Glioblastoma is frequently refractory to radiation, chemotherapy, and immunotherapy. The authors show in genetic mouse models that the mutational profile of glioblastoma influences the tumor immune microenvironment and offers a pharmacological means to sensitize glioblastoma to checkpoint immunotherapy.
Abstract
The composition of the tumor immune microenvironment (TIME) is considered a key determinant of patients’ response to immunotherapy. The mechanisms underlying TIME formation and development over time are poorly understood. Glioblastoma (GBM) is a lethal primary brain cancer for which there are no curative treatments. GBMs are immunologically heterogeneous and impervious to checkpoint blockade immunotherapies. Utilizing clinically relevant genetic mouse models of GBM, we identified distinct immune landscapes associated with expression of EGFR wild-type and mutant EGFRvIII cancer driver mutations. Over time, accumulation of polymorphonuclear myeloid-derived suppressor cells (PMN-MDSC) was more pronounced in EGFRvIII-driven GBMs and was correlated with resistance to PD-1 and CTLA-4 combination checkpoint blockade immunotherapy. We determined that GBM-secreted CXCL1/2/3 and PMN-MDSC–expressed CXCR2 formed an axis regulating output of PMN-MDSCs from the bone marrow leading to systemic increase in these cells in the spleen and GBM tumor-draining lymph nodes. Pharmacologic targeting of this axis induced a systemic decrease in the numbers of PMN-MDSC, facilitated responses to PD-1 and CTLA-4 combination checkpoint blocking immunotherapy, and prolonged survival in mice bearing EGFRvIII-driven GBM. Our results uncover a relationship between cancer driver mutations, TIME composition, and sensitivity to checkpoint blockade in GBM and support the stratification of patients with GBM for checkpoint blockade therapy based on integrated genotypic and immunologic profiles.
Introduction
Immune checkpoints blockade can achieve remarkable treatment outcomes for patients with cancer. Although mutational burden and dysfunction in mismatch repair genes positively correlate with efficacy of checkpoint blockade, in general, factors that predict positive therapeutic response remain poorly understood. Moreover, it is currently unknown whether cancer driver mutations influence the tumor immune microenvironment (TIME) and how that might have a role in therapeutic response to checkpoint blockade. Such an unaddressed question is of paramount mechanistic value in the field of tumor immunology.
Myeloid-derived suppressor cells (MDSC) represent major immunosuppressive populations of the tumor microenvironment. Generated by cancer-triggered emergency myelopoiesis (1), MDSCs are immature forms of neutrophils and monocytes, and are referred to as polymorphonuclear- (PMN-) and monocytic- (M-) MDSCs, respectively (2). MDSCs exert their immunosuppressive function through many mechanisms (1, 3) and can hinder the anticancer activities of chemotherapy, checkpoint blockade immunotherapy, and cancer vaccines (4).
Glioblastoma (GBM) is an incurable malignant primary brain cancer with a short (15 months) median survival. It is characterized by aberrant overexpression and amplification of EGFR in the majority (65%) of patients, and many of these patients also have an intragenic in-frame deletion mutant that encodes for a constitutively activated, ligand-independent receptor known as EGFRvIII (5–7). The current standard of care for patients with GBM is debulking surgical resection, fractionated irradiation concomitant with the DNA alkylating agent temozolomide (TMZ), followed by adjuvant TMZ. The recent success of checkpoint blockade immunotherapies in cancer has not translated in GBMs partly because GBMs have (i) a low incidence of intratumoral lymphocytes with immunologic features of elevated T-cell exhaustion, (ii) a high infiltration of MDSCs and immunosuppressive bone marrow–derived macrophages (BMDM), (iii) an impermeable blood brain barrier, (iv) a low abundance and/or ineffective antigen-presenting cells (APC), and (v) a low frequency of neoantigens and mutational burden (8, 9). GBMs develop asymptomatically until clinical manifestation of late disease stages is observed, which curtails detailed studies on the longitudinal evolution of GBM development using patient samples. In addition, GBM's genomic heterogeneity challenges correlative studies of driver mutations on tumor-microenvironment composition and function in patients. To overcome these caveats, we leveraged our genetically engineered mouse models of EGFR wild-type (WT)- and EGFRvIII-driven de novo GBM (10–13) to study the evolution of the TIME during disease progression, and to investigate how driver-gene mutations dictate immune composition and sensitivity to checkpoint blockade.
Here, we report that EGFR-WT and EGFRvIII-mutant GBMs activate distinct signaling pathways that correlate with different cytokine profiles and TIME, which in turn determine sensitivity to PD-1 and CTLA-4 combination immune checkpoint blockade. CXCL2 and CXCL3 produced by EGFRvIII-mutant GBM-induced CXCR2-dependent local and systemic recruitment of PMN-MDSCs, which correlated with resistance to PD-1 and CTLA-4 checkpoint inhibition, whereas CXCR2 antagonism resulted in systemic decrease of PMN-MDSC, enhanced the efficacy of combination immune checkpoint blockade and prolongation of survival.
Materials and Methods
EGFR conditional GBM mouse model and procedures
All mouse procedures were performed in accordance with Beth Israel Deaconess Medical Center's Institutional Animal Care and Use Committee. Lox-stop-lox (LSL)/Cre-mediated conditional expression of human EGFR WT, mutant EGFRvIII, firefly luciferase cDNAs and conditional deletion of Pten was achieved as described elsewhere (10, 14–16). Cohorts of LSL-EGFR-WT;Cdkn2a−/−;PTEN2lox;LSL-Luc and LSL-EGFRvIII;Cdkn2a−/−;PTEN2lox;LSL-Luc mice were injected with pTyf-TGFα-IRES-iCre or pTyf-iCre lentiviruses respectively and were imaged by bioluminescence imaging (BLI) for tumor staging as previously described in details (10, 17). Briefly, BLI was initiated 3 weeks post-lentivirus injection by intraperitoneal injection of Luciferin (Perkin-Elmer, #122799) into animals for 10 minutes prior to imaging using an IVIS xenogen imager (Perkin-Elmer). Animals were imaged every 7 days until they became moribund, at which point the experiments were terminated. BLI outputs of 10E07–10E08 (p/s/cm2/sr) were arbitrarily labeled as early-stage GBMs, whereas BLI outputs of >10E08 (p/s/cm2/sr) were considered late-stage tumors since the lag time between an animal that reaches >10E08 and moribund status is historically short (<10 days).
Histopathologic analysis
Histopathologic processing of mice and brain tissues has been described elsewhere (10–13, 18). For IHC, staining was performed on glass slides containing microtome-cut (10-μm-thick) sections of formalin-fixed paraffin-embedded tumor-bearing mouse brain sections from control animals, EGFR-WT or EGFRvIII-driven GBMs, control or treated with the indicated therapies. The following primary antibodies were used: cleaved caspase-3 (CC3; Cell Signaling Technology, #9661, 1:200) and Ki67 (NCL-ki67, 1:1,000, Novocastra). Further details on the antibodies and the secondary antibodies can be found in Supplementary Table S1. Ki-67 and CC3 staining was quantified using ImageJ (ver. 1.53t; https://imagej.net/ij/index.html) by two independent observers who were blind to the images. Images were obtained on a bright field Olympus BX43 microscope with a DP72 digital camera using a 20× objective and processed using the cellSens Entry (ver. 1.3) imaging software. A minimum of three fields of view per image, three images per tumors, and a minimum of n = 3 tumors were analyzed per condition.
In vivo reagents and treatment
GBM initiation and growth development was monitored by BLI as described above and animals that reached >1 × 107 p/s/cm2/sr were randomly enrolled in treatment groups. All antibodies were diluted with sterile 0.9% Saline (Hospira). Mice were injected intraperitoneally with 200 μg of anti–PD-1 (clone RMP 1–14, BioXcell) and/or 200 μg anti-CTLA-4 (clone 9D9, BioXcell), or the isotype control IgG2a mAb (clone 2A3, BioXcell), every 3 days for 3 doses total. For the immunodepletion experiments, mice were injected with 200 μg of anti-CD8 (clone 53.6.7, BioXcell) daily for 3 days prior to initiation of immune checkpoint blockade or with 200 μg anti-Ly6G (clone 1A8, BioXcell) 1 day prior to initiation of immune checkpoint blockade and depletion treatment was continued every 3 days for three treatments total. AZD-5069 (MedKoo Biosciences, #206473) was resuspended in Ora-Plus (Paddock) and administered daily by oral gavage at a dose of 100 mg/kg body weight for 12 days.
Tissue processing and flow cytometry
Tumor-bearing EGFR-WT and EGFRvIII-mutant GBM mice at either symptomatic moribund stage or 3 days after completion of the third and final dose of indicated treatment were perfused transcardiacally with 10mL PBS, brains were harvested, and cerebellum removed. Brain, tumor draining lymph nodes (TDLN), and spleen were minced and resuspended in 1.5 mg/mL Collagenase type IV (Gibco, #17–104–019) containing DNase I (Sigma, #D4263) in HBSS with calcium and magnesium and incubated rotating at 37°C for 45 minutes, with occasional gentle dissociation with a P1000 pipette and filtered through a 100 μm (brain) or 70 μm (TDLN and spleen) mesh filter and diluted with HBSS. All washes were pelleted at 400 × g for 5 minutes. Single-cell suspensions of brain tissues were resuspended in 30% Percoll (Sigma, #GE17–0891–02) in PBS for myelin removal by centrifugation at 700 × g for 15 minutes with low deceleration brake. Myelin layer was carefully aspirated off, and cells were diluted in PBS and pelleted at 400 × g for 5 minutes. Red blood cells (RBC) were lysed using RBC lysis buffer according to manufacturer protocol (BioLegend, #420302). Single-cell suspensions were blocked with FC block (BioLegend, #101301) for 5 minutes and stained with antibodies and viability dye (Zombie NIR Fixable Viability Kit, BioLegend, #423105; or Zombie Yellow Fixable Viability Kit, BioLegend, #423103). Cell surface antigens were stained 30 minutes at 4°C in the dark. Cells were washed 2× in PBS and fixed using a FoxP3 Intracellular Staining Kit according to manufacturer protocol (eBioscience, #00–5523–00), then permeabilized and stained with intracellular antibodies for 30 minutes at 4°C in the dark. Cell suspensions were washed 2× in permeabilization buffer and resuspended in PBS for analysis. A minimum of 20,000 events were collected on a Beckman Coulter Gallios flow cytometer or BD LSRFortessa and analyzed using FlowJo (version 10). Compensation was performed using Ultracomp ebeads Compensation Beads (Invitrogen, #01–2222–42), which were stained with appropriate antibody and analyzed on the same voltage and settings. Antibody combinations against the following target antigens were used to define these cell types: BMDM, CD45hiCD11b+Ly6C−Ly6G−; microglia, CD45loCD11b+Ly6C−Ly6G−; PMN-MDSCs CD45+CD11b+Ly6CloLy6G+; M-MDSCs, CD45+CD11b+Ly6ChiLy6G−; CD8+ T cells, CD45+CD3+CD8+CD4−; CD4+ T cells, CD45+CD3+CD4+CD8−; regulatory T cells (Treg cells), CD45+CD3+CD4+CD8−Foxp3+, tumor cells, CD45−hEGFR+. Antibodies details are listed in Supplementary Table S1.
MDSC suppression assay
MDSC-mediated suppression was assessed using previously established methodology (19). Briefly, splenic MDSCs were isolated from the spleens of tumor-bearing EGFR-WT and EGFRvIII-mutant GBM mice using the EasySep Mouse MDSC (CD11b+GR1+) Isolation Kit (StemCell Technologies, #19867). MDSC–splenocyte co-cultures were incubated in DMEM containing 5% FCS (R&D systems, S11150), 2 mmol/L glutamine (Corning, #25–005-Cl), 100 units/mL penicillin–streptomycin (Gibco, #15–140–122), 10 mmol/L HEPES (Corning, #25–060-Cl), and 20 μmol/L β-mercaptoethanol (Gibco, #31350010). Serial dilutions of MDSCs (2 × 105, 1 × 105, 1 × 104) were plated in RPMI1640 (Corning, #15–040-CM) in flat bottom 96-well plates with 2 × 105 splenocytes per well isolated from OTI-TCR transgenic mice [The Jackson Laboratory, C57BL/6-Tg(TcraTcrb)1100Mjb/J, stock No: 003831] and 0.1 nmol/L of ovalbumin peptide (OVA257–264; Sigma, #S7951–1MG) with 10 ng/mL of IL2 (Peprotech, #212–12) for 72 hours. As control, OTI splenocytes were incubated with OVA peptide (OVA257–264) and IL2 without MDSC. 3H-thymidine (Perkin-Elmer, 1 mCI, Catalog No. NET027W001MC1) was added for the last 16 hours of a 72-hour culture, and thymidine incorporation was measured by MicroBeta plate counter (Perkin-Elmer).
Bone marrow isolation
Symptomatic moribund tumor-bearing EGFR-WT and EGFRvIII-mutant GBM mice were sacrificed followed by dissection of tibias. Tibias were placed in ice cold RPMI1640 (Corning, #15–040-CM) to remove muscles and were washed in 70% ethanol 5 minutes, and ice-cold PBS 5 minutes. Bone marrow was flushed with PBS, cells pelleted at 1,200 rpm for 8 minutes followed by RBC lysis with ACK lysis buffer (Gibco, #A1049201) for 1 minute at room temperature. Lysis was stopped using RPMI1640+10% FBS (R&D systems, S11150). Cells were then counted using trypan blue. Common myeloid progenitors (CMP; Lin−Sca1−CD127−c-kit+CD16/CD32−) and granulocyte monocyte progenitors (GMP; Lin−Sca1−CD127−c-kit+CD16/CD32+) myeloid precursors were identified on a flow cytometer as described above.
RT-qPCR analysis of cytokine expression
Brain single-cell suspensions obtained as above were stained with anti-CD45 and Zombie Yellow fixable viability dye and immediately sorted on a FACS Aria Cell Sorter for viable CD45− and CD45+ populations and immediately resuspended in Qiagen RLT buffer. RNA was isolated using the Qiagen RNeasy Micro Kit (#74004) as per manufacture protocol and eluted using 14 μL of RNAse free water. Thirty-five nanograms of RNA was used for reverse-transcriptase reaction using the SuperScript III cDNA Synthesis Kit (Invitrogen, #11–752–050). Primers (Supplementary Table S2) were found in the literature as cited or from Primerbank (20–22). qPCR was performed using Sybr Green (Life Technologies, #4367659) on a Stratagene Mx3000p (Agilent Technologies). Expression level was determined using the ddCt method comparing CD45+ to CD45− expression and normalizing to GAPDH.
GBM primary cultures
Isolation of EGFR-driven mouse GBM primary cell cultures have been described previously (10–12, 18). Primary cultures of tumors were established as follows: tumor tissue from brains of moribund tumor-bearing EGFR-WT and EGFRvIII-mutant GBM mice were excised and minced in 0.25% trypsin (wt/vol) 1 mmol/L EDTA and allowed to disaggregate for 15 minutes at 37°C. The resulting cell suspension was then strained through a 100-μm cell strainer. The single-cell suspension was washed in PBS twice and plated on 0.2% gelatin-coated tissue culture plates. Cells were cultured with media that consisted of DMEM supplemented with 10% heat-inactivated FBS and antibiotics.
Human TCGA deconvolution analysis
Cell state fraction deconvolution analyses were performed on the processed TCGA glioma (GBMLGG) RNA sequencing (RNA-seq) dataset obtained from GDAC FireHose (RNAseqV2, RSEM). Cell state fractions were calculated as previously described (23) using CIBERSORTx (24) input with gene signatures derived from a pan-glioma single-cell RNA-seq dataset (25). Within the TCGA dataset, patients were stratified into EGFR mutation groups based on results from a prior publication (5). Patients with EGFRvIII mutations were defined as those with high expression (transcript allele frequency ≥10%) of EGFR containing an exon 1 to 8 junction (deletion of exons 2–7). Patients with EGFR mutations were those that had any non EGFRvIII mutation that exhibited either an allele frequency or transcript allele frequency >10%. Patients with WT EGFR were not part of the EGFRvIII and EGFR mutation groups and lacked focal amplifications of EGFR. Comparisons of T-cell fraction differences between these groups were performed using the Wilcoxon rank-sum test.
Quantitative proteomics and phosphoproteomics
Biological triplicates of primary cell cultures isolated from EGFR-WT and EGFRvIII GBMs were harvested, lysed, and processed for quantitative isobaric label-based Tandem Mass Tag (TMT) mass spectrometry as follows. Cell pellets were harvested and lysed with a buffer containing 50 mmol/L HEPES (pH 8.5), 8M urea, 150 mmol/L NaCl, protease inhibitors (mini-Complete EDTA-free; Roche, #04693159001), and phosphatase inhibitors (PhosSTOP; Roche, #4906845001). Cells were passed through a 22-gauge needle and syringe, 15 times for mechanical lysis. Lysates were cleared through centrifugation and protein concentrations were determined using a BCA assay (Thermo Fisher Scientific, #23225). Equal amounts of protein (4 mg) were reduced for 45 minutes at 37°C with 5 mmol/L DTT, alkylated with 15 mmol/L IAA for 30 minutes at room temperature in the dark, before final reduction with 5 mmol/L DTT for 15 minutes at room temperature. Protein contents were extracted through methanol–chloroform precipitation, before resuspension in 50 mmol/L HEPES, 8 mol/L urea, and 150 mmol/L NaCl. For proteolytic digestion LysC (Thermo Fisher Scientific, #NC9223464) was added at a substrate:enzyme ratio of 100:1 and incubated for 3 hours at 37°C. Samples were then diluted to 1.5 mmol/L urea with 50 mmol/L HEPES and digest overnight with Trypsin at room temperature with a substrate:enzyme ratio of 50:1. The peptide solutions were then acidified before solid-phase extraction via SepPak (Waters, #WAT020515). Peptide samples were resuspended in 1 mL 50% ACN, 2 mol/L lactic acid, and 100 μg of each sample was removed, desalted, and saved for protein-level measurements. Phosphopeptide enrichment was performed according to ref. 26.
Nonphosphorylated peptides saved prior to enrichment and enriched phosphopeptides were then suspended in 100 μL of 200 mmol/L EPPS pH 8.5 before the addition of 30 μL of anhydrous acetonitrile, and 10 μL of a 20 μg/μL stock of TMT reagent. Samples were incubated for 1 hour at room temperature before the addition of 10 μL 5% hydroxylamine. A small portion of each sample was mixed, desalted, and analyzed to determine relative analyte abundance in each sample. The remaining sample was then mixed to ensure equal loading of peptide and phosphopeptide content and acidified before solid-phase extraction via SepPak. Following isobaric labeling, enriched phosphopeptides were enriched again for phospho-tyrosine (pTyr) containing peptides. Enriched phosphopeptides were resuspended in 450 μL of immuno-affinity purification (IAP) buffer (50 mmol/L MOPS/NaOH pH 7.2, 10 mmol/L Na2PO4, and 50 mmol/L NaCl). A phospho-tyrosine specific antibody (P-Tyr-1000, Cell Signaling Technology) was incubated with protein A agarose beads (Roche) overnight at 4°C in 1% PBS to bind the antibody to the beads. Subsequently, the antibody-bead mixture was washed 3× with IAP before incubation with enriched phosphopeptides for 1 hour at room temperature to enable capture of pTyr containing peptides. The supernatant, containing enriched phosphopeptides, was removed, de-salted using a SepPak, and saved for offline fractionation. The beads were washed 1× with IAP and 1× with H2O before performing two elutions using 75 μL of 100 mmol/L formic acid. Enriched pTyr peptides were desalted and resuspended in 1% formic acid prior to nLC/MS-MS analysis. Nonphosphorylated and phosphorylated peptides were fractionated via basic-pH reversed-phase liquid chromatography (27). Nonphosphorylated samples were resuspended in 5% ACN, 1% formic acid, and phosphorylated peptides were resuspended in 1% formic acid before nLC/MS-MS analysis.
MS analysis
MS analyses were performed on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) coupled to an Easy-nLC 1200 ultra-high pressure LC pump (Thermo Fisher Scientific). Peptides were separated at 300 nL/min using an analytical column (75 μm inner diameter) that was packed self-packed with 0.5 cm of Magic C18 resin (5 μm, 100 Å; Michrom Bioresources) followed by 35 cm of Sepax Technologies GP-C18 resin (1.8 μm, 120 Å). LC buffers consisted of 0.1% formic acid (buffer A) and 80% ACN with 0.1% formic acid and LC gradients were optimized to ensure equal elution of peptides throughout the analysis. Survey scans (MS1) were performed in the Orbitrap (AGC target 1e6, 120,000 resolution, 100 milliseconds maximum injection time) and used to select the 10 most abundant features for MS-MS (MS2) analysis. Candidate peaks were filtered on the basis of charge sate ≥2 and monoisotopic peak assignment, and dynamic exclusion (60 second ± 10 ppm) was enabled. For nonphosphorylated peptide analysis, only one charge state was selected for each precursor. Precursor ions were isolated (AGC target = 2.5 × 104) at a width of 0.5 Th using a quadrupole mass filter and fragmented with collision-induced dissociation (CID, 35 NCE) in the ion trap with distinct maximum injection time settings for nonphosphorylated (150 milliseconds) and phosphorylated (200 milliseconds) peptides. To alleviate the effects of precursor ion interference (28), multiple fragment ions were isolated (29) using synchronous precursor selection (SPS) prior to HCD (55 NCE, SPS notches = 8, AGC target = 2.2 × 105, maximum injection time of 150 or 300 milliseconds for nonphosphorylated and phosphorylated peptides, respectively) MS3 fragmentation and Orbitrap analysis (50,000 resolution).
A compilation of in-house software was used to convert Thermo “.raw” mass spectrometric data to mzXML format, as well as to correct monoisotopic m/z measurements and erroneous peptide charge state assignments (30). The SEQUEST algorithm was used to assign MS-MS spectra to a peptide identification (31). Static modifications included TMT (229.16293 Da) on both the n-terminus of peptides and lysine residues and carbamidomethylation of cysteine residues (57.02146 Da). Phosphorylation (79.96633 Da) was included for phosphopeptide experiments. Peptide spectral matches were filtered to 1% FDR using the target-decoy strategy (32), before being grouped into proteins which were then filtered to 1% FDR at the protein level (30). Phosphorylation sites were localized with a modified version of the AScore algorithm and phosphorylation sites with an AScore >13 (P < 0.05) were considered localized (33). Proteins and phosphorylation isoforms were quantified according to ref. 27. “Relative abundance” expression values for each analyte (protein or phosphorylation isoform) and represent the signal-to-noise value of each sample divided by the sum of all samples for each analyte normalized to 100. For phosphorylated peptides, the quantitative values were normalized to the relative abundance of the protein, to account for changes in protein abundance upon treatment. All data analysis was performed using R (http://www.R-project.org).
Gene ontology analysis
The proteomics data described above was used to identify genes enriched in biological process (BP), molecular function (MF), and cellular component (CC), we utilized the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v7.0 (https://david.ncifcrf.gov; refs. 34, 35) with GOTERMs BP, MF, and CC. All terms with a P value (Benjamini or Benjamini–Hochberg adjusted) less than 0.05 were considered significant and ranked by the number of genes identified in the group.
Statistical analyses
Statistical analyses were performed using GraphPad Prism 9 (GraphPad Software). Values are given as mean ± SEM or SD as indicated. Numbers of experimental replicates are given in the figure legends. When two groups were compared, significance was determined using an unpaired two-tail t test. When multiple groups were compared, significance was determined using one-way ANOVA with Tukey's correction for multiple comparisons. Significance for survival analyses was determined by the log rank (Mantel–Cox) test. A P-value <0.05 is considered as statistically significant.
Data availability
The data generated in this study are available in the article and its supplementary data files or upon request from the corresponding authors.
Results
EGFR-WT and EGFRvIII GBMs have distinct cancer signaling and immune composition profiles
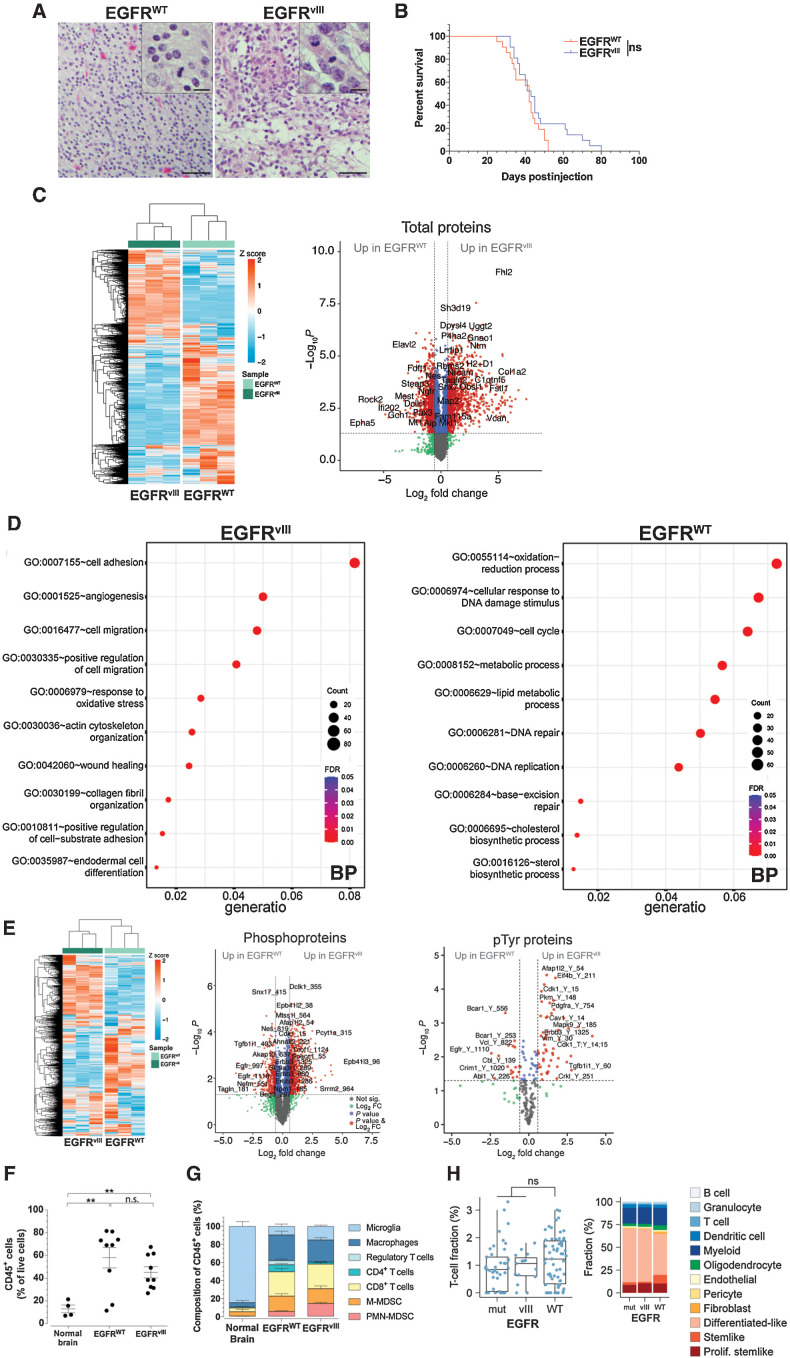
EGFR-WT and EGFRvIII utilize unique and overlapping pathways to convey oncogenic signaling inputs (36). We previously demonstrated that mice genetically engineered to overexpress human EGFR-WT or EGFRvIII in the context of loss of both Cdkn2a and Pten tumor suppressor genes in the CNS develop de novo GBMs (10–13). In this study, EGFR-WT (n = 8) and EGFRvIII (n = 9) mouse GBMs were blindly evaluated for neuropathologic features. Both had frequent mitoses, moderate to dense cellularity, brain infiltration, vascular proliferation, and necrosis, features consistent with high-grade gliomas (Fig. 1A). In addition, EGFR-WT GBMs had small, round, slightly hyperchromatic nuclei with minimal cytoplasm (Fig. 1A), histologic features highly reminiscent of small cell GBMs, which have a high frequency of EGFR amplification (70%) and PTEN loss (>95%; ref. 37). In contrast, EGFRvIII GBM cells had classical astrocytic morphology with readily identifiable glial processes, and moderate nuclear atypia (Fig. 1A). These differing features, however, had no influence on overall survival of GBM-bearing mice (median survival: 42 and 43 days for EGFR-WT and EGFRvIII, respectively; Fig. 1B).
Figure 1.
Distinctive histopathological, signaling and immune characteristics of EGFRvIII and EGFR-WT GBMs. A, Representative H&E sections of mouse GBMs. Scale bar = 50 μm; inset = 10 μm. B, Kaplan–Meier analysis of GBM mice. P = 0.1118; n.s., not significant, log-rank (Mantel–Cox), n = 21 each EGFR-WT and EGFRvIII. C, Unsupervised clustering heat map and volcano plot of quantitative proteomics from EGFR-WT and EGFRvIII GBM-derived cells. n = 3 from biological replicates for each line. D, GO analysis of significantly upregulated proteins in EGFR-WT and EGFRvIII GBM cells. BP, biological processes. E, Unsupervised clustering heat map and volcano plots of quantitative phosphoproteomics from EGFR-WT and EGFRvIII GBM-derived cells. n = 3 from biological replicates for each line. F, Flow cytometry of CD45+ cells in normal brain (n = 4), EGFR-WT (n = 9), and EGFRvIII (n = 9) GBMs. G, Flow cytometry of immune cells in normal brain (n = 4), EGFR-WT (n = 10), and EGFRvIII (n = 9) GBMs. H, CIBERSORTx cell state fraction deconvolution analysis performed on human TCGA glioma RNA-seq dataset from EGFR-WT (n = 62), EGFRvIII (n = 15), and mutated EGFR (mut) other than vIII (n = 41). Left, plot of T-cell fraction as percent of all cells, right panel, stacked percent fraction of all immune cells determined by CIBERSORTx. Mean ± SEM of biological replicates. **, P < 0.01; ns; not significant, unpaired t test, two-tailed (F, G).
To better characterize EGFR-WT and EGFRvIII GBMs, we performed an unbiased, comprehensive quantitative isobaric label-based TMT MS analysis to measure expression of proteins and phospho-isoforms (pTyr, pSer, and pThr) from mouse primary cell cultures derived from EGFR-WT and EGFRvIII GBMs (Fig. 1C–E). We identified 7,851 proteins across all conditions that clustered separately (Fig. 1C). Gene Ontology (GO) analysis of the significantly upregulated proteins demonstrated enrichment for cell oxidation–reduction process, lipid metabolic process, cell cycle, DNA repair and replication in EGFR-WT cells and cell adhesion, cell migration, actin cytoskeleton organization, and angiogenesis in EGFRvIII cells (Fig. 1D). To measure changes in signaling in EGFR-WT and EGFRvIII GBMs, we performed successive phospho-peptide enrichment and pTyr immunopurification and identified 6,054 phospho-peptides (210 pTyr) belonging to 1,807 unique proteins and computed Log2 fold change (FC) for each phospho-peptide between EGFR-WT and EGFRvIII. In EGFRvIII GBM cells, higher levels of Erbb3 pY1286 and pY1325 were observed (Fig. 1E). Erbb3 is a ligand-binding, kinase-deficient EGFR family member, which obligatory heterodimerizes with other ErbB members for signaling (38), and has been shown to activate oncogenic EGFRvIII signaling (39). Our results suggest that EGFRvIII may dimerize with ERBB3 for oncogenic activity in GBM. In addition, higher levels of PDGFRA pY754 were observed in EGFRvIII cells, corroborating our previous results demonstrating that EGFRvIII requires PDGFRA activity for oncogenic signaling (13). On the other hand, phosphorylation of the EGFR autophosphorylation site Y1110 and of the E3 ubiquitin ligase Cbl at Y139 were upregulated in EGFR-WT (Fig. 1E). Although observed in unrelated studies, the functional role of Cbl pY139 has not been elucidated. Phosphorylation of EGFR pY1110 increases kinase activity and plays a role in receptor trafficking and stability (40, 41). These results suggest that differences in histopathologic features in EGFR-WT and EGFRvIII GBMs are associated with distinct signaling events.
Increasing experimental evidence demonstrates that molecular features of cancer have a direct impact on the TIME (42). Using flow cytometry to characterize the TIME (Supplementary Figs. S1A and S1B), we observed significantly elevated levels of CD45+ cells in EGFR-WT (4.5 ± 0.71-fold) and EGFRvIII (3.5 ± 0.63 fold) GBMs when compared with normal brain, but no statistically significant differences between EGFR-WT and EGFRvIII GBMs (Fig. 1F). Using CD45lo and CD45hi to differentiate microglia from BMDMs (43), we observed that the majority of CD45+ cells of normal brains were CD45lo microglia (84.7±10.1% of total cells), whereas GBMs had markedly lower fractions of microglia and a significant increase in other immune cell types with differences in composition between EGFR-WT and EGFRvIII GBMs (Fig. 1G; Table 1). Compared with EGFR-WT GBMs, EGFRvIII GBMs had significantly reduced numbers of CD4+ T cells (7.63% ± 1.5 vs. 2.21% ± 0.43, P = 0.0042) and Treg cells (4.78% ± 1.69 vs. 0.00021% ± 0.0001, P = 0.0159) and elevated PMN-MDSCs (5.46% ± 1.01 vs. 14.33% ± 1.51, P = 0.0001; Fig. 1G; Table 1). These differences extended to cervical TDLNs and spleen of GBM-bearing animals (Supplementary Figs. S1C and S1D). We did not observe significant differences in CD8+ T cells, M-MDSCs, and BMDMs between EGFR-WT and EGFRvIII GBMs (Fig. 1G; Table 1). In TDLNs, significant increases in CD8+ T cells, PMN-MDSCs, M-MDSCs, and BMDMs and decreases in CD4+ T cells in EGFRvIII compared with EGFR-WT were observed (Supplementary Figs. S1C and S1D). The spleens of EGFRvIII-mutant GBM-bearing mice displayed higher fractions of CD8+ T cells and BMDMs and decreases in CD4+ T cells, compared with EGFR-WT GBM-bearing mice (Supplementary Figs. S1C and S1D). In contrast, splenic M-MDSCs and, more prominently, PMN-MDSCs were lower in EGFRvIII GBM-bearing mice compared with EGFR-WT GBM-bearing mice (Supplementary Figs. S1C and S1D). Together these findings indicate that the two types of GBM induced distinct systemic immunologic alterations and that EGFRvIII GBM displayed a systemic redistribution of PMN-MDSCs, including tumor site and TDLNs.
Table 1.
Immune composition of normal brain and EGFR glioblastomas.
| Percent of CD45+ cells ± SEM | P values | |||||
|---|---|---|---|---|---|---|
| Normal brain | EGFR WT | EGFR vIII | WT vs. Normal | vIII vs. Normal | WT vs. vIII | |
| CD8+ T cells | 4.12 ± 1.35 | 27.03 ± 3.68 | 27.29 ± 1.5 | 0.0025 | <0.0001 | n.s. |
| CD4+ T cells | 1.12 ± 0.59 | 7.63 ± 1.5 | 2.21 ± 0.43 | 0.0214 | n.s. | 0.0042 |
| Regulatory T cells | n.d. | 4.78 ± 1.69 | 0.00021 ± 0.0001 | n.d. | n.d. | 0.0159 |
| PMN-MDSCs | 0.33 ± 0.15 | 5.46 ± 1.01 | 14.33 ± 1.51 | 0.0088 | <0.0001 | 0.0001 |
| M-MDSCs | 4.91 ± 2.44 | 17.02 ± 2.72 | 16.46 ± 3.04 | 0.0228 | 0.0387 | n.s. |
| BMDMs | 4.79 ± 2.44 | 28.3 ± 4.14 | 24.3 ± 2.69 | 0.005 | 0.001 | n.s. |
| Microglia | 84.72 ± 5.03 | 9.76 ± 2.33 | 15.41 ± 1.03 | <0.0001 | <0.0001 | 0.0477 |
Note: Flow cytometry analysis of intratumoral CD45+ cells for the indicated cell type (percent). Data are mean ± SEM of biological replicates, P values from unpaired t test, two-tailed.
Abbreviation: ns, not significant.
We validated these findings in patients by conducting CIBERSORTx deconvolution analysis (44–46) on GBM TCGA data. We identified neoplastic cells (differentiated-like, stemlike, and proliferating stemlike), nontumorigenic cells and immune cells in EGFR-WT amplified and EGFR mutated/EGFRvIII human GBMs and we observed a trend for reduced T-cell infiltration in EGFRvIII GBMs (Fig. 1H). Together these results demonstrate that the immune cell composition of symptomatic EGFR-WT and EGFRvIII GBMs are distinctly different, both in mice and human patients.
Acquisition of immunosuppressive features during progression
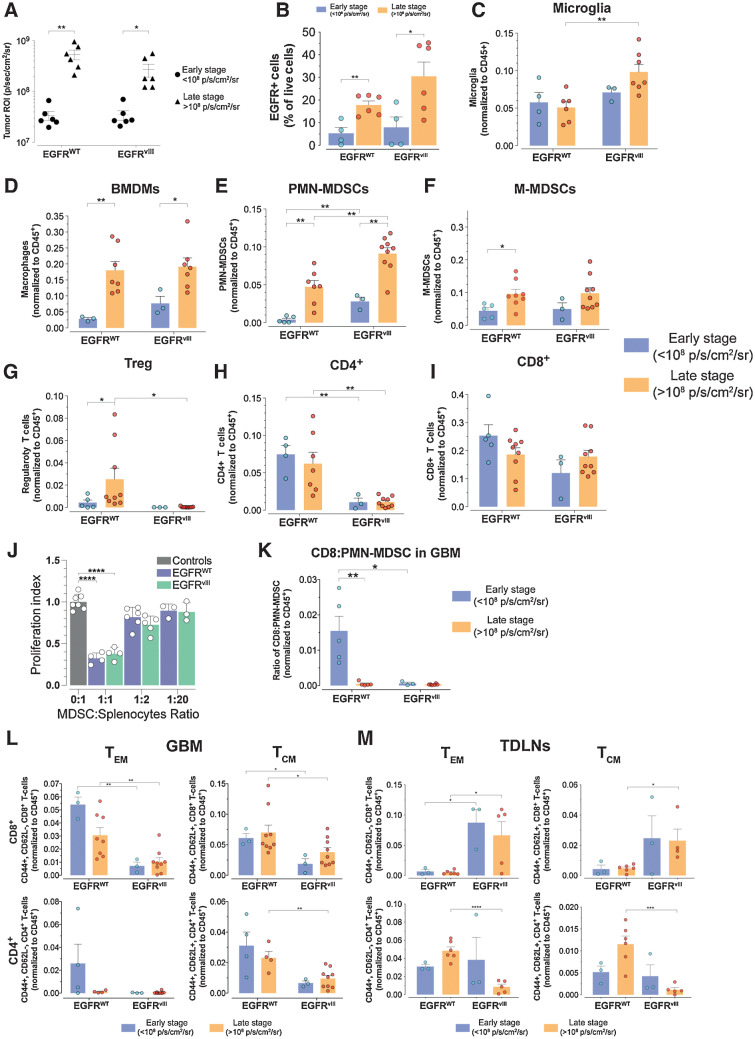
In patients, GBMs are diagnosed in symptomatic, advanced stages, which precludes studies on the evolution of the TIME during the gliomagenesis process. We leveraged our models to understand the development of the immune landscape during GBM progression. We analyzed the immune compositions of early- and late-stage GBMs as defined by BLI outputs (Fig. 2A; Supplementary Fig. S2A; ref. 47), which positively correlated with the number of EGFR+ GBM cells (Supplementary Fig. S2B). The number of intratumoral CD45+ cells remained unchanged during progression (Supplementary Fig. S2C), whereas the number of EGFR+ cells significantly increased over time in both GBM types (late vs. early GBMs, EGFR-WT 3.35 ± 1.00-fold, EGFRvIII 3.85 ± 1.26-fold) consistent with tumor growth (Fig. 2B).
Figure 2.
Distinct immune profile evolution of EGFR-WT and EGFRvIII GBMs during progression. A, BLI of GBMs at early- and late-stage tumor progression. n = 6 each EGFR-WT and EGFRvIII early and late stage. B, Relative number of EGFR+ GBM cells during early- and late-stage tumor progression. n = 4,6 and 4,6 for EGFR-WT early and late stage and EGFRvIII early and late stage, respectively. C–I, Flow cytometry longitudinal assessment of the indicated cell types during early- and late-stage tumor progression. J, Suppression of proliferation of T cells by MDSCs isolated from EGFR-WT and EGFRvIII GBM bearing mice. n = 4 for each control, EGFR-WT, and EGFRvIII mice. K, Ratio of CD8:PMN-MDSC from early- and late-stage GBMs. L, and M, Flow cytometry analysis of relative CD8+ and CD4+ TEM and TCM cell subset from GBM (L) and TDLNs (M) tissues of early- and late-stage GBMs. All data are mean ± SEM of biological replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; unpaired t test, two-tailed.
Although no detectable changes were observed in microglia (Fig. 2C), all other myeloid subsets displayed significant increases during progression (Fig. 2D–F). Concomitantly, there was an increase in Treg cells in EGFR-WT GBM, whereas in EGFRvIII GBM Treg cells remained undetectable at both early and late disease stages (Fig. 2G) and the numbers of CD4+ and CD8+ T cells did not change during tumor progression (Fig. 2H and I). The numbers of PMN-MDSCs were higher in EGFRvIII GBMs compared with EGFR-WT GBMs at both stages of tumor progression (Fig. 2E). Concordantly, the levels of CD4+ T cells were significantly lower in EGFRvIII than in EGFR-WT GBMs both in early- and late-stage tumors (Fig. 2H). To determine the suppression capacity of MDSCs in EGFRvIII and EGFR-WT GBM bearing mice, we performed suppression assays in vitro. We observed significant reduction (EGFR-WT MDSC 70.9% ± 4.3, EGFRvIII MDSC 60.4% ± 4.2) in antigen-specific responses of splenocytes from OTI TCR-transgenic mice at 1:1 MDSC/splenocyte ratio (Fig. 2J), indicating that MDSCs from EGFR-WT and EGFRvIII GBM mice displayed similar suppressive capacity. Together these findings suggest that the high levels of PMN-MDSCs in EGFRvIII GBMs might play a role in mediating an immunosuppressive TIME by inhibiting the expansion of CD4+ T cells.
Flow cytometry analyses of TDLNs and spleens demonstrated similar quantitative changes in key immune cell populations (Supplementary Fig. S2D), confirming an influence of EGFRvIII GBM on systemic immunity. Analysis of T-cell effector function showed no significant changes in the number of Granzyme B+ CD8+ T cells or the levels of Granzyme B protein in both EGFR-WT and EGFRvIII GBM and TDLNs (Supplementary Fig. S2E). However, the ratio of CD8+ T cells to PMN-MDSCs, a measure of immunosuppression, decreased during tumor progression in EGFR-WT GBM but was constitutively low in EGFRvIII GBM (Fig. 2K). Contrary to previously reported observations in syngeneic glioma mouse models (48), we did not detect sexual dimorphism in PMN- or M-MDSC infiltration (Supplementary Fig. S2F).
We observed significantly reduced proportions of CD8+ effector memory (TEM) and central memory (TCM) T cells in EGFRvIII compared with EGFR-WT GBMs in both early- and late-stage disease (Fig. 2L). There were also lower fractions of CD4+ TCM in late EGFRvIII compared with EGFR-WT GBMs (Fig. 2L). In TDLNs of EGFRvIII GBM mice, we found significantly higher relative numbers of CD8+ TEM and TCM cells than in EGFR-WT (Fig. 2M) but a significant reduction in CD4+ TEM and TCM in late-stage disease when compared with EGFR-WT (Fig. 2M). Collectively, these results demonstrate that EGFRvIII GBMs have a highly immunosuppressive TIME characterized by elevated fractions of PMN-MDSCs and suppressed lower relative numbers of effector and memory T cells.
In tumor-bearing mice and patients with cancer, MDSCs are produced during cancer-driven emergency myelopoiesis, characterized by an increased output of undifferentiated myeloid cells from the bone marrow in response to cancer (49). To determine if the increase in PMN-MDSC observed in EGFRvIII GBMs was associated with a distinct profile of myeloid progenitors and mature myeloid cells in the bone marrow, we analyzed bone marrow from control nontumor-bearing mice, and EGFR-WT and EGFRvIII GBM-bearing mice. In EGFRvIII GBM-bearing mice, we observed a significant expansion of Lin− populations compared with control and EGFR-WT GBM-bearing mice (Supplementary Fig. S2G). This was not due to an increase of GMP or CMP, indicating a predominant expansion at earlier stages of myeloid progenitors and a more rapid output of Lin+ cells from the bone marrow, as determined by the increased fractions of CD45+CD11b+ cells in the spleen of EGFRVIII GBM-bearing mice (Supplementary Fig. S2H). Consistent with these data, the fractions of Lin+ cells in EGFRvIII GBM-bearing mice were significantly lower. In contrast, in bone marrow from EGFR-WT GBM-bearing mice, the Lin+ fractions dominated whereas the frequencies of Lin− populations, including GMP, were lower compared with control and EGFRvIII GBM mice. In addition, the spleens of EGFR-WT GBM mice had lower fractions of CD45+CD11b+ cells compared with EGFRvIII GBM (Supplementary Fig. S2H). Together these results indicate a higher production of myeloid progenitors and output of myeloid cells in EGFRvIII GBM-bearing mice.
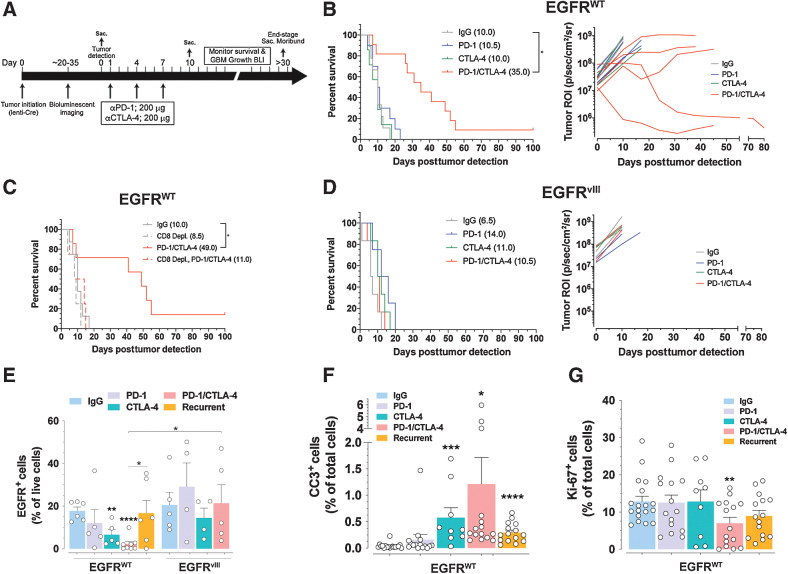
Checkpoint blockade treatment prolongs survival of EGFR-WT GBM mice
The immunosuppressive milieu of GBM is considered to be a major impediment to the clinical efficacy of immune checkpoint blockade therapy (8, 9). We leveraged the divergent immune compositions of EGFR-WT and EGFRvIII GBMs to analyze the efficacy of checkpoint blockade therapy. We used BLI to track GBM growth and initiation of treatment (Fig. 3A; Supplementary Fig. S3A) and mice were randomly enrolled into a four-arm treatment study with (i) control IgG, (ii) anti–PD-1, (iii) anti-CTLA-4, or (iv) anti–PD-1/anti-CTLA-4 combination treatment, after which response was followed by BLI and animals were monitored for survival (Fig. 3A). The majority of EGFR-WT GBM mice treated with combination therapy displayed responses consistent with sustained growth arrest and tumor regression, whereas single-agent treatments yielded no response by imaging (Fig. 3B; Supplementary Fig. S3A). Combination immune checkpoint blockade translated into a significant prolongation of survival in EGFR-WT GBM-bearing mice (Fig. 3B), with 10% of the treated animals surviving >100 days and tumor-free at termination. This survival benefit was dependent on CD8+ T cells because anti-CD8–mediated depletion eliminated the therapeutic effect (Fig. 3C; Supplementary Fig. S3B). Similar treatment of EGFRvIII GBM mice showed no inhibition of tumor growth (Fig. 3D; Supplementary Fig. S3A) and no significant prolongation of survival (Fig. 3D). We harvested GBMs, cervical TDLNs, and spleens from EGFR-WT and EGFRvIII GBM mice at the end of the third cycle of checkpoint blockade treatment, and upon recurrence of GBM growth, and analyzed their immune compositions by flow cytometry. Treatment of EGFR-WT mice with anti-CTLA-4 monotherapy and combination anti–PD-1/anti-CTLA-4 significantly decreased the number of EGFR+ tumor cells compared with IgG control (2.7 ± 1.2-fold and 7.9 ± 0.8-fold, respectively; Fig. 3E). This paralleled increases in apoptotic index (Fig. 3F; Supplementary Fig. S3C) and decreases in proliferative indexes (Fig. 3G; Supplementary Fig. S3C), cellular features that were annulled upon tumor recurrence (Fig. 3E–G). There were no decreases in EGFR+ tumor cells in EGFRvIII GBMs similarly treated, reflecting the absence of therapeutic benefit in GBMs bearing EGFRvIII mutation (Fig. 3E). These outcomes were independent from levels of expression of PD-1 and/or CTLA-4 on CD8+ T cells in GBMs, TDLNs, and spleens (Supplementary Fig. S3D).
Figure 3.
Combination PD-1/CTLA-4 checkpoint blockade promotes CD8+ T cell–dependent therapeutic response in EGFR-WT but not in EGFRvIII GBM. A, Treatment scheme of GBM mice. B, Left, Kaplan–Meier analysis of EGFR-WT GBM mice treated with indicated antibodies. Median survival (ms) in days. *, P = 0.0158, log-rank (Mantel–Cox) test. Right, BLI outputs over time of a subset of mice from left. C, Kaplan–Meier analysis of EGFR-WT mice treated with indicated antibodies. *, P = 0.0001, log-rank (Mantel–Cox) test. D, Left, Kaplan–Meier analysis of EGFRvIII GBM mice treated with indicated antibodies. Right, BLI outputs over time of a subset of mice from left. E, Flow cytometry of relative amounts of EGFR+ tumor cells in GBMs. Unbracketed; compared with IgG control. F and G, Relative levels of cleaved caspase 3 CC3; (F) and Ki-67 proliferative index (G) on GBM tissue from the indicated treatments. n = 3 GBMs and >3 serial sections stained for CC3 or Ki-67. Mean ± SEM of biological replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; unpaired t test, two-tailed.
Taken together with our findings of significantly lower numbers of CD8+ TEM and TCM in EGFRvIII GBM (Fig. 2K), this demonstrates that EGFRvIII creates a highly immunosuppressive TIME that negates the ability of combined PD-1 and CTLA-4 immune checkpoint blockade to induce antitumor immunity. Conversely, EGFR-WT GBM has a less immunosuppressive TIME that is sensitive to combination PD-1/CTLA-4 immune checkpoint blockade, capable of inducing a CD8+ T cell–dependent antitumor response.
Checkpoint blockade treatment reduces intratumoral PMN-MDSCs and BMDMs
To determine the cellular underpinnings of our results, we analyzed the immune profiles of GBMs, TDLNs, and spleens during treatment (Fig. 4A). In EGFR-WT GBMs, the proportions of PMN-MDSCs were decreased by CTLA-4 and combination PD-1/CTLA-4 immune checkpoint blockade (Fig. 4B). In EGFRvIII GBMs, the fractions of PMN-MDSCs were decreased by PD-1 and combination PD-1/CTLA-4 immune checkpoint blockade (Fig. 4B). The relative numbers of splenic PMN-MDSCs in EGFR-WT but not in EGFRvIII GBM-bearing mice were also significantly reduced by PD-1, CTLA-4, and combination PD-1/CTLA-4 immune checkpoint blockade (Fig. 4C), whereas no changes were observed in TDLNs (Supplementary Fig. S4A). Despite these decreases in PMN-MDSC in both GBM types, their relative numbers in EGFRvIII GBMs remained significantly higher than those in EGFR-WT GBMs (Fig. 4B).
Figure 4.
Changes in PMN-MDSCs and BMDMs parallels checkpoint blockade efficiency. A, Flow cytometry of the indicated immune cells in GBMs, TDLNs, and spleens of mice treated as indicated. EGFR-WT n = 10, 7, 6, 8 and EGFRvIII n = 9, 4, 4, 5 for IgG, PD-1, CTLA-4, and PD-1/CTLA-4 checkpoint blockade, respectively and n = 6 for recurrent EGFR-WT. B–D, Flow cytometry of PMN-MDSCs and BMDMs from GBM and spleen of mice treated as indicated. E, Ratio of CD8:PMN-MDSCs from GBMs treated as indicated. F and G, Flow cytometry of relative CD8+ and CD4+ TEM and TCM cell subset from GBM (F) and TDLNs (G) tissues of mice treated as indicated. Nonbracketed comparisons with IgG controls (B–G). Mean ± SEM of biological replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; unpaired t test, two-tailed.
Combination PD-1/CTLA-4 immune checkpoint blockade decreased relative numbers of BMDMs in EGFR-WT and EGFRvIII GBM (Fig. 4D); no effects in BMDM fractions were observed in the spleens or TDLN in any treatment group (Supplementary Figs. S4B and S4C). No changes were observed in CD8+, CD4+ T cells, Treg cells, M-MDSCs, and microglia in any treatment group (Supplementary Fig. S4D). Despite the lack of changes in CD8+ T cells, the decrease of PMN-MDSCs induced by combination PD-1/CTLA-4 immune checkpoint blockade in EGFR-WT mice translated into an increase in CD8:PMN-MDSCs ratio compared with control (Fig. 4E), suggesting a more immunologically active TIME after this combination therapy. This was not observed in EGFRvIII mice (Fig. 4E), which retained a strongly immunosuppressed TIME (high levels of PMN-MDSCs).
In EGFR-WT GBMs, the relative numbers of PMN-MDSCs and BMDMs increased upon recurrence (Fig. 4B–D), whereas no significant effects were observed in spleen or TDLN (Fig. 4C; Supplementary Figs. S4A–S4C). These changes translated into a decrease in the ratio of CD8:PMN-MDSCs in the tumor site (Fig. 4E). PD-1 and combination PD-1/CTLA-4 immune checkpoint blockade did not change the number of intratumoral GranzymeB+CD8+ T cells in both GBM subtypes (Supplementary Fig. S4E). In TDLN of EGFR-WT mice, we observed low fractions of GranzymeB+CD8+ T cells, which increased after combination PD-1/CTLA-4 treatment (Supplementary Fig. S4F). In contrast, in TDLNs of EGFRvIII mice, GranzymeB+CD8+ T cells were readily detected and were modestly increased by combination PD-1/CTLA-4 immune checkpoint blockade treatment (Supplementary Fig. S4F). Notably, GranzymeB+CD8+ T cells remained higher in EGFRvIII GBM than in EGFR-WT (Supplementary Figs. S4E and S4F), suggesting that CD8+ T cells are primed by tumor-associated antigens expressed in EGFRvIII GBM, although functional antitumor immunity is restrained.
As a result of these findings, we further investigated changes imposed on T-cell activation and expansion by GBM and checkpoint immunotherapy. Assessment of GBMs showed that EGFR-WT GBMs had proportionally low numbers of CD4+ and CD8+ TEM cells and these numbers readily increased in response to combination PD-1/CTLA-4 immune checkpoint blockade (CD8+ TEM 5.06 ± 0.64-fold and CD4+ TEM 42.6 ± 0.98-fold increases; Fig. 4F). In the TDLNs of these mice, the relative numbers of CD8+ TEM and TCM cells were upregulated, although to a lesser extent by combination PD-1/CTLA-4 immune checkpoint blockade (CD8+ TEM 2.26 ± 0.69-fold and CD8+ TCM 2.28 ± 0.7-fold increases; Fig. 4G). Similar increases were also observed in CD4+ TEM cells at the GBM site (Fig. 4F) as well as CD4+ TEM and TCM cells at the TDLN (Fig. 4G). Together these results suggest that in EGFR-WT GBM, after priming in TDLN, tumor-reactive TEM cells translocated to the tumor where they were able to expand after immunotherapy. In contrast, in EGFRvIII GBM, TDLN contained a high fraction of CD8+ TEM and TCM cells before treatment which further increased after immunotherapy (Fig. 4G). TDLN CD4+ TEM and TCM cells also relatively increased in number after PD-1 or combination PD-1/CTLA-4 immune checkpoint blockade (Fig. 4G). In these mice, intratumoral CD4+ and CD8+ TEM cells were sparce, and there was no increase under any treatment conditions (Fig. 4F), consistent with a TIME that imposes suppression on T-cell expansion.
These observations are consistent with enhanced T-cell priming in TDLN, indicative of a higher immunogenicity in EGFRvIII compared with EGFR-WT GBM. However, although CD4+ and CD8+ TEM and TCM cells were induced in TDLN in response to immunotherapy, they were significantly suppressed in the GBM microenvironment (Fig. 4F and G). The paucity of TEM and TCM cells and the lack of response to combination PD-1/CTLA-4 immune checkpoint blockade correlated with high levels of PMN-MDSC in EGFRvIII GBMs compared with EGFR-WT GBM in both early- and late-stage disease (Fig. 2E).
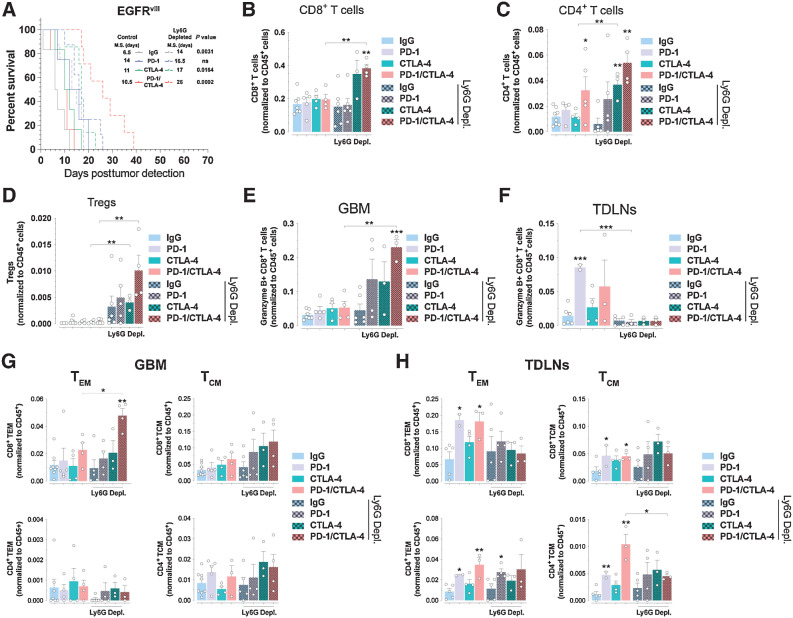
PMN-MDSC depletion sensitizes EGFRvIII GBM to checkpoint blockade treatments
We investigated whether PMN-MDSC were actively involved in the resistance to immune checkpoint blockade therapy in EGFRvIII GBMs. Anti-Ly6G immunodepletion in EGFRvIII mice (Supplementary Figs. S5A and S5B) did not change the levels of intratumoral CD45+ cells (Supplementary Fig. S5C) but improved survival in control, CTLA-4, and combination PD-1/CTLA-4 immune checkpoint blockade arms (Fig. 5A). Flow cytometry analysis showed no changes in the numbers of CD8+ T cells, CD4+ T cells, Treg cells, M-MDSCs, microglia, and BMDMs in control IgG-treated mice (Fig. 5B–D; Supplementary Fig. S5D). In contrast, we observed significant increases in the numbers of CD8+ and CD4+ T cells during combination PD-1/CTLA-4 immune checkpoint blockade (Fig. 5B–D), and increase in the numbers of CD4+ T cells after CTLA-4 blockade (Fig. 5C). PMN-MDSC depletion led to selective enrichment of intratumoral GranzymeB+CD8+ T cells with no changes in TDLNs (Fig. 5E and F). Depletion of PMN-MDSCs increased the numbers of intratumoral CD8+ TEM cells following combination immune checkpoint blockade (Fig. 5G), whereas no changes in the TDLN CD8+ and CD4+ T-cell subsets were observed (Fig. 5H). Collectively, these results indicate that PMN-MDSCs compromise the efficacy of combination PD-1/CTLA-4 immune checkpoint blockade by preventing the intratumoral expansion of primed T cells. Thus, elimination of PMN-MDSCs might represent a promising therapeutic option to sensitize GBMs to immune checkpoint blockade.
Figure 5.
PMN-MDSCs depletion sensitizes EGFR-driven GBMs to PD-1/CTLA-4 checkpoint blockade treatments. A, Kaplan–Meier analysis of EGFRvIII GBM mice treated as indicated. P values log-rank (Mantel–Cox) test. B–D, Relative numbers of CD8+ (B), CD4+ (C), and (D) regulatory T cells. E–H, Relative numbers of Granzyme B+ CD8+ T cells in GBMs (E) and TDLNs (F) and CD8+ and CD4+ T-cell subset from GBMs (G) and TDLNs (H) of PMN-MDSC (anti-Ly6G) depleted EGFRvIII mice treated as indicated. Nonbracketed comparisons with IgG controls (B–H). Mean ± SEM of biological replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; unpaired t test, two-tailed.
Distinct cytokine landscapes of EGFR-WT and EGFRvIII GBMs
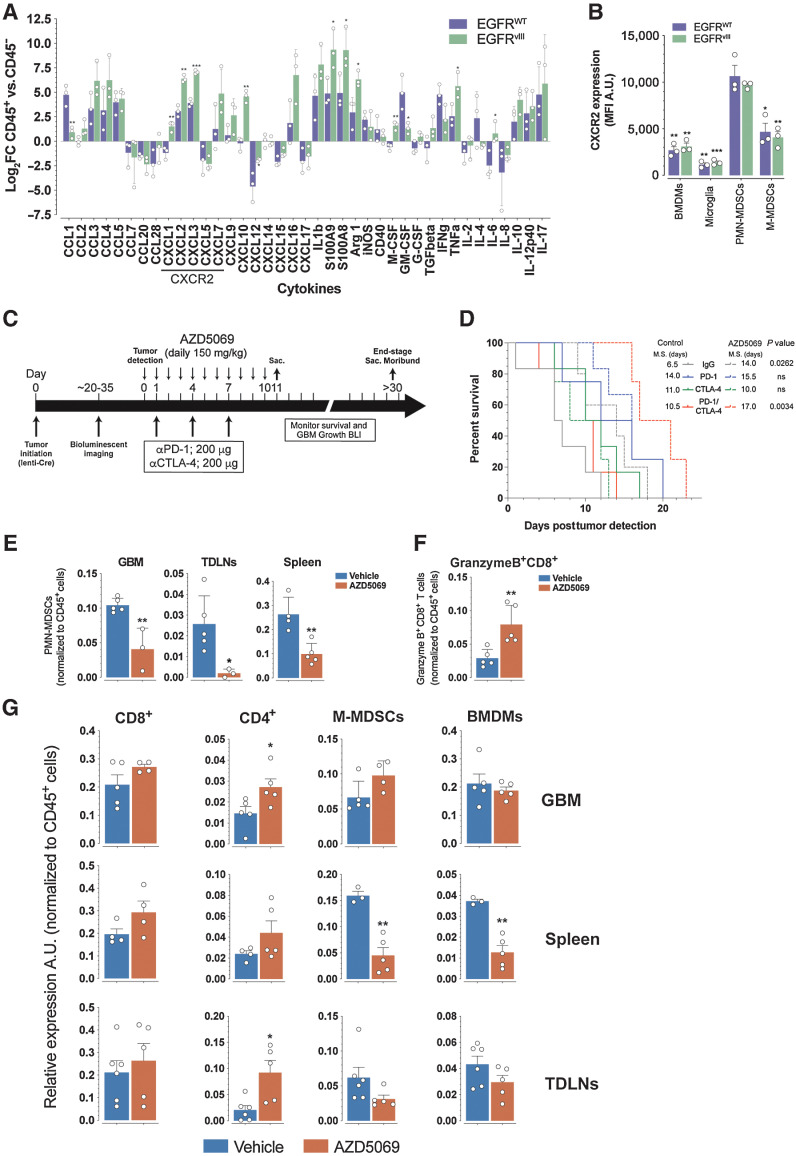
Because immune cells constitute ∼30% of GBM mass and represent an important source of cytokine production, we analyzed a panel of cytokines by RT-qPCR from EGFR-WT and EGFRvIII GBMs sorted CD45+ and CD45− cells. We found that the majority (27/39) of chemokines/cytokines were expressed at higher levels within the CD45+ immune fraction than the CD45− fraction in both GBM subtypes (Fig. 6A; Supplementary Fig. S6A). In particular, we observed significantly higher levels of CXCL1, CXCL2, and CXCL3 expression in EGFRvIII CD45+ cells when compared with EGFR-WT CD45+ cells (Fig. 6A; Supplementary Fig. S6A). CXCL1, CXCL2, and CXCL3 are ligands for the chemo-attractant neutrophil receptor CXCR2 (50). Flow cytometry analysis of CXCR2 expression in EGFR-WT and EGFRvIII GBM immune cells showed highest expression of CXCR2 on PMN-MDSCs when compared with other myeloid populations including BMDMs, microglia, and M-MDSCs (Fig. 6B; Supplementary Fig S6B). Combined with the higher expression of CXCL1, CXCL2, and CXCL3 in EGFRvIII (Fig. 6A; Supplementary Fig. S6A), these results offer a mechanistic basis for the higher number of PMN-MDSCs observed in EGFRvIII GBMs that provide these tumors with their intrinsic resistance to immune checkpoint blockade.
Figure 6.
EGFR WT and EGFRvIII GBMs have distinct cytokine profiles. A, Log2 fold change (FC) of CD45+/CD45− ratio from qRT PCR of the indicated cytokines from flow-sorted CD45+ and CD45− GBMs. B, Expression (mean fluorescent intensity MFI) of CXCR2 on the indicated cell types from GBMs. Comparisons of BMDMs, microglia, and M-MDSCs are to PMN-MDSCs. C, AZD5069 treatment schema of EGFRvIII mice. D, Kaplan–Meier analysis of EGFRvIII GBM mice treated with indicated treatments with or without AZD5069. M.S., median survival. P values log-rank (Mantel–Cox) test. E, Quantification of PMN-MDSCs in GBM, spleen, and TDLNs. F, Quantification of GranzymeB+CD8+ T cells in GBMs of control and AZD5069-treated mice. G, Quantification of the indicated cell types in GBM, spleen, and TDLNs from control and AZD5069 treated mice. Data are mean ± SEM of biological replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; unpaired t test, two-tailed.
Next, we pharmacologically targeted CXCR2 with AZD5069 (51, 52) in EGFRvIII GBM mice (Fig. 6C). In both control and combination PD-1/CTLA-4 immune checkpoint blockade treated animals, AZD5069 prolonged survival (Fig. 6D). In parallel, we observed no changes in the number of intratumoral CD45+ cells between AZD5069 treated and control animals (Supplementary Fig. S6C) while the levels of GBM PMN-MDSCs were reduced in AZD5069 treated (Fig. 6E). However, AZD5069 treatment markedly reduced PMN-MDSCs in TDLNs and to a lesser extent in the spleen of GBM-bearing mice (Fig. 6E), indicating a systemic decrease of PMN-MDSC, consistent with the established role of CXCR2 in regulating neutrophil homeostasis, as well as mobilization and trafficking from the bone marrow (53, 54). Concomitantly, AZD5069 treatment increased the fraction of intratumoral GranzymeB+CD8+ T cells (Fig. 6F), indicating that reduction of PMN-MDSC correlated with enhanced generation of cytolytic CD8+ TEF cells in response to immune checkpoint blockade. Importantly, we observed a significant increase in the relative number of intratumoral CD4+ T cells (Fig. 6G), which have a key role in supporting the survival, expansion, and function of cytolytic CD8+ T cells. There was also a clear trend for increased fractions of total intratumoral CD8+ T cells (Fig. 6G). AZD5069 treatment also induced a systemic decrease in the proportions of M-MDSC and BMDM detectable in the spleen of treated GBM EGFRvIII-tumor bearing mice, whereas no changes of these cell populations were observed at the tumor area including GBM and TDLN (Fig. 6G). Collectively, these results demonstrate that inhibition of PMN-MDSC recruitment from the bone marrow by AZD5069 treatment in EGFRvIII GBM mice caused a significant reduction in the relative numbers of PMN-MDSCs systemically and at the tumor region, namely TDLN, combined with a concomitant increase in the fractions of CD4+ and CD8+ T cells (Fig. 6G), and an increase of intratumoral cytolytic CD8+ T cells (Fig. 6F). Under these conditions, combination PD-1/CTLA-4 immune checkpoint blockade could induce antitumor immunity and extend survival.
Discussion
Despite recent advances in GBM multi-omics, technical limitations prevent using patient material to decode mechanisms underlying the development of the TIME. Among the challenges, human GBMs are almost universally diagnosed at advanced stages, thus preventing assessment of the immunologic landscape and evaluation of therapeutic efficacy of immune checkpoint inhibition at early versus late stages of human GBM. Here, we used genetic mouse models to overcome this limitation and obtain knowledge about GBM immune composition, its longitudinal evolution, and its role in shaping response to immunotherapy. By comparing TIME in early and late stages of GBM, we observed increases in myeloid infiltration over time, especially changes in PMN-MDSCs and BMDMs, revealing a gradual rise in immunosuppressive immune components that precluded T-cell activation and CTL differentiation. These findings unequivocally support the conclusion that at the time of diagnosis in patients, the GBM immune microenvironment is highly immunosuppressive and has already established a dysfunctional CD8+ T-cell state. Under these conditions, efficacy of immune checkpoint therapy is impaired, explaining why this treatment modality has only limited success in GBM, in contrast to several other cancer types, in which it is highly effective.
We observed discernable histopathologic characteristics and distinct signaling properties between EGFR-WT and EGFRvIII GBMs. We determined that EGFRvIII GBM induced a highly immunosuppressive environment primarily due to the accumulation of PMN-MDSC. Under these conditions, combination PD-1/CTLA-4 immune checkpoint blockade was unable to drive T-cell activation and expansion or confer protective antitumor immunity. Depleting PMN-MDSCs released the therapeutic effect of immune checkpoint blockade that was restrained by these immunosuppressive cells, and resulted in a prolongation of survival in EGFRvIII GBM. Our results set a paradigm of two-fold clinical relevance. First, our studies provide evidence that distinct cancer driver mutations, in the same cancer type, differentially alter the TIME in a manner that has a decisive impact on the outcome of checkpoint immunotherapy. Second, our findings reveal that tumors resistant to immune checkpoint blockade can be rendered amenable to the therapeutic benefit of T-cell intrinsic immune checkpoint blockade by concomitant interventions to release additional breaks mediated by cellular checkpoint components of the TIME, such as MDSC, which impose cell-extrinsic inhibitory effects on T-cell expansion.
An unexpected finding was that EGFR-WT and EGFRvIII GBMs not only had distinct intratumoral distribution of activated T cells, but also had TDLN distribution of activated T cells. In EGFR-WT GBMs, tumors contained mostly CD8+ cells with a TEM and TCM phenotype, whereas these T-cell subsets were very low in relative number in TDLN. In contrast, in EGFRvIII GBM, TDLN tumors contained a high fraction of CD8+ TEM and TCM cells, whereas these cell populations were sparce in the intratumoral space. These findings are indicative of a high ability of EGFRvIII GBM to induce T-cell priming in TDLN, where DCs induce presentation of tumor antigen and cross-priming of tumor-specific T cells (55). This may be mediated by the EGFRvIII mutation, which might serve as a neoantigen itself, or as an inducer of other neoantigens thereby leading to oligoclonal or polyclonal expansion of activated tumor-specific CD8+ T cells in TDLN. The high numbers of activated CD8+ T cells in the TDLN and the limited fractions of these activated CD8+ T cells in the intratumoral area are consistent with a TIME that imposes suppression of T effector–cell survival or expansion. Indeed, our studies revealed that, in both early- and late-stage disease, EGFRvIII-bearing GBM contained higher fractions of PMN-MDSC, a well-known immunosuppressive population, than EGFR-WT GBM. The higher numbers of PMN-MDSC in EGFRvIII GBM correlated with low numbers of CD4+ T cells, which have an instrumental role in supporting the survival and function of CD8+ cytolytic T cells. Intriguingly, the paucity of CD4+ and CD8+ TEM and TCM cells makes the GBM TIME reminiscent of a “cold” tumor because the targets of immune checkpoint blockade are missing from this microenvironment.
Our results showed that combination PD-1/CTLA-4 but not single immune checkpoint blockade induced CD8+ T-cell activation and effector differentiation alongside reduction in the fractions of PMN-MDSCs. The mechanisms underlying the effects of immune inhibition of PD-1 and CTLA-4 are inherently different (56, 57) and act on specific cells in anatomically defined locations. CTLA-4 blockade is thought to act mainly in lymphoid organs during T-cell receptor (TCR) engagement by recognition of tumor antigens and initiation of immune activation. In contrast, PD-1 blockade is thought to act predominantly on preactivated T cells, to promote Ras and PI3K/Akt signaling, cell-cycle progression, cytokine release, and metabolic reprogramming (58–63). In addition, PD-1 regulates differentiation, fate commitment, and output of myeloid cells from the bone marrow during emergency myelopoiesis (19). Consistent with the distinct but synergistic roles of these checkpoint inhibitors, our results showed that combination PD-1/CTLA-4 but not single immune checkpoint blockade induced reduction in the relative numbers of PMN-MDSCs alongside with CD8+ T-cell activation and effector differentiation. Notably, we achieved prolongation of survival using combination PD-1/CTLA-4 immune checkpoint blockade in EGFR-WT but not in EGFRvIII because of a lesser immunosuppressive environment in the former.
The higher immunosuppressive nature of the EGFRvIII GBM microenvironment was due to the greater infiltration by PMN-MDSCs in both early and late disease stages. The central role of PMN-MDSC in shaping the immunosuppressive TIME of EGFRvIII GBM is supported by our finding that depletion of PMN-MDSC induced significant prolongation of survivals.
Our studies also demonstrated that the increased numbers of PMN-MDSCs in EGFRvIII GBMs are associated with higher expression of CXCR2 ligands, namely CXCL1, CXCL2, and CXCL3. We documented a role for the CXCL1/2/3:CXCR2 axis in the immunosuppressive properties of EGFRvIII GBM by pharmacologic inhibition of CXCR2, which diminished the systemic levels of PMN-MDSC and sensitized GBMs to the therapeutic benefit of checkpoint blockade.
It should be noted that we only observed a modest decrease in the relative numbers of intratumoral PMN-MDSC during CXCR2 inhibition, perhaps reflecting a requirement for a longer treatment schedule, but we did observe an expansion of intratumoral CD4+ and cytotoxic CD8+ T cells. Together these results indicate that CXCR2 antagonism diminished the immunosuppressive TIME of EGFRvIII GBM. These data are consistent with prior reports that inhibition of CXCR2 reduces tumor MDSC infiltration and tumor growth in cancer models (64–67) and improved the efficacy of PD-1 immune checkpoint blockade (68, 69).
Relationships between neoplastic cell somatic mutations and alterations in TIME composition in glioma are slowly emerging (43, 70) and might have a decisive impact on the efficacy of checkpoint immunotherapy. Our present work revealed a previously unappreciated relationship among EGFR mutation status, GBM TIME composition, and response to combination immune checkpoint blockade, pointing to patient stratification for checkpoint blockade therapy based on integrated genotypic and immunological profiles, and to combinatorial targeting of TIME and checkpoint inhibitors for the treatment of GBM.
Supplementary Material
supplementary figures & legends
Supplementary Table 1
Supplementary table 2
Acknowledgments
NIH grants R01 CA 185137 (to S.P. Gygi, A. Charest), R01 CA229784 (to V.A. Boussiotis, A. Charest), and R01 CA238263 (to V.A. Boussiotis).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Contributions
A.T. Yeo: Conceptualization, formal analysis, investigation, writing–original draft. R. Shah: Formal analysis, investigation. K. Aliazis: Formal analysis, investigation. R. Pal: Investigation. T. Xu: Formal analysis, investigation, methodology. P. Zhang: Formal analysis, investigation. S. Rawal: Investigation. C.M. Rose: Investigation. F.S. Varn: Investigation. V.A. Appleman: Investigation. J. Yoon: Formal analysis. H. Varma: Formal analysis. S.P. Gygi: Supervision. R.G. Verhaak: Formal analysis. V.A. Boussiotis: Formal analysis, supervision, funding acquisition, validation, methodology, writing–original draft, writing–review and editing. A. Charest: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Veglia F, Sanseviero E, Gabrilovich DI. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol 2021;21:485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bronte V, Brandau S, Chen S-H, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun 2016;7:12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grover A, Sanseviero E, Timosenko E, Gabrilovich DI. Myeloid-derived suppressor cells: a propitious road to clinic. Cancer Discov 2021;11:2693–706. [DOI] [PubMed] [Google Scholar]
- 4. Weber R, Fleming V, Hu X, Nagibin V, Groth C, Altevogt P, et al. Myeloid-derived suppressor cells hinder the anti-cancer activity of immune checkpoint inhibitors. Front Immunol 2018;9:1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155:462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis M, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang EJ, Chen JS, Jain S, Morshed RA, Haddad AF, Gill S, et al. Immunotherapy resistance in glioblastoma. Front Genet 2021;12:750675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jackson CM, Choi J, Lim M. Mechanisms of immunotherapy resistance: lessons from glioblastoma. Nat Immunol 2019;20:1100–9. [DOI] [PubMed] [Google Scholar]
- 10. Zhu H, Acquaviva J, Ramachandran P, Boskovitz A, Woolfenden S, Pfannl R, et al. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc Natl Acad Sci U S A 2009;106:2712–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jun HJ, Acquaviva J, Chi D, Lessard J, Zhu H, Woolfenden S, et al. Acquired MET expression confers resistance to EGFR inhibition in a mouse model of glioblastoma multiforme. Oncogene 2012;31:3039–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Acquaviva J, Jun HJ, Lessard J, Ruiz R, Zhu H, Donovan M, et al. Chronic activation of wild-type epidermal growth factor receptor and loss of Cdkn2a cause mouse glioblastoma formation. Cancer Res 2011;71:7198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yeo AT, Jun HJ, Appleman VA, Zhang P, Varma H, Sarkaria JN, et al. EGFRvIII tumorigenicity requires PDGFRA co-signaling and reveals therapeutic vulnerabilities in glioblastoma. Oncogene 2021; 40:2682–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lesche R, Groszer M, Gao J, Wang Y, Messing A, Sun H, et al. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis 2002;32:148–9. [DOI] [PubMed] [Google Scholar]
- 15. Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell 1996;85:27–37. [DOI] [PubMed] [Google Scholar]
- 16. Woolfenden S, Zhu H, Charest A. A Cre/LoxP conditional luciferase reporter transgenic mouse for bioluminescence monitoring of tumorigenesis. Genesis 2009;47:659–66. [DOI] [PubMed] [Google Scholar]
- 17. Charest A, Wilker EW, McLaughlin ME, Lane K, Gowda R, Coven S, et al. ROS fusion tyrosine kinase activates a SH2 domain-containing phosphatase-2/phosphatidylinositol 3-kinase/mammalian target of rapamycin signaling axis to form glioblastoma in mice. Cancer Res 2006;66:7473–81. [DOI] [PubMed] [Google Scholar]
- 18. Jun HJ, Appleman VA, Wu HJ, Rose CM, Pineda JJ, Yeo AT, et al. A PDGFRalpha-driven mouse model of glioblastoma reveals a stathmin1-mediated mechanism of sensitivity to vinblastine. Nat Commun 2018;9:3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Strauss L, Mahmoud MAA, Weaver JD, Tijaro-Ovalle NM, Christofides A, Wang Q, et al. Targeted deletion of PD-1 in myeloid cells induces antitumor immunity. Sci Immunol 2020;5:eaay1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, Seed B. A comprehensive collection of experimentally validated primers for polymerase chain reaction quantitation of murine transcript abundance. BMC Genomics 2008;9:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res 2010;38(Database issue):D792–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res 2003;31:e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Varn FS, Johnson KC, Wade TE, Malta TM, Sabedot TS, Barthel FP, et al. Longitudinal analysis of diffuse glioma reveals cell state dynamics at recurrence associated with changes in genetics and the microenvironment. Biorxiv 2021:2021.05.03.442486. [Google Scholar]
- 24. Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37:773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson KC, Anderson KJ, Courtois ET, Gujar AD, Barthel FP, Varn FS, et al. Single-cell multimodal glioma analyses identify epigenetic regulators of cellular plasticity and environmental stress response. Nat Genet 2021;53:1456–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Erickson BK, Jedrychowski MP, McAlister GC, Everley RA, Kunz R, Gygi SP. Evaluating multiplexed quantitative phosphopeptide analysis on a hybrid quadrupole mass filter/linear ion trap/orbitrap mass spectrometer. Anal Chem 2015;87:1241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Isasa M, Rose CM, Elsasser S, Navarrete-Perea J, Paulo JA, Finley DJ, et al. Multiplexed, proteome-wide protein expression profiling: yeast deubiquitylating enzyme knockout strains. J Proteome Res 2015;14:5306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods 2011;8:937–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McAlister GC, Nusinow DP, Jedrychowski MP, Wuhr M, Huttlin EL, Erickson BK, et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem 2014;86:7150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 2010;143:1174–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 1994;5:976–89. [DOI] [PubMed] [Google Scholar]
- 32. Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 2007;4:207–14. [DOI] [PubMed] [Google Scholar]
- 33. Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol 2006;24:1285–92. [DOI] [PubMed] [Google Scholar]
- 34. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 35. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. An Z, Aksoy O, Zheng T, Fan QW, Weiss WA. Epidermal growth factor receptor and EGFRvIII in glioblastoma: signaling pathways and targeted therapies. Oncogene 2018;37:1561–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. B WCoTE. Central Nervous System Tumours WHO Classification of Tumours, 5th Edition, Volume6. Lyon: IARC Publications; 2021. [Google Scholar]
- 38. Black LE, Longo JF, Carroll SL. Mechanisms of receptor tyrosine-protein kinase ErbB-3 (ERBB3) action in human neoplasia. Am J Pathol 2019;189:1898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kancha RK, von Bubnoff N, Duyster J. Asymmetric kinase dimer formation is crucial for the activation of oncogenic EGFRvIII but not for ERBB3 phosphorylation. Cell Commun Signal 2013;11:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jiang X, Huang F, Marusyk A, Sorkin A. Grb2 regulates internalization of EGF receptors through clathrin-coated pits. Mol Biol Cell 2003;14:858–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sorkin A, Helin K, Waters CM, Carpenter G, Beguinot L. Multiple autophosphorylation sites of the epidermal growth factor receptor are essential for receptor kinase activity and internalization. Contrasting significance of tyrosine 992 in the native and truncated receptors. J Biol Chem 1992;267:8672–8. [PubMed] [Google Scholar]
- 42. Wellenstein MD, de Visser KE. Cancer-Cell-Intrinsic Mechanisms Shaping the Tumor Immune Landscape. Immunity 2018;48:399–416. [DOI] [PubMed] [Google Scholar]
- 43. Chen Z, Herting CJ, Ross JL, Gabanic B, Puigdelloses Vallcorba M, Szulzewsky F, et al. Genetic driver mutations introduced in identical cell-of-origin in murine glioblastoma reveal distinct immune landscapes but similar response to checkpoint blockade. Glia 2020;68:2148–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 2018;1711:243–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Steen CB, Liu CL, Alizadeh AA, Newman AM. Profiling cell type abundance and expression in bulk tissues with CIBERSORTx. Methods Mol Biol 2020;2117:135–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 2017;32:42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yeo AT, Rawal S, Delcuze B, Christofides A, Atayde A, Strauss L, et al. Single-cell RNA sequencing reveals evolution of immune landscape during glioblastoma progression. Nat Immunol 2022;23:971–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bayik D, Zhou Y, Park C, Hong C, Vail D, Silver DJ, et al. Myeloid-derived suppressor cell subsets drive glioblastoma growth in a sex-specific manner. Cancer Discov 2020;10:1210–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol 2012;12:253–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gouerou H, Crespon B, Favriel JM, Debray C. [Endoscopic retrograde duodenocholangiopancreatography. An effective and relatively safe diagnostic method]. Nouv Presse Med 1978;7:2876. [PubMed] [Google Scholar]
- 51. Kirsten AM, Forster K, Radeczky E, Linnhoff A, Balint B, Watz H, et al. The safety and tolerability of oral AZD5069, a selective CXCR2 antagonist, in patients with moderate-to-severe COPD. Pulm Pharmacol Ther 2015;31:36–41. [DOI] [PubMed] [Google Scholar]
- 52. Cullberg M, Arfvidsson C, Larsson B, Malmgren A, Mitchell P, Wahlby Hamren U, et al. Pharmacokinetics of the oral Selective CXCR2 antagonist AZD5069: a summary of eight Phase I studies in healthy volunteers. Drugs R D 2018;18:149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mei J, Liu Y, Dai N, Hoffmann C, Hudock KM, Zhang P, et al. Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. J Clin Invest 2012;122:974–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kohler A, De Filippo K, Hasenberg M, van den Brandt C, Nye E, Hosking MP, et al. G-CSF-mediated thrombopoietin release triggers neutrophil motility and mobilization from bone marrow via induction of Cxcr2 ligands. Blood 2011;117:4349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dammeijer F, van Gulijk M, Mulder EE, Lukkes M, Klaase L, van den Bosch T, et al. The PD-1/PD-L1-checkpoint restrains T cell immunity in tumor-draining lymph nodes. Cancer Cell 2020;38:685–700. [DOI] [PubMed] [Google Scholar]
- 56. Boussiotis VA, Charest A. Immunotherapies for malignant glioma. Oncogene 2018;37:1121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ott PA, Hodi FS, Kaufman HL, Wigginton JM, Wolchok JD. Combination immunotherapy: a road map. J Immunother Cancer 2017;5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Riley JL. PD-1 signaling in primary T cells. Immunol Rev 2009;229:114–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ahn E, Araki K, Hashimoto M, Li W, Riley JL, Cheung J, et al. Role of PD-1 during effector CD8 T cell differentiation. Proc Natl Acad Sci U S A 2018;115:4749–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Patsoukis N, Brown J, Petkova V, Liu F, Li L, Boussiotis VA. Selective effects of PD-1 on Akt and Ras pathways regulate molecular components of the cell cycle and inhibit T cell proliferation. Sci Signal 2012;5:ra46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Patsoukis N, Li L, Sari D, Petkova V, Boussiotis VA. PD-1 increases PTEN phosphatase activity while decreasing PTEN protein stability by inhibiting casein kinase 2. Mol Cell Biol 2013;33:3091–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Patsoukis N, Bardhan K, Chatterjee P, Sari D, Liu B, Bell LN, et al. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat Commun 2015;6:6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Boussiotis VA, Patsoukis N. Effects of PD-1 signaling on immunometabolic reprogramming. Immunometabolism 2022;4:e220007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Taki M, Abiko K, Baba T, Hamanishi J, Yamaguchi K, Murakami R, et al. Snail promotes ovarian cancer progression by recruiting myeloid-derived suppressor cells via CXCR2 ligand upregulation. Nat Commun 2018;9:1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bezzi M, Seitzer N, Ishikawa T, Reschke M, Chen M, Wang G, et al. Diverse genetic-driven immune landscapes dictate tumor progression through distinct mechanisms. Nat Med 2018;24:165–75. [DOI] [PubMed] [Google Scholar]
- 66. Di Mitri D, Mirenda M, Vasilevska J, Calcinotto A, Delaleu N, Revandkar A, et al. Re-education of tumor-associated macrophages by CXCR2 blockade drives senescence and tumor inhibition in advanced prostate cancer. Cell Rep 2019;28:2156–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang G, Lu X, Dey P, Deng P, Wu CC, Jiang S, et al. Targeting YAP-dependent MDSC infiltration impairs tumor progression. Cancer Discov 2016;6:80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Highfill SL, Cui Y, Giles AJ, Smith JP, Zhang H, Morse E, et al. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci Transl Med 2014;6:237ra67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Najjar YG, Rayman P, Jia X, Pavicic PG Jr, Rini BI, Tannenbaum C, et al. Myeloid-derived suppressor cell subset accumulation in renal cell carcinoma parenchyma is associated with intratumoral expression of IL1beta, IL8, CXCL5, and Mip-1alpha. Clin Cancer Res 2017;23:2346–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Magod P, Mastandrea I, Rousso-Noori L, Agemy L, Shapira G, Shomron N, et al. Exploring the longitudinal glioma microenvironment landscape uncovers reprogrammed pro-tumorigenic neutrophils in the bone marrow. Cell Rep 2021;36:109480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary figures & legends
Supplementary Table 1
Supplementary table 2
Data Availability Statement
The data generated in this study are available in the article and its supplementary data files or upon request from the corresponding authors.