Abstract
It was recently found that a mixture of nine amino acids down-regulate Clostridium difficile toxin production when added to peptone yeast extract (PY) cultures of strain VPI 10463 (S. Karlsson, L. G. Burman, and T. Åkerlund, Microbiology 145:1683–1693, 1999). In the present study, seven of these amino acids were found to exhibit a moderate suppression of toxin production, whereas proline and particularly cysteine had the greatest impact, on both reference strains (n = 6) and clinical isolates (n = 28) of C. difficile (>99% suppression by cysteine in the highest toxin-producing strain). Also, cysteine derivatives such as acetylcysteine, glutathione, and cystine effectively down-regulated toxin expression. An impact of both cysteine and cystine but not of thioglycolate on toxin yield indicated that toxin expression was not regulated by the oxidation-reduction potential. Several metabolic pathways, including butyric acid and butanol production, were coinduced with the toxins in PY and down-regulated by cysteine. The enzyme 3-hydroxybutyryl coenzyme A dehydrogenase, a key enzyme in solventogenesis in Clostridium acetobutylicum, was among the most up-regulated proteins during high toxin production. The addition of butyric acid to various growth media induced toxin production, whereas the addition of butanol had the opposite effect. The results indicate a coupling between specific metabolic processes and toxin expression in C. difficile and that certain amino acids can alter these pathways coordinately. We speculate that down-regulation of toxin production by the administration of such amino acids to the colon may become a novel approach to prophylaxis and therapy for C. difficile-associated diarrhea.
Clostridium difficile is the etiologic agent of pseudomembranous colitis and a major cause of antibiotic-associated diarrhea (14). Its key virulence factors are two toxins, A and B, which are members of the large clostridial cytotoxin family and thus have high molecular weight and conserved protein domains and enzymatic function (21). The onset of C. difficile growth in the large intestine, followed by C. difficile-associated diarrhea (CDAD), is thought to be caused by the reduction of protective colonic microbiota, especially by antibiotic treatment (14). Toxin production by C. difficile has been demonstrated to be dependent on the nutrient level of the growth medium (7, 10, 12, 18, 24, 25). The type and amount of nutrients present have been shown to affect growth of C. difficile in a continuous culture system containing a complex microflora (23, 26), suggesting that competition for nutrients in the colon plays a role in colonization by the pathogen and in the development of CDAD. Toxin expression may differ 100,000-fold between toxin-positive isolates of C. difficile in vitro (14) but the underlying reason is not understood. Previous studies have shown that regulation of toxin production in C. difficile may be affected by amino acid levels, as demonstrated in defined media during biotin starvation (25) and in complex media (12, 18). High toxin levels are observed in complex media deprived of glucose (4, 12). We recently found that during such conditions, toxin yields from strain VPI 10463 were reduced approximately 100-fold by adding the nine amino acids cysteine, glycine, isoleucine, leucine, methionine, proline, threonine, tryptophan, and valine, whereas a mix of eight other amino acids (alanine, arginine, aspartic acid, histidine, lysine, phenylalanine, serine, and tyrosine) had no effect (12). The aims of this study were (i) to test if the down-regulation of toxin production by these nine amino acids is a general response, i.e., medium independent and present in both reference strains and clinical isolates of C. difficile; (ii) to evaluate whether any of these nine amino acids are more potent than others in suppressing toxin production; (iii) to search for analogues or derivatives of amino acids having a similar effect; and (iv) to study the apparent connection between C. difficile metabolism and toxin production. The last was done by identifying differentially expressed proteins and by assaying the metabolic end products during high and low toxin production.
MATERIALS AND METHODS
Strains, growth conditions, and media.
Strains of C. difficile were obtained from the Culture Collection, University of Göteborg, Göteborg, Sweden (CCUG no. 4938, 8884, 9004, 9018, 19126 [VPI 10463], and 20309), and from Huddinge Hospital, Huddinge, Sweden (28 clinical isolates). Sterilized peptone yeast extract (PY) and brain heart infusion broth (BHI) were purchased from Karolinska Hospital, Stockholm, Sweden. The media were heated to 80°C and purged with a gas mixture (10% CO2, 10% H2, 80% N2) for 20 min, and aliquoted into tubes (Bellco glass). Where not otherwise indicated, the amounts of amino acids added to the media were as follows (gram/liter): cysteine, 0.5; glycine, 0.1; isoleucine, 0.3; leucine, 0.4; methionine, 0.2; proline, 0.3; threonine, 0.2; tryptophan, 0.1; and valine, 0.3. The defined medium SDM was made as described previously (12). Where indicated, butanol or butyric acid was added to the media. C. difficile cultures were obtained by 106-fold dilution of overnight cultures and were incubated at 37°C on a horizontal shaker for 10 to 48 h prior to harvesting. One milliliter of the harvested C. difficile cultures was sonicated and kept at −20°C until the total level of toxin was determined by enzyme immunoassay (EIA). For details see reference 12.
EIA of toxins.
Toxins A and B were measured using the Ridascreen C. difficile toxin A/B kit (r-Biopharm) according to the manufacturer's instructions. Thawed samples were diluted in buffer (r-Biopharm), and 50-μl aliquots were added to the wells. A microtiter plate reader (Labsystems Multiscan MCC/340) was used to monitor the absorbance at 450 nm (A450). An A450 value of 1.0 was defined to correspond to 1 U of toxin.
Measurements of oxidation-reduction potential.
Measurements of the oxidation-reduction potential (Eh) of cultures were carried out using a Blueline 31 RX platinum electrode connected to a CG 840 pH-meter (Schott). The Eh of C. difficile cultures grown 48 h was assayed in cell-free supernatant obtained after centrifugation. The probe was submerged in the medium and left for a 5-min equilibration at 22°C before the Eh value was recorded. Control experiments showed that the probe recorded a stable Eh value over at least 1 h, indicating that the immediate handling of the cultures did not affect the measurements.
Analysis of metabolic end products.
C. difficile grown in triplicates was harvested at 10, 14, 18, 24, and 36 h after inoculation, and optical density was measured at 600 nm (OD600). Bacteria and culture medium were separated by centrifugation and stored at −20°C. The cell pellets were suspended in sterile water to the original sample volume and were sonicated, after which the intracellular toxin yields were determined by EIA. For analysis of short-chain fatty acids (SCFAs), 1 ml of harvested medium was mixed with an equal volume of distilled water containing 3 mM 2-ethylbutyric acid as an internal standard and 0.5 ml of 0.5 mM H2SO4. The mixture was vacuum distilled according to the method of Zijlistra et al. (27) modified by Høverstad et al. (8), and the distillate was analyzed with gas-liquid chromatography on 10% SP-1200–1% H3PO4 (Chromosorb) at 120°C (Perkin-Elmer Autosystem XL with Turbochrome 4 automatic analyzer system). The injector and detector temperatures were 180 and 190°C, respectively, and the carrier gas was N2 (flow rate, 60 ml/min). Each series of analyses (10 to 15 samples) was started and finished with injection of a standard solution. For analysis of acetone and alcohols, the harvested medium was mixed with an equal volume of distilled water and was vacuum distilled using the same equipment as described above, without the internal standard. Instead, the results were compared with analyses performed in parallel with known amounts of acetone or each alcohol on a 5% Carbowax 20M glass column (Supelco) at 85°C. The injector and detector temperatures were 160°C, and the flow rate of the carrier gas N2 was 50 ml/min.
Two-dimensional polyacrylamide gel electrophoresis (2-D PAGE).
Forty microliters of cell extract was mixed with 160 μl of buffer containing 9.9 M urea, 4% (vol/vol) Igepal CA630, 2.2% (vol/vol) Pharmalytes 3-10, 100 mM dithiothreitol, and 2% (wt/vol) CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate) and was stored at −70°C. Proteins were focused at 20°C on 180-mm IPG Drystrip (pH 4 to 7) (Amersham Pharmacia Biotech) using the Multiphor II 2-D gel kit according to the manufacturer's instructions. The second dimension was run on sodium dodecyl sulfate–12% PAGE gels, and proteins were visualized by silver staining. Chemicals were obtained from Sigma except for Pharmalytes (Amersham Pharmacia Biotech). Proteins were transferred to polyvinylidene fluoride membranes and stained with Coomassie brilliant blue, and spots of interest were excised and N terminal sequenced.
RESULTS
Cysteine is a potent toxin down-regulator in C. difficile.
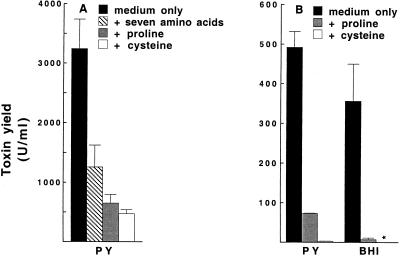
It was recently reported that toxin production in C. difficile VPI 10463 grown in PY medium was reduced 100-fold by the addition of a group of nine amino acids, whereas a mix of eight other amino acids had no effect (12). To test the generality of this observation, we studied six reference strains as well as 28 clinical isolates of C. difficile. In nonsupplemented PY there was a large variation of the toxin yields among the strains, and the highest yield was found for one clinical isolate (13,300 U/ml), followed by the reference strain CCUG 20309 (3,710 U/ml). The nine amino acids markedly reduced the toxin yield in both the reference strains and the clinical isolates; for the two highest toxin producers, the toxin yields were lowered by approximately 99 and 96%. Of the nine amino acids, cysteine had the strongest toxin-suppressing impact, followed by proline, while the remaining seven amino acids together showed a moderate effect in strain VPI 10463 grown in PY (Fig. 1A). Cysteine was also more potent than proline in reducing toxin yields in a clinical isolate grown in PY or BHI (Fig. 1B). A marked impact of cysteine on toxin yield was also apparent in the other 27 clinical isolates (data not shown).
FIG. 1.
Impact of amino acids on toxin production in C. difficile. Toxin yield (U/ml) in 48-h cultures of (A) strain VPI 10463 grown in PY or PY supplemented with cysteine, proline, or the seven amino acids glycine, isoleucine, leucine, methionine, threonine, tryptophan, and valine and (B) a clinical isolate of C. difficile grown in PY or BHI with or without cysteine or proline. Amino acid concentrations are given in Materials and Methods. The mean and standard error of three experiments are shown. The asterisk indicates that the toxin yield was below the detection limit of 0.2 U/ml.
Cysteine and cysteine derivatives down-regulate toxin production in a dose-dependent manner.
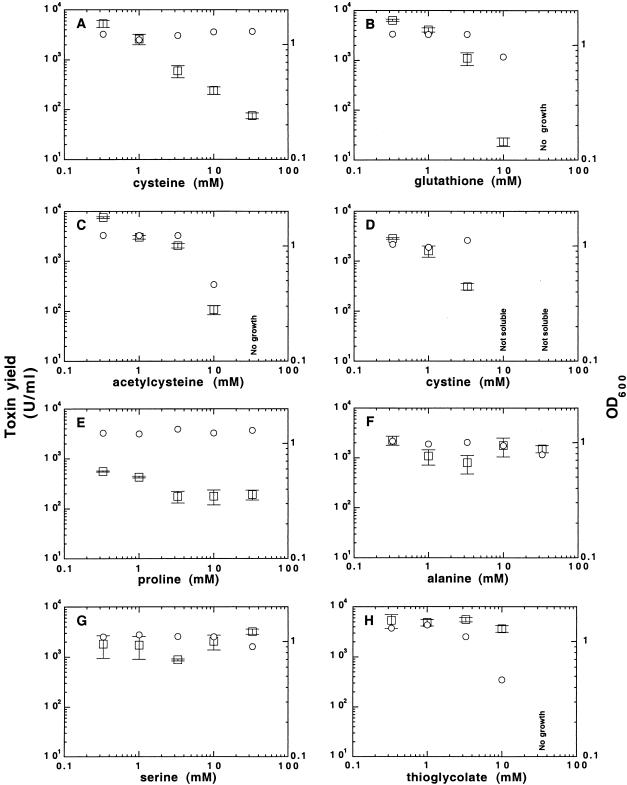
Cysteine showed a clear dose-response effect on toxin expression in strain VPI 10463 during growth in PY; the highest toxin yield (5,000 U/ml) was found at 0.33 mM and the lowest (80 U/ml) at an added level of 33 mM (Fig. 2A). The OD600 values of the 48-h cultures were approximately the same regardless of cysteine concentrations, showing that the observed reduction of toxin yields was not caused by growth inhibition. Control experiments in which cysteine (33 mM) was added to sonicated culture samples showed no changes in toxin levels over 72 h at 37°C; i.e., the toxins were stable and the EIA measurements were not affected by cysteine. Dose-response experiments with cysteine in a defined medium (SDM) were difficult to interpret since growth was supported only within a narrow range of cysteine concentrations (3.3 to 10.0 mM), and the toxin yields were generally low (12).
FIG. 2.
Impact of amino acids and reducing agents on toxin expression. Toxin yield (U/ml; left scale and squares) and cell yield (OD600; right scale and circles) in C. difficile VPI 10463 grown for 48 h in PY with different concentrations of various amino acids, thioglycolate, and cysteine derivatives added. Where indicated, the compound inhibited growth or was not soluble. The mean and standard error of three experiments are shown.
Other molecules containing cysteine residues, such as glutathione (γ-Glu-Cys-Gly), acetylcysteine, or cystine, also down-regulated toxin production in C. difficile VPI 10463 in a dose-dependent manner similar to cysteine (Fig. 2B to D). Supplementing PY with 10 mM taurine, a common metabolite of cysteine formed by its carboxylation (9), had no effect, however, on toxin production in C. difficile VPI 10463 or in a clinical isolate (data not shown). Proline showed a dose-dependent effect on toxin production up to 3.3 mM but not at higher concentrations (Fig. 2E). Alanine and serine belonging to the group of amino acids not affecting toxin production (cf. above and reference 12) and included as negative controls had no effect (Fig. 2F and G).
Eh does not affect toxin production.
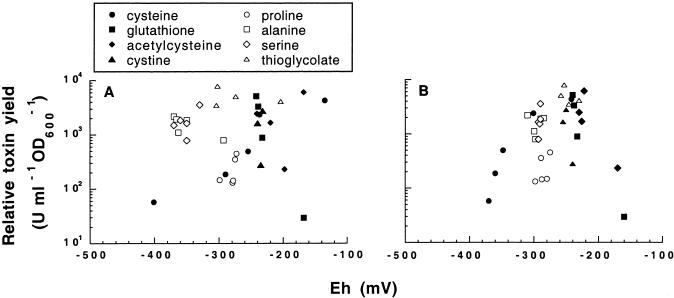
The Eh of the growth medium has been shown to influence the extracellular toxin levels in C. difficile (17). We speculated that the lowered Eh of the medium due to the addition of cysteine could be the cause of altered toxin expression. Increasing cysteine concentrations (0.33 to 33 mM added) successively reduced the Eh of the PY medium from −100 mV to −400 mV (data not shown), which correlated with lowered toxin yields in VPI 10463 cultures (Fig. 2A). By contrast, the toxin yields were unaffected by the reducing agent thioglycolate (Fig. 2H), despite the fact that it lowered the Eh of the medium from −200 mV at 0.33 mM to −300 mV at 10 mM (data not shown; levels above 10 mM inhibited growth). The lack of influence on toxin expression by Eh was further supported by the finding that cystine (i.e., the oxidized dimer of cysteine and thus Eh inactive) caused a reduction of toxin yields comparable to that of cysteine (Fig. 2D; cystine was not soluble at ≥10 mM). The absence of an overall correlation between Eh and toxin production is depicted in Fig. 3, where the data from Fig. 2 were plotted as relative toxin yield at 48 h versus the Eh of the medium measured at inoculation (Fig. 3A) and after 48 h of growth (Fig. 3B).
FIG. 3.
Eh and toxin production in C. difficile strain VPI 10463. Relative toxin yield (U ml−1 OD600−1) after 48 h of growth in PY was plotted against the Eh of the medium at inoculation (A) and 48 h (B). Relative yields were used to adjust for the variations in cell yield. The values were obtained from the experiment depicted in Fig. 2.
Enzymes involved in butyric acid and butanol production and one-carbon transfer are down-regulated by cysteine.
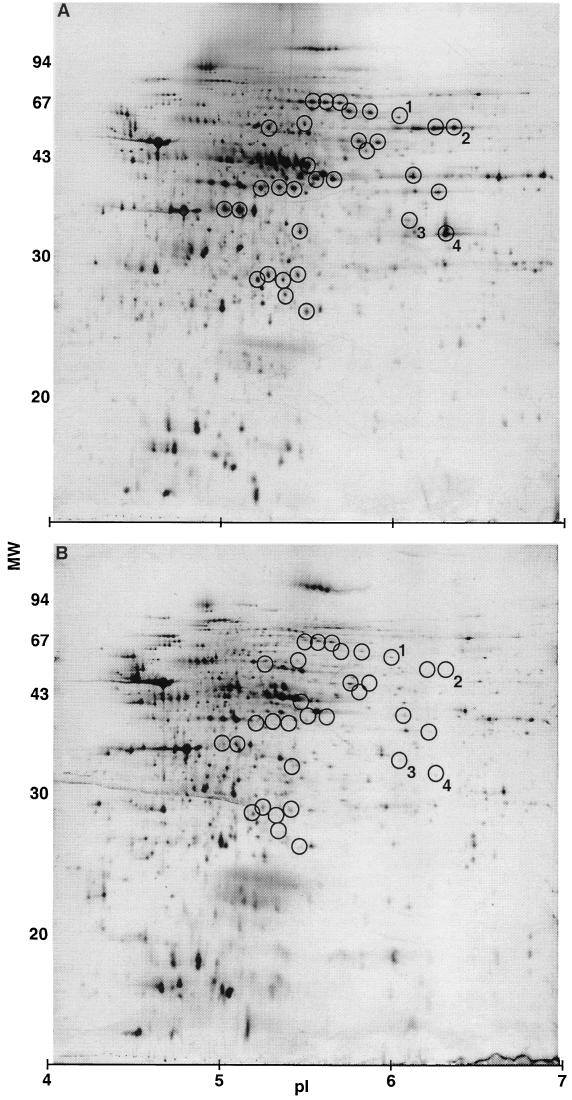
It was recently found that several other proteins were down-regulated together with the C. difficile toxins in PY cultures supplemented with nine amino acids (12). Here we investigated whether cysteine causes a similar change in the protein expression pattern. In cell extracts of PY cultures, approximately 500 proteins were visualized by 2-D PAGE (Fig. 4A). More than 30 proteins were markedly down-regulated or absent in PY cultures with 30 mM cysteine added (Fig. 4B, circled spots). Several of these proteins corresponded to those previously found to be down-regulated by adding the nine amino acids (data not shown). Four of the proteins (Fig. 4, no. 1 to 4) were here N-terminal amino acid sequenced and were identified in the genome sequence database for C. difficile strain 630 at the Sanger Center. Protein spot no. 1 matched 4-hydroxyphenylacetate-3-hydroxylase (Table 1). Adjacent to this gene on the C. difficile chromosome, we found a gene encoding 4-hydroxybutyryl-coenzyme A (CoA) transferase, an enzyme in the butyric acid and butanol production pathway. Spot no. 2 matched formate tetrahydrofolate dehydrogenase, an enzyme involved in one-carbon transfer and important for synthesis of, e.g., amino acids. Spot no. 3 matched indolepyruvate ferredoxin oxidoreductase, which catalyzes the ferredoxin-dependent oxidative decarboxylation of arylpyruvates. The open reading frame (ORF) next to this gene encodes butyrate kinase, which catalyzes the final step in butyric acid production. Spot no. 4 was identified as 3-hydroxybutyryl-CoA dehydrogenase, a key regulatory enzyme involved in the formation of butyric acid and butanol.
FIG. 4.
Impact of cysteine on protein expression in C. difficile VPI 10463. Sonicates of 14-h cultures in PY (A) or PY plus 30 mM cysteine (B) were analyzed by 2-D PAGE. Protein (15 μg) was loaded onto each gel. Circles highlight proteins consistently found to be down-regulated by cysteine in three experiments (between 3- and >100-fold difference in spot intensity), and the numbered protein spots were subjected to N-terminal sequencing (Table 1). Molecular weights in thousands (left) and pI (bottom) are indicated.
TABLE 1.
Identification of the N-terminal-sequenced proteins down-regulated by cysteine in C. difficile VPI 10463 cultures
| Spot no.a | N-terminal sequenceb | Identityc |
|---|---|---|
| 1 | AXMTG AQYIE SLRKL | 4-Hydroxyphenylacetate-3-hydroxylase |
| 2 | GFKSD IEIAQ EAKPQ | Formate tetrahydrofolate dehydrogenase |
| 3 | MKQLM TGNEA IARGA | Indolepyruvate ferredoxin oxidoreductase |
| 4 | MKLAV IGSGT MGSGI VQTFA | 3-Hydroxybutyryl-CoA dehydrogenase |
Numbers refer to circled spots in Fig. 4.
X denotes unidentified amino acid.
Genes were identified by searching for a high-score match of the N-terminal sequence in the C. difficile strain 630 sequence database (http://www.sanger.ac.uk/Projects/C_difficile/) using the BLAST algorithm. These sequence data were produced by the Clostridium difficile Sequencing Group at the Sanger Center and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/cd/CD.dbs. The found genome segments were further analyzed using the ORF finder at http://www.ncbi.nlm.nih.gov/, and the ORFs in the contigs were identified using the BLAST algorithm.
Toxin expression correlates with butyric acid production.
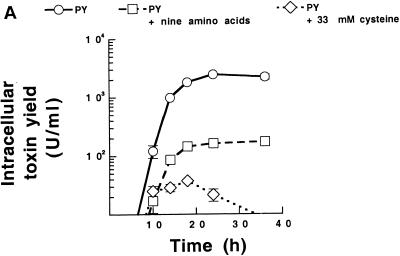
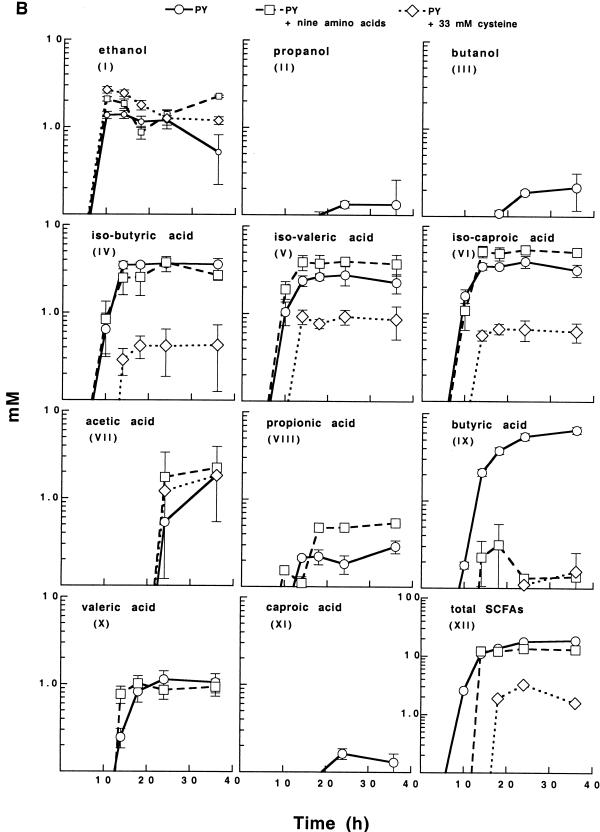
The results described above indicated that certain amino acids and especially cysteine concomitantly down-regulate toxin expression and butyrate and butanol production in C. difficile. To study this in greater detail, we monitored the metabolic end products (SCFAs, acetone, and alcohols) produced by strain VPI 10463 grown in PY, PY supplemented with the nine amino acids, or PY with only cysteine added. As expected, the toxin yield was high in PY cultures, reduced by over 90% in PY supplemented with nine amino acids, and reduced by about 99% in PY supplemented with 33 mM cysteine (Fig. 5A). The end-product pattern was complex, and the final levels ranged from 0.1 to 10.0 mM (Fig. 5B). The overall result was that the addition of cysteine lowered the yields of all end products except ethanol (Fig. 5B-I) and acetic acid (Fig. 5B-VII) and reduced the total yield of SCFAs by approximately 90% (Fig. 5B-XII). The production of one particular SCFA, butyric acid, was markedly down-regulated by both cysteine and the nine amino acids (Fig. 5B-IX). Taken together, the results showed that amino acids, and especially cysteine, dramatically down-regulate metabolic pathways and that there is a correlation between the expression of toxins and butyric acid and butanol production.
FIG. 5.
Intracellular toxin yield (A) and metabolic end products (B) in culture medium during growth of C. difficile VPI 10463 in PY, PY with nine amino acids added, and PY with cysteine added (30 mM). End products are given after subtraction of levels present in noninoculated medium. Acetone, 2-propanol, and 2-butanol were not found in any culture. The mean and standard error of three experiments are shown.
Butyric acid and butanol have opposite impact on toxin production.
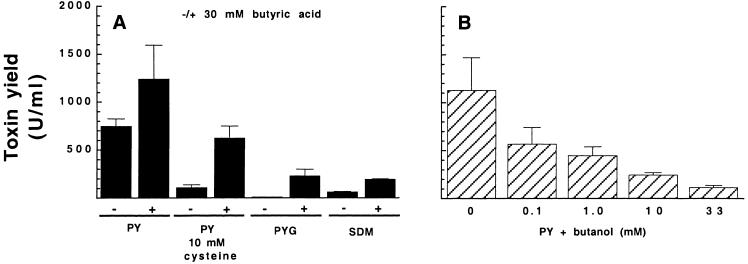
In Clostridium acetobutylicum, the addition of acetic and butyric acid or methyl viologen to cultures induces solventogenesis, i.e., the conversion of acetic acid and butyric acid to acetone and butanol, respectively (see Discussion). Since butyric acid and butanol production and toxin expression correlated in C. difficile (see above), we wanted to test whether the solventogenesis-inducing agents could promote toxin production in C. difficile. Indeed, adding butyric acid to C. difficile cultures enhanced toxin yields in all PY media tested (Fig. 6A). The level of induction ranged from 1.5-fold (PY) to 30-fold (PY with glucose). Similarly, the addition of acetic acid, acetic acid plus butyric acid, or methyl viologen to PY cultures induced toxin production (results not shown). Butyric acid also enhanced toxin production in SDM, a defined medium that supports low toxin production (Fig. 6A). The addition of caproic acid to PY did not alter toxin production, but since high concentrations of this acid inhibited growth of C. difficile, this result was difficult to interpret. In SDM, which has a higher buffering capacity than PY, caproic acid elevated the toxin yield to a level similar to that of butyric acid in PY (data not shown). Interestingly, the addition of butanol, i.e., the product formed from butyric acid during solventogenesis, to PY cultures decreased the toxin yield in a dose-dependent manner (Fig. 6B). Thus, the metabolically related end products butyric acid and butanol had an opposite effect on toxin production in C. difficile.
FIG. 6.
Impact of butyric acid or butanol on toxin yield (U/ml) in C. difficile. Strain VPI 10463 was inoculated into (A) different media (PY, PY supplemented with 10 mM cysteine or with 0.9% glucose [PYG], and SDM), with or without 30 mM butyric acid added, and (B) PY supplemented with different concentrations of butanol. Toxin yield was measured after 48 h of growth. The mean and standard error of three experiments are shown.
DISCUSSION
The regulation of toxin production in C. difficile is not well understood, although amino acids, glucose, and biotin exogenously added to growth media influence toxin production (see the introduction). Here we studied the effects of amino acids on growth, toxin production, protein expression, and metabolic end product formation in various strains of C. difficile. Among the nine amino acids previously found to down-regulate toxin production, cysteine was found the most potent, followed by proline. Also, the cysteine-containing compounds cystine, acetylcysteine, and glutathione had a strong negative impact on toxin expression. We did not find any consistent correlation between the Eh and toxin production, although Eh may play a role in the release of toxin from the bacteria (17). A moderate toxin-suppressing effect of glycine, isoleucine, leucine, methionine, threonine, tryptophan, and valine was observed. Interestingly, these amino acids together with cysteine and proline are required for maximum cell yield of C. difficile in defined media (7, 10). Although these amino acids did not affect growth in PY, there was an inverse relationship between toxin production and the levels of these amino acids.
The published effects of amino acids on toxin production in C. difficile are complex. For example, it has been reported that both high and low levels of arginine promote toxin production (11, 18). Furthermore, each of the amino acids alanine, isoleucine, leucine, serine, threonine, and valine was shown to suppress toxin production below the detection level in tryptone yeast medium when added at 50 mM (18). In contrast, we did not observe any toxin-suppressing effect by alanine or serine in C. difficile strain VPI 10463 PY cultures within the concentration range 0.33 to 33 mM. These discrepancies are difficult to explain, but may be the result of differences in bacterial metabolism depending on the growth medium used. Furthermore, studies on toxin expression in different media are difficult to evaluate, since cysteine, here found to markedly influence toxin production, has been commonly used as a reducing agent during cultivation of C. difficile. When cysteine was added to the defined medium SDM, the lowest and highest concentrations did not support growth of C. difficile, making the results difficult to interpret. The low toxin level and the lack of impact of cysteine on toxin production in SDM cultures are likely to be due to the presence of both other amino acids and glucose, which suppress toxin production (see above). Recently, the VirR/VirS two-component system in Clostridium perfringens was shown to regulate both toxin production and cysteine synthase in an opposite manner (1). Thus, there may be a coordinate regulation of genes involved in virulence and amino acid metabolism. Whether a system similar to VirR/VirS affects toxin production in C. difficile is not known.
In addition to the C. difficile toxins, metabolic enzymes involved in carbon transfer, electron transport, and butyric acid and butanol production were down-regulated by cysteine. The large number of proteins in C. difficile regulated by cysteine indicates that there is a switch of a global regulator. The enzyme 3-hydroxybutyryl-CoA dehydrogenase was among the most prominent cysteine-regulated proteins. This enzyme is involved in fatty acid metabolism, particularly the production of butyric acid and butanol (solventogenesis) in C. acetobutylicum (5, 22). Solventogenesis is a process in C. acetobutylicum cultures in which certain SCFAs become converted to ketones and alcohols. Solventogenesis is linked to sporulation in this organism, and mutants unable to convert, e.g., butyric acid to butanol sporulate poorly (15). Conversion of accumulated acids (acidogenesis) to solvents (solventogenesis) is probably induced to detoxify the acidic environment, and this process enables the bacteria to complete endospore formation and survive (5). Solventogenesis can thus be considered to be a stress response, and, in parallel with solvent-producing enzymes, heat shock proteins are induced (19). Three genes encoding enzymes in the solventogenesis pathway in C. acetobutylicum (3-hydroxybutyryl-CoA dehydrogenase, thiolase, and crotonase) are clustered on the C. difficile chromosome (16), and the homologous genes encoding 3-hydroxybutyryl-CoA dehydrogenase and thiolase in Bacillus subtilis have been shown to be regulated by the global regulator sigma factor E (3). The molecular mechanism that governs the switch from butyric acid to butanol production in C. acetobutylicum is not known. Solventogenesis can be induced by the addition of acetic acid, butyric acid, or methyl viologen to the culture medium (6, 20). The elevation of toxin production in C. difficile cultures by these compounds is thus a response similar to the induction of solventogenesis in C. acetobutylicum, although the levels of butyric acid and butanol produced were 10- to 100-fold lower in C. difficile than in C. acetobutylicum. Our finding that the addition of butanol suppressed toxin production was intriguing and further indicates that toxin and butyric acid and butanol production share the same regulatory control.
The large intestine is a milieu essentially lacking free glucose (2), and the availability of amino acids is likely to be crucial to the growth of C. difficile and other bacteria in vivo. In continuous cultures containing human feces, significant competition for amino acids between C. difficile and other microorganisms of the biota was shown (26). The apparent association between amino acid limitation and toxin production may be of clinical relevance in that protein malnutrition may be a risk factor for CDAD. This could explain why patients on chronic dialysis due to renal insufficiency and elderly hospitalized individuals are at particular risk of developing CDAD when exposed to antimicrobial agents (13). The specific therapy for CDAD is currently to give metronidazole or vancomycin, further disrupting the bowel flora and predisposing to relapse. Thus, there is a need for novel strategies for prophylaxis and treatment. We hypothesize that down-regulation of toxin production by the administration of amino acids to the colon may become such an alternative.
ACKNOWLEDGMENTS
This work was supported by SBL Vaccin AB, Stockholm, Sweden, and by grant V96230 from the Vårdal Foundation, Sweden.
Protein data were obtained at the Protein Analysis Center, Karolinska Institute, Sweden. The sequence data were produced by the Clostridium difficile Sequencing Group at the Sanger Center. We are grateful for the excellent technical support provided by Anna-Karin Persson.
REFERENCES
- 1.Banu S, Ohtani K, Yaguchi H, Swe T, Cole S T, Hayashi H, Shimizu T. Identification of novel VirR/VirS-regulated genes in Clostridium perfringens. Mol Microbiol. 2000;35:854–864. doi: 10.1046/j.1365-2958.2000.01760.x. [DOI] [PubMed] [Google Scholar]
- 2.Bond J H, Levitt M D. Fate of soluble carbohydrates in the colon of rats and man. J Clin Investig. 1976;57:1158–1164. doi: 10.1172/JCI108383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryan E M, Beall B W, Morgan C P., Jr A ςE-dependent operon subject to catabolite repression during sporulation in Bacillus subtilis. J Bacteriol. 1996;178:4778–4786. doi: 10.1128/jb.178.16.4778-4786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dupuy B, Sonenshein A L. Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol. 1998;27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- 5.Dürre P, Fischer R-J, Kuhn A, Lorenz K, Schreiber W, Stürzenhofecker B, Ullman S, Witzer K, Sauer U. Solventogenic enzymes of Clostridium acetobutylicum: catalytic properties, genetic organization and transcriptional regulation. FEMS Microbiol Rev. 1995;17:251–262. doi: 10.1111/j.1574-6976.1995.tb00209.x. [DOI] [PubMed] [Google Scholar]
- 6.Gottschal J C, Morris J G. The induction of acetone and butanol production of Clostridium acetobutylicum by elevated concentrations of acetate and butyrate. FEMS Microbiol Lett. 1981;12:385–389. [Google Scholar]
- 7.Haslam S C, Ketley J M, Mitchel T J, Stephen J, Burdon D W, Candy D C A. Growth of Clostridium difficile and production of toxins A and B in complex and defined media. J Med Microbiol. 1986;21:293–297. doi: 10.1099/00222615-21-4-293. [DOI] [PubMed] [Google Scholar]
- 8.Høverstad T, Fausa O, Bjørneklett A, Bøhmer T. Short-chain fatty acids in the normal faeces. Scand J Gastroenterol. 1984;19:375–381. [PubMed] [Google Scholar]
- 9.Jacobsen J G, Smith L H. Biochemistry and physiology of taurine and taurine derivatives. Physiol Rev. 1968;48:424–511. doi: 10.1152/physrev.1968.48.2.424. [DOI] [PubMed] [Google Scholar]
- 10.Karasawa T, Ikoma S, Yamakawa K, Nakamura S. A defined medium for Clostridium difficile. Microbiology. 1995;141:371–375. doi: 10.1099/13500872-141-2-371. [DOI] [PubMed] [Google Scholar]
- 11.Karasawa T, Maegawa T, Nojiri T, Yamakawa K, Nakamura S. Effect of arginine on toxin production by Clostridium difficile in defined medium. Microbiol Immunol. 1997;41:581–585. doi: 10.1111/j.1348-0421.1997.tb01895.x. [DOI] [PubMed] [Google Scholar]
- 12.Karlsson S, Burman L G, Åkerlund T. Suppression of toxin production in Clostridium difficile VPI 10463 by amino acids. Microbiology. 1999;145:1683–1693. doi: 10.1099/13500872-145-7-1683. [DOI] [PubMed] [Google Scholar]
- 13.Karlström O, Fryklund B, Tullus K, Burman L G. A prospective nationwide study of Clostridium difficile-associated diarrhea in Sweden. Clin Infect Dis. 1998;26:141–145. doi: 10.1086/516277. [DOI] [PubMed] [Google Scholar]
- 14.Lyerly D M, Krivan H C, Wilkins T D. Clostridium difficile: its disease and toxins. Clin Microbiol Rev. 1988;1:1–18. doi: 10.1128/cmr.1.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattson D M, Rogers P. Analysis of Tn916-induced mutants of Clostridium acetobutylicum altered in solventogenesis and sporulation. J Ind Microbiol. 1994;13:258–268. doi: 10.1007/BF01569758. [DOI] [PubMed] [Google Scholar]
- 16.Mullany P, Clayton C L, Pallen M J, Slone R S, Al-Saleh A, Tabaqchai S. Genes encoding homologues of three consecutive enzymes in the butyrate/butanol-producing pathway of Clostridium acetobutylicum are clustered on the Clostridium difficile chromosome. FEMS Microbiol Lett. 1994;124:61–68. doi: 10.1111/j.1574-6968.1994.tb07262.x. [DOI] [PubMed] [Google Scholar]
- 17.Onderdonk A B, Lowe B R, Bartlett J G. Effect of environmental stress on Clostridium difficile toxins levels during continuous cultivation. Appl Environ Microbiol. 1979;38:637–641. doi: 10.1128/aem.38.4.637-641.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osgood D P, Wood N P, Sperry J F. Nutritional aspects of cytotoxin production by Clostridium difficile. Appl Environ Microbiol. 1993;59:3985–3988. doi: 10.1128/aem.59.12.3985-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pich A, Narberhaus F, Bahl H. Induction of heat shock proteins during initiation of solvent formation in Clostridium acetobutylicum. Appl Microbiol Biotechnol. 1990;33:697–704. [Google Scholar]
- 20.Rao G, Mutharasan R. Alcohol production by Clostridium acetobutylicum induced by methyl viologen. Biotechnol Lett. 1986;8:893–896. [Google Scholar]
- 21.von Eichel-Streiber C, Boquet P, Sauerborn M, Thelestam M. Large clostridial cytotoxins—a family of glycosyltransferase modifying small GTP-binding proteins. Trends Microbiol. 1996;4:375–382. doi: 10.1016/0966-842X(96)10061-5. [DOI] [PubMed] [Google Scholar]
- 22.Wilkinson S R, Young D I, Morris J G, Young M. Molecular genetics and the initiation of solventogenesis in Clostridium beijerinckii (formerly Clostridium acetobutylicum) NCIMB 8052. FEMS Microbiol Rev. 1995;17:275–285. doi: 10.1111/j.1574-6976.1995.tb00211.x. [DOI] [PubMed] [Google Scholar]
- 23.Wilson K H, Perini F. Role of competition for nutrients in suppression of Clostridium difficile by the colonic microflora. Infect Immun. 1988;56:2610–2614. doi: 10.1128/iai.56.10.2610-2614.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamakawa K, Kamiya S, Meng X Q, Karasawa T, Nakamura S. Toxin production by Clostridium difficile in a defined medium with limited amino acids. J Med Microbiol. 1994;41:319–323. doi: 10.1099/00222615-41-5-319. [DOI] [PubMed] [Google Scholar]
- 25.Yamakawa K, Karasawa T, Ohta T, Hayashi H, Nakamura S. Inhibition of enhanced toxin production by Clostridium difficile in biotin-limited conditions. J Med Microbiol. 1998;47:767–771. doi: 10.1099/00222615-47-9-767. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto-Osaki T, Kamiya S, Sawamura S, Kai M, Ozawa A. Growth inhibition of Clostridium difficile by intestinal flora of infant faeces in continuous flow culture. J Med Microbiol. 1994;40:179–187. doi: 10.1099/00222615-40-3-179. [DOI] [PubMed] [Google Scholar]
- 27.Zijlistra J B, Beukema J, Wolgers B G, Byrne B M, Groen A, Dankert J. Pretreatment methods prior to gas chromatographic analysis of volatile fatty acids from faecal samples. Clin Chim Acta. 1977;78:243–250. doi: 10.1016/0009-8981(77)90312-6. [DOI] [PubMed] [Google Scholar]