Abstract
Next‐generation sequencing of AML has identified specific genetic mutations in AML patients. Hematologic Malignancies (HM)‐SCREEN‐Japan 01 is a multicenter study to detect actionable mutations using paraffin‐embedded bone marrow (BM) clot specimens rather than BM fluid in AML patients for whom standard treatment has not been established. The purpose of this study is to evaluate the presence of potentially therapeutic target gene mutations in patients with newly diagnosed unfit AML and relapsed/refractory AML (R/R‐AML) using BM clot specimens. In this study, 188 patients were enrolled and targeted sequencing was undertaken on DNA from 437 genes and RNA from 265 genes. High‐quality DNA and RNA were obtained using BM clot specimens, with genetic alterations successfully detected in 177 patients (97.3%), and fusion transcripts in 41 patients (23.2%). The median turnaround time was 13 days. In the detection of fusion genes, not only common fusion products such as RUNX1‐RUX1T1 and KMT2A rearrangements, but also NUP98 rearrangements and rare fusion genes were observed. Among 177 patients (72 with unfit AML, 105 with R/R‐AML), mutations in KIT and WT1 were independent factors for overall survival (hazard ratio = 12.6 and 8.88, respectively), and patients with high variant allele frequency (≥40%) of TP53 mutations had a poor prognosis. As for the detection of actionable mutations, 38% (n = 69) of patients had useful genetic mutation (FLT3‐ITD/TKD, IDH1/2, and DNMT3A R822) for treatment selection. Comprehensive genomic profiling using paraffin‐embedded BM clot specimens successfully identified leukemic‐associated genes that can be used as therapeutic targets.
Keywords: actionable mutation, AML, FoundationOne Heme, ineligible for standard chemotherapy, paraffin‐embedded bone marrow clot
Comprehensive genomic profiling was undertaken using paraffin‐embedded bone marrow clot specimens. High accuracy mutation results (97.3%) and fusion transcripts (23.2%) in newly diagnosed unfit AML and relapsed/refractory AML were obtained.
Abbreviations
- BM
bone marrow
- CI
confidence interval
- F1H
FoundationOne Heme
- HM‐SCREEN‐Japan 01
Hematologic Malignancies‐SCREEN‐Japan 01
- HR
hazard ratio
- NR
not reached
- OS
overall survival
- R/R‐AML
relapsed/refractory AML
- TMB
tumor mutation burden
- VAF
variant allele frequency
1. INTRODUCTION
Genomic and chromosomal abnormalities are known to be crucial factors in predicting prognosis and determining the WHO classification in patients with AML. 1 , 2 , 3 Recently, novel agents targeting specific gene mutations, such as FLT3 and IDH1/IDH2, as well as BCL‐2 inhibitors have made significant contributions to AML treatment strategies. 4 , 5 , 6 , 7 The iconic Beat AML basket trial suggested that comprehensive genomic screening and subsequent genomic alteration‐specific treatment could improve the prognosis of patients with AML. 8
With these successes in newly diagnosed AML, therapeutic strategies using precision medicine with a similar genetic approach are becoming more important in unfit AML or R/R‐AML, for which standard treatment has not been established. In R/R‐AML, it is especially important to identify the genetic mutation(s) to be treated, as leukemic clones are selected by previous therapy and might have different genetic backgrounds at the time of initial diagnosis. However, reassessment of genomic abnormalities in AML patients who are not eligible for standard treatment in real‐world practice is limited, and there are few comprehensive genomic studies that identify associations between these abnormalities and clinical outcomes.
We undertook a multicenter study of prospective genomic profiling in patients with AML named the HM‐SCREEN‐Japan 01 (UMIN000035233) using a hybrid capture‐based comprehensive genomic profiling assay, FoundationOne Heme (Foundation Medicine Inc.). This study aimed to evaluate genomic abnormalities in relation to clinical characteristics and outcome using paraffin‐embedded BM clot specimens in patients with R/R‐AML or newly diagnosed unfit AML who were ineligible for standard therapy. Detection of gene mutations and/or fusion transcripts using archival paraffin‐embedded BM clot specimens is expected to reveal genetic characteristics in R/R‐AML and newly diagnosed unfit AML and to identify molecular abnormalities that could be targets or candidates for novel therapeutic options.
2. METHOD
2.1. Study overview
A total of 17 institutions in Japan participated in this study, of which the National Cancer Center East was the authorized representative. The protocol was approved by each institutional review board and carried out in accordance with the Declaration of Helsinki. All patients provided written informed consent.
2.2. Study design
The HM‐SCREEN‐JAPAN 01 is a multicenter genomic profiling study with next‐generation sequencing by F1H. The inclusion criteria were patients with R/R‐AML or previously untreated AML who are ineligible for standard therapy (unfit AML). The available specimens were paraffin‐embedded BM clots, and submission of archival specimens was also allowed. Patients with FLT3 mutation were allowed to submit specimens at multiple time points. After submission, annotated genomic reports were returned to the participants. Clinical information such as age, sex, treatment technique, response, stem cell transplantation status, and survival information were collected.
2.3. Comprehensive genome profiling assay
FoundationOne Heme is a comprehensive genomic profile developed by Foundation Medicine Inc. to identify somatic genomic alterations in genes known to be clear drivers of hematologic malignancies using formalin‐fixed paraffin‐embedded specimens. Next‐generation sequencing was applied to a hybrid capture‐based target enrichment approach. Each profile simultaneously sequences the complete coding region of 406 genes and selected introns of 31 genes involved in rearrangements (Figure S1). FoundationOne Heme also examines RNA sequences of 265 commonly rearranged genes to better identify rare gene fusions. FoundationOne Heme can detect genomic alterations such as base substitutions, insertions/deletions, copy number changes, as well as TMB status. The TMB is determined by measuring the number of somatic mutations occurring in the sequenced genes on the F1H profile and extrapolating to the genome as a whole.
2.4. Statistical analysis
Clinical information was summarized as descriptive statistics. Overall survival, defined as the time from the date the patients enrolled in this study, was determined by the Kaplan–Meier method. Prognostic effects of basic characteristics (e.g., age, sex, and transplant status) and genomic alterations status were statistically calculated using the Cox regression model and summarized as HR of death.
3. RESULTS
3.1. Baseline patient characteristics and FIH results
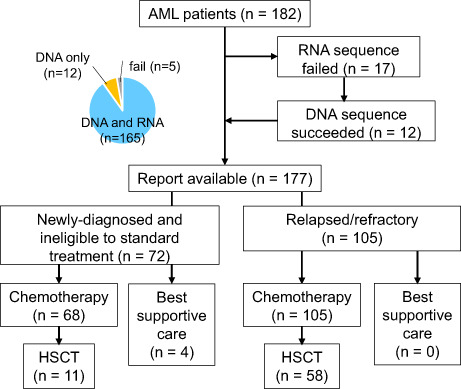
From January 2019 to April 2021, 182 patients with AML across 17 clinical sites consented to enrolment in HM‐SCREEN‐Japan 01. High‐quality DNA was obtained from 177 of 182 (97.3%) patients and high‐quality RNA suitable for further applications was obtained from 165 of 182 (90.6%) patients (Figure 1A). The main cause of insufficient DNA/RNA yields was a lack of leukemic cells in BM specimens. Mutation event was successfully obtained for all 177 patients and fusion transcripts were detected in 41 patients (23.2%). The mean number of mutations was approximately four, with no difference in the number of mutations among newly diagnosed unfit AML, R/R‐AML, and therapy‐related AML, or between groups with recurrent genetic abnormalities (Figure 1B). The median turnaround time from sample submission to results reporting was 13 days (25th to 75th percentile, 11–18 days) (Figure S2). Of the 177 samples, 72 (40.7%) were newly diagnosed and 105 (59.3%) were relapsed or refractory. The characteristics of the 177 eligible patients are shown in Table 1. The median age of the 72 patients with newly diagnosed unfit AML was 73 (61–78) years, and the median age of the 105 patients with R/R‐AML was 51 (40–69) years.
FIGURE 1.
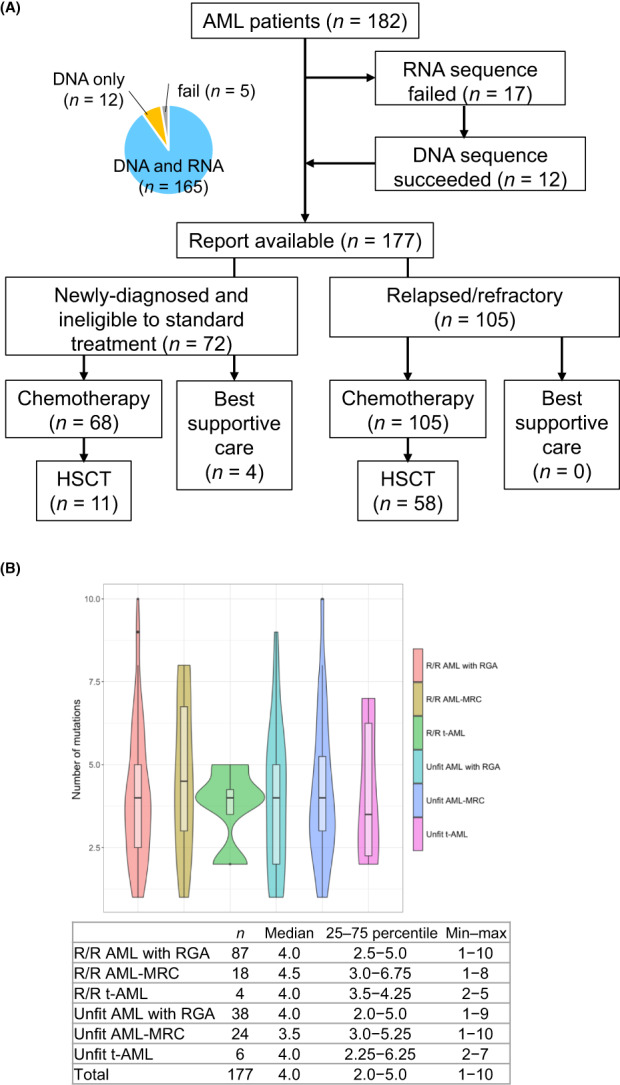
Extraction of DNA and RNA from bone marrow (BM) clot specimens from AML patients and mutation analysis. (A) Consort diagram and success rate of DNA and RNA yielded from BM clot samples. (B) Distribution of tumor mutation burden according to the WHO 2017 classification in relapsed refractory (R/R)‐AML and newly diagnosed unfit AML. HSCT, hematopoietic stem cell transplantation; MRC, myelodysplasia‐related changes; RGA, recurrent genetic abnormality; t‐AML, therapy‐related AML
TABLE 1.
Characteristics of 177 patients with AML
| Total | ND unfit | R/R | ||||
|---|---|---|---|---|---|---|
| n | 177 | Patients | 72 | Patients | 105 | Patients |
| Age (years) | 63 | 46–74 | 73 | 61–78 | 51 | 40–69 |
| Sex | ||||||
| F | 62 | 35.0% | 23 | 31.9% | 39 | 37.1% |
| M | 115 | 65.0% | 49 | 68.1% | 66 | 62.9% |
| Dx | ||||||
| Acute megakaryoblastic leukemia | 1 | 1.0% | 0 | 0.0% | 1 | 1.0% |
| Acute monoblastic/monocytic leukemia | 22 | 12.0% | 5 | 6.9% | 17 | 16.2% |
| Acute myelomonocytic leukemia | 13 | 7.0% | 5 | 6.9% | 8 | 7.6% |
| Acute panmyelosis with myelofibrosis | 1 | 1.0% | 1 | 1.4% | 0 | 0.0% |
| Acute promyelocytic leukemia with PML‐RARA | 1 | 1.0% | 1 | 1.4% | 0 | 0.0% |
| AML (megakaryoblastic) with t(1; 22)(p13.3; q13.3); RBM15‐MKL1 | 3 | 2.0% | 2 | 2.8% | 1 | 1.0% |
| AML with biallelic mutations of CEBPA | 3 | 2.0% | 0 | 0.0% | 3 | 2.9% |
| AML with inv (16)(p13.1q22) or t(16; 16)(p13.1; q22); CBFB‐MYH11 | 7 | 4.0% | 4 | 5.6% | 3 | 2.9% |
| AML with inv (3)(q21.3q26.2)or t(3; 3)(q21.3; q26.2); GATA2, MECOM(EVI1) | 3 | 2.0% | 0 | 0.0% | 3 | 2.9% |
| AML with maturation | 25 | 14.0% | 8 | 11.1% | 17 | 16.2% |
| AML with minimal differentiation | 6 | 3.0% | 3 | 4.2% | 3 | 2.9% |
| AML with mutated NPM1 | 9 | 5.0% | 2 | 2.8% | 7 | 6.7% |
| AML with myelodysplasia‐related changes | 42 | 24.0% | 26 | 36.1% | 16 | 15.2% |
| AML with t(8; 21)(q22; q22.1); RUNX1‐RUNX1T1 | 15 | 8.0% | 5 | 6.9% | 10 | 9.5% |
| AML with t(9; 11)(p21.3; q23.3); MLLT3‐KMT2A | 1 | 1.0% | 0 | 0.0% | 1 | 1.0% |
| AML without maturation | 13 | 7.0% | 4 | 5.6% | 9 | 8.6% |
| Provisional entity: AML with mutated RUNX1 | 1 | 1.0% | 0 | 0.0% | 1 | 1.0% |
| Pure erythroid leukemia | 1 | 1.0% | 0 | 0.0% | 1 | 1.0% |
| Therapy‐related myeloid neoplasms | 10 | 6.0% | 6 | 8.3% | 4 | 3.8% |
| Allo.SCT | ||||||
| N | 108 | 61.0% | 61 | 84.7% | 47 | 44.8% |
| Y | 69 | 39.0% | 11 | 15.3% | 58 | 55.2% |
Abbreviations: Allo.SCT, allogeneic stem cell transplantation; Dx, diagnosis; F, female; M, male; N, no; ND, newly diagnosed; R/R, relapsed/refractory; Y, yes.
3.2. Mutation and rearrangement analysis
In the entire cohort, the most common genetic alterations observed were FLT3 (27.7%), TP53 (20.9%), and RUNX1 (20.3%). Mutations in ASXL1 and TET2 were frequently observed in newly diagnosed unfit AML (Figure 2A), while mutations in FLT3, NPM1, and DNMT3A were observed more frequently in R/R‐AML (Figure 2B). Mutations in NPM1, which is known to be associated with a favorable prognosis, were detected in 24/109 (22%) in R/R‐AML. Compared to the more than 2000 AML patients who underwent F1H analysis in the United States and Europe, the Japanese cohort was similar except for a higher frequency of FLT3 and KIT mutations and a lower frequency of SRSF2 mutations (Figure 2C).
FIGURE 2.
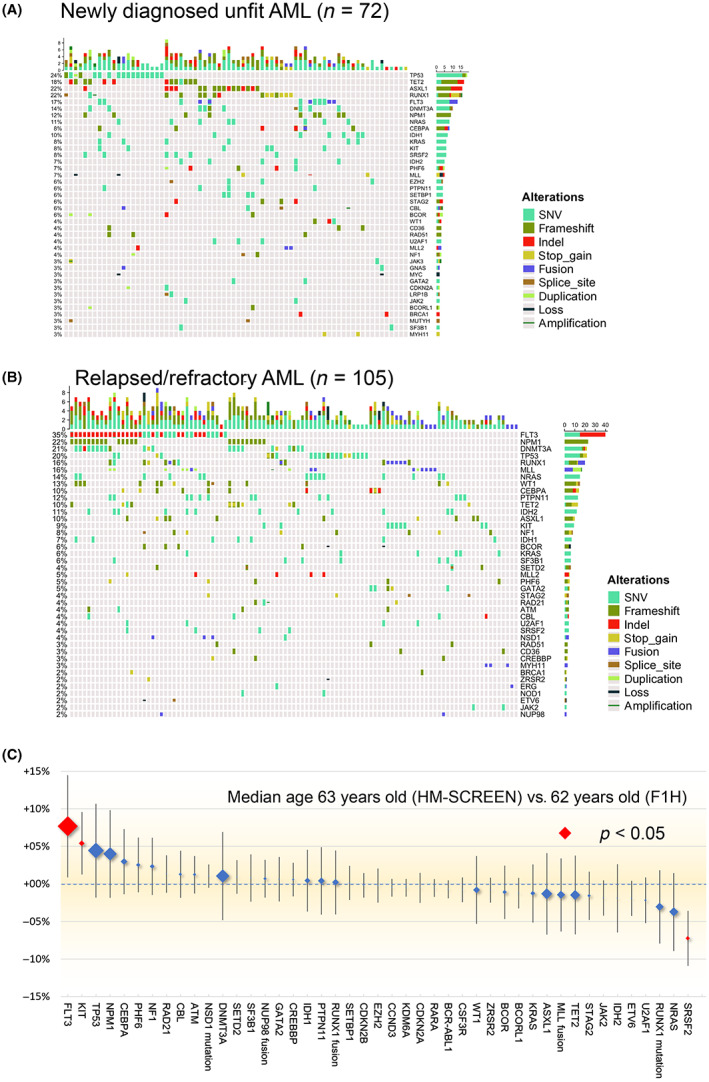
Landscape of mutations and mutational spectrum of the cohort of AML patients. (A) Gene mutation profile in newly diagnosed unfit AML. (B) Gene mutation profile in relapsed or refractory AML (R/R‐AML). (C) Differences in the prevalence of gene mutations. Comparison of mutation analysis using FoundationOne Heme between the 177 patient Japanese cohort and the 2247 patients in the cohort from the United States and Europe. The symbol size represents the number of cases. SNV, single nucleotide variation
3.3. RNA‐based fusion transcript analysis
The F1H analysis using BM clot samples successfully detected fusion gene transcripts in 41 patients. The concordance rate between the fusion gene from the extracted RNA and the karyotypic aberration from the clinical data was extremely high at 0.96 (κ = 0.86, Cohen's kappa) (Figure S3). The detected fusion genes transcripts are shown in Table 2. RUNX1‐RUNX1T1 fusion was most frequently observed (n = 12), followed by KMT2A rearrangement (n = 10), MYH11‐CBFB fusion (n = 5), and NUP98 rearrangement (n = 5). In addition to recurrent genetic abnormalities, rare fusion transcripts such as BCR‐ABL1, JAK3‐SYMPK, and NTRK3‐ETV1 were also detected. Among the fusion partners of the KMT2A rearrangement, AF9 and AF6 were frequently observed, but fusions with AF1Q, SEPT9, and ENL were also observed, indicating that the translocation partners are diverse (Table 2, Figure S4). In addition, HOXA9 and NSD1 were also detected as fusion partners of the NUP98 rearrangement.
TABLE 2.
Detected fusion genes
| Fusion transcript | Translocation partner | Number (N = 41) |
|---|---|---|
| RUNX1‐RUNX1T1 | 12 | |
| KMT2A rearrangement | 10 | |
| AF9 | (3) | |
| AF6 | (2) | |
| AF1Q | (1) | |
| AF10 | (1) | |
| SEPT9 | (1) | |
| ENL | (1) | |
| SEPT6 | (1) | |
| MYH11‐CBFB | 5 | |
| NUP98‐rearrangement | 5 | |
| HOXA9 | (3) | |
| NSD1 | (1) | |
| TNRC18 | (1) | |
| PML‐RARA | 1 | |
| BCR‐ABL1 | 1 | |
| JAK3‐SYMPK | 1 | |
| NTRK3‐ETV1 | 1 |
3.4. Patient survival and impact of genetic alterations
The median follow‐up of all 177 eligible patients was 7.4 months (range, 0.03–73.6 months), and the estimated median OS was 17.3 months (95% CI, 12.0–NR) in R/R‐AML and 10.6 months (95% CI, 6.9–NR) in newly diagnosed unfit AML (Figure 3A). In R/R‐AML, allogeneic stem cell transplantation was carried out in 58 patients (55%), and OS was significantly longer in transplant recipients (p < 0.001) (Figure 3B). Multivariate analysis of the impact on survival showed that clinical information indicated worse OS in age (HR 1.04; 95% CI, 1.03–1.06) and female patients (HR 1.92; 95% CI, 1.41–2.61). Multivariate analysis of the effect of each gene mutation on survival showed that patients with KIT, WT1, PHF6, and TP53 mutations had significantly inferior OS with HRs of 12.6 (95% CI, 6.89–23.11), 8.88 (95% CI, 5.11–15.44), 5.07 (95% CI, 2.02–6.32), and 3.14 (95% CI, 2.44–4.98) for death, respectively (Figure 3C). Of the 49 patients with FLT3 mutations, 18 (36.7%) received FLT3 inhibitors and had an improved risk of death. The group with a high VAF (≥40%) of TP53 clearly had a worse prognosis, suggesting that residual normal alleles correlate with prognosis (Figure 3D). In the comparison of single‐hit versus multi‐hit TP53 mutations, 31 cases were single‐hit and six cases were multi‐hit, with no difference in prognosis between the single‐hit and multi‐hit groups (Figure S5).
FIGURE 3.
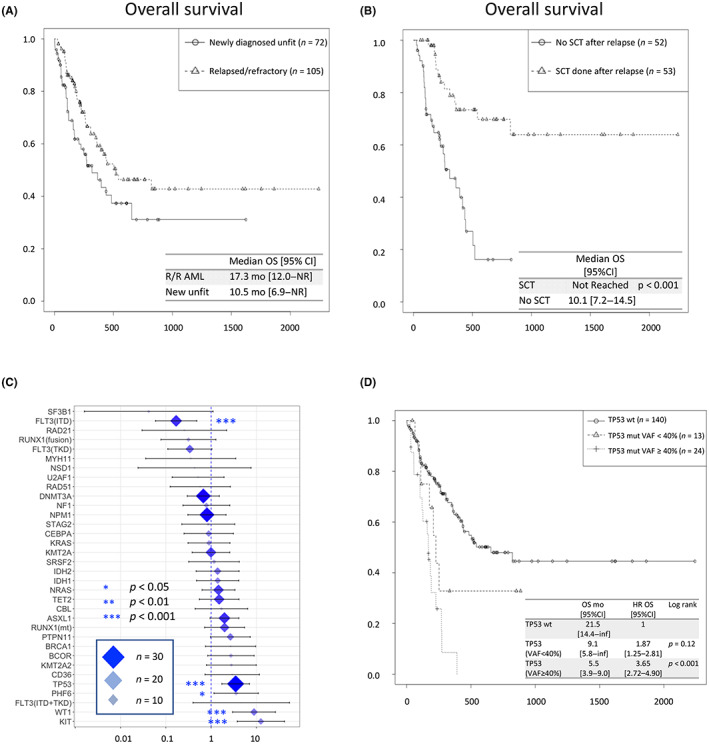
Overall survival (OS) and impact for death of the analysis cohort of AML patients. (A) Survival curves for newly diagnosed unfit AML and relapsed or refractory (R/R) AML. (B) Survival analysis with or without stem cell transplantation (SCT) in relapsed or refractory AML. Log–rank test was used in survival time analysis. (C) OS, hazard ratio (HR) of death by each mutation. Multivariable Cox proportional hazards regression model was used to estimate the effect of various predictor variables. *p < 0.05; **p < 0.01; ***p < 0.001. (D) Survival analysis according to TP53 valiant allele frequency (VAF). Log–rank test was used in survival time analysis. CI, confidence interval; mo, months; mOS, median overall survival; NR, not reached.
3.5. Detection of actionable mutation
Finally, we evaluated the extent to which we could detect actionable mutations that might be candidates for treatment in a group of AML patients for whom standard treatment has not been established. Overall, 38% (n = 69) of the cases possessed usable genetic alteration for selecting treatment options (Figure 4A). The detection rate of actionable mutations, FLT3‐ITD and FLT3‐TKD mutations, for which FLT3 inhibitors are therapeutic candidates, were found in 18% (n = 32) of all cases. The detection rate of IDH1or IDH2 mutations for which IDH inhibitors or venetoclax are therapeutic candidates was 17% in both newly diagnosed unfit AML (n = 12) and R/R‐AML (n = 18). The DNMT3A R822 mutation, which has been reported to be associated with anthracycline sensitivity, 9 was found in 11.8% (n = 21) of cases. When drugs in development (currently in clinical trials) targeting mutations in TP53, KIT D816 , KRAS G12C , NPM1, and KMT2A rearrangements become available, an additional 31.1% (n = 55) of cases are expected to receive selective treatment based on genetic information (Figure 4B). KRAS mutations were found in 12 cases, but no KRAS G12C mutations were found to be therapeutic indications. In summary, our results suggest that genetic analysis of BM clot specimens can detect genetic abnormalities associated with leukemia and could provide useful information for determining treatment strategies for unfit AML and R/R‐AML patients who are ineligible for standard therapy.
FIGURE 4.
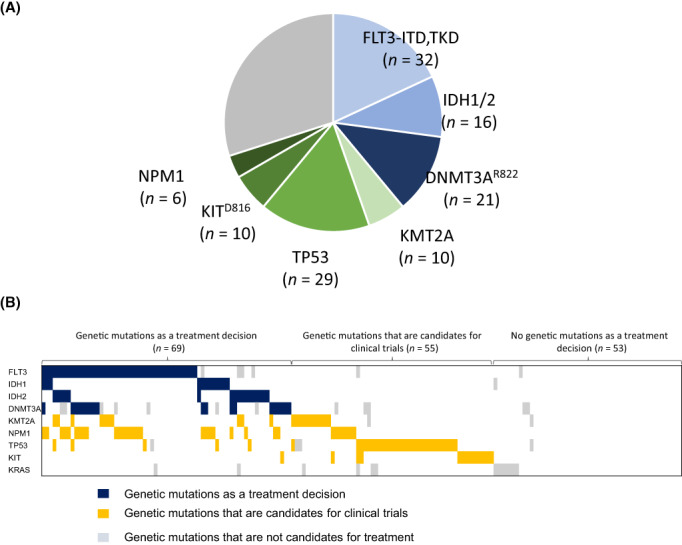
Number of detected genetic mutations in AML patients. (A) Detection of genetic mutations that affect treatment decisions. Actionable mutations include FLT3‐ITD/TKD, IDH2 R140Q/W,R172K, IDH1 R132, and DNMT3A R882C/H mutation. Overall, 38% (n = 69) of the cases possessed usable genetic alteration for selecting treatment options. (B) Distribution of therapeutic target genes in the analyzed cohort. Gene mutations that can be used to determine treatment include FLT3‐ITD/TKD, IDH2 R140Q/W,R172K, IDH1 R132, and DNMT3A R882C/H mutation. Candidate gene mutations for clinical trials include mutation of TP53, KIT D816 , KRAS G12C , NPM1, and KMT2A rearrangements
4. DISCUSSION
The HM‐SCREEN‐Japan 01 is a multicenter study focusing on actionable mutation profiling using paraffin‐embedded BM clot specimens from patients with newly diagnosed unfit AML or R/R‐AML who cannot be treated with intensive chemotherapy. This study demonstrates the utility and reliability of our approach in detecting genetic alterations and fusion genes derived from leukemia cells, as well as new possibilities for precision treatment of AML based on genetic profiling from BM clot specimens.
The purpose of this project was to: (i) assess the effectiveness of detection of genetic events (including fusion transcripts) using paraffin‐embedded clot samples, (ii) investigate the relationship between genetic alterations and prognosis in newly diagnosed unfit AML or R/R‐AML for which standard treatment has not been established, and (iii) evaluate the feasibility of selective treatment based on the results of genetic alterations. While most targeted sequencing protocols for gene panels have been analyzed using fresh BM aspiration liquid or peripheral blood, recently there have been reports on the utility of genetic analysis using fragmented DNA extracted from BM clot samples. 10 , 11 In this study, using the BM clot specimens, we were also able to obtain sufficient quality and quantity of DNA for sequencing in 97.3% (177/182) of patients and sufficient RNA samples for fusion gene detection in 90.1% (163/182) of patients. Furthermore, the average turnaround time was 13 days, indicating its utility in clinical practice. In general, genetic alterations in leukemia are examined in BM aspiration liquid, but this is difficult to perform on the initial examination, and is often omitted in elderly patients or those who have relapsed. The genetic analysis using paraffin‐embedded specimens that we have demonstrated is expected to be a very useful tool that can detect omitted cases, even during treatment and after remission, as long as the clots are available. Despite the convenience of being able to examine both DNA and RNA at any point in the clinical course of leukemia, there is a concern that the DNA is extracted from the BM clot sample, which contains whole blood components other than leukemic cells, resulting in lower VAF than conventional sequencing methods.
The genetic landscape of the AML cohort in Japan was almost the same as in the United States, except for the low frequency of SRSF2 mutations. The data from the United States and Europe were provided by the companies that undertook the FIH analysis, and although detailed clinical information is missing, the results were interesting for information on genetic alteration in AML in each region. The genetic events of newly diagnosed unfit AML and R/R‐AML appeared to differ, with the NPM1 mutation highly observed in R/R‐AML (22.2%), suggesting the presence of genetic abnormalities that affect outcomes other than FLT3‐ITD in NPM1 mutated AML. Neither unfit AML nor R/R‐AML are indicated for standard induction chemotherapy (so‐called 3 + 7), but have different mutation profiles and require different treatment strategies. At a minimum, patients who received SCT showed better prognosis, indicating that SCT is an effective option for R/R‐AML (Figure 3B), and younger patients have a better prognosis (Figure S6). Notably, this study was able to detect fusion transcripts and various partner genes in KMT2A‐related or NUP98‐related fusion transcripts using paraffin‐embedded clot specimens. The detection of various partner genes in KMT2A translocations indicates that chimeric screening of fusion genes is insufficient. However, F1H does not complement chromosome abnormality result from G‐banding. Because F1H uses probes that recognize specific sequences of extracted RNA, detection is limited by sequence variation, and the G‐banding is conversely more useful for detecting complex karyotypes. Regarding the relationship between gene mutations and prognosis, mutations in KIT, TP53, or WT1 were poor prognostic factors (Figure 3C). In our F1H cohort, TP53 mutations were more prevalent (21.4% overall, 20% in R/R‐AML, and 23.6% in newly diagnosed unfit AML) because the analysis was carried out in R/R‐AML or newly diagnosed unfit cases (i.e., older patients). Associations between VAF or allelic state and prognosis in TP53 mutations have been reported, 12 and in our cohort, survival was clearly inferior with VAF ≥ 40% compared to VAF < 40% (Figure 3D). Unfortunately, our analysis did not provide information on the status of the TP53 allele, only the VAF of the TP53 mutation. Most of the WT1 mutations were loss‐of‐function mutations (Figure S7), but in our study, WT1 expression tended to be higher in the WT1 mutant group (Figure S8). WT1 is known to act as an oncogene in AML, 13 , 14 and it was of interest that loss‐of‐function mutations were associated with WT1 expression, which is consistent with poor prognosis. 15 , 16
The purpose of this study was to evaluate the feasibility of selective treatment based on the results of genetic events. The newly diagnosed unfit AML and R/R‐AML that were the subject of our F1H analysis in this study are not eligible for standard 3 + 7 therapy, and it would be very useful to predict the possibility of novel therapy based on these genetic analyses. In our cohort, 17.1% (n = 18) of R/R‐AML cases were treated with selective inhibitors as FLT3 mutation‐positive AML because, at the time of analysis, venetoclax was not approved. The selective inhibitors currently available include FLT3 inhibitors, BCL2 inhibitors, IDH inhibitors (United States/Europe), and clinical trials are ongoing for new drugs such as APR‐246, a mutant p53 reactivator, menin‐MLL inhibitors, and anti‐CD47 Ab. 5 , 6 , 17 , 18 , 19 , 20 When these drugs become available, 31.1% of cases will have the opportunity to receive selective inhibitors, and it is strongly expected that these agents will be approved in the near future.
The HM‐SCREEN‐Japan 01 study successfully detected both the mutation profile and fusion transcripts of AML patients using paraffin‐embedded clot specimens. This is highly advantageous in that specimens can be examined retrospectively when fresh samples are not available. Mutation profiles differed between newly diagnosed unfit AML and R/R‐AML patients, and specific treatment strategies based on mutation profiles can be expected to yield better results. This study revealed the future applicability of genetic panels and the potential for targeted treatment of AML by genetic testing.
AUTHOR CONTRIBUTIONS
K.F., H.S., S.C., N.H., T.Y., S.K., A.G., M.E., T.M., R.O., T.Kon, M.Y., K.Y., T.Kob, J.K., K.U., Y.U., N.A., M.A., K.Iya., T.O., N.T., S.I., M.N., Y.N., S.F., K.Izu, N.Y., J.Y., and Y.M. contributed to data acquisition and interpretation, writing and reviewing the manuscript, and reviewing and approving the final manuscript. H.A. contributed to this paper as a representative of the Chugai Foundation Medicine, Inc. laboratory.
CONFLICT OF INTEREST STATEMENT
T.Y.: Otsuka, Pfizer, Abbie, Astellas, Daiichi Sankyo, Solasia Pharma (research funding); Ono Pharmaceutical, Pfizer, Chugai (honoraria). Y.M.: Bristol‐Myers Squibb, Novartis Pharma KK, Pfizer Japan, Inc., Takeda (honoraria). H.S.: Astellas, Teijin, Shionogi, Taiho, Eisai, Celgene, Ono, Takeda, Merck Sharp & Dohme, Sumitomo Dainippon, Nippon Shinyaku, Novartis, Janssen, Chugai, AbbVie (research funding); Eisai, Ono, Takeda, Sumitomo Dainippon, Nippon Shinyaku, Daiichi Sankyo, Novartis, Janssen, Chugai, Kyowa Kirin, Otsuka, Bristol‐Myers Squibb, Pfizer, Fujimoto, AbbVie, AstraZeneca, Sanofi, Mundi Pharma (honoraria); Eisai, Celgene, Chugai, AbbVie, AstraZeneca (membership on an entity's Board of Directors or advisory committees). K.Y.: AbbVie, Astra‐Zeneca, Bayer, Celgene, Chugai, Eisai, IQIVA/Incyte, Gilead Sciences, MSD, Mundipharma, Nippon Shinyaku, Novartis, Ono, Otsuka, Solasia Pharma, SymBio, Takeda, Yakult, Zenyaku (research funding); AbbVie, Bristol‐Myers Squibb, Celgene, Chugai, Eisai, IQIVA/HUYA, Janssen, Kyowa Kirin, Meiji Seika Pharma, Mochida, MSD, Mundipharma, Nippon Shinyaku, Novartis, Ono, Otsuka, Pfizer, Sanofi, Sumitomo Dainippon, Takeda (honoraria); AbbVie, Astra‐Zeneca, Celgene, Chugai, Eisai, Daiichi Sankyo, HUYA, Meiji Seika Pharma, MSD, Mundipharma, Ono, Otsuka, Stemline Therapeutics, Takeda (consultancy). J.K.: Bristol‐Myers Squibb, Chugai Pharmaceutical, Dainippon Sumitomo Pharma, Daiichi Sankyo, Sanofi, Kyowa Kirin, Otsuka Pharmaceutical, Astellas Pharma, Takeda, Celgene, MSD, Ono Pharmaceutical, Eisai, Sysmex, Pfizer, Nippon Shinyaku, Shionogi, Asahi Kasei, Taiho Pharmaceutical, Fujimoto Pharmaceutical (research funding); Bristol‐Myers Squibb, Chugai Pharmaceutical, Dainippon Sumitomo Pharma, Daiichi Sankyo, Sanofi, Kyowa Kirin, Otsuka Pharmaceutical, Astellas Pharma, Takeda, Celgene, Abbvie, Ono Pharmaceutical, Eisai, Pfizer, Nippon Shinyaku, Fujimoto Pharmaceutical (honoraria); Janssen Pharmaceutical KK, Bristol‐Myers Squibb, Sanofi, Celgene, Abbvie (consultancy). K.U.: Astellas, Abbvie, Gilead, Symbio, Daiichi Sankyo, Sumitomo Dainippon, Otsuka, Novartis, Bristol‐Myers Squibb, Ono, Janssen, Celgene, Takeda, Nippon Boehringer Ingelheim, Mundipharma, Astellas‐Amgen‐Biopharma, Nippon Shinyaku, Kyowa Kirin, Pfizer (research funding); Astellas, Symbio, Daiichi Sankyo, Otsuka, Novartis, Bristol‐Myers Squibb, Ono, Celgene, Nippon Shinyaku, Kyowa Kirin, Alexion, Eisai, MSD, Takeda, PharmaEssentia, Yakult (speakers bureau). A.G.: Eisai Co., Ltd., Ono Pharmaceutical Co., Ltd., Taiho Pharmaceutical Co., Ltd., Takeda Pharmaceutical Co., Ltd., Nippon Shinyaku Co., Ltd., Chugai Pharmaceutical Co., Ltd., MSD KK, Otsuka Pharmaceutical Co., Ltd., Sumitomo Dainippon Pharma Co., Ltd., Bayer Yakuhin, Ltd., Daiichi‐Sankyo Co., Ltd., and Nihon Pharmaceutical Co., Ltd. (research funding); Novartis Pharma KK, Alexion Pharmaceuticals, Inc., Eisai Co., Ltd., Ono Pharmaceutical Co., Ltd., Taiho Pharmaceutical Co., Ltd., Takeda Pharmaceutical Co., Ltd., Nippon Shinyaku Co., Ltd., Chugai Pharmaceutical Co., Ltd., Otsuka Pharmaceutical Co., Ltd., Sumitomo Dainippon Pharma Co., Ltd., Daiichi‐Sankyo Co., Ltd., Nihon Pharmaceutical Co., Ltd., Kyowa Kirin Co., Ltd., Janssen Pharmaceutical KK, Pfizer Japan Inc., Sanofi KK (honoraria); PharmaEssentia Japan KK, Chugai Pharmaceutical Co. (consultancy).
ETHICS STATEMENT
Approval of the research protocol by an institutional review board: This study was approved by the Research Ethics Committee of National Cancer Center Hospital East and was carried out in accordance with the Declaration of Helsinki.
Informed consent: Written informed consent was obtained from all donors who provided samples.
Registry and the registration no. of the study/trial: UMIN000035233.
Animal studies: N/A.
Supporting information
Figure S1–S8
ACKNOWLEDGMENTS
The authors thank all collaborators who participated in this project.
Hosono N, Chi S, Yamauchi T, et al. Clinical utility of genomic profiling of AML using paraffin‐embedded bone marrow clots: HM‐SCREEN‐Japan 01. Cancer Sci. 2023;114:2098‐2108. doi: 10.1111/cas.15746
Naoko Hosono and SungGi Chi contributed equally to this work.
REFERENCES
- 1. Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209‐2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ley TJ, Miller C, Ding L, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059‐2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424‐447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perl AE, Martinelli G, Cortes JE, et al. Gilteritinib or chemotherapy for relapsed or refractory FLT3‐mutated AML. N Engl J Med. 2019;381(18):1728‐1740. [DOI] [PubMed] [Google Scholar]
- 5. DiNardo CD, Stein EM, de Botton S, et al. Durable remissions with Ivosidenib in IDH1‐mutated relapsed or refractory AML. N Engl J Med. 2018;378(25):2386‐2398. [DOI] [PubMed] [Google Scholar]
- 6. Stein EM, DiNardo CD, Pollyea DA, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. 2017;130(6):722‐731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DiNardo CD, Jonas BA, Pullarkat V, et al. Azacitidine and Venetoclax in previously untreated acute myeloid leukemia. N Engl J Med. 2020;383:617‐629. [DOI] [PubMed] [Google Scholar]
- 8. Burd A, Levine RL, Ruppert AS, et al. Precision medicine treatment in acute myeloid leukemia using prospective genomic profiling: feasibility and preliminary efficacy of the beat AML master trial. Nat Med. 2020;26:1852‐1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guryanova OA, Shank K, Spitzer B, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med. 2016;22(12):1488‐1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bartels S, Schipper E, Kreipe HH, Lehmann U. Comprehensive molecular profiling of archival bone marrow trephines using a commercially available leukemia panel and semiconductor‐based targeted resequencing. PLos One. 2015;10(7):e0133930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fukuhara S, Oshikawa‐Kumade Y, Kogure Y. Feasibility and clinical utility of comprehensive genomic profiling of hematological malignancies. Cancer Sci. 2022;113(8):2763‐2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bernard E, Nannya Y, Hasserjian RP, et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat Med. 2020;26(10):1549‐1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y, Xiao M, Chen X, et al. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell. 2015;57(4):662‐673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang L, Han Y, Saurez Saiz F, Minden MD. A tumor suppressor and oncogene: the WT1 story. Leukemia. 2007;21:868‐876. [DOI] [PubMed] [Google Scholar]
- 15. Hou HA, Huang TC, Lin LI, et al. WT1 mutation in 470 adult patients with acute myeloid leukemia: stability during disease evolution and implication of its incorporation into a survival scoring system. Blood. 2010;115(25):5222‐5231. [DOI] [PubMed] [Google Scholar]
- 16. Nomdedéu JF, Hoyos M, Carricondo M, et al. Bone marrow WT1 levels at diagnosis, post‐induction and post‐intensification in adult de novo AML. Leukemia. 2013;27:2157‐2164. [DOI] [PubMed] [Google Scholar]
- 17. Perdrix A, Najem A, Saussez S, et al. PRIMA‐1 and PRIMA‐1(met) (APR‐246): from mutant/wild type p53 reactivation to unexpected mechanisms underlying their potent anti‐tumor effect in combinatorial therapies. Cancer. 2017;9(12):172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sallman DA, DeZern AE, Garcia‐Manero G, et al. Eprenetapopt (APR‐246) and Azacitidine in TP53‐mutant myelodysplastic syndromes. J Clin Oncol. 2021;39(14):1584‐1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fiskus W, Boettcher S, Daver N. Effective Menin inhibitor‐based combinations against AML with MLL rearrangement or NPM1 mutation (NPM1c). Blood Cancer J. 2022;12(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chao MP, Takimoto CH, Feng DD, et al. Therapeutic targeting of the macrophage immune checkpoint CD47 in myeloid malignancies. Front Oncol. 2020;9:1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1–S8


