Abstract
Ribosome biogenesis in the nucleolus is an important process that consumes 80% of a cell's intracellular energy supply. Disruption of this process results in nucleolar stress, triggering the activation of molecular systems that respond to this stress to maintain homeostasis. Although nucleolar stress was originally thought to be caused solely by abnormalities of ribosomal RNA (rRNA) and ribosomal proteins (RPs), an accumulating body of more current evidence suggests that many other factors, including the DNA damage response and oncogenic stress, are also involved in nucleolar stress response signaling. Cells reacting to nucleolar stress undergo cell cycle arrest or programmed death, mainly driven by activation of the tumor suppressor p53. This observation has nominated nucleolar stress as a promising target for cancer therapy. However, paradoxically, some RP mutations have also been implicated in cancer initiation and progression, necessitating caution. In this article, we summarize recent findings on the molecular mechanisms of nucleolar stress and the human ribosomal diseases and cancers that arise in its wake.
Keywords: nucleolar stress, p53, PICT1, ribosomal disease, RPL11, signal transduction
Disruption in ribosome biogenesis results in nucleolar stress, under which several nucleolar proteins, including ribosomal proteins, play roles in the activation of p53.

Abbreviations
- AAA
abdominal aortic aneurysm
- ALS
amyotrophic lateral sclerosis
- BCS
Bowen‐Cornadi syndrome
- CBS
cystathionine β‐synthase
- DBA
Diamond‐Blackfan anemia
- EMG1
EMG N1‐specific pseudouridine methyltransferase
- FTD
frontotemporal dementia
- GRWD1
glutamate‐rich WD repeat containing‐1
- MJD
Machado‐Joseph disease
- MPA
mycophenolic acid
- MYBBP1A
MYB‐binding protein‐1A
- NPM1
nucleophosmin‐1
- PD
Parkinson's disease
- PICT1
protein interacting with carboxyl terminus‐1
- Pol I
RNA polymerase I
- Pol III
RNA polymerase III
- PPAN
Peter Pan homolog
- pre‐rRNA
rRNA precursor
- RNP
ribonucleoprotein
- ROS
reactive oxygen species
- RP
ribosomal protein
- RPL
large ribosomal subunit protein
- RPS
small ribosomal subunit protein
- rRNA
ribosomal RNA
- SBDS
Shwachman‐Bodian‐Diamond syndrome ribosome maturation factor
- SDS
Schwachman‐Diamond syndrome
- SOD2
superoxide dismutase‐2
- TCOF1
treacle ribosome biogenesis factor‐1
- TCS
Treacher‐Collins syndrome
- IMPDH2
inosine monophosphate dehydrogenase‐2
1. INDUCTION OF NUCLEOLAR STRESS BY ABNORMAL RIBOSOME BIOGENESIS
Ribosome biogenesis initiates in the nucleolus through three critical steps (Figure 1): (i) A 47S rRNA precursor (pre‐rRNA) is transcribed by RNA polymerase I (Pol I) in the nucleolus, with a 5S rRNA being concurrently transcribed by Pol III outside the nucleolus.; (ii) The 47S pre‐rRNA is processed and modified to generate three types of rRNA molecules, the 28S, 18S, and 5.8S rRNAs. (iii) The 18S rRNA is assembled with 33 types of small ribosomal subunit proteins (RPS) to form the Pre‐40S ribosome particle, whereas the 5S, 28S, and 5.8S rRNAs are assembled with 48 types of large ribosomal subunit proteins (RPL) to form the Pre‐60S ribosome particle. These pre‐ribosomes exit the nucleolus and pass through the nuclear pore into the cytoplasm, where they combine to form the mature 80S ribosome capable of translating mRNA.
FIGURE 1.
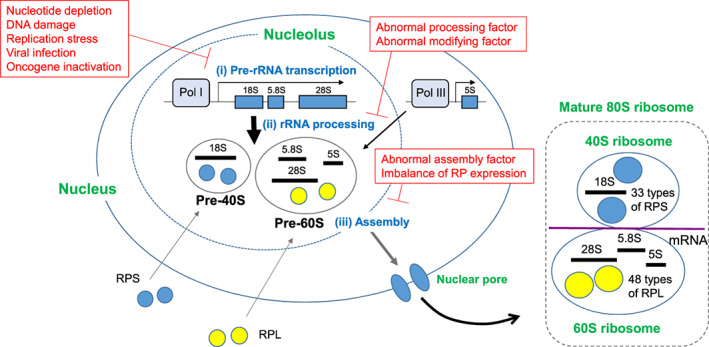
Mechanisms of ribosome biogenesis and triggers of nucleolar stress. Normal ribosome biogenesis starts in the nucleolus via three initial steps (shown in blue): (i) Pol I–mediated transcription of a large rRNA precursor (pre‐rRNA) and Pol III–mediated transcription of a smaller rRNA precursor that constitutes the 5S subunit; (ii) processing of the large rRNA precursor into 18S, 5.8S, and 28S mature rRNAs; (iii) assembly of the 18S subunit with 33 types of RPS to form the pre‐40S ribosome particle, and assembly of the 5.8S, 28S, and 5S subunits with 48 types of large ribosomal subunit protein (RPL) to form the pre‐60S ribosome particle. The pre‐40S and pre‐60S ribosome particles are then exported from the nucleolus through the nucleoplasm into the cytoplasm, where they combine to form mature 80S ribosomes. It is the 80S ribosomes that take up mRNA and translate it into protein. Insults that cause nucleolar stress, for example, those which interfere with steps (i)‐(iii) above during early ribosome biogenesis, are listed in red text
About 80% of a cell's intracellular energy supply is spent on the three steps of ribosome biogenesis occurring in the nucleolus. 1 , 2 This energy‐consuming process is constantly monitored, and if any one of these three steps fails, “nucleolar stress” (also called “ribosomal stress”) arises. The cell cycle is immediately stopped while the cell attempts to fix the defect. However, if the damage is so severe that it cannot be repaired, programmed cell death is induced. 3 , 4
There are many forms of disruption of steps (i)‐(iii) above that lead to nucleolar stress. 3 , 4 Inhibition of pre‐rRNA transcription can be caused by nucleotide depletion or DNA damage, as well as by viral infection 5 , 6 or oncogene inactivation. 7 , 8 , 9 , 10 , 11 Replication stress, which is caused by replication fork arrest and collapse, is also known to induce nucleolar stress. 4 Pharmacological interference with pre‐rRNA transcription can be achieved using a low concentration of actinomycin D to inhibit Pol I. 12 Similarly, many anticancer drugs that induce DNA damage block pre‐rRNA transcription. 13 , 14 Abnormalities in rRNA processing or modification or in ribosome assembly can also cause nucleolar stress. 15 Lastly, an imbalance in ribosomal protein (RP) expression levels (such as a deficiency of certain RPs) can lead to abnormal ribosome assembly and induce nucleolar stress. 16 , 17
2. NUCLEOLAR STRESS RESPONSE PATHWAYS
Cells exposed to nucleolar stress undergo p53 activation and other responses that lead to cell cycle arrest or cell death. 3 , 16 Most of these responses are triggered by the transfer of nucleolar proteins, including RPs, from the nucleolus to the nucleoplasm. In this section, we discuss the molecular mechanisms of these response pathways, which we have divided into two major categories based on their dependence on p53.
2.1. p53‐dependent pathways
In cells at rest, p53 exists mainly in an inactivated state imposed by polyubiquitination via the ubiquitin E3 ligase MDM2 (Figure 2). Proteasome‐mediated degradation of polyubiquitinated p53 can then follow. In contrast, in most cells under nucleolar stress, RPs (mainly RPL5, RPL11, and RPL23) migrate from the nucleolus to the nucleoplasm, where they bind to MDM2 and inhibit its activity. p53 is therefore stabilized and activated, inducing cell cycle arrest and/or cell death. 4 , 16 Transcriptional upregulation of p53 is thought to contribute only minimally to p53 activity triggered by nucleolar stress. 18
FIGURE 2.
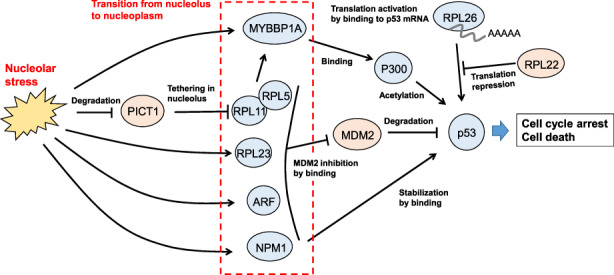
p53‐dependent nucleolar stress response pathways. Nucleolar stress induces the translocation of the nucleolar proteins indicated in the red dotted box into the nucleoplasm. These proteins then stabilize p53 by the various indicated processes (see main text for details). Cells with elevated p53 then undergo cell cycle arrest and/or cell death
Among RPs inhibiting MDM2, RPL11 is the best characterized. RPL11 forms a 5S RNP (ribonucleoprotein) complex with RPL5, 19 which then moves together with RPL11. It is now clear that the nucleolar protein interacting with carboxyl terminus‐1, also known as NOP53 or GLTSCR2 (PICT1) plays an important role in the RPL11‐mediated response pathway. 20 , 21 In resting cells, PICT1 binds to RPL11 and retains it in the nucleolus. However, in cells exposed to nucleolar stress, PICT1 is rapidly destabilized and degraded such that RPL11 translocates from the nucleolus to the nucleoplasm. Glutamate‐rich WD repeat containing‐1 (GRWD1), which may have PICT1‐like functions, may also participate in this nucleolar stress response. 22
Another RPL11‐dependent p53 activation pathway is mediated by MYB‐binding protein‐1A (MYBBP1A). 23 In resting cells, MYBBP1A is predominantly localized in the nucleolus through its binding to rRNA. However, upon a decrease in rRNA levels caused by nucleolar stress, MYBBP1A moves from the nucleolus to the nucleoplasm in an RPL11‐dependent manner. In the nucleoplasm, MYBBP1A forms a complex with acetyltransferase p300 and p53, promoting the acetylation of p53 by p300 to stabilize p53.
Other molecules that play important roles in p53 regulation by nucleolar stress are nucleophosmin‐1 (NPM1), the most abundant protein in the nucleolus, and p14ARF. 24 In resting cells, these proteins are localized in the nucleolus, but under nucleolar stress, they translocate to the nucleoplasm where they bind to and inhibit MDM2, leading to p53 stabilization.
Some RPs are known to contribute to p53 regulation, but it is not clear if these activities are linked to nucleolar stress. For example, loss of RPL22 specifically promotes p53 translation, 25 and RPL26 promotes p53 translation by binding to the 5'‐UTR of p53 mRNA. 26 The relationship of these actions to nucleolar stress remains elusive.
Despite the above detail on how nucleolar proteins activate p53, it is still largely a mystery how cells sense nucleolar stress in the first place and initiate the stress response pathway. For example, how does nucleolar stress induce PICT1 degradation? Does the RPL11 released from PICT1 binding in response to nucleolar stress participate in MYBBP1A translocation? Further studies are needed to elucidate the crosstalk between these pathways and determine the magnitude and significance of their contributions to p53 activation.
2.2. p53‐independent pathways
Cells lacking p53 can also undergo cell cycle arrest and death induced by nucleolar stress (Figure 3). 16 E2F‐1, MYC and NFκB are key players in these p53‐independent pathways.
FIGURE 3.
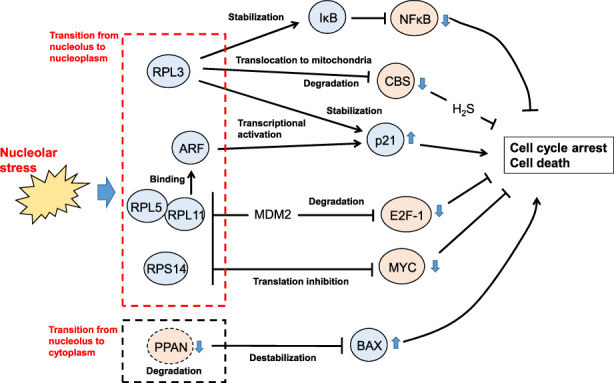
p53‐independent nucleolar stress response pathways. Nucleolar stress induces the translocation of the nucleolar proteins indicated in the dotted boxes into the nucleoplasm or cytoplasm, where they trigger cell cycle arrest and/or cell death through the inhibition of NFκB, CBS, E2F‐1, or MYC or the activation of BAX (see main text for details)
Like p53, E2F‐1 is regulated by MDM2‐mediated polyubiquitination followed by proteasomal degradation. However, unlike the case of p53, the binding of RPL11 to MDM2 induced by nucleolar stress promotes E2F‐1 polyubiquitination and degradation. The transcription of E2F‐1–dependent genes is thus repressed, thereby inducing cell death in a p53‐independent manner. 27 RPL11 also binds to p14ARF, triggering transcriptional activation of the CDK inhibitor p21 and p53‐independent cell cycle arrest. 28
MYC translation is repressed by the direct binding of RPL11, RPS14, and RPL5 to its mRNA. 28 , 29 MYC regulates ~15% of total cellular transcription, and MYC suppression leads to arrested cell proliferation and death induction. 30 As the transcriptional targets of MYC include almost all genes involved in ribosome biogenesis as well as Pol I–mediated rRNA transcription, this RP‐mediated MYC repression may suppress ribosome biogenesis, thereby enhancing the nucleolar stress response. 30
Like other RPs, RPL3 translocates from the nucleolus to the nucleoplasm in response to nucleolar stress, but it induces cell cycle arrest and death by a different mechanism. In the nucleoplasm, RPL3 both stabilizes p21 protein and activates p21 transcription by binding directly to the transcription factor Sp1; these activities lead to p21‐dependent cell cycle arrest. 31 On the other hand, RPL3 both represses transcription of cystathionine β‐synthase (CBS) and directly binds to CBS protein to promote its translocation into mitochondria, enhancing its degradation. This RPL3‐mediated CBS downregulation decreases levels of the CBS reaction product H2S and thereby induces mitochondria‐based apoptosis. 32 Finally, RPL3 stabilizes IκB, which inhibits the nuclear transfer and activation of NFκB and thereby suppresses its cell survival signaling. 33
In addition to these RP‐dependent pathways, the Peter Pan Homolog (PPAN) pathway functions in the nucleolar stress response in an RP‐ and p53‐independent manner. In resting cells, PPAN inhibits mitochondria‐based apoptosis by destabilizing BAX. Upon nucleolar stress, PPAN migrates from the nucleolus to the cytoplasm, where PPAN is degraded by caspases. PPAN thus loses its anti‐apoptotic effect, resulting in apoptotic cell death. 34
3. NUCLEOLAR STRESS AND HUMAN RIBOSOMAL DISEASES
Noncancerous pathological conditions linked to abnormal ribosome biosynthesis are called “ribosomal diseases” (Figure 4). Patients often present with anemia, developmental abnormalities, and/or degenerative disorders. In this section, we describe various ribosomal diseases grouped by their phenotypes, and introduce their molecular mechanisms. For more detail, we refer the reader to two excellent reviews. 35 , 36
FIGURE 4.
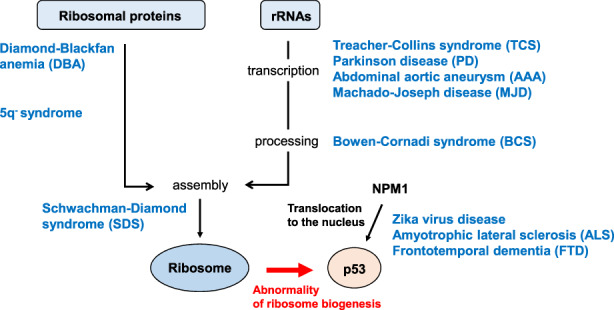
Nucleolar stress and human ribosomal diseases. During normal ribosome biogenesis, ribosomal proteins (RPs) are assembled with rRNAs (transcribed and processed) to form pre‐ribosomes, which then combine to produce mature ribosomes. Any abnormality in this sequence of events stabilizes and activates p53. In blue text are human diseases whose molecular defects affect ribosome biogenesis at the indicated stage. p53 is also stabilized when NPM1 abnormally translocates from the nucleolus into the nucleoplasm, an effect associated with Zika virus infection, amyotrophic lateral sclerosis (ALS), and frontotemporal dementia (FTD) (see main text for details)
3.1. Hematopoietic disorders and/or developmental abnormalities
Diamond‐Blackfan anemia (DBA) is characterized by anemia and malformations of the upper extremities, face and head (including microcephaly). DBA is caused mainly by RP mutation or reduced RP expression. Causal mutations affect genes encoding 19 RPs (particularly RPS19) and GATA binding protein‐1. 35 , 36 Analyses of mouse and zebrafish models have indicated that mutations in some of these RPs induce p53 stabilization and cause anemia and other DBA‐like symptoms. 37
“5q− syndrome” stands for “myelodysplastic syndromes associated with isolated del(5q) chromosome abnormality,” which is caused by a defect in a region of chromosome 5 containing RPS14. 5q− syndrome is characterized by anemia and increased platelets but normal body morphology. 38
Schwachman‐Diamond syndrome (SDS) features anemia and abnormal pancreatic exocrine secretion. Mutations in the Shwachman‐Bodian‐Diamond syndrome ribosome maturation factor (SBDS) gene, which is required for 80S ribosome assembly, occur in 90% of SDS patients. Loss of SBDS function is linked to RPL11‐dependent p53 activation. 39 , 40
Morphological (but not hematological) abnormalities characterize Treacher‐Collins syndrome (TCS) and Bowen‐Cornadi syndrome (BCS). Mutations of genes related to the Pol I transcription complex (such as treacle ribosome biogenesis factor‐1 [TCOF1]) cause TCS because the ensuing decreased transcription/processing of rRNA activates p53. 41 Mice or African clawed frogs lacking TCOF1 function show TCS‐like phenotypes, including skull dysplasia. 41 In contrast, BCS is caused by mutation of the EMG N1‐specific pseudouridine methyltransferase (EMG1) gene. EMG1 is degraded, which impairs pseudouridine rRNA modification and processing, and results in microcephaly. 42
Zika virus infection during pregnancy also causes microcephaly and may involve nucleolar stress. The viral ZIKV‐C protein binds to NPM1 in neural progenitors and forces its transfer from the nucleolus to the nucleoplasm. The resulting excessive p53 activation may induce neuronal cell cycle arrest and/or death leading to microcephaly. 43
3.2. Degenerative diseases
Degenerative diseases are characterized by exacerbation of symptoms over time. These disorders arise from impaired organ function caused partly by decreased cell proliferation or increased cell death. Degenerative diseases involving nucleolar stress include abdominal aortic aneurysm (AAA), amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), Machado‐Joseph disease (MJD), and Parkinson's disease (PD).
In AAA, decreased expression of Pol I transcription complex components reduces rRNA levels, triggering a nucleolar stress response involving p53 activation. 44 ALS and FTD patients also exhibit p53 activation but the molecular mechanism differs. 45 Despite exhibiting distinct neurological and psychiatric symptoms, ALS and FTD share a molecular pathway in which amplified GGGGCC repeats arise in the C9orf72‐SMCR8 complex subunit (C9orf72) gene. 46 The peptides generated by these repeats force NPM1 translocation from the nucleolus to the nucleoplasm, leading to p53 activation. 47
Machado‐Joseph disease is a spinocerebellar degenerative disease featuring RNA transcripts containing amplified CAG repeats within the ataxin‐3 (ATXN3) gene. 48 These transcripts bind to nucleolin and repress rRNA transcription, resulting in p53 activation via pathways involving RPL5, RPL11, and RPL23. 48 Reduced rRNA levels also lead to p53 activation in ~5%‐10% of PD patients with inactivation of Parkin, an E3 ubiquitin ligase. Loss of Parkin leads to accumulation of Parkin‐interacting substrate (PARIS; also called ZNF746), which represses rRNA transcription. 49
4. NUCLEOLAR STRESS AND CANCER
Cancer cells dramatically boost ribosome biosynthesis to promote cell growth and proliferation (Figure 5). In addition, de novo nucleotide biosynthesis is activated to supply sufficient nucleotides for increased rRNA production. 50 , 51 , 52 Thus, cancer cells should be sensitive to nucleolar stress. However, some RP mutations have been linked to tumor initiation and progression. 53 , 54 , 55 This section focuses on the association between nucleolar stress and cancer from two points of view. The first is the paradoxical phenomenon that, for some ribosomal diseases, the longer the nucleolar stress persists, the greater the risk of tumorigenesis. The second perspective concerns a potential new approach to cancer therapy based on manipulating nucleolar stress pathways mediating p53 activation.
FIGURE 5.
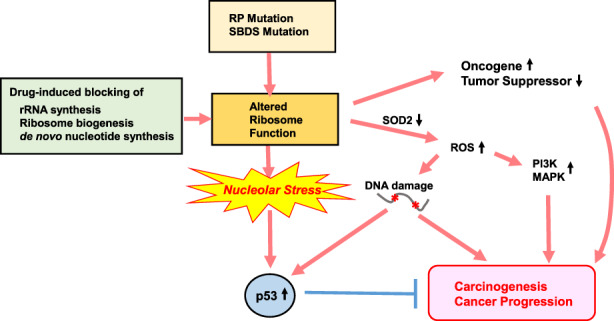
Nucleolar stress and cancer. Mutations in ribosomal proteins (RPs) and Shwachman‐Bodian‐Diamond syndrome ribosome maturation factor (SBDS) may alter ribosome function, which induces nucleolar stress and p53 activation. Although this p53 activation attempts to block tumorigenesis, the altered ribosome function may lead to increased translation of oncogenes and decreased translation of tumor suppressors, all of which promote cancer. These mutations also reduce superoxide dismutase‐2 (SOD2) activity and promote reactive oxygen species (ROS) production, which may activate PI3K/MAPK signaling supporting cancer development. ROS also trigger DNA damage, which contributes to tumor formation on one hand but also activates tumor suppression by p53 on the other hand. When these cell‐proliferative and deleterious events combine to overcome p53 signaling mediating tumor suppression, cancer starts to develop. In contrast, novel types of drugs that can inhibit ribosome biogenesis and induce nucleolar stress without triggering DNA damage or excessive ROS may activate p53 sufficiently to suppress tumorigenesis. These agents could represent a fresh avenue of anticancer therapy (see main text for details)
4.1. Increased risk of tumorigenesis
As noted above, in DBA, 5q− syndrome, and SDS, mutations in RPs and SBDS activate p53 via the typical nucleolar stress response pathway. However, despite this p53 activation, along with their anemia and malformations, DBA, 5q− syndrome, and SDS patients have an increased long‐term risk of developing various malignancies, especially blood cancers. 54 Furthermore, mutations in RPS15, RPL5, RPL10, and RPL22 occur in various solid tumors and blood cancers. 53 , 54 , 55 The mechanism by which defective ribosome biosynthesis increases cancer risk is not clear, but two possibilities have been raised.
The first possibility is that ribosome function is profoundly altered by an RP mutation. Ribosomes with RPS23 mutations are less accurate during translation, potentially compromising protein function. 56 Ribosomes with mutated RPL10 show enhanced BCL‐2 translation, 57 whereas ribosomes incorporating mis‐modified rRNAs due to processing errors exhibit reduced p53 and p27 translation. 58 , 59 If such translation anomalies affect molecules contributing to DNA repair or proliferation/survival signaling, a cell might be pushed toward oncogenesis.
The second possibility is that the reactive oxygen species (ROS) balance is altered by defective ribosome biogenesis, with RP mutations increasing ROS production. High levels of ROS are cytotoxic, but low levels of ROS enhance PI3K and MAPK signaling and cause DNA damage, which may initiate malignant transformation. 60 , 61 , 62 Increased ROS occur in lymphocytes of SDS patients and SDS‐deficient mice. 63 , 64 Loss of RPS19, RPL5, or RPL11, the genes involved in DBA, reduces the activity of superoxide dismutase‐2 (SOD2), 65 which is responsible for ROS degradation.
4.2. Nucleolar stress pathway as a target for cancer therapy
Cancer cells actively proliferate and so require vigorous ribosomal biosynthesis consuming large amounts of nucleotides, amino acids, and energy. p53 mutations arise in ~50% of human cancers, with the remaining 50% exhibiting normal p53. Therefore, drugs targeting ribosome biosynthesis should inhibit tumor cell growth in two ways: firstly, by reducing the number of ribosomes available for cancer cell growth; and secondly, by inducing a nucleolar stress response that activates p53 in cancer cells without p53 mutation.
Many conventional anticancer drugs inhibit DNA replication by intercalating into the DNA duplex. Cancer cells have a high rDNA copy number and strong rRNA transcriptional activity, making them susceptible to nucleolar stress when exposed to such drugs. Although this nucleolar stress should lead to the demise of the cancer cells, the DNA damage caused by drug intercalation may lead to the accumulation of mutations promoting oncogenesis. Thus, there is a need to develop drugs that more specifically inhibit rRNA transcription without causing DNA damage. 66
The novel agent CX‐5461 was developed as a Pol I–specific inhibitor and indeed induces a significant decrease in rRNA transcription. However, subsequent studies have revealed that CX‐5461 acts as a DNA intercalator and stabilizes guanine quadruplexes, which induce DNA replication stress. 67 Clinical trials of this agent are ongoing and should reveal if the benefits of its effects on rRNA outweigh the drawbacks of its effects on DNA. BMH‐21 is another drug that was developed as a Pol I inhibitor and functions as a DNA intercalator, but this agent does not appear to cause DNA damage. 68 , 69
Another strategy for inducing nucleolar stress to activate p53 in cancer cells relies on inhibition of nucleotide biosynthesis, which ultimately decreases pre‐rRNA transcription. 51 , 70 Mycophenolic acid (MPA) inhibits inosine monophosphate dehydrogenase‐2 (IMPDH2), a key enzyme in the de novo pathway of purine nucleotide biosynthesis. Glioblastoma cells treated in vitro with MPA show markedly impaired growth. 50 PICT1 may be another interesting target for cancer therapy because it plays roles in both the nucleolar stress response and rRNA processing. PICT1 depletion triggers p53 activation in cancer cells in an RPL11‐MDM2–dependent manner 20 and also represses cancer cell proliferation by disrupting rRNA processing. 71
5. CONCLUSION
It was discovered about 50 years ago that abnormal ribosome biosynthesis causes cell cycle arrest. The molecular basis of this phenomenon, originally named “ribotoxic stress” and now known as nucleolar stress, was elucidated in the early 2000s. In the nearly 20 years since, much has been revealed about the underlying mechanisms, but some major issues remain to be addressed: (i) How does a cell detect nucleolar stress, and what are the sensing molecules?; (ii) Why do so many diverse nucleolar stress response pathways exist?; (iii) How do we explain the tissue specificity of ribosomal diseases despite the ubiquitous presence of ribosomes?; and (iv) Why do patients with ribosomal diseases show an increased risk of cancer development? We expect that future research will answer these questions one by one and hope that the findings will one day be applied to both the establishment of therapies for ribosomal diseases and the generation of anticancer drugs targeting the nucleolar stress pathway.
CONFLICT OF INTEREST STATEMENT
A.S. is a current member of the Editorial Board of Cancer Science. The other authors have no conflict of interest.
ETHICS STATEMENT
Approval of the research protocol by an Institutional Reviewer Board: not applicable (N/A). Informed Consent: N/A. Registry and the Registration No. of the study/trial: N/A. Animal Studies: N/A.
ACKNOWLEDGMENTS
We are grateful for funding provided by the Japan Agency for Medical Research and Development (AMED P‐PROMOTE; Grant Number 22ama221117h0001 to A.S.); the Japan Society for the Promotion of Science (JSPS KAKENHI, Grant‐in‐Aid for Scientific Research [A] Grant Number 21H04806, and Grant‐in‐Aid for Scientific Research on Innovative Areas Grant Number 20H04905, all to A.S.); Nanken‐Kyoten, Tokyo Medical and Dental University (TMDU) (to A.S.); and the Project Mirai Cancer Research Grants (to A.S.).
Maehama T, Nishio M, Otani J, Mak TW, Suzuki A. Nucleolar stress: Molecular mechanisms and related human diseases. Cancer Sci. 2023;114:2078‐2086. doi: 10.1111/cas.15755
Contributor Information
Tomohiko Maehama, Email: tmaehama@med.kobe-u.ac.jp.
Akira Suzuki, Email: suzuki@med.kobe-u.ac.jp.
REFERENCES
- 1. Sollner‐Webb B, Tower J. Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem. 1986;55:801‐830. [DOI] [PubMed] [Google Scholar]
- 2. Schmidt EV. The role of c‐myc in cellular growth control. Oncogene. 1999;18:2988‐2996. [DOI] [PubMed] [Google Scholar]
- 3. Yang K, Yang J, Yi J. Nucleolar stress: hallmarks, sensing mechanism and diseases. Cell Stress. 2018;2:125‐140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lindstrom MS, Bartek J, Maya‐Mendoza A. p53 at the crossroad of DNA replication and ribosome biogenesis stress pathways. Cell Death Differ. 2022;29:972‐982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matthews DA. Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J Virol. 2001;75:1031‐1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Westdorp KN, Sand A, Moorman NJ, Terhune SS. Cytomegalovirus late protein pUL31 alters pre‐rRNA expression and nuclear organization during infection. J Virol. 2017;91:e00593‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poortinga G, Hannan KM, Snelling H, et al. MAD1 and c‐MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J. 2004;23:3325‐3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grandori C, Gomez‐Roman N, Felton‐Edkins ZA, et al. c‐Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311‐318. [DOI] [PubMed] [Google Scholar]
- 9. Arabi A, Wu S, Ridderstrale K, et al. c‐Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303‐310. [DOI] [PubMed] [Google Scholar]
- 10. Zhao J, Yuan X, Frodin M, Grummt I. ERK‐dependent phosphorylation of the transcription initiation factor TIF‐IA is required for RNA polymerase I transcription and cell growth. Mol Cell. 2003;11:405‐413. [DOI] [PubMed] [Google Scholar]
- 11. Chan JC, Hannan KM, Riddell K, et al. AKT promotes rRNA synthesis and cooperates with c‐MYC to stimulate ribosome biogenesis in cancer. Sci Signal. 2011;4:ra56. [DOI] [PubMed] [Google Scholar]
- 12. Perry RP, Kelley DE. Inhibition of RNA synthesis by actinomycin D: characteristic dose‐response of different RNA species. J Cell Physiol. 1970;76:127‐139. [DOI] [PubMed] [Google Scholar]
- 13. Burger K, Muhl B, Harasim T, et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem. 2010;285:12416‐12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068‐6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holzel M, Orban M, Hochstatter J, et al. Defects in 18S or 28S rRNA processing activate the p53 pathway. J Biol Chem. 2010;285:6364‐6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Russo A, Russo G. Ribosomal proteins control or bypass p53 during nucleolar stress. Int J Mol Sci. 2017;18:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luan Y, Tang N, Yang J, et al. Deficiency of ribosomal proteins reshapes the transcriptional and translational landscape in human cells. Nucleic Acids Res. 2022;50:6601‐6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. James A, Wang Y, Raje H, Rosby R, DiMario P. Nucleolar stress with and without p53. Nucleus. 2014;5:402‐426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sloan KE, Bohnsack MT, Watkins NJ. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 2013;5:237‐247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sasaki M, Kawahara K, Nishio M, et al. Regulation of the MDM2‐P53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nat Med. 2011;17:944‐951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Suzuki A, Kogo R, Kawahara K, et al. A new PICTure of nucleolar stress. Cancer Sci. 2012;103:632‐637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ichikawa MK, Saitoh M. Direct and indirect roles of GRWD1 in the inactivation of p53 in cancer. J Biochem. 2022;171:601‐603. [DOI] [PubMed] [Google Scholar]
- 23. Kuroda T, Murayama A, Katagiri N, et al. RNA content in the nucleolus alters p53 acetylation via MYBBP1A. EMBO J. 2011;30:1054‐1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gjerset RA, Bandyopadhyay K. Regulation of p14ARF through subnuclear compartmentalization. Cell Cycle. 2006;5:686‐690. [DOI] [PubMed] [Google Scholar]
- 25. Anderson SJ, Lauritsen JP, Hartman MG, et al. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53‐dependent checkpoint. Immunity. 2007;26:759‐772. [DOI] [PubMed] [Google Scholar]
- 26. Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49‐63. [DOI] [PubMed] [Google Scholar]
- 27. Wang HT, Chen TY, Weng CW, Yang CH, Tang MS. Acrolein preferentially damages nucleolus eliciting ribosomal stress and apoptosis in human cancer cells. Oncotarget. 2016;7:80450‐80464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Donati G, Montanaro L, Derenzini M. Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer Res. 2012;72:1602‐1607. [DOI] [PubMed] [Google Scholar]
- 29. Challagundla KB, Sun XX, Zhang X, et al. Ribosomal protein L11 recruits miR‐24/miRISC to repress c‐Myc expression in response to ribosomal stress. Mol Cell Biol. 2011;31:4007‐4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301‐309. [DOI] [PubMed] [Google Scholar]
- 31. Russo A, Esposito D, Catillo M, Pietropaolo C, Crescenzi E, Russo G. Human rpL3 induces G(1)/S arrest or apoptosis by modulating p21 (waf1/cip1) levels in a p53‐independent manner. Cell Cycle. 2013;12:76‐87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pagliara V, Saide A, Mitidieri E, et al. 5‐FU targets rpL3 to induce mitochondrial apoptosis via cystathionine‐beta‐synthase in colon cancer cells lacking p53. Oncotarget. 2016;7:50333‐50348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen J, Stark LA. Insights into the relationship between nucleolar stress and the NF‐kappaB pathway. Trends Genet. 2019;35:768‐780. [DOI] [PubMed] [Google Scholar]
- 34. Pfister AS, Keil M, Kuhl M. The Wnt target protein Peter pan defines a novel p53‐independent nucleolar stress‐response pathway. J Biol Chem. 2015;290:10905‐10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lafita‐Navarro MC, Conacci‐Sorrell M. Nucleolar stress: from development to cancer. Semin Cell Dev Biol. 2022;136:64‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sulima SO, Kampen KR, De Keersmaecker K. Cancer biogenesis in Ribosomopathies. Cells. 2019;8:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McGowan KA, Mason PJ. Animal models of diamond Blackfan anemia. Semin Hematol. 2011;48:106‐116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ebert BL, Pretz J, Bosco J, et al. Identification of RPS14 as a 5q‐ syndrome gene by RNA interference screen. Nature. 2008;451:335‐339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boocock GR, Morrison JA, Popovic M, et al. Mutations in SBDS are associated with Shwachman‐diamond syndrome. Nat Genet. 2003;33:97‐101. [DOI] [PubMed] [Google Scholar]
- 40. Hao Q, Wang J, Chen Y, et al. Dual regulation of p53 by the ribosome maturation factor SBDS. Cell Death Dis. 2020;11:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gal Z, Nieto B, Boukoura S, Rasmussen AV, Larsen DH. Treacle sticks the nucleolar responses to DNA damage together. Front Cell Dev Biol. 2022;10:892006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Armistead J, Khatkar S, Meyer B, et al. Mutation of a gene essential for ribosome biogenesis, EMG1, causes Bowen‐Conradi syndrome. Am J Hum Genet. 2009;84:728‐739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Slomnicki LP, Chung DH, Parker A, Hermann T, Boyd NL, Hetman M. Ribosomal stress and Tp53‐mediated neuronal apoptosis in response to capsid protein of the zika virus. Sci Rep. 2017;7:16652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang W, Cheng W, Parlato R, et al. Nucleolar stress induces a senescence‐like phenotype in smooth muscle cells and promotes development of vascular degeneration. Aging. 2020;12:22174‐22198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weishaupt JH, Hyman T, Dikic I. Common molecular pathways in amyotrophic lateral sclerosis and frontotemporal dementia. Trends Mol Med. 2016;22:769‐783. [DOI] [PubMed] [Google Scholar]
- 46. DeJesus‐Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p‐linked FTD and ALS. Neuron. 2011;72:245‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tao Z, Wang H, Xia Q, et al. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation‐induced cytotoxicity. Hum Mol Genet. 2015;24:2426‐2441. [DOI] [PubMed] [Google Scholar]
- 48. Tsoi H, Lau TC, Tsang SY, Lau KF, Chan HY. CAG expansion induces nucleolar stress in polyglutamine diseases. Proc Natl Acad Sci U S A. 2012;109:13428‐13433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kang H, Shin JH. Repression of rRNA transcription by PARIS contributes to Parkinson's disease. Neurobiol Dis. 2015;73:220‐228. [DOI] [PubMed] [Google Scholar]
- 50. Kofuji S, Hirayama A, Eberhardt AO, et al. IMP dehydrogenase‐2 drives aberrant nucleolar activity and promotes tumorigenesis in glioblastoma. Nat Cell Biol. 2019;21:1003‐1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lafita‐Navarro MC, Venkateswaran N, Kilgore JA, et al. Inhibition of the de novo pyrimidine biosynthesis pathway limits ribosomal RNA transcription causing nucleolar stress in glioblastoma cells. PLoS Genet. 2020;16:e1009117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hubackova S, Davidova E, Boukalova S, et al. Replication and ribosomal stress induced by targeting pyrimidine synthesis and cellular checkpoints suppress p53‐deficient tumors. Cell Death Dis. 2020;11:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. De Keersmaecker K, Atak ZK, Li N, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T‐cell acute lymphoblastic leukemia. Nat Genet. 2013;45:186‐190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sulima SO, Hofman IJF, De Keersmaecker K, Dinman JD. How ribosomes translate cancer. Cancer Discov. 2017;7:1069‐1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sulima SO, Patchett S, Advani VM, De Keersmaecker K, Johnson AW, Dinman JD. Bypass of the pre‐60S ribosomal quality control as a pathway to oncogenesis. Proc Natl Acad Sci U S A. 2014;111:5640‐5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Paolini NA, Attwood M, Sondalle SB, et al. A Ribosomopathy reveals decoding defective ribosomes driving human dysmorphism. Am J Hum Genet. 2017;100:506‐522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kampen KR, Sulima SO, Verbelen B, et al. The ribosomal RPL10 R98S mutation drives IRES‐dependent BCL‐2 translation in T‐ALL. Leukemia. 2019;33:319‐332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene‐induced senescence and p53 translational control in X‐linked dyskeratosis congenita. EMBO J. 2010;29:1865‐1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yoon A, Peng G, Brandenburger Y, et al. Impaired control of IRES‐mediated translation in X‐linked dyskeratosis congenita. Science. 2006;312:902‐906. [DOI] [PubMed] [Google Scholar]
- 60. Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24:R453‐R462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ishikawa K, Takenaga K, Akimoto M, et al. ROS‐generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661‐664. [DOI] [PubMed] [Google Scholar]
- 62. Woo DK, Green PD, Santos JH, et al. Mitochondrial genome instability and ROS enhance intestinal tumorigenesis in APC(min/+) mice. Am J Pathol. 2012;180:24‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ravera S, Dufour C, Cesaro S, et al. Evaluation of energy metabolism and calcium homeostasis in cells affected by Shwachman‐diamond syndrome. Sci Rep. 2016;6:25441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zambetti NA, Ping Z, Chen S, et al. Mesenchymal inflammation drives genotoxic stress in hematopoietic stem cells and predicts disease evolution in human pre‐leukemia. Cell Stem Cell. 2016;19:613‐627. [DOI] [PubMed] [Google Scholar]
- 65. Kapralova K, Jahoda O, Koralkova P, et al. Oxidative DNA damage, inflammatory signature, and altered erythrocytes properties in diamond‐Blackfan anemia. Int J Mol Sci. 2020;21:9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ferreira R, Schneekloth JS Jr, Panov KI, Hannan KM, Hannan RD. Targeting the RNA polymerase I transcription for cancer therapy comes of age. Cell. 2020;9:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xu H, Di Antonio M, McKinney S, et al. CX‐5461 is a DNA G‐quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat Commun. 2017;8:14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jacobs RQ, Huffines AK, Laiho M, Schneider DA. The small‐molecule BMH‐21 directly inhibits transcription elongation and DNA occupancy of RNA polymerase I in vivo and in vitro. J Biol Chem. 2022;298:101450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Colis L, Peltonen K, Sirajuddin P, et al. DNA intercalator BMH‐21 inhibits RNA polymerase I independent of DNA damage response. Oncotarget. 2014;5:4361‐4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Huang M, Ji Y, Itahana K, Zhang Y, Mitchell B. Guanine nucleotide depletion inhibits pre‐ribosomal RNA synthesis and causes nucleolar disruption. Leuk Res. 2008;32:131‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tafforeau L, Zorbas C, Langhendries JL, et al. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre‐rRNA processing factors. Mol Cell. 2013;51:539‐551. [DOI] [PubMed] [Google Scholar]


