Abstract
G‐proteins are intracellular partners of G‐protein‐coupled receptors. As a member of the G‐protein family, GNB1 has been shown to play a pro‐cancer role in lung cancer and breast cancer. However, the biological function and detailed mechanisms of GNB1 in hepatocellular carcinoma progression are unclear. In this study, we investigated the effects of GNB1 and its possible mechanism of action in hepatocellular carcinoma (HCC). The clinical significance of GNB1 was evaluated in a large cohort of HCC patients, showing that GNB1 was overexpressed in HCC compared to adjacent normal liver tissues, and increased GNB1 expression was associated with poor prognosis. We also demonstrated that GNB1 enhances cell proliferation, colony formation, and cell migration and invasion in vitro and promotes the epithelial‐to‐mesenchymal transition process in HCC cells. Tumor xenograft model assay confirmed the oncogenic role of GNB1 in tumorigenicity in nude mice. Activation of P38 signaling was found in the GNB1 overexpressed HCC cells. Further intervention of P38 confirmed it as an important signaling pathway for the oncogenic role of GNB1 in HCC. Moreover, co‐immunoprecipitation followed by liquid chromatograph‐mass spectrometry identified that GNB1 exerted oncogenic functions via the interaction of BAG2 and activated P38 signaling pathway. Together, our results reveal that GNB1 plays a pivotal oncogenic role in HCC by promoting the P38 pathway via cooperating with BAG2. GNB1 may serve as a prognostic biomarker for patients with HCC.
Keywords: BAG2, GNB1, hepatocellular carcinoma, metastasis, P38, proliferation
Our study revealed that the expression of GNB1 in HCC tissue was significantly upregulated and closely associated with patient prognosis. Overexpression of GNB1 stimulates a repertoire of downstream tumorigenic responses, including proliferation, migration, invasion, and EMT. Mechanistically, GNB1 interactes with BAG2 and facilitates its expression. This thereby activates the P38/MAPK signaling pathway, leading to the development of HCC.
Abbreviations
- AFP
Alpha‐fetoprotein
- ALT
Alanine aminotransferase
- AST
Aspartate aminotransferase
- BAG2
The Bcl‐2 associated athanogene
- CCK8
Cell counting kit‐8
- COIP
Co‐immunoprecipitation
- DFS
Disease‐free survival
- EMT
Epithelial–mesenchymal transition
- GPCR
G protein‐coupled receptors
- HBV
Hepatitis B virus
- HCC
Hepatocellular carcinoma
- IHC
Immunohistochemistry
- IOD
Integrated option density
- LC–MS
Liquid chromatograph‐mass spectrometry
- OS
Overall survival
- shRNA
Short hairpin RNA
- siRNA
Small interfering RNA
- TCGA
The Cancer Genome Atlas
1. INTRODUCTION
Liver cancer is the sixth most common and fourth most fatal cancer worldwide. 1 Hepatocellular carcinoma (HCC) is the most common form of liver cancer, accounts for approximately 90% of cases. HCC is a highly heterogeneous disease with multiple risk factors and etiology, among which Hepatitis B virus (HBV) infection is still the most prominent risk factor for developing HCC, accounting for approximately 50% of cases. 2 Half of HCC patients are diagnosed in advanced stages, when liver resection and transplantation are not feasible. 3 Although drugs such as sorafenib have been used in clinical practice, patients with advanced hepatocellular carcinoma still have a mortality rate of 80%, median survival of <1 year, and a 5‐year survival rate of <20% due to its high recurrence, invasion, and metastasis rates. 4 , 5 Consequently, it is important to uncover the molecular mechanisms of HCC progression to identify new therapeutic targets and seek new strategies for treatment.
Guanine nucleotide‐binding proteins (G proteins), mediating intracellular responses to various extracellular stimuli, is a ubiquitous signaling mechanism found in many receptor‐dependent pathways. 6 There are three known types of G proteins, called guanine nucleotide‐binding polypeptides α, β, and γ (Gα, Gβ, and Gγ). GNB1 (Gβ1) is one of the isoforms of Gβ that have been identified. 6 Some G protein‐coupled receptors (GPCRs) are associated with tumorigenesis and metastasis, while GPCR is coupled to a heterotrimeric G protein consisting of Gα, Gβ, and Gγ subunits. 7 Therefore, the role of G proteins in tumors has attracted the attention of many researchers. In past studies, GNB1 has been shown to be involved in the development or drug resistance of several kinds of cancer, such as acute lymphoblastic leukemia, 8 ETV6‐ABL1 positive leukemia, 9 lung cancer, 10 cervical squamous cell carcinoma, 11 clear cell renal cell carcinoma, 12 and breast cancer. 13 In addition, bioinformatics analysis has identified GNB1 as a potential therapeutic target and prognostic marker for ovarian cancer and colorectal cancer, which is also a digestive system tumor. 14 , 15 However, the function and regulatory mechanism of GNB1 in HCC remain unknown.
The mitogen‐activated protein kinases (MAPKs), which are well known to be activated by GPCRs in various systems, have an important role for cellular activities, such as gene expression, cell proliferation, survival, and migration. 16 , 17 Because of the important role in regulating these cellular processes, the MAPK signaling pathway is frequently found to be dysregulated in various types of human tumors, including HCC. 18 Recently, BAG2 was found to be involved in the regulation of the MAPK pathway. 19 The Bcl‐2 associated athanogene (BAG) family, a group of proteins that prevent cell death by interacting with Bcl‐2, is involved in regulating physiological activities from apoptosis to tumorigenesis. 20 , 21 As a member of the BAG family, BAG2 has been shown to be associated with the progression and prognosis of hepatocellular carcinoma by bioinformatic‐based analysis. 22 In this study, we focused on the role of GNB1 in HCC and found its increased expression, leading to poor prognosis. Further experiments confirmed its cancer‐promoting effect. Based on the fact that BAG proteins interact with many proteins and participate in different cellular processes, 21 we demonstrated that GNB1 interacts with BAG2 and is jointly involved in the activation of the MAPK pathway, thus promoting the development of HCC.
2. MATERIALS AND METHODS
2.1. Clinical samples
Human HCC tissues were obtained from 105 patients at Tongji Hospital of Huazhong University of Science and Technology between 2012 and 2016. Of these, 91 tissues with complete clinical information were included in this study. Data including clinical information and pathological features were approved by the institutional ethics committee of Huazhong University of Science and Technology. Written informed consent for data analysis was obtained from all patients before operation.
2.2. Plasmids and reagents and antibodies
GNB1 cDNA was obtained from NCBI and cloned into the pLenti 3*Flag‐CMV‐GFP vector to generate a GNB1 expression plasmid, which was confirmed by sequencing. The target sequences in the pLKO.1‐GNB1 shRNA vector against human GNB1 were 5′‐CGAGCAACTTAAGAACCAGAT‐3′, 5′‐GCCATTTGCTTCTTTCCAAAT‐3′, and 5′‐GCTTGTGATGCTTCAGCCAAA‐3′. PLenti 3*Flag‐CMV‐GFP and PLKO.1 were purchased from Addgene. SB202190, P38 MAPK inhibitor, was from MedChemExpress. PEI was from Shanghai FuShen Biotechnology. DMEM was from Hyclone. Opti‐MEM reduced serum media was from Gibco. FBS was from QmSuero. Polybrene and puromycin were from Biosharp. Primary antibodies used in this study include GNB1 (Proteintech, 10247‐2‐AP), BAG2 (Santa, sc‐390262), tubulin (Proteintech, 10094‐1‐AP), p‐P38 (CST, 9211s), P38 (CST, 9212s), p‐ERK (CST, 4370s), ERK (CST, 9102s), p‐JNK (CST, 4668s), and JNK (CST, 9252s).
2.3. Cell culture, GNB1 overexpression and knockdown
HL‐7702, HepG2, Hep3B, Huh7, HLF MHCC‐97H, and 293T cell lines were obtained from Hepatobiliary Surgery Laboratory, Huazhong University of Science and Technology. Cells were cultured in DMEM (Hyclone) supplemented with 4.5 g/L glucose and 10% FBS (QmSuero). Transfection was performed using PEI according to the manufacturer's instructions. Viral packaging was performed in HEK‐293T cells after co‐transfection of pLenti‐GNB1 and plko.1‐GNB1 with the packaging plasmid psPAX2 and the envelope plasmid pMD2.G (Addgene) using PEI. Virus was harvested 48 h after transfection. Freshly made virus supernatants supplemented with 8 μg/mL polybrene (Biosharp) were added to exponentially growing HLF, Huh7, or 97H cells. After 24 h, fresh medium was added. GNB1 stable overexpression or knockdown cells were achieved by 1‐week puromycin (4 μg/mL, Biosharp) selection.
2.4. Immunohistochemical staining
Immunohistochemistry (IHC) was conducted using antibodies against GNB1. Briefly, sections were deparaffinized with xylene and ethanol and rehydrated before antigen retrieval by heating to just below boiling temperature in Tris/EDTA buffer (pH 9.0) for 20 min in a microwave oven. Images were acquired using the 3DHIESTECH scanning system and software. The mean integrated option density (IOD) of GNB1‐expressing in each sample was analyzed using Image‐Pro Plus 6.0 software (Media Cybernetics). The samples were divided into two groups for further analysis using X‐tile: 23 , 24 , 25 , 26 , 27 a high expression group and a low expression group.
2.5. Western blotting
Cells or tissues were lysed in RIPA buffer. The protein from each group was separated by 10% SDS‐PAGE and transferred onto a PVDF membrane (Bio‐Rad Laboratory). The membranes were blocked in protein‐free rapid‐blocking buffer (Epizyme). The membrane was then incubated with specific antibodies overnight at 4°C and probed with appropriate secondary antibodies. The signals were visualized using Bio‐Rad ChemiDoc MP. Band intensities were quantified using ImageLab. Tubulin was used as the loading control.
2.6. RNA isolation and quantitative RT‐PCR
The total RNA from the human cells or tissues was extracted using TRIzol Reagent, as per the kit's instructions. Then a reverse transcription system kit (Servicebio) was applied to generate the first cDNA strand. Thereafter, the qRT‐PCR technique was implemented using the Universal Blue SYBR Green qPCR Master Mix (Servicebio). The qRT‐PCR parameters were as follows: 30 s at 95°C, then 40 cycles (15 s at 95°C, 10 s at 60°C, 30 s at 72°C). Here, β‐actin was used as the control. Then, the relative fold change was determined using the 2−ΔΔCT technique in triplicate. Table S1 shows the list of primers used.
2.7. Cell proliferation and colony formation assays
Cell proliferation was estimated with a Cell Counting Kit‐8 (CCK8), and cells in the logarithmic phase were seeded onto 96‐well plates (3000 cells/well). CCK8 solution (10 μL, Biosharp) was incubated with cells for 1 h. The OD450 was measured using a multimode plate reader (PerkinEimer), which reflects the number of viable cells, on days 1, 2, and 3. Lentivirus‐infected cells were inoculated in a six‐well plate with 1000 cells/well/2 mL DMEM with 10% FBS for colony formation assay. Medium was exchanged every 2–3 days. Cell clones were fixed and stained by 4% paraformaldehyde and Giemsa (Servicebio), respectively. The numbers of colonies were counted.
2.8. Wound‐healing assay
Cells in six‐well plates were allowed to reach confluence, and wounds were scratched using sterile tips. Wound closure was recorded every 24 h using a microscope.
2.9. Transwell assay
Cells in serum‐free medium were seeded in the upper chamber of a transwell apparatus (8 mmol/L pore size) or on a Matrigel‐coated transwell insert (BD Biosciences). The lower chamber contained DMEM supplemented with 10% FBS as a chemoattractant. After 48 h of incubation, the cells in the upper chamber were gently removed. The remaining cells were fixed and stained using 4% paraformaldehyde and Giemsa solution. The cells were counted in five randomly selected visual fields.
2.10. Co‐immunoprecipitation
Co‐immunoprecipitation (Co‐IP) was performed to confirm protein–protein binding properties. 97H cells were grown in 10‐cm dishes, and approximately 1 × 107 cells were lysed by sonication in 1 mL NP‐40 lysis buffer (P0013F, Beyotime) supplemented with Cocktail and PhosSTOP Cocktail Tablets. After sonication and centrifugation, agarose beads were added to remove nonspecific adsorption. Supernatants were incubated with corresponding antibodies overnight, then incubated with protein A/G beads for 4 h at 4°C on a rotating platform. Immunoprecipitates (IPs) were then subjected to western blot analyses.
2.11. In vivo growth assays
Animal experiments were approved by the Institutional Review Board of Renmin Hospital of Wuhan University (WDRM 22008). All animal experiments were performed in the Animal Experiment Center of Renmin Hospital of Wuhan University in accordance with standard guidelines. HLF cells (4 × 106) bearing GNB1‐overexpression or vector‐control constructs were suspended in 100 μL of a mixture of PBS and subcutaneously injected into nude mice. The pharmaceutical formulation consisting of the SB202190 was successively dissolved in 5% DMSO, 40% PEG300, 5%Tween‐80, and 45% saline, and the mixed solution was intraperitoneally injected into mice daily. The mice were anesthetized using isoflurane inhalation before sample collection. Death was confirmed using cervical dislocation.
2.12. Statistical analysis
Data are presented as mean ± SEM and analyzed by SPSS 26.0 (IBM, U.S.A.) and Prism 8.0 software (GraphPad, USA). An unpaired Student's t test was used to evaluate the differences between two groups. p < 0.05 was considered statistically significant.
3. RESULTS
3.1. GNB1 is highly expressed in hepatocellular carcinoma tissues and predicts poor prognosis
Through TCGA cohort analyses, upregulation of GNB1 in HCC was found (Figure 1A). In addition, GNB1 is shown to be upregulated in most of the tumors, including bladder urotheliar carcinoma, invasive breast carcinoma, cholangiocarcinoma, esophageal carcinoma, glioblastoma multiforme, kidney chromophobe, prostate adenocarcinoma, stomach adenocarcinoma, thyroid carcinoma, and uterine carcinosarcoma (Figure S1A). Then, we detected the expression levels of GNB1 in 91 pairs of tumorous tissues and nontumorous adjacent liver tissues from HCC patients by IHC staining. We found that GNB1 was significantly overexpressed in HCC compared to adjacent normal liver tissues (Figure 1C,D). Upregulation of GNB1 was further validated by western blot and real‐time PCR (Figure 1B,E).
FIGURE 1.
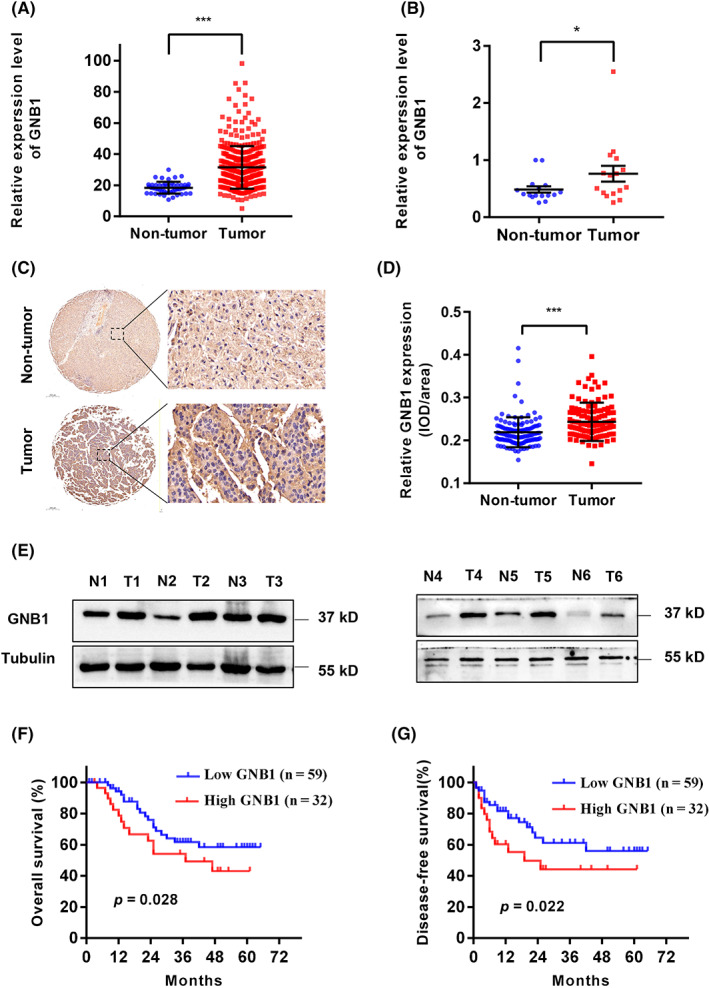
GNB1 is highly expressed in hepatocellular carcinoma (HCC) tissues and predicts poor prognosis. (A) The expression of GNB1 mRNA in the TCGA‐OV dataset was analyzed. (B) The expression of GNB1 mRNA was verified by RT‐PCR in 16 pairs of HCC and adjacent normal tissues. (C) Representative images of IHC staining for GNB1 in 91 paired HCC tumor and adjacent normal tissues. Scale bar, 200 μm (left panel) or 20 μm (right panel). (D) Dot density plot shows the distribution of average integrated option density (IOD) for GNB1 level in the samples from (C). ***p < 0001, paired t test. (E) The GNB1 protein levels in six pairs of HCC and adjacent normal tissues was analyzed by western blot. (F) Kaplan–Meier curves of the overall survival rate of 91 HCC patients with GNB1 high or low expression levels. p = 0.028, log‐rank test. (G) Kaplan–Meier curves of the disease‐free survival rate of 91 HCC patients with GNB1 high or low expression levels. p = 0.022, log‐rank test.
To determine the clinical significance of GNB1 upregulation in HCC tissues, the 91 HCC patients were divided into two groups based on the results of IHC analysis: a high GNB1 expression group (n = 33) and a low GNB1 expression group (n = 58). Kaplan–Meier survival analysis showed that HCC patients with high GNB1 expression levels had worse overall survival (OS) (p = 0.028) and disease‐free survival (p = 0.022) than patients with low GNB1 levels (Figure 1F,G). This prognosis effect was also observed in TCGA (n = 324) cohort (Figure S1B). In addition, the larger sample size in TCGA revealed the correlation between GNB1 and HCC grading as well as clinical staging (Figure S1C,D). The clinical data from 91 HCC patients only revealed the correlation between GNB1 expression levels and Child–Pugh class (Table 1), suggesting that a larger sample size is needed for further study.
TABLE 1.
The correlation between GNB1 expression and clincopathological features in hepatocellular carcinoma (HCC)
| Clinical variables | Number of patients | GNB1 expression level | p value | |
|---|---|---|---|---|
| n = 91 | Low (n = 59) | High (n = 32) | ||
| Age (years) | 0.476 | |||
| <60 | 72 | 48 (66.7%) | 24 (33.3%) | |
| ≥60 | 19 | 11 (57.9%) | 8 (42.1%) | |
| Gender | 0.307 | |||
| Male | 76 | 51 (67.1%) | 25 (32.9%) | |
| Female | 15 | 8 (53.3%) | 7 (46.7%) | |
| HBsAg | 0.428 | |||
| Positive | 75 | 50 (66.7%) | 25 (33.3%) | |
| Negative | 16 | 9 (56.3%) | 7 (43.7%) | |
| ALT (U/L) | 0.907 | |||
| ≤40 | 59 | 38 (64.4%) | 21 (35.6%) | |
| >40 | 32 | 21 (65.6%) | 11 (34.4%) | |
| AST (U/L) | 0.926 | |||
| ≤40 | 62 | 40 (64.5%) | 22 (35.5%) | |
| >40 | 29 | 19 (65.5%) | 10 (34.5%) | |
| AFP (ng/mL) | 0.735 | |||
| <400 | 49 | 31 (63.3%) | 18 (36.7%) | |
| ≥400 | 42 | 28 (66.7%) | 14 (33.3%) | |
| Child–Pugh class | 0.014 | |||
| A | 81 | 49 (60.5%) | 32 (39.5%) | |
| B | 10 | 10 (100%) | 0 (0%) | |
| Liver cirrhosis | 0.170 | |||
| No | 25 | 19 (76.0%) | 6 (24.0%) | |
| Yes | 66 | 40 (60.6%) | 26 (39.4%) | |
| Tumor size (cm) | 0.396 | |||
| ≤5 | 29 | 17 (58.6%) | 12 (41.4%) | |
| >5 | 62 | 42 (67.7%) | 20 (32.3%) | |
| Tumor number | 0.582 | |||
| Single | 74 | 47 (63.5%) | 27 (36.5%) | |
| Multiple | 17 | 12 (70.6%) | 5 (29.4%) | |
| Tumor encapsulation | 0.890 | |||
| Yes | 56 | 36 (64.3%) | 20 (35.7%) | |
| No | 35 | 23 (65.7%) | 12 (34.3%) | |
| Vascular invasion | 0.712 | |||
| Yes | 18 | 11 (61.1%) | 7 (38.9%) | |
| No | 73 | 48 (65.8%) | 25 (34.2%) | |
| Tumor differentiation | 0.105 | |||
| Well | 53 | 38 (71.7%) | 15 (28.3%) | |
| Poor | 38 | 21 (55.3%) | 17 (44.7%) | |
| TNM stage | 0.579 | |||
| I–II | 65 | 41 (63.1%) | 24 (36.9%) | |
| III–IV | 26 | 18 (69.2%) | 8 (30.8%) | |
3.2. GNB1 promotes hepatocellular carcinoma cell growth in vitro.
To explore the biological function of GNB1 in HCC cells, we established stable models of GNB1 overexpression in HLF cell lines as well as knockdown in Huh7 and 97H cell lines based on the expression of GNB1 in HCC cell lines (Figure 2A). In cultured HLF cells, GNB1 overexpression increased cell viability, while GNB1 knockdown showed the opposite (Figure 2B,C). In addition, overexpression of GNB1 promoted clone formation, while knockdown of GNB1 inhibited clone formation (Figure 2D,E).
FIGURE 2.
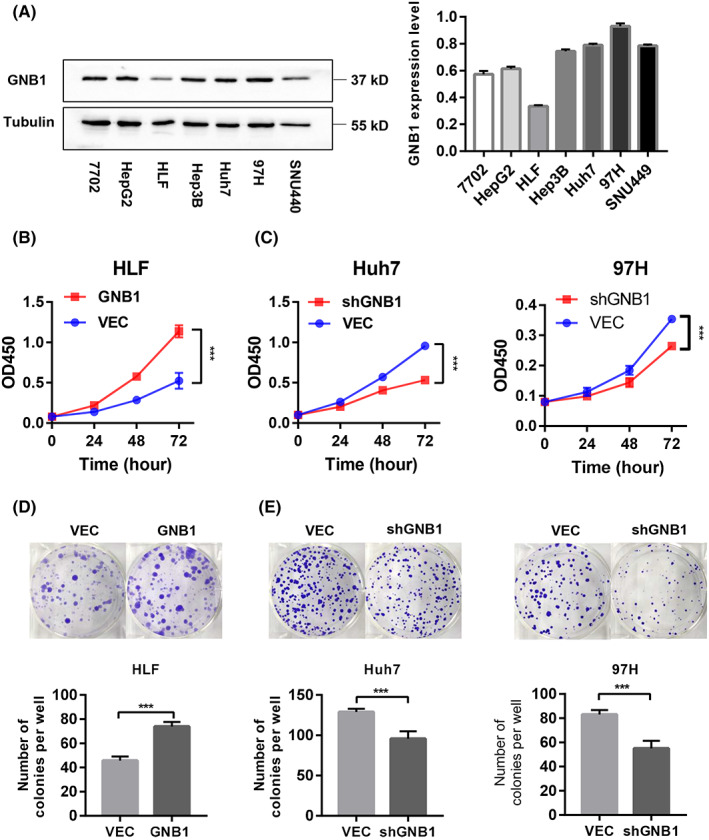
GNB1 promotes hepatocellular carcinoma (HCC) cell growth in vitro. (A) GNB1 expression levels in various HCC cells were detected by western blotting. (B, C) The cell viability of indicated stable cells was measured by CCK‐8 assay at different time points. ***p < 0.001, Student t test. (D, E) The anchorage‐depend colony formation assay was performed to assess the effects of GNB1 knockdown or overexpression on the clone formation ability of HCC cells. Representative images of colonies are shown (upper panel), and the number of colonies have been counted (lower panel). **p < 0.01, ***p < 0.001, Student t test.
3.3. GNB1 overexpression induces epithelial‐to‐mesenchymal transition of hepatocellular carcinoma cells and promotes metastasis
To determine the effect of GNB1 on cell metastasis, wound‐healing assays were performed to show that overexpression of GNB1 increases the migration of HCC cells, whereas GNB1 knockdown represses the cell mobility (Figure 3A,B). The transwell assays also demonstrated that GNB1 promotes cell invasion and migration (Figure 3C,D). Given that epithelial‐to‐mesenchymal transition (EMT) is one of the hallmarks of elevated cell mobility and metastasis, the expression of several EMT markers was tested. As shown in Figure 3E,F, GNB1 knockdown suppressed the mesenchymal marker (N‐cadherin) and upregulated the epithelial markers (occludin) in Huh7 and 97H cells. In contrast, overexpression of Flag‐tagged GNB1 exhibited the opposite effects in HLF cells. Altogether, these results demonstrate that GNB1 is the crucial factor regulating cell migration and invasion, inducing the EMT process in HCC cells.
FIGURE 3.
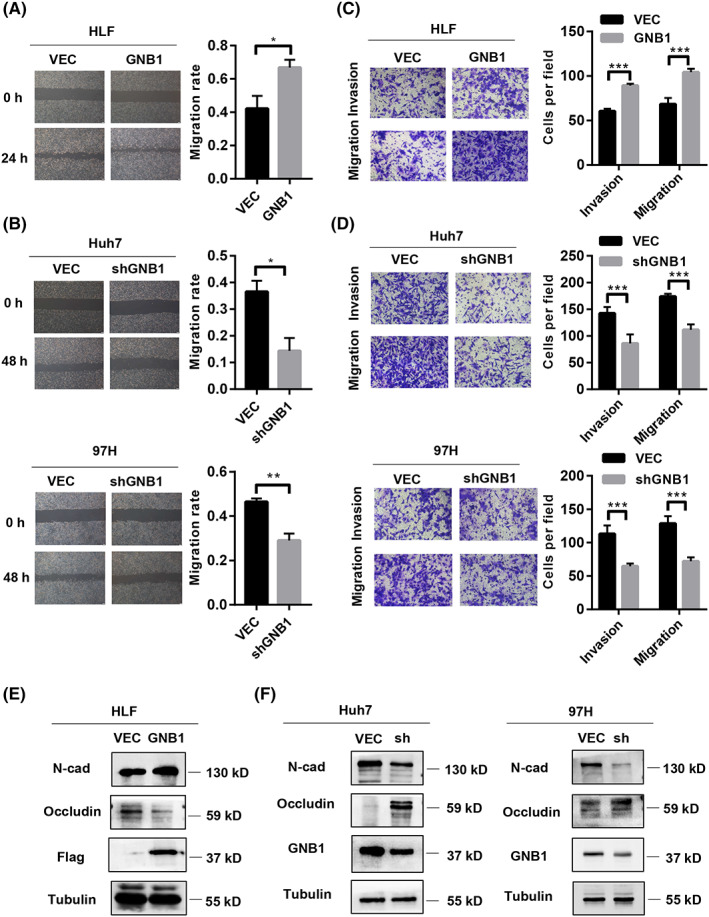
GNB1 overexpression induces epithelial‐to‐mesenchymal transition (EMT) of hepatocellular carcinoma (HCC) cells and promotes metastasis. (A) Cells migration at 24 h after scratching (0 h) in HLF cells with GNB1 overexpression were determined by wound‐healing assays. The scale bar represents 200 μm. *p < 0.05. (B) Cell migration at 48 h after scratching (0 h) in Huh7 and 97H cells with GNB1 knockdown were determined by wound‐healing assays. The scale bar represents 200 μm. **p < 0.01. (C) Cell migration and invasion abilities in HLF cells with GNB1 overexpression were determined by transwell assay. Representative image (left panel) and summary bar chart (left panel) are shown. Scale bar, 50 μm. ***p < 0.001. (D) Cell migration and invasion abilities in 97H and Huh7 cells with GNB1 knockdown were determined by transwell assay. Representative image (left panel) and summary bar chart (left panel) are shown. Scale bar, 50 μm. ***p < 0.001, Student t test. (E and F) Expression of EMT markers, N‐cadherin and occludin in indicated cells were examined by western blotting.
3.4. GNB1 activates the mitogen‐activated protein kinases pathway in hepatocellular carcinoma cells
In investigating the mechanisms underlying GNB1 abnormal expressed‐induced HCC progression, gene set enrichment analysis (GSEA) of the RNA‐seq data revealed significant overlaps in MAPK signaling pathway‐targeted genes (Figure 4A, Table S2). Further, we confirmed the protein expression of total and phosphorylation levels of ERK, JNK, and P38 by western blotting. P38 phosphorylation (p‐P38) and ERK1/2 phosphorylation (p‐ERK1/2) were markedly reduced in GNB1 knockdown cells in comparison to vector‐infected cells. There was a significant increase in GNB1‐overexpressing cells in comparison to the vector‐infected cells. However, JNK phosphorylation (p‐JNK) showed no significant change, indicating that P38 and ERK‐dependent signaling, rather than JNK‐dependent signaling pathway, played a major role in GNB1‐enhanced HCC proliferation and metastasis (Figure 4B,C). Previous studies have confirmed the activating effect of GNB1 on ERK. 11 In this study, P38 inhibitor SB202190 was applied, and the inhibition of P38 resulted in diminished cell proliferation, migration, and invasion, suggesting that the promotion of GNB1 on hepatocellular carcinoma progression is dependent on the activation of P38 (Figure 4C–E).
FIGURE 4.
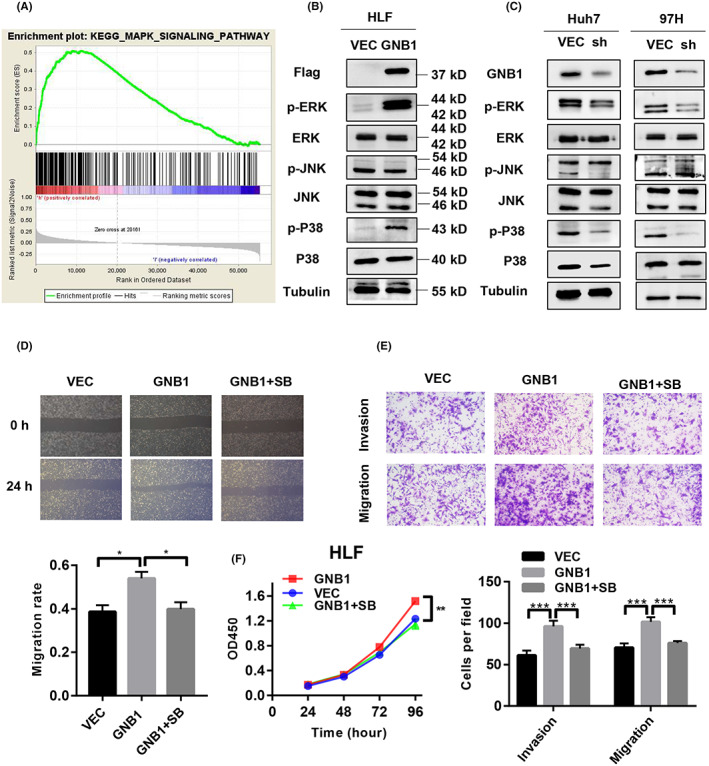
GNB1 activates the mitogen‐activated protein kinase (MAPK) pathway in hepatocellular carcinoma (HCC) cells. (A) GSEA of the RNA‐seq data revealed significant overlaps among MAPK pathway‐targeted genes. (B) Important molecules in the MAPK pathway, such as p‐ERK, ERK, p‐P38, P38, p‐JNK, and JNK, were determine by western blotting in GNB1 overexpression cell lines. Stable overexpression of GNB1 enhanced the phosphorylation of P38 and ERK in HLF cells. (C) Important molecules in the MAPK pathway levels were determined by western blotting in GNB1 knockdown cell line. Knockdown of GNB1 inhibited the phosphorylation of P38 and ERK in Huh7 and 97H cells. (D) HLF cells stably overexpressing GNB1 were treated with SB202190 (10 μmol/L). Wound‐healing assay was performed to assess cell migrative abilities. SB202190 exposure significantly attenuated the enhancement in cell migration induced by GNB1 overexpression. (E) Transwell assay was performed to assess cell invasive and migrative abilities. SB202190 exposure significantly attenuated the enhancement in cell migration and invasion induced by GNB1 overexpression. (F) Cell viability of indicated cells were measured by CCK‐8 assay. SB202190 treatment reversed the increased proliferation induced by GNB1 overexpression in HLF cells. *p < 0.05; **p < 0.01; ***p < 0.001, Student t test.
3.5. GNB1 regulates the phosphorylation of P38 through BAG2
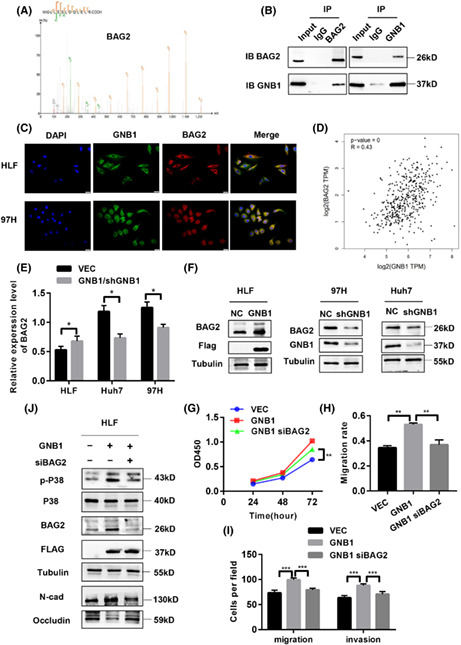
To further determine the downstream target of GNB1, we performed Co‐IP followed by liquid chromatograph‐mass spectrometry (LC–MS). GNB1‐binding candidates were then identified by comparing the anti‐GNB1‐Flag with anti‐IgG IP products of GNB1 (Flag)‐overexpressed cells. Identified candidate genes included TRIM33, BAG2, and MANBA (Figure 5A). Among them, BAG2 has been shown to be associated with the progression and prognosis of HCC. 22 The 91 HCC patients were divided into two groups based on the results of IHC analysis: a high BAG2 expression group (n = 51) and a low BAG2 expression group (n = 40) (Figure S2A). Kaplan–Meier survival analysis showed that HCC patients with high BAG2 expression levels had worse (OS) (p = 0.038) and disease‐free survival (p = 0.029) than patients with low BAG2 levels (Figure S2B,C). High BAG2 expression correlates with high levels of AST and AFP, and worse TNM stage and may predict the presence of tumor encapsulation (Table 2). Otherwise, high expression of BAG2 in tumor tissues was confirmed by western blotting (Figure S2D). Co‐IP assays were performed in the 97H cells to validate the interaction between GNB1 and BAG2 (Figure 5B). The localization of the two proteins was further confirmed by microscopy that GNB1 co‐localized with BAG2 both in the cytoplasm of HLF and 97H cells (Figure 5C). Through the GEPIA2 website, we found that the mRNA level of GNB1 was positively correlated with BAG2 (Figure 5D). The mRNA and protein expression of BAG2 was upregulated by GNB1 overexpression, while low expression of GNB1 led to reduced protein and mRNA levels of BAG2 in HCC cells (Figure 5E,F). To investigate the effect of BAG2 on the GNB1‐mediated cell proliferation and metastasis, HLF cells stably overexpressed GNB1 were co‐transfected with siRNA against BAG2 (Figure 5J). BAG2 knockdown significantly abolished the promoting effect of GNB1 on cell viability, migration, and invasion abilities in HLF cells (Figures 5G–I, S3B,C). We further examined whether GNB1 activated the P38 signaling pathway through mediating BAG2. As expected, BAG2 knockdown inhibited P38 phosphorylation (Figure 5J). To investigate the effect of the combination of GNB1 and BAG2 on the prognosis of clinical patients, we divided 91 patients’ hepatocellular carcinoma tissues into groups with low expression of both GNB1 and BAG2 and other groups. Analysis of overall survival and disease‐free survival showed that patients in the group with low expression of both GNB1 and BAG2 had a better prognosis (Figure S3D,E). The analysis for TCGA reveals the same conclusion (Figure S3F). These results collectively suggested that the oncogenic role of GNB1 and activating P38 signaling are at least in part dependent on BAG2 expression in HCC cells.
FIGURE 5.
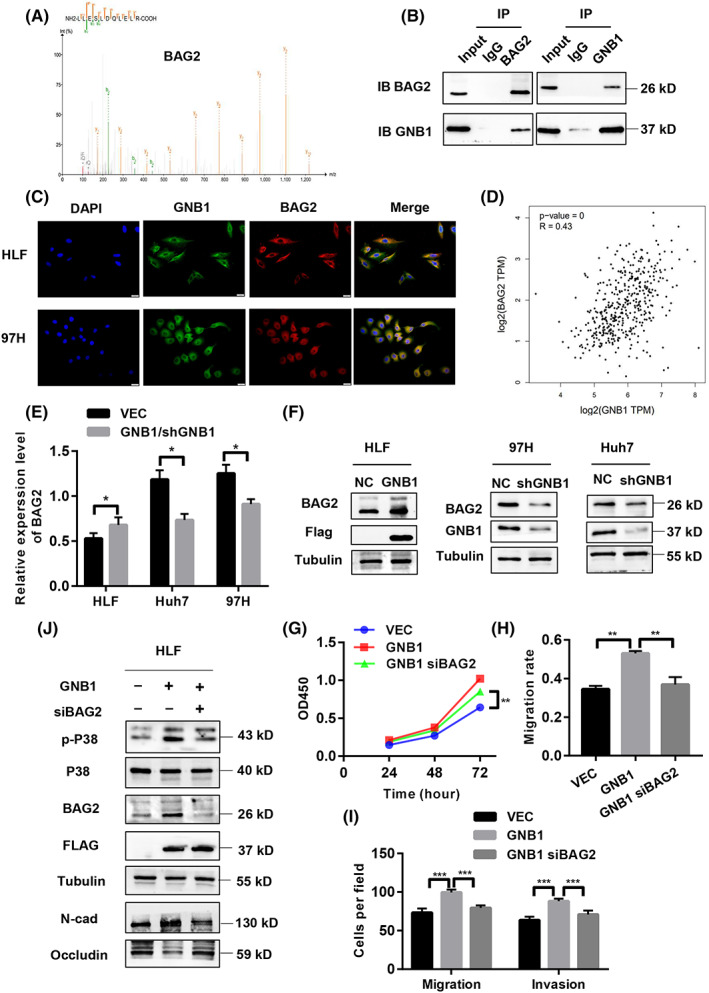
GNB1 regulate the phosphorylation of P38 through BAG2. (A) Co‐immunoprecipitation (Co‐IP) followed by liquid chromatography‐mass spectrometry (LC–MS) identified BAG2 to be a GNB1‐binding protein. (B) Co‐IP followed by western blot analyses confirmed the binding between GNB1 and BAG2 in 97H cells. (C) GNB1 and BAG2 are mainly co‐localized in cytoplasm, as demonstrated by confocal immunofluorescence analysis in HLF and 97H cells. (D) The correlation analysis in GEPIA2 shows that GNB1 and BAG2 expression levels are positively correlated. (E) The mRNA level of BAG2 was upregulated in HLF cells stably overexpressing GNB1, while 97H and Huh7cells with GNB1 knockdown showed lower BAG2 mRNA levels. (F) The protein level of BAG2 was upregulated in HLF cells stably overexpressing GNB1, while 97H and Huh7cells with GNB1 knockdown showed lower BAG2 expression levels. (G) HLF cells stably overexpressing GNB1 were transiently transfected siRNA targeting BAG2. Cell viability of indicated cells were measured by CCK‐8 assay. BAG2 knockdown reversed the increased proliferation induced by GNB1 overexpression in HLF cells. **p < 0.01, Student t test. (H, I) Wound‐healing and transwell assays were performed to assess cell migrative and invasion abilities. SiRNA targeting BAG2 treatment significantly attenuated the enhancement in cell migration and invasion induced by GNB1 overexpression. **p < 0.01, ***p < 0.001, Student t test. (J) Expression of p‐P38, P38 as well as EMT markers, N‐cadherin, and Occludin in indicated cells were examined by western blotting.
TABLE 2.
The correlation between BAG2 expression and clincopathological features in hepatocellular carcinoma (HCC)
| Clinical variables | Number of patients | BAG2 expression level | p value | |
|---|---|---|---|---|
| n = 91 | Low (n = 40) | High (n = 51) | ||
| Age (years) | 0.736 | |||
| <60 | 72 | 31 (43.1%) | 41 (56.9%) | |
| ≥60 | 19 | 9 (47.4%) | 10 (52.6%) | |
| Gender | 0.364 | |||
| Male | 76 | 35 (46.1%) | 41 (53.9%) | |
| Female | 15 | 5 (33.3%) | 10 (66.4%) | |
| HBsAg | 0.985 | |||
| Positive | 75 | 33 (44%) | 42 (56%) | |
| Negative | 16 | 7 (43.8%) | 9 (56.2%) | |
| ALT (U/L) | 0.632 | |||
| ≤40 | 59 | 27 (45.8%) | 32 (54.2%) | |
| >40 | 32 | 13 (40.6%) | 19 (59.4%) | |
| AST (U/L) | 0.009 | |||
| ≤40 | 62 | 33 (53.2%) | 29 (46.8%) | |
| >40 | 29 | 7 (24.1%) | 22 (75.9%) | |
| AFP (ng/mL) | 0.021 | |||
| <400 | 49 | 27 (55.1%) | 22 (44.9%) | |
| ≥400 | 42 | 13 (31.0%) | 29 (69.0%) | |
| Child–Pugh class | 0.789 | |||
| A | 81 | 36 (44.4%) | 45 (55.6%) | |
| B | 10 | 4 (40.0%) | 6 (60.0%) | |
| Liver cirrhosis | 0.347 | |||
| No | 25 | 9 (36.0%) | 16 (64.0%) | |
| Yes | 66 | 31 (47.0%) | 35 (53.0%) | |
| Tumor size (cm) | 0.307 | |||
| ≤5 | 29 | 15 (51.7%) | 14 (48.3%) | |
| >5 | 62 | 25 (40.3%) | 37 (59.7%) | |
| Tumor number | 0.060 | |||
| Single | 74 | 36 (48.6%) | 38 (51.4%) | |
| Multiple | 17 | 4 (23.5%) | 13 (76.5%) | |
| Tumor encapsulation | 0.015 | |||
| Yes | 56 | 19 (33.9%) | 37 (66.1%) | |
| No | 35 | 21 (60.0%) | 14 (40.0%) | |
| Vascular invasion | 0.311 | |||
| Yes | 18 | 6 (33.3%) | 12 (66.7%) | |
| No | 73 | 34 (46.6%) | 39 (53.4%) | |
| Tumor differentiation | 0.247 | |||
| Well | 53 | 26 (49.1%) | 27 (50.9%) | |
| Poor | 38 | 14 (36.8%) | 24 (63.2%) | |
| TNM stage | 0.003 | |||
| I–II | 65 | 35 (53.8%) | 30 (46.2%) | |
| III–IV | 26 | 5 (19.2%) | 21 (80.8%) | |
3.6. GNB1 promotes tumorigenicity in nude mice
The oncogenic role of GNB1 was further investigated with an in vivo model. As shown in Figure 6A,B, GNB1 markedly promoted the growth of tumor volume and increased tumor weight in subcutaneous xenograft models, while the P38 inhibitor SB202190 could abolished the promoting effect. The efficient ectopic expression of GNB1 in xenograft tumors was confirmed by immunohistochemistry. Ki‐67 staining showed that GNB1 significantly promoted cell proliferation as compared to controls. Moreover, the expression of BAG2 and n‐cad was markedly increased in GNB1‐overexpressed xenografts, while the expression of occludin was reduced by IHC and western blotting. All of the above can be reversed by SB202190 (Figure 6C,D), validating the molecular mechanisms identified in vitro.
FIGURE 6.
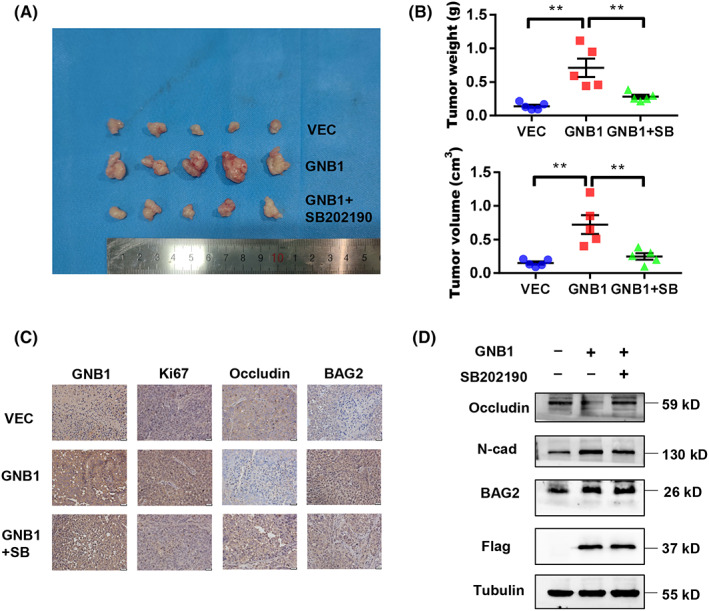
GNB1 promotes tumorigenicity in nude mice. (A) HLF cells stably transfected with control or GNB1 were injected into the flank of nude mice. After the tumors grew to 3–5 mm in diameter, mice with GNB1 overexpression HLF cells were treated with SB202190 (5 mg/kg, daily for 10–12 days). Photos for xenograft tumors isolated from nude mice 21 days post‐inoculation. (B) Tumor volume and weight were compared between the three groups at the end of the experiment. **p < 0.01, one‐way ANOVA. (C) The expression of GNB1, Ki67, occludin and BAG2 in indicated subcutaneous xenografts was determined by immunohistochemistry (IHC). Scale bar, 50 μm. (D) The levels of GNB1, N‐cadherin, occludin, and BAG2 were analyzed by western blotting from three representative xenograft tumors in each group.
4. DISCUSSION
Since its discovery more than 40 years ago, Gβ protein has been investigated extensively. 28 Both mutations and aberrant expression are biological factors that cause heterotrimeric G proteins to lose their normal function and acquire pro‐oncogenic capacities. 29 Mutations in Gα or Gβ have been discovered in, for instance, melanomas, congenital hemangiomas, B cell neoplasms, and myeloid neoplasms. 30 , 31 , 32 Their abnormal expression is also associated with the progression of a variety of tumors. The latest research shows that malignant progression of HCC progenitor cells requires increased K28‐acetylated and cytoplasm‐translocated GαS. 33 To our knowledge, this is the first study to comprehensively identify the biological function and mechanism of GNB1 in HCC. The results of the current study indicated that GNB1 was extensively upregulated in HCC tissues, and the overexpression of GNB1 was correlated with the prognosis of patients. Next, we undertook a series of in vitro and in vivo experiments to systematically investigate the effects of GNB1 on HCC progression. Our data showed that the knockdown of GNB1 inhibited HCC cell proliferation, colony formation, migration, invasion, and tumorigenicity and induced EMT. In contrast, the opposite results were witnessed in GNB1 overexpression assays. Therefore, our study first indicated that GNB1 enhanced cancer proliferation and invasion abilities and might be a novel therapeutic target for HCC.
Subsequently, to further elucidate the underlying mechanism by which GNB1 exerted its functions in HCC, we combined IP and LC–MS/MS to screen out the target proteins that interacted with GNB1. BAG2 is characterized by the BAG domain, which is a conserved region located at the C‐terminus of the BAG‐family proteins that binds to the ATPase domain of Hsc70 and has general nucleotide exchange activities toward Hsc70. Therefore, proteins containing the BAG domain often function as a co‐chaperone protein. 34 To date, BAG2 has been shown to be involved in the development of several kinds of cancer, such as gastric cancer, 19 colorectal cancer, 35 breast cancer, 36 , 37 esophageal squamous cell carcinoma, 38 gliomagenesis, 39 and thyroid carcinoma, 40 and some researchers have found that it is associated with the progression and prognosis of oral cancer and HCC. 22 , 41 However, how BAG2 promotes the progression of HCC remains to be elucidated. As extensively reported, BAG2 was upregulated in HCC, and high expression of this gene was associated with an unfavorable prognosis. BAG2 promoted cell proliferation, blocked apoptosis, and facilitated tumor invasion in HCC22. Because of its relative high abundance in the interactome and definite oncogenic properties, BAG2 was selected as the target that GNB1 interacted with. In this study, we confirmed that GNB1 and BAG2 are co‐localized and can interact with each other. As a downstream molecule of GNB1, BAG2 plays a pro‐cancer role in HCC. Previous studies have respectively revealed the activation of ERK by GNB1 and BAG2. In our study, P38, another important molecule in the MAPK signaling pathway, was shown to play an important role in the pro‐cancer effects of GNB1 and BAG2.
Xu et al. suggested that the BAG2 C‐terminal domain, with its atypical dimeric structure, should be considered a unique Hsp70/Hsc70–NEF and proposed that it be defined as the brand new BAG (BNB) domain. 42 Meanwhile, BAG2 N‐terminal domain (NTD) forms a dimer and adopts a folded conformation annotated in the Pfam or SMART domain databases. 43 Our future research will focus on the domain where GNB1 binds to BAG2. In addition, it would be of great interest to further investigate whether GNB1 regulates the protein stability of BAG2, such as ubiquitinated degradation, for the reason that GSEA of the RNA‐seq data revealed significant overlaps in ubiquitin mediated proteolysis (Table S2). Owing to the target proteins screened out by LC–MS/MS, including more than 30 cancer‐related proteins, further work is required to elaborate the interaction network of GNB1 that is involved in the HCC tumorigenesis. The upstream molecules that lead to the overexpression of GNB1 are also worth exploring.
In conclusion, our study revealed that the expression of GNB1 in HCC tissue was significantly upregulated and closely associated with patient prognosis. Overexpression of GNB1 stimulates a repertoire of downstream tumorigenic responses, including proliferation, migration, invasion, and EMT. Mechanistically, GNB1 interactes with BAG2 and facilitates its expression. This thereby activates the P38/MAPK signaling pathway, leading to the development of HCC.
AUTHOR CONTRIBUTIONS
Xin Zhang conceived and designed the research studies, conducted experiments, acquired and analyzed data, and wrote the manuscript. Keshuai Dong designed and conducted experiments and analyzed data. Jiacheng Zhang designed and conducted experiments and prepared figures. Xin Zhang conducted experiments and wrote the manuscript. Tianrui Kuang and Yiyun Luo acquired data and reviewed the manuscript. Weixing Wang, Jia Yu and Jinming Yu conceived and designed the research studies.
FUNDING INFORMATION
This work was supported by grants from the National Natural Science Foundation of China (No. 82003063) and the Natural Science Foundation of Hubei Province, China (No. 2020CFB213).
CONFLICT OF INTEREST STATEMENT
There are no competing financial interests among the authors.
ETHICAL APPROVAL
This study was approved by the Ethics Committee of Renmin Hospital of Wuhan University.
Informed Consent: Written informed consent was obtained from all patients to participate in the study.
Registry and the registration number of the study/trial: N/A.
Animal Studies: Animal experiments were approved by the Institutional Review Board of Renmin Hospital of Wuhan University (WDRM 22008).
Supporting information
Figure S1
Table S1
Table S2
ACKNOWLEDGMENTS
We would like to acknowledge the reviewers for their helpful comments on this paper.
Zhang X, Dong K, Zhang J, et al. GNB1 promotes hepatocellular carcinoma progression by targeting BAG2 to activate P38/MAPK signaling. Cancer Sci. 2023;114:2001‐2013. doi: 10.1111/cas.15741
Xin Zhang and Keshuai Dong contributed equally to this work.
Contributor Information
Jia Yu, Email: yogaqq116@whu.edu.cn.
Jinming Yu, Email: sdyujinming@163.com.
Weixing Wang, Email: sate.llite@163.com.
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this published article.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209‐249. [DOI] [PubMed] [Google Scholar]
- 2. Akinyemiju T, Abera S, Ahmed M, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and National Level: results from the global burden of disease study 2015. JAMA Oncol. 2017;3:1683‐1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dai Z, Wang X, Peng R, et al. Induction of IL‐6Rα by ATF3 enhances IL‐6 mediated sorafenib and regorafenib resistance in hepatocellular carcinoma. Cancer Lett. 2022;524:161‐171. [DOI] [PubMed] [Google Scholar]
- 4. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589‐604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu L‐C, Hsu C‐H, Hsu C, Cheng A‐L. Tumor heterogeneity in hepatocellular carcinoma: facing the challenges. Liver Cancer. 2016;5:128‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McCudden CR, Hains MD, Kimple RJ, Siderovski DP, Willard FS. G‐protein signaling: back to the future. Cell Mol Life Sci. 2005;62:551‐577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rasheed SAK, Subramanyan LV, Lim WK, Udayappan UK, Wang M, Casey PJ. The emerging roles of Gα12/13 proteins on the hallmarks of cancer in solid tumors. Oncogene. 2022;41:147‐158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brett M, Lai AHM, Ting T‐W, et al. Acute lymphoblastic leukemia in a child with a de novo germline gnb1 mutation. Am J Med Genet A. 2017;173:550‐552. [DOI] [PubMed] [Google Scholar]
- 9. Zimmermannova O, Doktorova E, Stuchly J, et al. An activating mutation of GNB1 is associated with resistance to tyrosine kinase inhibitors in ETV6‐ABL1‐positive leukemia. Oncogene. 2017;36:5985‐5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fan Z, Bai Y, Zhang Q, Qian P. CircRNA circ_POLA2 promotes lung cancer cell stemness via regulating the miR‐326/GNB1 axis. Environ Toxicol. 2020;35:1146‐1156. [DOI] [PubMed] [Google Scholar]
- 11. Cao Y, Li J, Jia Y, Zhang R, Shi H. CircRNA circ_POLA2 promotes cervical squamous cell carcinoma progression via regulating miR‐326. Front Oncol. 2020;10:959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen C, Chi H, Min L, Junhua Z. Downregulation of guanine nucleotide‐binding protein beta 1 (GNB1) is associated with worsened prognosis of clearcell renal cell carcinoma and is related to VEGF signaling pathway. J BUON. 2017;22:1441‐1446. [PubMed] [Google Scholar]
- 13. Wazir U, Jiang WG, Sharma AK, Mokbel K. Guanine nucleotide binding protein β1: a novel transduction protein with a possible role in human breast cancer. Cancer Genomics Proteomics. 2013;10:69‐73. [PubMed] [Google Scholar]
- 14. Zhang J, Huang S, Quan L, et al. Determination of potential therapeutic targets and prognostic markers of ovarian cancer by bioinformatics analysis. Biomed Res Int. 2021;2021:8883800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hameed Y, Usman M, Liang S, Ejaz S. Novel diagnostic and prognostic biomarkers of colorectal cancer: capable to overcome the heterogeneity‐specific barrier and valid for global applications. PLoS One. 2021;16:e0256020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim EK, Choi E‐J. Compromised MAPK signaling in human diseases: an update. Arch Toxicol. 2015;89:867‐882. [DOI] [PubMed] [Google Scholar]
- 17. Jain R, Watson U, Vasudevan L, Saini DK. ERK activation pathways downstream of GPCRs. Int Rev Cell Mol Biol. 2018;338:79‐109. [DOI] [PubMed] [Google Scholar]
- 18. Min L, He B, Hui L. Mitogen‐activated protein kinases in hepatocellular carcinoma development. Semin Cancer Biol. 2011;21:10‐20. [DOI] [PubMed] [Google Scholar]
- 19. Sun L, Chen G, Sun A, et al. BAG2 promotes proliferation and metastasis of gastric cancer via ERK1/2 signaling and partially regulated by miR186. Front Oncol. 2020;10:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee JH, Takahashi T, Yasuhara N, Inazawa J, Kamada S, Tsujimoto Y. Bis, a Bcl‐2‐binding protein that synergizes with Bcl‐2 in preventing cell death. Oncogene. 1999;18:6183‐6190. [DOI] [PubMed] [Google Scholar]
- 21. Kabbage M, Dickman MB. The BAG proteins: a ubiquitous family of chaperone regulators. Cell Mol Life Sci. 2008;65:1390‐1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang X, Zhang J, Liu Y, Li J, Tan J, Song Z. Bcl‐2 associated Athanogene 2 (BAG2) is associated with progression and prognosis of hepatocellular carcinoma: a bioinformatics‐based analysis. Pathol Oncol Res. 2021;27:594649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Camp RL, Dolled‐Filhart M, Rimm DL. X‐tile: a new bio‐informatics tool for biomarker assessment and outcome‐based cut‐point optimization. Clin Cancer Res. 2004;10:7252‐7259. [DOI] [PubMed] [Google Scholar]
- 24. Zhang C, Zhao S, Wang X. Analysis of the risk factor of insufficient examined lymph nodes in stage II colon cancer from the perspective of stage migration: a retrospective study combined with external validation. Int J Surg. 2022;101:106628. [DOI] [PubMed] [Google Scholar]
- 25. Tan Z, Ma G, Yang H, Zhang L, Rong T, Lin P. Can lymph node ratio replace pn categories in the tumor‐node‐metastasis classification system for esophageal cancer? J Thorac Oncol. 2014;9:1214‐1221. [DOI] [PubMed] [Google Scholar]
- 26. Zhang J‐X, He W‐L, Feng Z‐H, et al. A positive feedback loop consisting of C12orf59/NF‐κB/CDH11 promotes gastric cancer invasion and metastasis. J Exp Clin Cancer Res. 2019;38:164. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27. Zhang Y, Cao Y, Zhang J, et al. Lymph node ratio improves prediction of overall survival in esophageal cancer patients receiving neoadjuvant chemoradiotherapy: a National Cancer Database Analysis. Ann Surg. 2022. doi: 10.1097/SLA.0000000000005450. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Northup JK, Sternweis PC, Smigel MD, Schleifer LS, Ross EM, Gilman AG. Purification of the regulatory component of adenylate cyclase. Proc Natl Acad Sci USA. 1980;77:6516‐6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chaudhary PK, Kim S. An insight into GPCR and G‐proteins as cancer drivers. Cell. 2021;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191‐2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ayturk UM, Couto JA, Hann S, et al. Somatic activating mutations in GNAQ and GNA11 are associated with congenital hemangioma. Am J Hum Genet. 2016;98:789‐795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yoda A, Adelmant G, Tamburini J, et al. Mutations in G protein β subunits promote transformation and kinase inhibitor resistance. Nat Med. 2015;21:71‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou Y, Jia K, Wang S, et al. Malignant progression of liver cancer progenitors requires lysine acetyltransferase 7‐acetylated and cytoplasm‐translocated G protein GαS. Hepatology. 2022. doi: 10.1002/hep.32487. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yue X, Zhao Y, Liu J, et al. BAG2 promotes tumorigenesis through enhancing mutant p53 protein levels and function. Elife. 2015;4:e08401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tu R, Kang W, Kang Y, et al. c‐MYC‐USP49‐BAG2 axis promotes proliferation and chemoresistance of colorectal cancer cells in vitro. Biochem Biophys Res Commun. 2022;607:117‐123. [DOI] [PubMed] [Google Scholar]
- 36. Yoon C‐I, Ahn S‐G, Cha Y‐J, et al. Metastasis risk assessment using BAG2 expression by cancer‐associated fibroblast and tumor cells in patients with breast cancer. Cancers (Basel). 2021;13:4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang K‐M, Bae E, Ahn SG, et al. Co‐chaperone BAG2 determines the pro‐oncogenic role of cathepsin B in triple‐negative breast cancer cells. Cell Rep. 2017;21:2952‐2964. [DOI] [PubMed] [Google Scholar]
- 38. Hong Y‐C, Wang Z, Peng B, Xia L‐G, Lin L‐W, Xu Z‐L. BAG2 overexpression correlates with growth and poor prognosis of esophageal squamous cell carcinoma. Open Life Sci. 2018;13:582‐588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu H, Ding J, Zhu H, et al. LOXL1 confers antiapoptosis and promotes gliomagenesis through stabilizing BAG2. Cell Death Differ. 2020;27:3021‐3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang HQ, Zhang HY, Hao FJ, Meng X, Guan Y, Du ZX. Induction of BAG2 protein during proteasome inhibitor‐induced apoptosis in thyroid carcinoma cells. Br J Pharmacol. 2008;155:655‐660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu Y‐S, Wei B. Over‐expression of Bcl2‐associated athanogene 2 in oral cancer promotes cellular proliferation and is associated with poor prognosis. Arch Oral Biol. 2019;102:164‐170. [DOI] [PubMed] [Google Scholar]
- 42. Xu Z, Page RC, Gomes MM, et al. Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2. Nat Struct Mol Biol. 2008;15:1309‐1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Page RC, Xu Z, Amick J, Nix JC, Misra S. Crystallization and preliminary X‐ray crystallographic analysis of the Bag2 amino‐terminal domain from Mus musculus. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68:647‐651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Table S1
Table S2
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


