alveolar bone of each individual mouse was calculated by subtracting the CEJ-to-ABC distance (total of 14 sites) for that mouse from the mean CEJ-to-ABC distance of groups of sham-infected mice. Since the distance from the CEJ to the ABC increases as bone is resorbed, this calculation produces negative values for total millimeters of change in bone when bone loss has occurred.
Statistics.
Differences between the infected and sham groups and between infected mice of pairs of strains were evaluated by two-tailed, unpaired t tests (Stat View; Abacus Concepts, Inc.).
RESULTS
To determine the effects of oral infection with P. gingivalis on alveolar bone, the distance from the CEJ to the ABC at 7 buccal sites per side (14 buccal sites per mouse) was measured after 47 days. Figure 1 shows one side of an upper jaw from a BALB/cByJ mouse orally infected with P. gingivalis (Fig. 1B) and from an age- and sex-matched mouse sham infected with carboxymethylcellulose rather than the bacterial preparation (Fig. 1A). Clear evidence of bone resorption was observed at several buccal sites, with the distance from the CEJ to the ABC greatly increased in the infected mouse compared to that in the sham-infected mouse. In contrast, infected C57BL/6J mice failed to show bone loss compared with their sham controls (Fig. 1D and C, respectively).
As can also be seen in Fig. 1, the alveolar anatomy differs among mouse strains (compare Fig. 1A and C). The CEJ-to-ABC distance in infected mice of different strains therefore cannot be compared directly, showing why it is important to compare levels in infected mice to those in sham-infected controls of the same strain. By subtracting the CEJ-to-ABC distance in an infected mouse from the mean CEJ-to-ABC distance in sham-infected mice, we can calculate the exact millimeter change in bone that can be attributed to the P. gingivalis infection.
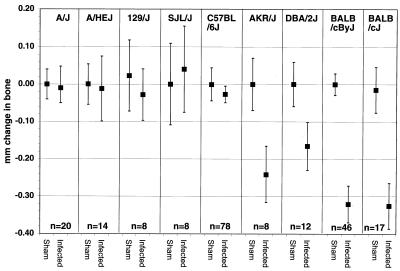
Results comparing nine inbred mouse strains are summarized in Fig. 2. A/J, A/HeJ, 129/J, SJL/J, and C57BL/6J mice were more resistant to bone loss as a consequence of P. gingivalis infection than were AKR/J, DBA/2J, BALB/cByJ, or BALB/cJ mice.
FIG. 2.
Effect of P. gingivalis oral infection on alveolar bone levels in different strains of SPF mice. The values shown are the total millimeters of change at 14 sites (mean ± standard error). Negative values of millimeter change in bone indicate bone loss. Infected AKR/J, DBA2/J, BALB/cByJ, and BALB/cJ mice lost bone compared to sham controls (P = 0.03 for AKR/J, P = 0.04 for DBA/2J, P = 2 × 10−7 for BALB/cByJ, and P = 0.006 for BALB/cJ). In the other mouse strains, bone levels were not different in sham controls and infected mice (P > 0.05). n, number of mice tested. An equivalent number of P. gingivalis-infected and sham mice were tested per strain. When n is greater than 8, results are pooled data from 8 to 10 infected and control mice per experiment.
However, it is important to note that in our experience, the amount of bone lost varies from one experiment to another, and resistant strains sometimes lose bone after infection, but they do so at fewer sites and in fewer mice than more susceptible strains (3). A/J appears to be more strongly resistant than C57BL/6J; in no experiment, has bone loss been initiated after P. gingivalis infection of A/J mice. When susceptible strains were run in the same experiment as resistant strains (as in the results shown below), the susceptible strains always lost more bone than the resistant strains after infection.
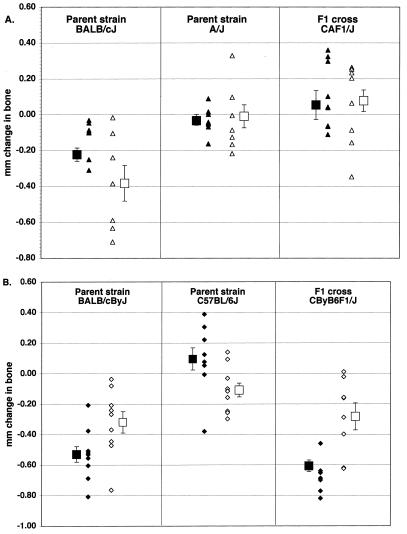
To determine whether resistance or susceptibility demonstrated Mendelian properties, F1 mice from matings of susceptible strains with resistant strains were analyzed. The results are shown in Fig. 3, which shows the data from individual mice as well as the calculated mean values. When BALB/cJ mice (susceptible) were mated with A/J mice (resistant), their F1 offspring were highly resistant to bone loss after infection with P. gingivalis (Fig. 3A). In contrast, F1 progeny of BALB/cByJ mice mated to the slightly less resistant strain (C57BL/6J) were susceptible (Fig. 3B).
FIG. 3.
Bone loss in P. gingivalis-infected F1 mice from two different crosses. Each symbol represents one infected mouse. Data from sham-infected mice are not shown. Open symbols and solid symbols are results from independent experiments. Squares with brackets represent the mean ± standard error of the respective experiments. (A) BALB/cJ × A/J cross. Infected mice of one parent strain (BALB/cJ) lost bone (different from BALB/cJ sham controls at P = 0.02 in one experiment P = 0.01 in the other, and P = 0.0006 for pooled data); infected mice of the other parent strain (A/J) and infected F1 mice (CAF1/J) did not lose bone (not different from sham controls; P > 0.05). Values from infected BALB/c mice differed from those from infected A/J mice (P = 0.031 in one trial, P = 0.028 in the other, and P = 0.0024 for pooled data). Infected BALB/cJ mice also differed from infected CAF1 mice (P = 0.0049 in one experiment, P = 0.0041 in the other, and P = 6 × 10−6 for pooled data). (B) BALB/cByJ × C57BL/6J cross. Infected mice of one parent strain (BALB/cByJ) and infected F1 mice (CByB6F1/J) lost bone. (Infected BALB/cByJ mice were different from BALB/cByJ sham controls at P = 7 × 10−6 in one experiment, P = 0.01 in the other, and P = 9 × 10−7 for pooled data; infected CByB6F1 mice were different from CByB6F1 sham controls at P = 5 × 10−7 in one trial, P = 0.04 in the other, and P = 2 × 10−5 for pooled data.) Infected mice of the other parent strain (C57BL/6J) mice did not lose bone (not different from sham controls, P > 0.05). Values from infected BALB/c mice differed from those from infected C57BL/6J mice (P = 2 × 10−7 in one trial, P = 0.04 in the other, and P = 3 × 10−7 for pooled data). Infected CByB6F1/J mice also differed from infected C57BL/6J mice (P = 3 × 10−8 in one trial, P = 0.05 in the other, and P = 3 × 10−6 for pooled data).
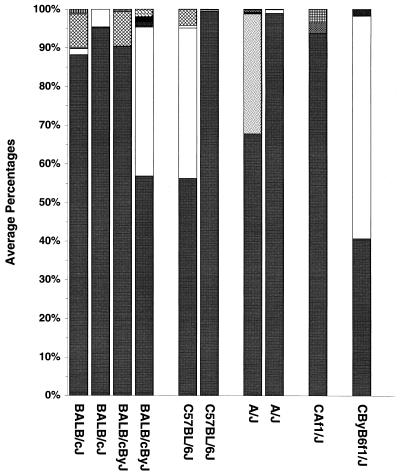
Differences in bone loss could not be explained by differences in the background oral flora, either prior to superinfection with P. gingivalis or at the termination of the experiments. The data shown in Fig. 4 represent the oral anaerobic microflora as sampled by paper points immediately prior to superinfection with P. gingivalis (i.e., mice had been on antibiotics 10 days and off antibiotics 3 days). Based on colony morphology and gram stain, mice had a mixed oral flora of gram-positive and gram-negative species. P. gingivalis was never present prior to infection. One species predominated in every experiment except one, and this was a large gram-negative coccobacillus. There were generally low numbers of two to three other species present. There was more variation between experiments than there was among mouse strains. The mouse strains that were resistant to bone loss (C57BL/6J and A/J) had essentially the same background flora as did the bone loss-susceptible BALB/cJ and BALB/cByJ strains. Resistant CAF1 mice had essentially the same flora as both the susceptible BALB/cJ and resistant A/J parent strains from the same experiment, as did susceptible CByB6F1 mice and the susceptible BALB/cByJ and resistant C57BL/6J parent strains from the same experiment. Aerobic preinfection cultures revealed an additional two to three species of bacteria, all gram-positive cocci, which did not differ between mouse strains (data not shown). Aerobic or anaerobic cultures from paper point sampling done at the time of termination revealed a somewhat more complex flora, but again there were no differences among mouse strains (data not shown). The presence or percentage, either preinfection or at termination, of any bacterial species other than P. gingivalis did not correlate with bone loss in any mouse strain.
FIG. 4.
Viable anaerobic microflora from various strains of mice immediately prior to infection with P. gingivalis. Duplicate bars for the same mouse strain are from two different experiments. Each pattern or tone of gray is a different bacterial species; the proportion of the bar shows the mean percentage of the total flora represented by that species.
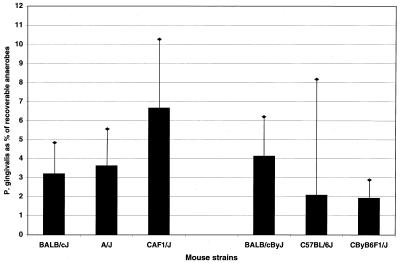
The presence of P. gingivalis at termination of the experiments correlated with bone loss in susceptible strains. The lack of bone loss in resistant strains could not, however, be attributed to smaller amounts of recoverable P. gingivalis. At termination, the percentage of P. gingivalis in the total recoverable anaerobic flora (Fig. 5) and the number of P. gingivalis colonies per plate (data not shown) did not differ significantly between the susceptible (BALB/cJ) and resistant (A/J or C57BL/6J) mouse strains. The strain which tended to have the largest amounts of P. gingivalis was the resistant F1 strain CAF1/J, while the susceptible F1 strain CByB6F1/J tended to have the smallest amounts of P. gingivalis, although again, differences between strains were not statistically significant (Fig. 5).
FIG. 5.
Recovery of P. gingivalis from different mouse strains. P. gingivalis is shown as a percentage of the total anaerobic flora at the termination of the experiments. The first three mouse strains (BALB/cJ, A/J, and CAF1/J) correspond to the strains in Fig. 3A, while the second three (BALB/cByJ, C57BL/6J, and CByB6F1/J) are from the experiment shown in Fig. 3B. Bars represent the means from 10 mice ± standard error. No groups were significantly different (P > 0.05).
DISCUSSION
Studies of the genetic basis of susceptibility and resistance to infectious disease are difficult to perform in humans. Mouse models are playing an increasingly prominent role in this endeavor. Common inbred strains often show differences in genetic predisposition to infectious diseases, and the mode of inheritance can be elucidated by genetic techniques regardless of whether the resistance or susceptibility trait is unigenic or polygenic (11, 23, 26).
Our results suggest that susceptibility to alveolar bone loss in mice as a consequence of exposure to P. gingivalis is a genetically determined trait. Bone loss was readily initiated in AKR/J, DBA/2J, BALB/cByJ, and BALB/cJ mice, while A/J, A/HeJ, 129/J, SJL/J, and C57BL/6J mice were more resistant. Early studies showed that mouse strains differ in their tendency toward spontaneous alveolar bone loss (1, 2). These studies differed from the present model in two important ways. Mice were not infected with P. gingivalis; uninfected mice of different strains were compared. Additionally, the mice were examined only at termination at 16 months. In the much shorter time course of our experiments, sham-infected mice showed no change in the CEJ-to-ABC distance (4). Sham-infected animals of different strains with different anatomy were not compared to each other, but were compared only to P. gingivalis-infected mice of the same strain. Thus, although there may also be genetic differences in bone breakdown over time, our model examined genetic influences on bone loss induced specifically by oral infection. Interestingly, mice of strain DBA/2A, which are resistant to spontaneous alveolar bone resorption (1, 2), are susceptible to P. gingivalis-induced bone resorption (Fig. 2).
Whether these results would only be induced by infection with P. gingivalis is not known at this time. The presence of other bacterial species did not correlate with bone loss (Fig. 4); however, Gilbert and Sofaer have shown differences in mouse strains in alveolar bone loss after infection with another species of oral bacteria, Actinomyces viscosus (12). In their studies, bone levels in infected mice were compared to those in sham-infected mice of the same strain. Bone loss was maximal at a lower bacterial inoculum size than we used and was not induced by either higher or lower inocula. We have not seen this dependence on inoculum size (M. Dixon, unpublished observations). At the inoculum size which produced maximal bone loss in Gilbert and Sofaer's experiments, BALB/c mice lost more bone than did C57BL/6J mice, which is in agreement with our results.
Differences that we saw in bone loss among mouse strains are not likely to be due to differences in background flora (Fig. 4) or to P. gingivalis load (Fig. 5), because these did not show significant differences between resistant and susceptible mouse strains. We cannot entirely rule out these possibilities, however, due to the limits of the sensitivity of the paper point sampling method (N. Price, P. Baker, and U. Wikesjö, Int. Assoc. Dent. Res., abstr. 129, 1992).
In the experiments shown here, the results with the two F1 crosses indicate that the genetic influences are likely to be complex. F1 progeny were resistant or susceptible, depending on the cross. In a cross of BALB/cJ (susceptible) mice with A/J (resistant) mice, the F1 mice were resistant, suggesting that resistance is dominant in this cross. However, when BALB/cByJ mice were mated with another resistant strain, C57BL/6J, the F1 mice were susceptible, suggesting that resistance is recessive in this combination. Further crosses (F2 and backcrosses) will be needed to establish any pattern of dominance. Quantitative trait locus analysis in conjunction with genome markers can be used to unravel genetic contributions whether they are unigenic or polygenic (9, 20). Our ability to measure bone loss as the millimeters of change in bone after infection makes this model amenable to quantitative trait locus analysis.
At this point, we do not know what specific loci may be involved. Susceptibility or resistance to many infectious diseases is dependent on genetically controlled differences in inflammatory responses, cytokine secretion, or T-cell recruitment after exposure to the pathogen (11, 26, 29). It is possible that genetically determined differences in immune regulation or in homeostatic bone remodeling are also important to the outcome of periodontal disease (14, 19). Our previous studies have shown that targeted knockout mice that are genetically deficient in CD4+ T cells or in two T-cell cytokines (gamma interferon or interleukin-6) lose less bone in response to P. gingivalis infection than do immune-competent mice, while those that are deficient in the adhesion molecules ICAM-1 and P-selectin lose more bone (5, 6). Whether these are the traits that account for the observed differences between strains will become apparent during quantitative trait locus analysis.
Our findings of a heritable genetic basis for P. gingivalis-induced alveolar bone loss in mice provides a potentially important model to track the loci that confer susceptibility and resistance to alveolar bone loss that accompanies periodontal disease.
ACKNOWLEDGMENTS
We thank Teresa Hopkins and Tom Sproule for contributions to this project and Dave Serreze for critical reading of the manuscript.
This work was supported by Public Health Service grants R29 DE10728 (to P.J.B.) and R01 AI24544 (to D.C.R.) from the National Institutes of Health, by a grant to Bates College from the Howard Hughes Medical Institute, and by CORE grant CA34196.
REFERENCES
- 1.Baer P N, Lieberman J E. Observations on some genetic characteristics of the periodontium of three strains of inbred mice. Oral Surg Oral Med Oral Pathol. 1959;12:820–829. doi: 10.1016/0030-4220(59)90031-3. [DOI] [PubMed] [Google Scholar]
- 2.Baer P N, Crittenden L B, Jay G E, Lieberman J E. Studies on periodontal disease in the mouse. II. Genetic and maternal effects. J Dent Res. 1961;40:23–33. doi: 10.1177/00220345610400011901. [DOI] [PubMed] [Google Scholar]
- 3.Baker P J, Evans R T, Roopenian D C. Oral infection with Porphyromonas gingivalis and induced alveolar bone loss in immunocompetent and severe combined immunodeficient mice. Arch Oral Biol. 1994;39:1035–1040. doi: 10.1016/0003-9969(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 4.Baker P J, Carter S, Dixon M, Evans R T, Roopenian D C. Serum antibody response to oral infection precedes but does not prevent Porphyromonas gingivalis-induced alveolar bone loss in mice. Oral Microbiol Immunol. 1999;14:194–196. doi: 10.1034/j.1399-302x.1999.140309.x. [DOI] [PubMed] [Google Scholar]
- 5.Baker P J, DuFour L, Dixon M, Roopenian D C. Adhesion molecule deficiencies increase Porphyromonas gingivalis-induced alveolar bone loss in mice. Infect Immun. 2000;68:3103–3107. doi: 10.1128/iai.68.6.3103-3107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker P J, Dixon M, Evans R T, Dufour L, Johnson E, Roopenian D C. CD4+ T cells and the proinflammatory cytokines gamma interferon and interleukin-6 contribute to alveolar bone loss in mice. Infect Immun. 1999;67:2804–2809. doi: 10.1128/iai.67.6.2804-2809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown L J, Loë H. Prevalence, extent, severity and progression of periodontal disease. Periodontology 2000. 1993;2:57–71. doi: 10.1111/j.1600-0757.1993.tb00220.x. [DOI] [PubMed] [Google Scholar]
- 8.Corey L A, Nance W E, Hofstede P, Schenkein H A. Self-reported periodontal disease in a Virginia twin population. J Periodontol. 1993;64:1205–1208. doi: 10.1902/jop.1993.64.12.1205. [DOI] [PubMed] [Google Scholar]
- 9.Frankel W N. Taking stock of complex genetic traits. Trends Genet. 1995;11:471–477. doi: 10.1016/s0168-9525(00)89155-6. [DOI] [PubMed] [Google Scholar]
- 10.Genco R J, van Dyke T E, Levine M J, Nelson R D, Wilson M E. Molecular factors influencing neutrophil defects in periodontal disease. J Dent Res. 1986;65:1379–1391. doi: 10.1177/00220345860650120201. [DOI] [PubMed] [Google Scholar]
- 11.Gervais F, Stevenson M, Skamene E. Genetic control of resistance to Listeria monocytogenes: regulation of leukocyte inflammatory responses by the Hc locus. J Immunol. 1984;132:2078–2083. [PubMed] [Google Scholar]
- 12.Gilbert A D, Sofaer J A. Host genotype pathogenic challenge and periodontal bone loss in the mouse. Arch Oral Biol. 1988;33:855–861. doi: 10.1016/0003-9969(88)90013-1. [DOI] [PubMed] [Google Scholar]
- 13.Hart T C. Genetic considerations of risk in human periodontal disease. Curr Opin Periodontol. 1994;1994:3–11. [PubMed] [Google Scholar]
- 14.Hopps R M, Sismey-Durrant H J. Mechanisms of alveolar bone loss in periodontal disease. In: Hamada S, Holt S C, McGhee J R, editors. Periodontal disease: pathogens and host immune response. Tokyo, Japan: Quintessence Publishing Co., Ltd.; 1991. pp. 307–320. [Google Scholar]
- 15.Ismail A J, Morrison E C, Burt B A, Caffesee R G, Kavanagh M T. Natural history of periodontal disease in adults: findings from the Tecumseh Periodontal Disease Study, 1959–1987. J Dent Res. 1990;69:430–435. doi: 10.1177/00220345900690020201. [DOI] [PubMed] [Google Scholar]
- 16.Johnson N W. Detection of high-risk groups and individuals for periodontal disease. Int Dent J. 1989;39:33–47. [PubMed] [Google Scholar]
- 17.Johnson N W, Griffith G S, Wilson J M A, Maiden M F, Curtis M A, Gillett I R, Wilson D T, Sterne J A. Detection of high-risk groups and individuals for periodontal diseases: evidence of existence of high-risk groups and individuals and approaches to their detection. J Clin Periodontol. 1988;15:276–282. doi: 10.1111/j.1600-051x.1988.tb01584.x. [DOI] [PubMed] [Google Scholar]
- 18.Klausen B, Evans R T, Sfintescu C. Two complementary methods of assessing periodontal bone level in rats. Scand J Dent Res. 1989;97:494–499. doi: 10.1111/j.1600-0722.1989.tb00922.x. [DOI] [PubMed] [Google Scholar]
- 19.Kornman K S, Crane A, Wang H-Y, di Giovine F S, Newman M G, Pirk F W, Wilson T G, Higginbottom F L, Duffy G W. The interleukin-1 genotype as a severity factor in adult periodontal disease. J Clin Periodontol. 1997;24:72–77. doi: 10.1111/j.1600-051x.1997.tb01187.x. [DOI] [PubMed] [Google Scholar]
- 20.Lander E S, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindhe J, Haffajee A D, Socransky S S. Progression of periodontal disease in adult subjects in the absence of periodontal therapy. J Clin Periodontol. 1983;10:433–442. doi: 10.1111/j.1600-051x.1983.tb01292.x. [DOI] [PubMed] [Google Scholar]
- 22.Loë H, Anerud A, Boysen H, Morrison E. Natural history of periodontal disease in man. Rapid, moderate and no loss of attachment in Sri Lankan laborers 14–46 years of age. J Clin Periodontol. 1986;13:431–446. doi: 10.1111/j.1600-051x.1986.tb01487.x. [DOI] [PubMed] [Google Scholar]
- 23.Malo D, Skamene E. Genetic control of host resistance to infection. Trends Genet. 1994;10:365–371. doi: 10.1016/0168-9525(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 24.Michalowicz B S. Genetic and heritable risk factors in periodontal disease. J Periodontol. 1994;65:479–488. doi: 10.1902/jop.1994.65.5s.479. [DOI] [PubMed] [Google Scholar]
- 25.Savit E D, Socransky S S. Distribution of certain subgingival microbial species in selected periodontal conditions. J Periodontal Res. 1984;19:111–115. doi: 10.1111/j.1600-0765.1984.tb00800.x. [DOI] [PubMed] [Google Scholar]
- 26.Skamene E. Inflammatory vs. protective host responses. Immunobiology. 1994;191:451–460. doi: 10.1016/S0171-2985(11)80451-1. [DOI] [PubMed] [Google Scholar]
- 27.Slots J, Listgarten M A. Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitans in human periodontal diseases. J Clin Periodontol. 1988;15:85–93. doi: 10.1111/j.1600-051x.1988.tb00999.x. [DOI] [PubMed] [Google Scholar]
- 28.Sofaer J A. Genetic approaches in the study of periodontal diseases. J Clin Periodontol. 1990;17:401–408. doi: 10.1111/j.1600-051x.1990.tb02337.x. [DOI] [PubMed] [Google Scholar]
- 29.Stevenson M M, Tam M-F. Differential induction of helper T cell subsets during blood-stage Plasmodium chabaudi AS infection in resistant and susceptible mice. Clin Exp Immunol. 1993;92:77–83. doi: 10.1111/j.1365-2249.1993.tb05951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanner A C R, Haffer C, Bratthall G T, Visconti R A, Socransky S S. A study of the bacteria associated with advancing periodontitis in man. J Clin Periodontol. 1979;6:278–307. doi: 10.1111/j.1600-051x.1979.tb01931.x. [DOI] [PubMed] [Google Scholar]
- 31.van Winkelhoff A J, van Steenbergen T J M, de Graaff J. Occurrence and association with disease. In: Shah H N, Mayrand D, Genco R J, editors. Biology of the species Porphyromonas gingivalis. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 33–42. [Google Scholar]
- 32.Williams R C. Periodontal disease. N Engl J Med. 1990;322:373–382. doi: 10.1056/NEJM199002083220606. [DOI] [PubMed] [Google Scholar]