Abstract
Pneumococcal surface protein A (PspA) is a serologically variable protein of Streptococcus pneumoniae. Twenty-four diverse alleles of the pspA gene were sequenced to investigate the genetic basis for serologic diversity and to evaluate the potential of diversity to have an impact on PspA's use in human vaccination. The 24 pspA gene sequences from unrelated strains revealed two major allelic types, termed “families,” subdivided into clades. A highly mosaic gene structure was observed in which individual mosaic sequence blocks in PspAs diverged from each other by over 20% in many cases. This level of divergence exceeds that observed for blocks in the penicillin-binding proteins of S. pneumoniae or in many cross-species comparisons of gene loci. Conversely, because the mosaic pattern is so complex, each pair of pspA genes also has numerous shared blocks, but the position of conserved blocks differs from gene pair to gene pair. A central region of pspA, important for eliciting protective antibodies, was found in six clades, which each diverge from the other clades by >20%. Sequence relationships among the 24 alleles analyzed over three windows were discordant, indicating that intragenic recombination has occurred within this locus. The extensive recombination which generated the mosaic pattern seen in the pspA locus suggests that natural selection has operated in the history of this gene locus and underscores the likelihood that PspA may be important in the interaction between the pneumococcus and its human host.
Streptococcus pneumoniae is a formidable human pathogen responsible for a major portion of over 3 million deaths worldwide of children from pneumonia and meningitis (22). Recent increases in the rate of isolation of pneumococci resistant to antibiotics promote the expectation that morbidity and mortality caused by this pathogen will increase in the near future. The impact on health is greatest in very young children, elderly individuals, sickle cell patients, and immunocompromised persons of all ages (4). Currently available vaccines are based on immunity to capsular polysaccharides of the pneumococcus, of which 90 serotypes exist. A 23-valent polysaccharide vaccine, recommended for use in adults, is not effective for a large number of the at-risk individuals, including children less than 2 years of age, who are not yet capable of responding adequately to polysaccharide antigens (14, 29). A 7-valent conjugated polysaccharide vaccine was recently licensed for use in children (53), but nonvaccine serotypes will be likely to cause substantial pneumococcal disease even in vaccinated individuals (26, 27). Pneumococcal proteins either in addition to polysaccharides or as stand-alone vaccines have the potential to better protect those for whom the current vaccine is ineffective (5, 7, 45). PspA, which has been shown to effect antibody-mediated protection in mouse models of pneumococcal disease (38) and to be safe in administration to humans (43), is one such candidate.
PspA is an important virulence factor of the pneumococcus (63) that influences bacterium-host interactions through interference with the fixation of complement C3 (61). PspA also binds human lactoferrin (24). PspA is present on all pneumococcal strains and is serologically variable (15). Mouse models have shown that cross-reactive anti-PspA antisera are also cross-protective (reviewed in references 8, 9, 60). Human antisera from a phase I vaccine trial were also competent for protecting mice from pneumococcal infection by challenge strains of various PspA types (5a).
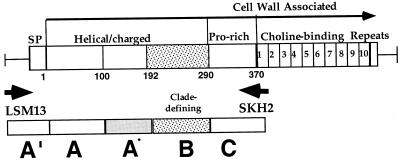
Basic information about protein structural domains in PspA comes from the DNA sequence of pspA/Rx1 (gene/strain name [described below]) and pspA/EF5668 (28, 36, 62, 63). There are five domains (see Fig. 1), including (i) a signal peptide, (ii) an α-helical and charged domain that bears a strong 7-residue periodicity typical of coiled-coil proteins (amino acids 1 to 288), (iii) a proline-rich region (amino acids 289 to 370), (iv) a choline-binding domain consisting of 10 20-amino-acid repeats (amino acids 371 to 571), and (v) a C-terminal 17-amino-acid tail (amino acids 572 to 589). The large choline-binding repeat domain is required for the attachment of PspA on the pneumococcal cell surface via interaction with choline in membrane-associated lipoteichoic acid (64). This orientation results in the α-helical or charged domain of PspA being exposed on the surface and thus available to interact with the human host (8, 20).
FIG. 1.
Modular pspA gene showing windows of the sequence analyzed. Major domains of PspA are indicated above the line drawing. Within the α-helical or charged domain, a clade-defining region of the PspA molecule is indicated by the stippled box. Choline-binding repeats are numbered 1 to 10. The small numbers directly below the gene are equivalent to amino acid residue numbers based on PspA/Rx1, the prototype PspA molecule previously sequenced (62). Arrows LSM13 and SKH2 indicate two primers used for PCR. Boxes A′, A, A*, B, and C indicate windows of sequence for analysis.
The combinatorial serological diversity of monoclonal antibody (MAb)-detected epitopes on PspA proteins from different strains (15) coupled with the presence of one major chromosomal locus encoding PspA suggested that PspA proteins might be mosaics. Mosaic gene alleles in bacteria are formed by recombination following horizontal gene transfer (34, 35). The term “mosaic” derives from the pattern of interspersed blocks of nucleotide sequence which have different evolutionary histories, but are found combined in the resulting gene allele subsequent to recombination events (41). Recombination follows the horizontal transfer of gene segments mediated by transformation, transduction, conjugation, or other means; the recombined segments can be derived from other strains in the same species or from other more distant bacterial relatives (35, 40).
The most intensively studied mosaic alleles in S. pneumoniae have been those encoding several penicillin-binding proteins (PBPs), including 1a, 2x, and 2b, in which the mosaic blocks have often been identified as resulting from an interspecies DNA transfer event (18, 23, 52, 54, 65; M. C. Maiden, B. Malorny, and M. Achtman, Letter, Mol. Microbiol. 21:1297–1298, 1996). If pneumococci are capable of between-species horizontal transfer, they, in all likelihood, undergo even more frequent within-species horizontal gene transfer that could contribute to the development of mosaic alleles. The transfer of capsule cassettes (16) is one example of an intraspecies horizontal gene transfer, and these events have recently been documented in vivo (12, 13, 31). Both interspecies and intraspecies horizontal gene transfers are facilitated in the pneumococci because of the widespread capability for natural transformation within this species (49, 51).
In this study, the pspA alleles examined were from a group of 24 diverse clinical pneumococcal isolates. These alleles were sequenced, revealing that pspA genes and PspA proteins have a highly complex mosaic structure. This mosaic diversity was examined in the light of its potential cross-reactive immunogenicity for use in protein-based vaccines to protect children and adults from this pathogen.
MATERIALS AND METHODS
DNA isolation, primers, and gene-specific PCR.
Pneumococcal strains (see Table 1 and Results for description) were inoculated from agar plates containing 5% sheep blood into 15-ml cultures of Todd-Hewitt broth with 0.5% added yeast extract, and cells were harvested after only a few hours of growth. Chromosomal DNA for each strain was isolated by a modification of the genomic DNA procedure of Promega.
TABLE 1.
Strains chosen for pspA sequencing
| Strain namea | Capsule type | PspA typeb | Yr isolated | Site of origin | Accession no. |
|---|---|---|---|---|---|
| Clade 1 | |||||
| BG9739 | 4 | 26 | 1988 | Alabama | AF071804 |
| DBL6A | 6A | 19 | <1980 | Unknownc | AF071805 |
| L81905 | 4 | 23 | 1984 | Alaska | AF071809 |
| BG8743 | 23 | NDe | 1987 | Alabama | AF071803 |
| AC94 | 9L | 0 | 1987 | Canada | AF071802 |
| BG6692 | 33 | 16 | 1984 | Alabama | AF071808 |
| BG8838 | 6 | 27 | 1987 | Alabama | AF071807 |
| DBL1 | 6B | 0 | <1980 | Unknownc | AF071806 |
| Clade 2 | |||||
| Rx1 | Rough | 25 | 1940s | United States | M74122 |
| E134 | 23 | ND | 1994 | Alabama | AF071811 |
| EF10197 | 3 | 18 | <1985 | Sweden | AF071812 |
| EF6796 | 6A | 1 | <1985 | Sweden | AF071813 |
| BG9163 | 6B | 21 | 1987 | Alabama | AF071815 |
| DBL5 | 5 | 33 | <1980 | Unknownd | AF071810 |
| WU2 | 3 | 1 | 1966 | Missouri | AF071814 |
| Clade 3 | |||||
| EF3296 | 4 | 20 | <1985 | Sweden | AF071816 |
| BG8090 | 19 | 16 | 1986 | Alabama | AF071817 |
| AC122 | 9V | 18 | 1984 | Canada | AF071818 |
| Clade 4 | |||||
| EF5668 | 4 | 12 | <1985 | Sweden | U89711 |
| BG7561 | 15 | ND | 1985 | Alabama | AF071824 |
| BG7817 | 12 | ND | 1986 | Alabama | AF071826 |
| BG11703 | 18 | ND | 1992 | Alabama | AF071821 |
| Clade 5 | |||||
| ATCC6303 | 3 | 7 | 1940s | Unknown | AF071820 |
| Clade 6 | |||||
| BG6380 | 37 | ND | 1984 | Alabama | AF071823 |
PspA clade designations are from this paper.
Based on typing with MAbs (15).
From B. Gray, origin unknown.
From S. Szu, origin unknown.
ND, not determined.
By using two oligonucleotide primers, LSM13 (in the promoter region of Rx1) and SKH2 (in the first of the 10 C-terminal repeats), a DNA fragment of about 1,200 to 1,800 bp corresponding to the pspA gene was amplified from nearly all pneumococcal strains (Fig. 1). Because the site for LSM13 is in the region of DNA immediately upstream of pspA, which is highly conserved in all pneumococcal strains (59), this particular PCR-generated fragment derives from the pspA chromosomal locus. The sequence of primer LSM13 is 5′-GCAAGCTTATGATATAGAAATTTGTAAC-3′, and that of primer SKH2 is 5′-CCACATACCGTTTTCTTGTTTCCAGCC-3′.
PCRs were carried out in a standard PCR mixture of 50 μl containing 2.5 mM MgCl2, 200 μM (each) deoxynucleoside triphosphates (dNTPs), 50 pmol of each primer, and 2.5 U of Taq DNA polymerase. Cycling consisted of 95°C for 1 min, 62°C for 1 min, and 72°C for 3 min, repeated 30 times. Before sequencing, PCR products were purified from agarose gels by using Microcon gel nebulizer filters or were treated with shrimp alkaline phosphatase and exonuclease I (25).
Automated DNA sequence analysis and DNA sequencing strategy.
DNA sequencing was performed by directly sequencing the LSM13 and SKH2 PCR-generated DNA fragments. Automated sequencing reactions used dye terminator chemistry and were run on an Applied Biosystems model ABI Prism 377 sequencer. Initial sequence runs for each strain used one of the two primers from the initial PCR amplification of the pspA gene—either LSM13 or SKH2. The sequence was extended and also confirmed by sequence runs in the opposite direction through the use of additional primers that were designed based on the initial sequence data for each gene. Approximately 40 additional primers were necessary to fully assemble the sequence of the 24 distinct pspA genes (sequences of primers are available upon request). The total length of the sequence data examined for each gene was >1,100 bp.
DNA sequence alignment.
Sequence data for each strain's pspA PCR-generated fragments were assembled and edited by using Sequencher (GeneCodes, Inc.). Further editing, alignment, and additional analysis were performed with MacVector DNA sequence analysis software (Oxford Molecular). The protein alignment presented was generated by the Clustal W algorithm by using the Blosum30 amino acid-scoring matrix. Distance calculations given in Fig. 2 and 4 were those calculated by MacVector, but equivocal distances were found for alternative alignments in other programs. Because variation is high, simple distances were used. The graph of the distribution of distances was generated by using Microsoft Excel.
FIG. 2.
Pairwise comparisons among pspA genes and PspA proteins. Below the diagonal, values represent percent DNA identity. DNA comparisons are highlighted white for values less than 60%, light gray for values between 60 and 75%, and dark gray for values greater than 75%. Above the diagonal, values represent percent protein identity. Protein comparisons are highlighted white for values less than 55%, light gray for values between 55 and 73%, and dark gray for values greater than 73%. Black boxes along the diagonal indicate the 100% identity for each gene when compared with itself. Indications of the groups forming clades 1 to 6 and families 1 to 3 as described in the text are given along the left axis and across the top.
FIG. 4.
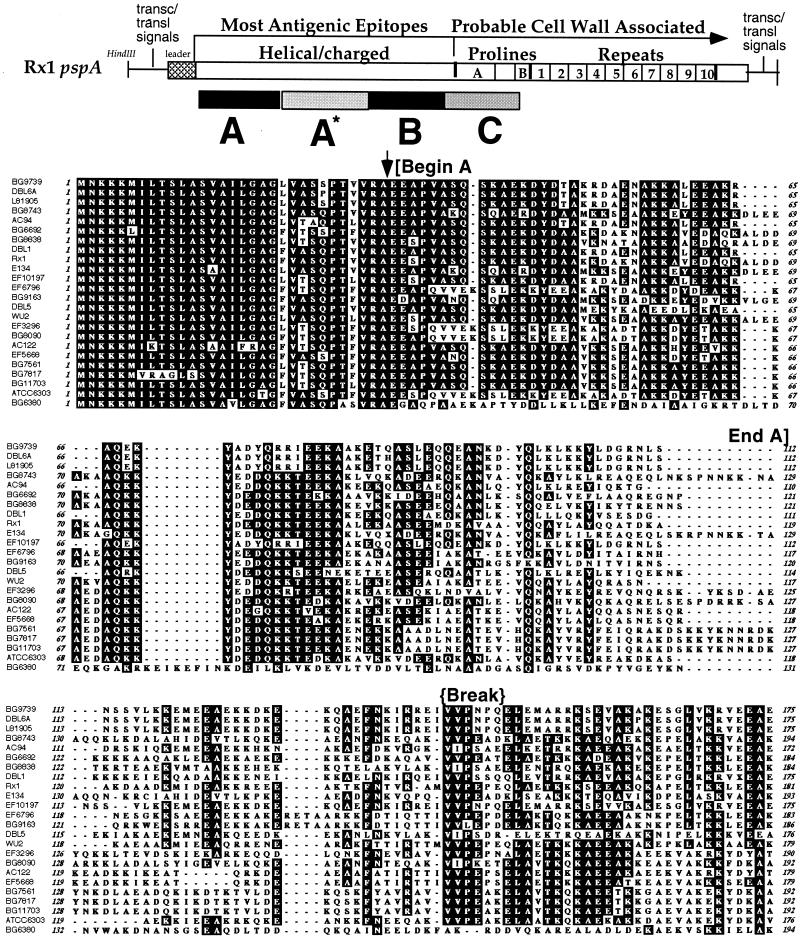
Alignment of all 24 PspAs by the Clustal W algorithm and the Blosum30 amino acid scoring matrix in MacVector. The printed output shows amino acids common to over 51% of the group as darkened boxes. Regions used for window A, B, and C analyses are marked on the figure by brackets at the beginning and end. The alignment is ordered based on the clade groupings identified in the B window. Line breaks under strain names indicate clades 1 to 6. Indications above the sequence mark the start of the mature protein (amino acid [AA] 32), regions of break in coiled-coil (approximately from AA 143 to 148), and, in window C, the location of an non-proline sequence block (approximately from AA 368 to 395) that is optionally present or absent in this region of PspA molecules (Fig. 5).
Nucleotide sequence accession numbers.
The pspA sequences have been submitted to GenBank and assigned the accession no. AF071802 to AF071827 as indicated beside each strain in Table 1.
RESULTS
Selection of strains and nomenclature.
Twenty-four independent clinical isolates representing 13 different capsular and 17 serologic PspA types (based on an earlier typing scheme using a MAb [15]) were chosen for sequencing in order to evaluate the patterns of diversity present at the pspA chromosomal locus in pneumococcal strains. An effort was made to choose strains which would span the range of diversity among strains available to us. No two of the pspA genes examined came from strains either suspected or known to be clonally related. An initial group of 19 strains were chosen from strains of the most diverse PspA types we could find based on the serologic typing with seven MAbs that detected different combinations of epitopes in the α-helical or charged region of PspA (Fig. 1). In reviewing additional data on the reactivity of over 50 MAbs (R. Becker, unpublished data) with a panel of >50 additional strains, we identified 5 additional PspAs for sequencing that failed to react with any of the 7 original MAbs and that had unusual patterns of reactivity with the larger group of 50 MAbs. Thus, these 24 isolates are expected to exhibit a greater diversity than the PspAs in any random selection of pneumococcal isolates. The serologic properties, dates, and places of origin of the S. pneumoniae strains are given in Table 1. These isolates are from four main geographic sites: Alabama, Sweden, Alaska, and Canada. Because each gene sequenced differs from every other gene at multiple positions, each pspA gene was designated pspA/strain name and, similarly, the protein encoded was designated PspA/strain name (e.g., PspA/Rx1 and pspA/Rx1 from strain Rx1, an unencapsulated derivative of D39).
Identification of PspA families.
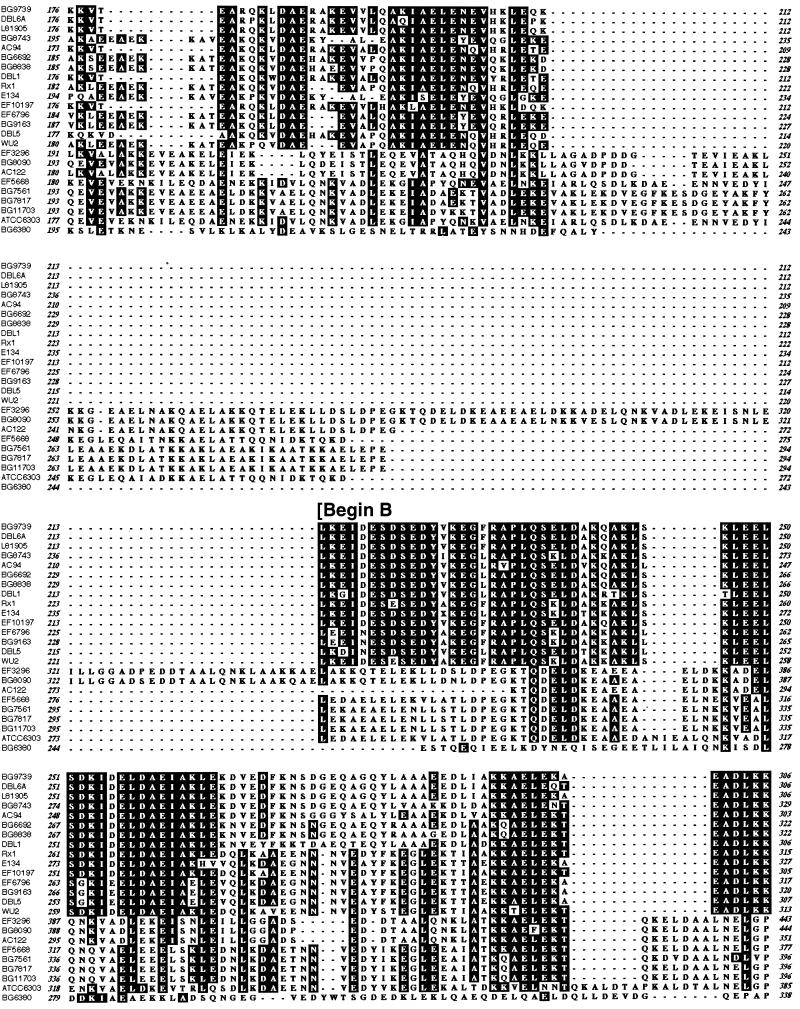
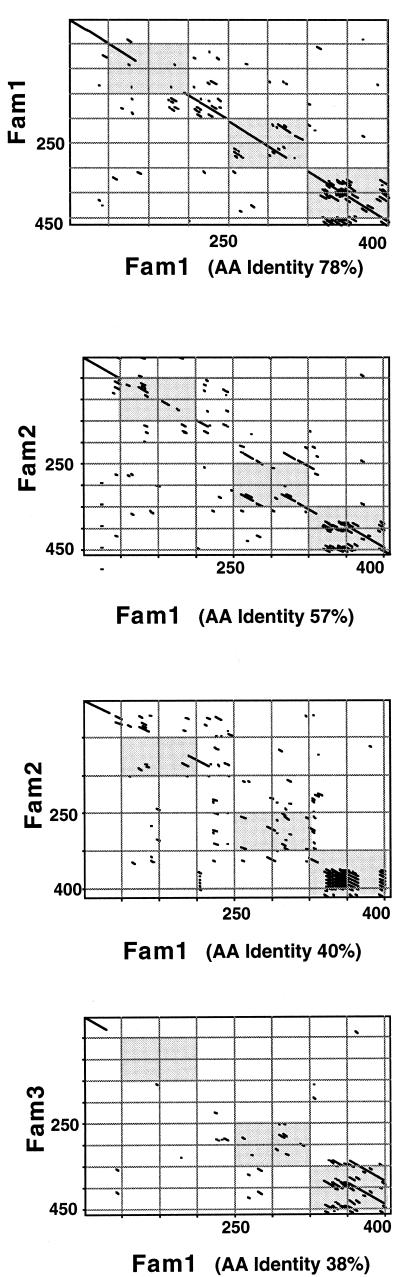
Using the specific PCR primers LSM13 and SKH2, we have found that virtually every isolate of S. pneumoniae has a pspA gene (S. K. Hollingshead and D. E. Briles, unpublished data). We amplified and sequenced the complete α-helical portion of each of the 24 genes from strains in Table 1. The repeat region encoding the carboxy-terminal choline-binding domain was not sequenced, since previous studies have indicated that it is relatively invariant (10, 36). In pairwise comparisons, the genes and the proteins were exceptionally diverse in their α-helical regions. Figure 2 gives the percent identity values for nucleotide-nucleotide and protein-protein comparisons. When the pairwise identity was ≥60%, the sequences could be aligned with minimal gaps. When a pairwise comparison showed less than 60% identity, the alignment required the introduction of multiple gaps. By using the 40% nucleotide divergence as a cutoff value, two major families of PspA proteins were identified, with a single PspA falling into a third family. The requirement for multiple gaps in the alignments of proteins in different families, but not in the alignment of proteins within families, indicates a much weaker phylogenic relationship between PspAs in different families than between the PspAs within a single family. Thus, all 24 pspA genes may not descend from a common ancestral gene over their length, and each PspA family may have a distinct ancestor. For the data set of 24 genes, base substitutions, replacement blocks, and small insertions or deletions were not clustered in one or more gene regions, but were distributed throughout the entire gene. Sample dot plots showing both same-family and cross-family comparisons indicate the variation throughout the genes (Fig. 3).
FIG. 3.
Dot matrix analysis of representative PspA pairs indicating the regions where variance is found. In each case, the x axis is family 1 (Fam1) and the y axis is either family 1, family 2, or family 3. The protein comparison begins with the signal peptide and continues to around amino acid 400. Comparisons were made by the method of Pustell and Kafatos (50) with the default values of a window size of 8 and a minimum percent score of 60 by using the pam250 matrix and a hash value of 2. Specific proteins used in this analysis were PspA/Rx1 (family 1, x axis), PspA/BG6692 (family 1, y axis), PspA/EF5668 (family 2, first), PspA/EF3296 (family 2, second), and PspA/BG6380 (family 3). Three gray boxes in each graph represent windows A, B, and C. The percent amino acid (AA) identity values are given for each case.
Several alignments of the 24 pspA genes were considered. One generated by the Clustal W algorithm and later adjusted to align portions of the proteins that are structurally the same and thus more likely to be homologous is presented in Fig. 4. These anchor regions included the mature N terminus of the protein, the transition zone between the α-helical region and the proline-rich regions of PspA, and one or more breakpoints in the coiled-coil structure as noted previously (28, 36, 62). The breakpoints serve to anchor the alignment even as diversity increases, necessitating the insertion of the many gaps in the overall alignment.
The low sequence identities between pspA genes in different families suggest that at least two different parental ancestral genes contributed to the current diversity in the pspA locus. This being the case, nonsynonymous sites tend to be saturated for some genes and not for others, and their examination is not valid for the cases in which ancestral sequences differ. The comparison of divergent sequences potentiates a high risk for nonhomologous alignment, especially when the overall lengths of the sequences compared differ. For this reason, we focused the initial examination of this diverse group of pspA genes on several windows within the gene that had the maximum potential to be correctly aligned. The three windows, A, B, and C, used for analysis are depicted in Fig. 1.
Alignment.
The aligned amino acid sequences show the diversity of PspAs over the different windows (Fig. 4). The sequence differences in windows A to C are analyzed further in Fig. 5. Window A′ includes approximately 200 nucleotides of DNA sequence upstream of each gene, including the region encoding the signal peptide. A′ was highly conserved, ranging from 95 to 100% nucleotide identity (data not shown). The conservation of the DNA sequence in A′ provided confirmation that the DNA fragment amplified and sequenced came from the pspA gene locus and not a related paralogous gene, such as that of pspC. This A′ window is not included in the Fig. 5 analyses because it was so highly conserved and because it contains both coding and noncoding sequences.
FIG. 5.
Distribution of pairwise comparisons of sequence distance among PspA proteins in windows A, B, and C. The y axis in each graph represents the number of pairwise comparisons out of 276 total comparisons which fell within a range of the percent amino acid identity given on the x axis. To the right of each graph is a dendrogram representing the relationships between the PspAs for the sequence obtained in window A, B, or C as described in the text. The numbers at the indicated nodes indicate the cutoff values for DNA (above) or amino acid (below) identity that define the branches extending from that node. Clade groups defined in the text based on region B are indicated by arrows in the center of the figure.
Window A encodes the first 100 amino acids (300 nucleotides) of each PspA molecule, beginning with the first amino acid of the mature protein. The conserved region of signal peptides extends into the first 50 amino acids, with the majority of comparisons exhibiting greater than 50% amino acid identity in A. The PspA molecules are much less conserved over the second half of window A, where the sequences begin to diverge and fall into groups (Fig. 4).
Window A* is highly diverse (Fig. 4). The distribution values for this region are also not included in Fig. 5, because the lengths of the sequences in this region are so variable that meaningful alignments are not possible in this context. Inspection of this region, however, shows it to be a transition zone between the picture shown by window A and that shown by window B. The N-terminal end of A* is hypervariable, and the C-terminal end is characterized for the most part by the same sequence groups that are identified in window B.
Window B shows six sequence groups that are quite distinct (Fig. 4). These will be discussed further below.
Window C contains the proline-rich region of the PspA molecules. This region of PspA is quite repetitive, with many imperfect repeats of the sequence PAPAP (Fig. 4). The sequence-to-sequence distances in this window largely reflect the variation in the iterations of this repetitive sequence. A second factor influencing the distribution of sequence distances in this window is the presence or the absence of a 27-amino-acid non-proline-rich block of the consensus amino acid sequence EKS/TADQQAEEDYARRSEEEYNRLTQQQ, which is present in this region in 14 of 24 of the sequences and absent from the others (Fig. 4 and 5).
Distribution of pairwise comparisons.
The A-to-C windows are of approximately equal lengths, and each is proximal to the transition between one domain of the protein and another (Fig. 1). As was observed in the overall comparison of genes (Fig. 2), when individual sequence windows A to C were analyzed, any two PspA molecules were found to share as few as 14% or as many as 100% of the amino acids in a given window (data not shown). Although a similar range of distance comparisons was observed for each of the three windows, a closer examination reveals that the pattern of pspA diversity differs within each window (Fig. 5).
The 24×24 gene comparisons per window resulted in 276 distance calculations. The distribution of the 276 distances by percentile was plotted for each window. Figure 5 shows the distribution of these pairwise comparisons and indicates some striking differences between the variations seen at windows A, B, and C. Window A shows a normal distribution around the median of 70% amino acid identity. The pairwise comparisons at window B exhibit two modes. One peak in the distribution reflected pairwise comparisons of quite similar proteins (60 to 70% amino acid identity), and the second peak reflected pairwise comparisons of quite divergent proteins (20 to 30% identity). This biphasic distribution represents the distance profiles between genes of the same PspA sequence “type” versus distances between PspAs of differing sequence “types.” The two major types here correspond to PspA families as defined previously. The deviation of this profile from a normal distribution is significant (P < 0.0001; χ2 test). Window C showed yet a third type of distribution in which the frequencies of pairwise comparisons were nearly equivalent over each distribution range sampled, giving a relatively broad flat curve with more spread. Hence, almost the same number of pairwise comparisons yielded identities in the 31 to 40, 41 to 50, 51 to 60, 61 to 70, 71 to 80, 81 to 90, and 91 to 100% ranges, with only a slight peak in the 51 to 60% range.
The normal distribution for sequence comparisons in the A region results in a deep branching dendrogram for the A region in Fig. 5, which reflects the equidistant relationship among the proteins in this window. The strong group structure (B region) forming the two major families drives the biphasic distribution of the pairwise comparisons (Fig. 4 and 5) and the relatively tight branches or clusters seen in the dendrogram in the B window. The six groups seen in this window represent major PspA lineages and are discussed further below. The dendrogram for window C is mainly explained by the presence or the absence of the non-proline-rich block.
Clade-defining region of PspA.
The region of sequence including window B was previously found to be an important protection-eliciting region in PspA/Rx1, making it critical to the development of PspA as a protein-based vaccine (37). Among the 24 PspAs, there are six groups that are mutually distinct at differences of >20% of amino acid positions in this B region. We are defining these groups as PspA clades, a term coming from the science of cladistics and meaning monophyletic group. On the phenogram generated from the amino acid sequence in this region, each monophyletic group that was separated from the others with bootstrap values of 100% was considered a clade (Fig. 5B and 6C). The clades were numbered from 1 to 6. The assignment of the individual pspA genes to clade groups is given in Table 1, Fig. 5B, and Fig. 6C.
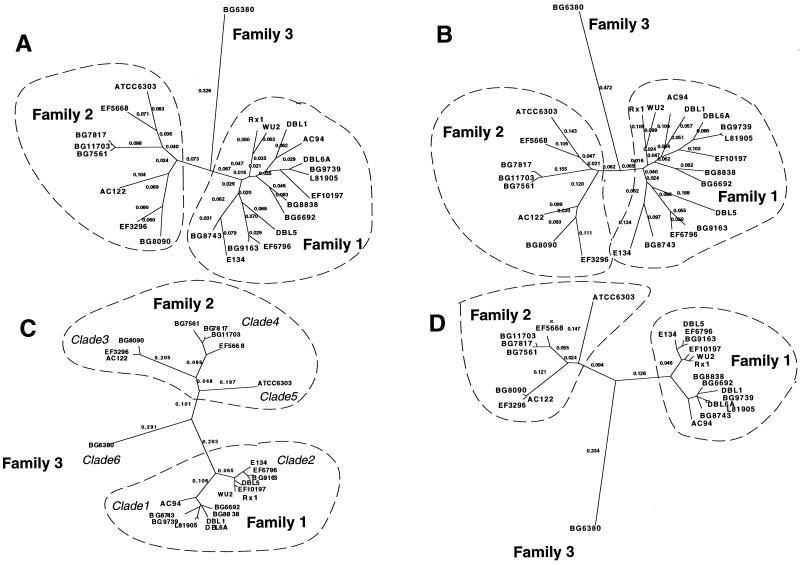
FIG. 6.
Relationship between PspA sequences over window B and over the entire sequence. Shown are four unrooted phylograms generated from mean distances by the neighbor-joining method in the program PAUP 4.0B, as follows: A, proteins, all; B, DNA, all; C, proteins, B region; D, DNA, B region. The numbers on the tree indicate the distances along the branch lengths as calculated by PAUP with the terminal branch distances suppressed. Clades and families as defined in the text are listed near the clusters or branches that they encompass.
The clades of the 24 sequences are depicted in a tree that represents the different groupings as clusters or branch points on the tree (Fig. 6C, protein; 6D, DNA). Proteins within the same clade are greater than 90% identical in this B region. A higher branching division, which corresponds to the family division described earlier, is also shown on the tree (Fig. 2 and 6). Each of the two major families contains more than one clade, and family 3 has only a single clade (Fig. 6). The three families diverge from each other at >45% of amino acid positions in this B region. The family structure is also evident in distance trees constructed from the whole sequences (Fig. 6A, protein; 6B, DNA). The clade structure is blurred somewhat in whole gene trees due to the discordant contribution of main recombinant blocks within the A and C regions.
PspA proteins in the same clade appear to share a recent ancestral sequence for the B window or “clade-defining” region because they share their monophyletic position. Variation in this case is found to be restricted to single-amino-acid substitutions (Fig. 4). PspA proteins in the same family are also related in ancestry, but may represent products of parallel lineages from each other over the B window if they are in different clades. Variation in this case sometimes involves small blocks of divergent amino acid substitutions (Fig. 4). PspA proteins in different families are quite divergent, indicating a significant separation in ancestry of the respective pspA genes over the B window. Variation, in this third case, is the norm, and few identifiable blocks of conserved sequence are present.
Evidence for recombination in pspA genes.
Strong evidence for past recombination events is apparent from the discordance of the trees for windows A, B, and C. Figure 5 shows the dendrograms generated based on sequence similarities for each of the three windows. The three dendrograms are noncongruent; the relationship for a strain-to-strain comparison is dependent upon the window or block of sequence used for assigning the relationship. For example, in window B, PspA/EF6796 is only 15% diverged from PspA/Rx1, and they both fall into clade 2; PspA/Rx1 is 74% diverged from PspA/EF3296. In window A, the situation is reversed; PspA/EF6796 is 65% diverged from PspA/Rx1 in this window and only 33% diverged from PspA/EF3296. Numerous examples of such crossovers exist within this limited data set of 24 PspA molecules. Evidence for recombination may also be viewed in Fig. 2 in the isolated shaded blocks as indications of the uneven distribution of homology and in numerous blocks along the alignment in Fig. 4.
DISCUSSION
The alignment of 24 diverse PspA proteins provides a snapshot of a highly complex mosaic gene pattern (Fig. 4). Mosaic genes are formed by recombination. In this data set, there are numerous instances in which individual pspA sequences may be very similar in sequence to certain pspA sequences in one location and very similar in sequence to other pspA sequences in another location within the alignment. Examples are present throughout both the DNA and protein alignments (Fig. 4). At the level of the encoded proteins, the impression is formed that nature has been shuffling the deck of epitopes through repeated recombination events over the pspA locus. This may largely explain the previously noted complex combinatorial patterns of MAb reactivity found for PspAs (15).
The discordant trees in sequence windows A, B, and C emphasize that recombination has been an important contributor to diversity in the pspA gene locus (Fig. 5 and 6). Despite extensive recombination, we have been able to identify families and clades (Fig. 5 and 6) that depict lineages within the pspA genes and the encoded proteins. For this purpose, the B window region was used because it was previously shown to be a site important to protective immune responses (37). The division of PspAs into families by the criteria used here has turned out to be especially important, because it reflects differences in PspAs readily detectable with polyclonal antisera to PspA (S. K. Hollingshead and D. E. Briles, unpublished data).
The level of divergence seen in the pspA gene locus is generally greater than that in previously studied mosaic genes in S. pneumoniae or other bacteria. For instance, the divergence within a mosaic block of the pbp2x gene is only 3% when comparing DNA in sensitive alleles, but was 18 to 23% for the same block between sensitive and resistant alleles (32). Other PBP mosaic genes have similar values for mosaic blocks (18). PspAs are so diverse that the boundaries of the numerous recombinant blocks are difficult to enumerate from this data set, so a sliding window comparison was used to initially examine diversity. The pspA genes from different pneumococcal isolates showed at least 5 to 15% divergence between even those pspA genes that are very similar for a particular window comparison. Divergence was sometimes >70% for those genes that are the least similar within the window. When the divergence between two PspAs is >70% in a particular window, the two proteins would be expected to be products from different genes if it were not clear from the upstream regions that these sequences are from the same gene locus.
Mosaic blocks are distributed throughout the pspA locus (Fig. 3 and 4). In other known mosaic gene families, discrete conserved blocks are often found to be interspersed among the variable divergent blocks (e.g., intimin in Escherichia coli [39] and vacA in Helicobacter pylori [2, 3]). For PspA, the only regions conserved among all pspA genes seemed to be at the 5′ end of the gene encoding the N terminus of the protein and the 3′ end of the gene encoding the choline-binding domain (36). The variant mosaic sequences seemed to be distributed along the locus as a whole (Fig. 4). Each pair of pspA genes shares a number of different sequence blocks, but the location of the shared blocks differs, depending on the pair under examination (Fig. 3).
This extraordinary magnitude of the pspA gene mosaicism is also reflected in the large number of nucleotide positions that are polymorphic. For example, the average interclade distance is about 28% when the genes compared are in the same family and usually exceeds 50% when the genes are from different PspA families (Fig. 2). This level of polymorphism is as great as that often observed when comparing orthologs from the genomes of distinct bacterial species. For example, the hyaluronate lyase gene from S. pneumoniae diverges from the hyaluronate lyase gene of group B streptococci at 50% of nucleotide sites and 49% of amino acid sites (33). Similarly, the immunoglobulin A2 (IgA2) protease gene from S. pneumoniae diverges from the IgA2 protease gene in Streptococcus sanguis at 38% of nucleotide sites and 41% of amino acid sites (48).
Source of mosaic blocks.
Recent studies in a number of laboratories investigating both Neisseria and streptococci have suggested the possibility of “global” gene pools (19, 52, 65; Maiden et al., Letter) based on documented interspecies transfer of gene segments (18, 32, 56, 57). For the most part, these global gene pools operate at the genus-wide level. For S. pneumoniae, the closest relatives among other streptococci appear to be Streptococcus oralis and Streptococcus mitis based on 16S rRNA, 23S rRNA, and other factors (30, 58). Indeed, transfers in pbp alleles of S. pneumoniae have most frequently been traced to these very closely related oral commensal species (11, 17, 52). IgA1 protease alleles of S. oralis and S. mitis are also shared (47). Although not extensively studied, there are gene loci whose alleles are found to be species restricted as well. The presence of the pspA gene is not demonstrable in the nearest relatives of S. pneumoniae, with the possible exception of a few S. mitis-like organisms (46). Thus, a source for interspecies transfer of pspA blocks is missing. Second, both family 1 and family 2 PspA proteins are prevalent among recent clinical isolates of pneumococci of all capsular serotypes, a finding which argues that they are not recent acquisitions from another streptococcal species (Hollingshead and Briles, unpublished data).
Although interspecies horizontal transfer of pspA is possible, it seems likely that the vast majority of recombination in the pspA locus comes from intraspecies horizontal transfer. The pneumococcus is carried in the nasopharynx, and frequently more than one strain can be carried at the same time (21, 55). S. pneumoniae is well known for its special capacity to take up DNA from its environment and incorporate it into its chromosome. Because other pneumococci share variant pspA loci that still have enough homology to allow efficient recombination, the neighboring strains of pneumococci are likely to serve as the most common donors.
Implications for vaccine development.
One aim of sequencing this many pspA genes was to address the breadth and depth of the genetic diversity that was present at this locus. It was felt that this information might be an aid in understanding the serological diversity of pspA and thus shed light on the appropriate composition of a PspA-based pneumococcal vaccine for humans. Although at first glance the level of diversity appears daunting for this purpose, PspA is remarkably immunogenic and cross-reactive. The data from cross-protective studies in mice (36, 39) show that a PspA molecule in clades 1 to 4 can elicit protective antibody responses to pneumococci which differ from the immunogen at >50% of their amino acid positions in the α-helical portion of the molecule (5, 60). Immunization of healthy adults with a single recombinant PspA stimulated cross-reactive antibodies to heterologous PspAs in different clades or families which were cross-protective in mice (43; Briles et al., submitted). In producing a broadly protective vaccine, the cross-reactive responses that have been documented could easily be further encouraged by the inclusion of PspA molecules from each of the families and clades in the vaccine.
The verification from sequence data that virtually all PspAs share significant short stretches of amino acids helps our understanding of the nature of PspA cross-reactivity. Cross-reactive antibodies may recognize the amino acid identities that exist in small stretches depicted as short lines (Fig. 3) and indicating a region scoring above the cutoff value in a dot plot of pspA genes or PspA proteins. These short stretches of matching amino acids are present in comparing any two PspA proteins when using default cutoff values with a window size of 8, minimal percentage score of 60%, and hash value of 2. If the short conserved sequences are of sufficient length and structure to comprise an epitope, then their presence along the α-helical or charged region of PspA may mark encoded cross-reactive sites. The cross-reactive sites are the residues of the repeated recombination that has exchanged sequence blocks and generated new chimeric sites within the pspA gene. The scattered distribution of cross-reactive sites explains in part the different combinations of epitopes in PspA that were previously detected by MAbs (15).
Paradoxically, the extraordinary degree of mosaicism exhibited by pspA genes may indicate the importance of this surface protein as a natural target for host defense against the pneumococcus. Surface proteins in bacteria and antigen receptor proteins of T and B cells in eukaryotes are often those proteins which exhibit the greatest divergence in species-to-species comparisons (42). This divergence often results from the processes that create mosaic proteins. The presence of mosaicism and, in particular, the complex pattern of mosaicism seen in the pspA gene locus could indicate that pspA is often the target of positive or negative selection in the interaction between the pneumococcus and its human host. The antibodies that PspA elicits clearly play an important role in protection in mouse models (8), and the protein appears to interfere with complement fixation in vivo in mice and is likely to do so in humans as well (1, 6, 44, 61). Boosting the level of natural antibodies to PspA may bolster the host's ability to resist the pneumococcus.
ACKNOWLEDGMENTS
We gratefully acknowledge the technical support of Xinping Wu and Terri Readdy and are thankful to Alexis Brooks-Walter, Elliot Lefkowitz, and D. Ashley Robinson for suggestions throughout the course of this work and Sylvie Rodriguez for reading the manuscript.
The work was supported in part by grants AI21548, AI40645, and HL54818 from the National Institutes of Health (NIH) and by a contract from Aventis Pasteur. The DNA Sequencing Core Facilities were supported by a grant from NIH to the Center for Aids Research (AI27767), by a grant from the Tennessee Valley Authority to the Department of Microbiology at the University of Alabama at Birmingham, and by the Howard Hughes Foundation to the University of Alabama at Birmingham Medical School.
REFERENCES
- 1.Abeyta M. Ph.D. dissertation. Birmingham: University of Alabama at Birmingham; 1999. [Google Scholar]
- 2.Atherton J C, Cao P, Peek R M, Jr, Tummuru M K, Blaser M J, Cover T L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 3.Atherton J C, Sharp P M, Cover T L, Gonzalez-Valencia G, Peek R M, Jr, Thompson S A, Hawkey C J, Blaser M J. Vacuolating cytotoxin (vacA) alleles of Helicobacter pylori comprise two geographically widespread types, m1 and m2, and have evolved through limited recombination. Curr Microbiol. 1999;39:211–218. doi: 10.1007/s002849900447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breiman R, Butler J C, Tenover F C, Elliot J, Facklam R R. Emergence of drug-resistant pneumococcal infections in the United States. JAMA. 1994;271:1831–1835. [PubMed] [Google Scholar]
- 5.Briles D E, Hollingshead S, Brooks-Walter A, Nabors G S, Ferguson L, Schilling M, Gravenstein S, Braun P, King J, Swift A. The potential to use PspA and other pneumococcal proteins to elicit protection against pneumococcal infection. Vaccine. 2000;18:1707–1711. doi: 10.1016/s0264-410x(99)00511-3. [DOI] [PubMed] [Google Scholar]
- 5a.Briles, D. E., S. K. Hollingshead, J. E. King, A. Swift, P. Braun, L. M. Ferguson, M. Nahm, and G. S. Nabors. Immunization of humans with rPspA elicits antibodies which passively protect mice from fatal infection with Streptococcus pneumoniae expressing heterologous PspA molecules. J. Infect. Dis., in press. [DOI] [PubMed]
- 6.Briles D E, Hollingshead S K, Swiatlo E, Brooks Walter A, Szalai A, Virolainen A, McDaniel L S, Benton K A, White P, Prellner K, Hermansson A, Needleman C, Van Dijk H, Crain M J. PspA and PspC: their potential for use as pneumococcal vaccines. Microb Drug Resist. 1997;3:401–408. doi: 10.1089/mdr.1997.3.401. [DOI] [PubMed] [Google Scholar]
- 7.Briles D E, Swiatlo E, Edwards K. Vaccine strategies for S. pneumoniae. In: Stevens D L, editor. Streptococci. New York, N.Y: Oxford University Press; 2000. pp. 419–433. [Google Scholar]
- 8.Briles D E, Tart R C, Swiatlo E, Dillard J P, Smith P, Benton K A, Ralph B A, Brooks-Walter A, Crain M J, Hollingshead S K, McDaniel L S. Pneumococcal diversity: considerations for new vaccine strategies with emphasis on pneumococcal surface protein A (PspA) Clin Microbiol Rev. 1998;11:645–657. doi: 10.1128/cmr.11.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briles D E, Tart R C, Wu H-Y, Ralph B A, Russell M W, McDaniel L S. Systemic and mucosal protective immunity to pneumococcal surface protein A. Ann N Y Acad Sci. 1996;797:118–126. doi: 10.1111/j.1749-6632.1996.tb52954.x. [DOI] [PubMed] [Google Scholar]
- 10.Brooks-Walter A, Briles D E, Hollingshead S K. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect Immun. 1999;67:6533–6542. doi: 10.1128/iai.67.12.6533-6542.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffey T J, Dowson C G, Daniels M, Spratt B G. Horizontal spread of an altered penicillin-binding protein 2B gene between Streptococcus pneumoniae and Streptococcus oralis. FEMS Microbiol Lett. 1993;110:335–339. doi: 10.1111/j.1574-6968.1993.tb06345.x. [DOI] [PubMed] [Google Scholar]
- 12.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal transfer of multiple penicillin-binding protein genes, and capsule biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 13.Coffey T J, Enright M C, Daniels M, Morona J K, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 14.Cowan M J, Ammann A J, Wara D W, Howie V M, Schultz L, Doyle N, Kaplan M. Pneumococcal polysaccharide immunization in infants and children. Pediatrics. 1978;62:721–727. [PubMed] [Google Scholar]
- 15.Crain M J, Waltman II W D, Turner J S, Yother J, Talkington D E, McDaniel L S, Gray B M, Briles D E. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect Immun. 1990;58:3293–3299. doi: 10.1128/iai.58.10.3293-3299.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dillard J P, Vandersea M W, Yother J. Characterization of the cassette containing genes for type 3 capsular polysaccharide biosynthesis in Streptococcus pneumoniae. J Exp Med. 1995;181:973–983. doi: 10.1084/jem.181.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowson C, Hutchison A, Woodford N, Johnson A, George R, Spratt B. Penicillin-resistant viridans streptococci have obtained altered penicillin-binding protein genes from penicillin-resistant strains of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1990;87:5858–5862. doi: 10.1073/pnas.87.15.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dowson C G, Hutchinson A, Brannigan J A, George R C, Hansman D, Liñares J, Tomasz A, Maynard J, Spratt B G. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1989;86:8842–8846. doi: 10.1073/pnas.86.22.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fussenegger M, Rudel T, Barten R, Ryll R, Meyer T F. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae—a review. Gene. 1997;192:125–134. doi: 10.1016/s0378-1119(97)00038-3. [DOI] [PubMed] [Google Scholar]
- 20.Gray B M. Pneumococcal infections in an era of multiple antibiotic resistance. Adv Pediatr Infect Dis. 1995;11:55–100. [PubMed] [Google Scholar]
- 21.Gray B M, Converse III G M, Dillon H C. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J Infect Dis. 1980;142:923–933. doi: 10.1093/infdis/142.6.923. [DOI] [PubMed] [Google Scholar]
- 22.Greenwood B. The epidemiology of pneumococcal infection in children in the developing world. Philos Trans R Soc Lond B Biol Sci. 1999;354:777–785. doi: 10.1098/rstb.1999.0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hakenbeck R, König A, Kern I, van der Linden M, Keck W, Billot-Klein D, Legrand R, Schoot B, Gutmann L. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level β-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J Bacteriol. 1998;180:1831–1840. doi: 10.1128/jb.180.7.1831-1840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammerschmidt S, Bethe G, Remone P H, Chhatwal G S. Identification of pneumococcal surface protein A as a lactoferrin-binding protein of Streptococcus pneumoniae. Infect Immun. 1999;67:1683–1687. doi: 10.1128/iai.67.4.1683-1687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanke M, Wink M. Direct DNA sequencing of PCR-amplified vector inserts following enzymatic degradation of primer and dNTPs. BioTechniques. 1994;17:858–860. [PubMed] [Google Scholar]
- 26.Hausdorff W P, Bryant J, Kloek C, Paradiso P R, Siber G R. The contribution of specific pneumococcal serogroups to different disease manifestations: implications for conjugate vaccine formulation and use, part II. Clin Infect Dis. 2000;30:122–140. doi: 10.1086/313609. [DOI] [PubMed] [Google Scholar]
- 27.Hausdorff W P, Bryant J, Paradiso P R, Siber G R. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin Infect Dis. 2000;30:100–121. doi: 10.1086/313608. [DOI] [PubMed] [Google Scholar]
- 28.Jedrzejas M J, Hollingshead S K, Lebowitz J, Chantalat L, Briles D E, Lamani E. Production and characterization of the functional fragment of pneumococcal surface protein A. Arch Biochem Biophys. 2000;373:116–125. doi: 10.1006/abbi.1999.1544. [DOI] [PubMed] [Google Scholar]
- 29.Jernigan D B, Cetron M S, Breiman R F. Defining the public health impact of drug resistant Streptococcus pneumoniae: report of a working group. Morb Mortal Wkly Rep. 1996;45:1–20. [PubMed] [Google Scholar]
- 30.Kawamura Y, Hou X-G, Sultana F, Miura H, Ezaki T. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol. 1995;45:406–408. doi: 10.1099/00207713-45-2-406. . (Erratum, 45:882.) [DOI] [PubMed] [Google Scholar]
- 31.Kell C M, Jordens J Z, Daniels M, Coffey T J, Bates J, Paul J, Gilks C, Spratt B G. Molecular epidemiology of penicillin-resistant pneumococci isolated in Nairobi, Kenya. Infect Immun. 1993;61:4382–4391. doi: 10.1128/iai.61.10.4382-4391.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laible G, Spratt B G, Hakenbeck R. Interspecies recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1991;5:1993–2002. doi: 10.1111/j.1365-2958.1991.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 33.Lin B, Hollingshead S K, Coligan J E, Egan M L, Baker J R, Pritchard D E. Cloning and expression of the gene for group B streptococcal hyaluronate lyase. J Biol Chem. 1994;269:30113–30116. [PubMed] [Google Scholar]
- 34.Maynard Smith J. Population genetics: an introduction. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: American Society of Microbiology; 1996. pp. 2685–2690. [Google Scholar]
- 35.Maynard Smith J, Dowson C G, Spratt B G. Localized sex in bacteria. Nature. 1991;349:29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- 36.McDaniel L S, McDaniel D O, Hollingshead S K, Briles D E. Comparison of the PspA sequence from Streptococcus pneumoniae EF5668 to the previously identified PspA sequence from strain Rx1 and ability of PspA from EF5668 to elicit protection against pneumococci of different capsular types. Infect Immun. 1998;66:4748–4754. doi: 10.1128/iai.66.10.4748-4754.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDaniel L S, Ralph B A, McDaniel D O, Briles D E. Localization of protection-eliciting epitopes on PspA of Streptococcus pneumoniae between amino acid residues 192 and 260. Microb Pathog. 1994;17:323–337. doi: 10.1006/mpat.1994.1078. [DOI] [PubMed] [Google Scholar]
- 38.McDaniel L S, Sheffield J S, Delucchi P, Briles D E. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect Immun. 1991;59:222–228. doi: 10.1128/iai.59.1.222-228.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGraw E A, Li J, Selander R K, Whittam T S. Molecular evolution and mosaic structure of alpha, beta, and gamma intimins of pathogenic Escherichia coli. Mol Biol Evol. 1999;16:12–22. doi: 10.1093/oxfordjournals.molbev.a026032. [DOI] [PubMed] [Google Scholar]
- 40.Milkman R. Recombination and population structure in Escherichia coli. Genetics. 1997;146:745–750. doi: 10.1093/genetics/146.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milkman R, Stoltzfus A. Molecular evolution of the Escherichia coli chromosome. II. Clonal segments. Genetics. 1988;120:359–366. doi: 10.1093/genetics/120.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy P M. Molecular mimicry and the generation of host defense protein diversity. Cell. 1993;72:823–826. doi: 10.1016/0092-8674(93)90571-7. [DOI] [PubMed] [Google Scholar]
- 43.Nabors G S, Braun P A, Herrmann D J, Heise M L, Pyle D J, Gravenstein S, Schilling M, Ferguson L M, Hollingshead S K, Briles D E, Becker R S. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) stimulated cross-reactive antibodies to heterologous PspA molecules. Vaccine. 2000;18:1743–1754. doi: 10.1016/s0264-410x(99)00530-7. [DOI] [PubMed] [Google Scholar]
- 44.Neeleman C, Geelen S P M, Aerts P C, Daha M R, Mollnes T E, Roord J J, Posthuma G, van Dijk H, Fleer A. Resistance to both complement activation and phagocytosis in type 3 pneumococci is mediated by binding of complement regulatory protein factor H. Infect Immun. 1999;67:4517–4524. doi: 10.1128/iai.67.9.4517-4524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogunniyi A D, Folland R L, Briles D E, Hollingshead S K, Paton J C. Immunization of mice with combinations of pneumococcal virulence proteins elicits enhanced protection against challenge with Streptococcus pneumoniae. Infect Immun. 2000;68:3028–3033. doi: 10.1128/iai.68.5.3028-3033.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poulsen K, Kilian M. Proceedings of the ASM Conference on Streptococcal Genetics. Washington, D.C.: American Society for Microbiology; 1998. Genetic relationships between Streptococcus pneumoniae, Streptococcus mitis, and Streptococcus oralis; p. 81. [Google Scholar]
- 47.Poulsen K, Reinholdt J, Jespersgaard C, Boye K, Brown T A, Hauge M, Kilian M. A comprehensive genetic study of streptococcal immunoglobulin A1 proteases: evidence for recombination within and between species. Infect Immun. 1998;66:181–190. doi: 10.1128/iai.66.1.181-190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poulsen K, Reinholdt J, Kilian M. Characterization of the Streptococcus pneumoniae immunoglobulin A1 protease gene (iga) and its translation product. Infect Immun. 1996;64:3957–3966. doi: 10.1128/iai.64.10.3957-3966.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pozzi G, Masala L, Iannelli F, Manganelli R, Håvarstein L S, Piccoli L, Simon D, Morrison D A. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J Bacteriol. 1996;178:6087–6090. doi: 10.1128/jb.178.20.6087-6090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pustell J, Kafatos F C. A high speed, high capacity homology matrix: zooming through SV40 and polyoma. Nucleic Acids Res. 1982;10:4765–4782. doi: 10.1093/nar/10.15.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramirez M, Morrison D A, Tomasz A. Ubiquitous distribution of the competence related genes comA and comC among isolates of Streptococcus pneumoniae. Microb Drug Resist. 1997;3:39–52. doi: 10.1089/mdr.1997.3.39. [DOI] [PubMed] [Google Scholar]
- 52.Reichmann P, Konig A, Linares J, Alcaide F, Tenover F C, McDougal L, Swidsinski S, Hakenbeck R. A global gene pool for high-level cephalosporin resistance in commensal Streptococcus species and Streptococcus pneumoniae. J Infect Dis. 1997;176:1001–1012. doi: 10.1086/516532. [DOI] [PubMed] [Google Scholar]
- 53.Shinefield H R, Black S, Ray P, Chang I, Lewis N, Fireman B, Hackell J, Paradiso P R, Siber G, Kohberger R, Madore D V, Malinowski F J, Kimura A, Le C, Landaw I, Aguilar J, Hansen J. Safety and immunogenicity of heptavalent pneumococcal CRM197 conjugate vaccine in infants and toddlers. Pediatr Infect Dis J. 1999;18:757–763. doi: 10.1097/00006454-199909000-00004. [DOI] [PubMed] [Google Scholar]
- 54.Sibold C, Henrichsen J, Konig A, Martin C, Chalkley L, Hakenbeck R. Mosaic pbpX genes of major clones of penicillin-resistant Streptococcus pneumoniae have evolved from pbpX genes of a penicillin-sensitive Streptococcus oralis. Mol Microbiol. 1994;12:1013–1023. doi: 10.1111/j.1365-2958.1994.tb01089.x. [DOI] [PubMed] [Google Scholar]
- 55.Sluijter M, Faden H, de Groot R, Lemmens N, Goessens W H F, van Belkum A, Hermans P W M. Molecular characterization of pneumococcal nasopharynx isolates collected from children during their first 2 years of life. J Clin Microbiol. 1998;36:2248–2253. doi: 10.1128/jcm.36.8.2248-2253.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spratt B G. Hybrid penicillin-binding proteins in penicillin-resistant strains of Neisseria gonorrhoeae. Nature. 1988;332:173–176. doi: 10.1038/332173a0. [DOI] [PubMed] [Google Scholar]
- 57.Spratt B G. Resistance to antibiotics mediated by target alterations. Science. 1994;264:388–393. doi: 10.1126/science.8153626. [DOI] [PubMed] [Google Scholar]
- 58.Sultana F, Kawamura Y, Hou X G, Shu S E, Ezaki T. Determination of 23S rRNA sequences from members of the genus Streptococcus and characterization of genetically distinct organisms previously identified as members of the Streptococcus anginosus group. FEMS Microbiol Lett. 1998;158:223–230. doi: 10.1111/j.1574-6968.1998.tb12824.x. [DOI] [PubMed] [Google Scholar]
- 59.Swiatlo E, Brooks-Walter A, Briles D E, McDaniel L S. Oligonucleotides identify conserved and variable regions of pspA and pspA-like sequences of Streptococcus pneumoniae. Gene. 1997;188:279–284. doi: 10.1016/s0378-1119(96)00823-2. [DOI] [PubMed] [Google Scholar]
- 60.Tart R C, McDaniel L S, Ralph B A, Briles D E. Truncated Streptococcus pneumoniae PspA molecules elicit cross-protective immunity against pneumococcal challenge in mice. J Infect Dis. 1996;173:380–386. doi: 10.1093/infdis/173.2.380. [DOI] [PubMed] [Google Scholar]
- 61.Tu A-H, Fulgham R L, McCrory M A, Briles D E, Szalai A J. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect Immun. 1999;67:4720–4724. doi: 10.1128/iai.67.9.4720-4724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yother J, Briles D E. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J Bacteriol. 1992;174:601–609. doi: 10.1128/jb.174.2.601-609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yother J, Handsome G L, Briles D E. Truncated forms of PspA that are secreted from Streptococcus pneumoniae and their use in functional studies and cloning of the pspA gene. J Bacteriol. 1992;174:610–618. doi: 10.1128/jb.174.2.610-618.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yother J, White J M. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J Bacteriol. 1994;176:2976–2985. doi: 10.1128/jb.176.10.2976-2985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou J, Bowler L D, Spratt B G. Interspecies recombination, and phylogenetic distortions, within the glutamine synthetase and shikimate dehydrogenase genes of Neisseria meningitidis and commensal Neisseria species. Mol Microbiol. 1997;23:799–812. doi: 10.1046/j.1365-2958.1997.2681633.x. [DOI] [PubMed] [Google Scholar]