Abstract
Discovery of antibiotics acting against Gram-negative species is uniquely challenging due to their restrictive penetration barrier. BamA, which inserts proteins into the outer membrane, is an attractive target due to its surface location. Darobactins produced by Photorhabdus, a nematode gut microbiome symbiont, target BamA. We reasoned that a computational search for genes only distantly related to the darobactin operon may lead to novel compounds. Following this clue, we identified dynobactin A, a novel peptide antibiotic from Photorhabdus australis containing two unlinked rings. Dynobactin is structurally unrelated to darobactins, but also targets BamA. Based on a BamA-dynobactin co-crystal structure and a BAM-complex-dynobactin cryo-EM structure, we show that dynobactin binds to the BamA lateral gate, uniquely protruding into its β-barrel lumen. Dynobactin showed efficacy in a mouse systemic Escherichia coli infection. This study demonstrates the utility of computational approaches to antibiotic discovery and suggests that dynobactin is a promising lead for drug development.
The antimicrobial resistance crisis is fuelling a search for novel antibiotics1. Several promising drug leads have been described in the past several years, and new approaches to discovery are being developed2. In situ growth of uncultured bacteria has led to the discovery of teixobactin that binds bacterial cell wall precursors and kills Gram-positive pathogens3. Remarkably, resistance to this compound has not been detected. The current need for novel antibiotics is especially acute for multidrug-resistant Gram-negative species4. Their outer membrane restricts permeation of compounds5, making discovery challenging6,7. An additional major obstacle for synthetic and natural-product discovery is the enormous background of toxic molecules, such as membrane-acting or DNA intercalators. Differential screening, testing for activity against two different groups of bacteria, largely resolves this bottleneck8-11. A recent differential screen against Borreliella burgdorferi, the causative agent of Lyme disease, rediscovered hygromycin A11.
Reasoning that particular bacterial ecology may lead to drug-like compounds, we recently turned to Xenorhabdus and Photorhabdus—symbionts of the nematode gut microbiome. When nematodes infect insect larvae, these bacteria are released, and produce antimicrobials to fend off competitors12,13. The main competitors in that environment are Gram-negative bacteria. The antimicrobials these symbionts produce must be non-toxic to the nematode, and have systemic availability throughout the larvae, and thus inherently good pharmacokinetics. These features match our requirements for antibiotics. Screening a small library of Photorhabdus led to the discovery of darobactins, a novel class of antibiotics targeting an essential outer membrane chaperone and insertase, the protein BamA14. BamA is a β-barrel protein that inserts and folds β-barrel proteins, such as porins, into the outer membrane. Darobactin A has an unusual fold and is a 7mer peptide with two fused rings that produce a rigid, preformed β-strand. Darobactin forms a β-β interaction with the lateral gate of BamA, outcompeting native substrates15.
Computational approaches comprise another important area of antibiotic discovery. Genomes of producing bacteria contain multiple biosynthetic gene clusters (BGCs), and only a fraction express their products in the lab16. A computational search for BGCs with a particular rare feature, a Ca2+ binding domain, led to the discovery of malacidins17. These compounds bind lipid II and have topical activity against Gram-positive bacteria. Another computational approach based on machine learning used a training set of unrelated compounds with antimicrobial activity from a synthetic library, resulting in the identification of halicin18. This is a membrane-acting compound with activity against Gram-negative bacteria and is also a topical agent. These are encouraging examples that set the stage for advancing computational approaches to antibiotic discovery.
Here we combine several approaches: computational analysis of BGCs distantly related to the darobactins and differential screening, to search for novel antibiotics against Gram-negative pathogens. This enabled discovery of a new class of antibiotics, the dynobactins, which act against Gram-negative bacteria by targeting BamA. Dynobactin A is apparently the first exemplar of a novel systemically active antibiotic lead compound discovered through a computational approach.
Bioprospecting radical S-adenosyl-l-methionine enzymes
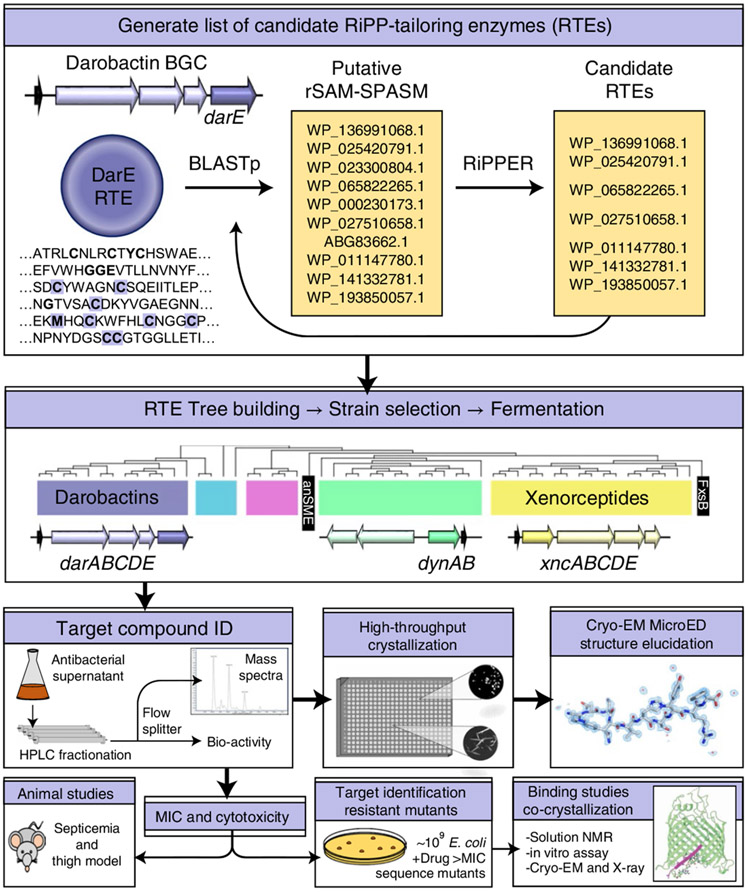
We reasoned that ribosomally synthesized and post-translationally modified peptides (RiPPs) distantly related to the darobactins could lead to novel antimicrobials (Fig. 1). We began with an iterative search using the darobactin radical SAM enzyme (DarE) as the starting point. First, the National Center for Biotechnology Information (NCBI) databases were queried using Basic Local Alignment Search Tool (BLAST)19 to generate a list of enzymes similar to DarE (Supplementary Table 1). This was curated by examining the genomic neighbourhood surrounding putative hits for propeptides with sequence similarity to the darobactins, or with the RiPP Precursor Peptide Enhanced Recognition (RiPPER) search tool, which compiles and scores prospective RiPP genes20. Upon identifying putative RiPP-tailoring enzymes (RTEs), these were leveraged for additional BLAST queries, expanding our list. This strategy yielded numerous additional darobactins (F–L) alongside many diverse RiPP operons (Supplementary Table 1).
Fig. 1 ∣. Schematic overview of the workflow.
The darobactin operon radical SAM enzyme, DarE, was used to query NCBI BLASTp. BLASTp hits were examined for associations with putative RiPPs in the surrounding genomic neighbourhood using the RiPPER search tool or manual curation, and were applied as further BLASTp queries in such cases. Producers of putatively identified RiPPs were fermented in media to screen for the presence of these RiPPs by biological activity assays or mass spectrometry. Once candidates were identified, these were purified and characterized through both biological assays to determine spectrum of action/cytotoxicity and structural elucidation approaches. A promising compound with low toxicity was then tested for efficacy in animal models of E. coli infection and its mechanism of action identified.
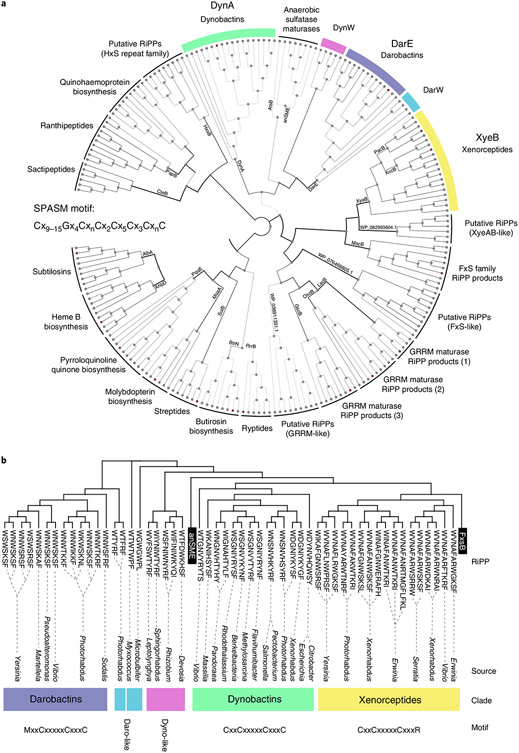
To differentiate putative RTEs, a multi-sequence alignment generated with ClustΩ21 was used to build a neighbour-joining phylogenetic tree (Fig. 2a). Outgroups of radical S-adenosyl-l-methionine enzymes (rSAM) enzymes containing SPASM and Twitch family domains, similar to DarE, but modifying a variety of substrates were selected22,23. Predicted clades had internally consistent operon structures (Fig. 2 and Extended Data Fig. 1), propeptide sequences and variations within their iron-coordinating SPASM domains (Fig. 2b and Supplementary Table 1). Utilizing the phylogenetic tree (Fig. 2), we examined SPASM motifs to guide our search for RiPPs. The canonical SPASM motif contains seven conserved cysteines that coordinate iron-sulfur clusters, Cx9–15Gx4CxnCx2Cx5Cx3CxnC22-25. Interestingly, the anaerobic sulfatase maturing enzyme SPASM group (anSME) fully coordinates their auxiliary iron-sulfur clusters26, with an additional cysteine upstream of the canonical motif. The DarE enzyme shares this irregular feature; however, DarE also contains a conserved methionine, exchanging a key cysteine (Fig. 2b). The DarE SPASM motif also terminates with an unusual cysteine pair. The relevance of these features are interesting topics for further enquiry, but for our purposes served as indicative patterns—a roadmap to unique RiPP-tailoring chemistry.
Fig. 2 ∣. Phylogenetic tree of rSAM-SPASM enzymes.
a, Neighbour-joining tree generated from ClustΩ multi-sequence alignment of distantly related SPASM enzymes. Putatively identified cyclophane RTEs were plotted alongside major characterized SPASM enzymes and their closest homologues (selected using NCBI BLASTp). RTE clades of note are highlighted. b, Neighbour-joining tree generated from ClustalΩ multi-sequence alignment of known RTEs (XyeB, DarE), close homologues and newly identified clades neighbouring DarE (DynA, DynW and DarW).
The largest clade neighbouring DarE bore striking resemblance to the anSME group. Uniquely, this RiPP-associated clade also contained eight cysteines. We named this enzyme group the DynA clade. Like both anSME and DarE, DynA enzymes contain the cysteine immediately upstream of the SPASM motif, an addendum to the motif we term the ‘tweak’ cysteine. Other characteristic features of DarE SPASM are absent in this group. Pairwise similarities of DynA and DarE proteins from a single Photorhabdus strain were compared, and despite high alignment coverage (~80%) and domain architecture, lower sequence conservation (25% identity, 47% similarity) indicated that these enzymes had substantially diverged. Other minor groups of putative RTE neighbouring the DarE and DynA clades on the tree were also identified to contain the tweak cysteine. We tentatively named these clades DynW and DarW (Supplementary Table 1).
Identification of dynobactin A
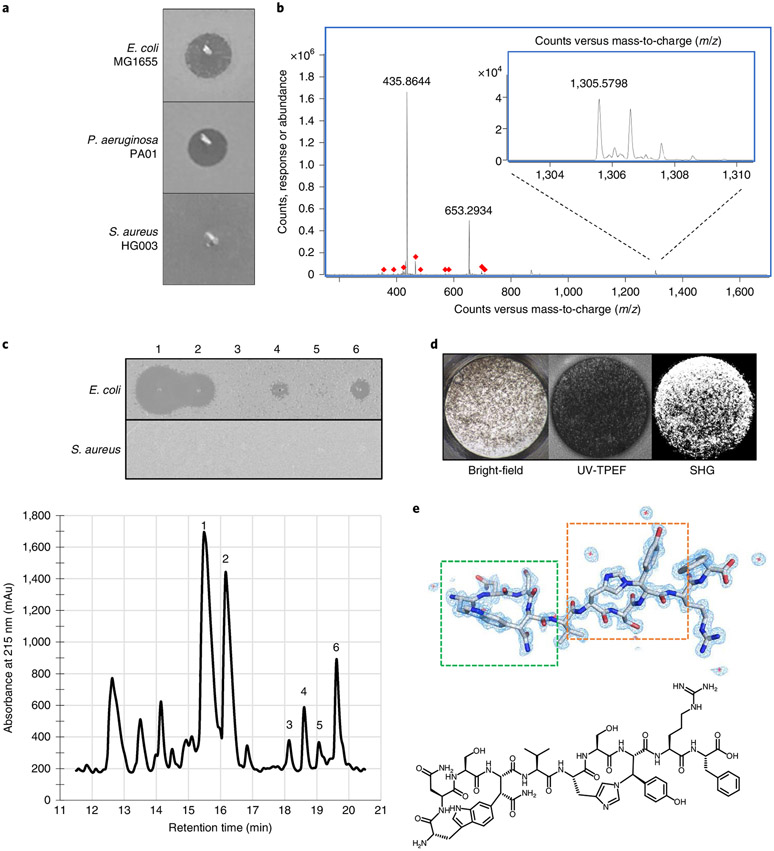
We assembled 14 strains containing RTEs (Extended Data Table 1) using our SPASM motif tree as a guide. These strains represented each of the diverse clades neighbouring DarE identified within the tree (Fig. 2). These strains were screened for antimicrobial activity as described previously14. Briefly, strains were fermented in tryptic soy broth for 8 d at 28 °C with aeration, and culture supernatant was concentrated 20-fold. This increases the likelihood of detecting poorly expressed compounds coded by silent operons. The resulting concentrates were spotted onto lawns of E. coli, P. aeruginosa and S. aureus to identify compounds acting selectively against Gram-negative bacteria. Photorhabdus australis displayed strong selective activity (Fig. 3a). In addition to DarE, the P. australis genome contains several RTEs depicted in our tree (Extended Data Fig. 1 and Table 1): the PacB Xye operon27, a DarW operon and a DynA operon.
Fig. 3 ∣. Identification of dynobactin A from P. australis.
a, Lawn bio-assay of 20x concentrated P. australis W34 supernatant. b, Mass spectra of dominant Gram-negative selective metabolite, exact mass (M) 1,304.57. c, Bottom: RP-HPLC chromatogram of XAD16N-extracted and SP-FF ion-exchanged P. australis supernatant. Top: isolated peaks assayed against lawns of E. coli and S. aureus. d, Well from high-throughput crystallization screen containing fluorescent crystals. e, Top: cryo-EM microED-generated 3D crystal structure of dynobactin A molecule. Dashed green box, carbon-carbon bond formed between the W1 C6 and the β-carbon of N4; dashed orange box, nitrogen-carbon linkage between the H6 imidazole Nε2 and the β-carbon of Y8. Bottom: accompanying 2D structure.
Table 1 ∣.
Dynobactin A minimum inhibitory concentrations
| MIC | |
|---|---|
| Gram-negative bacteria | μg ml−1 |
| Escherichia coli ATCC 25922 | 8 |
| +10% FBS | 8 |
| Escherichia coli K-12 MG1655 | 16 |
| Strain 1, BamA (G429V, G807V) | >128 |
| Strain 2, BamA (F394V, E435K, G443D) | 16 |
| Strain 3, BamA (T434A, Q445P, A705T) | 16 |
| Escherichia coli AR350 | 8 |
| Shigella sonnei ATCC 25931 | 4 |
| Shigella flexneri KLE 2512 | 8 |
| Salmonella enterica typhimurium LT2 ATCC 19585 | 8 |
| Salmonella enterica Enteritidis AR496 | 8 |
| Enterobacter cloaceae ATCC 13047 | 16 |
| Klebsiella pneumoniae AR347 | 64 |
| Pseudomonas aeruginosa PA01 | 8 |
| Pseudomonas aeruginosa PA14 | 16 |
| Acinetobacter baumannii ATCC 19606 | 16 |
| Moraxella catarrhalis ATCC 25238 | 2 |
| Yersinia pseudotuberculosis ATCC 6904 | 8 |
| Vibrio vulnificus KLE δ-1125 | 8 |
| Veillonella ratti KLE 2365 | 16 |
| Bacteroides fragilis KLE 2244 | >128 |
| Bacteroides stercoris KLE 2537 | >128 |
| Photorhabdus australis DSM 17609 | >128 |
| Gram-positive bacteria | |
| Staphylococcus aureus HG003 | >1000 |
| Bacillus subtilis 168 | >1000 |
| Limosilactobacillus reuteri LTH5448 | >128 |
| Clostridium bifermentans KLE 2329 | >128 |
| Human cell lines | |
| FaDu (Epithelium) | >1000 |
| HepG2 (Hepatocytes) | >1000 |
| HEK293 (Embryonic kidney) | >1000 |
| A549 (Lung epithelium) | >1000 |
MICs and cytotoxicity data for pathogens, human gut microbiome members and human cell lines. MICs were determined in biological triplicate.
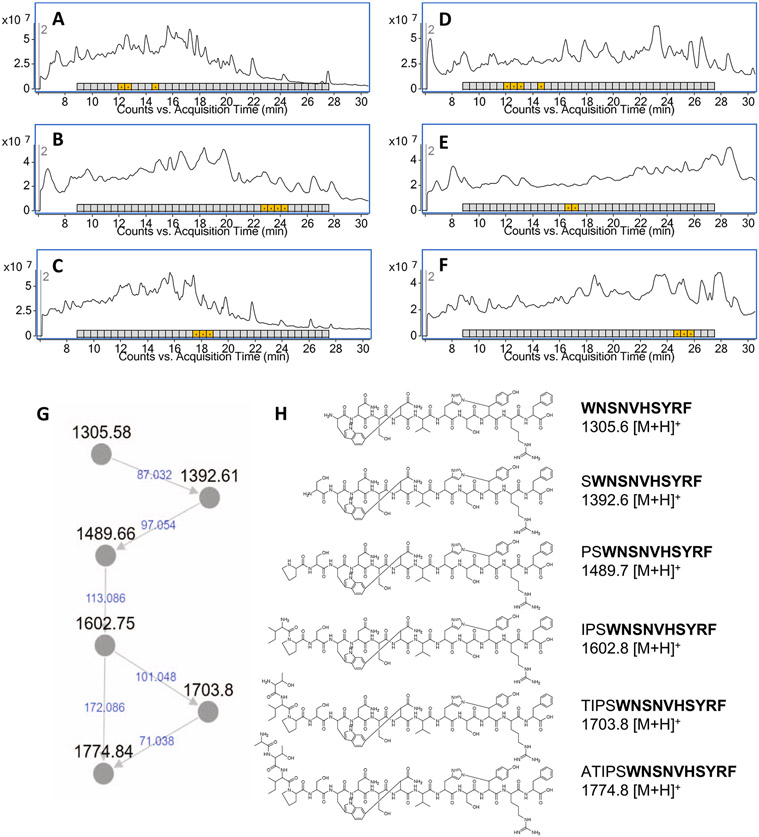
To rapidly dereplicate active compounds, concentrated culture supernatant was separated by high-performance liquid chromatography (HPLC), splitting flow between a fraction collector and the mass spectrometer (MS). Fractions simultaneously enter the MS and the well of a microtitre plate where E. coli is screened for growth inhibition. A small panel of analytical columns (C18, Phenyl and Pentafluorophenyl) were used in parallel with two different eluants (acetonitrile and methanol) (Extended Data Fig. 2a-f). From these data, candidate ions from antimicrobial fractions were compiled from the independent HPLC conditions and subsequently cross-referenced to rule out inactive co-eluants (Supplementary Table 2). This cross-referencing identified the dominant selectively active compound corresponding to an ion of exact mass (M) of 1,304.57 (Fig. 3b).
The dominant metabolite showed no activity against Gram-positive representatives and no detectable toxicity against four human cell lines at 1 mg ml−1. A larger panel was used to assess the spectrum of action (Table 1). The compound is effective against γ-Proteobacteria but did not harm Bacteroides, the main Gram-negative human gut symbionts. Interestingly, Veillonella, a Gram-negative Firmicute that borrowed the genes for an outer membrane from Proteobacteria long ago, also inherited susceptibility to this metabolite28,29.
Production was scaled up, combining resin extraction, cation exchange and HPLC. In addition to the dominant metabolite, further examination revealed others with selective activity, having exact masses of 1,488.65, 1,601.74, 1,702.79 and 1,773.82. These showed similar potency and selective activity (Fig. 3c and Supplementary Table 3). To ascertain whether these metabolites were structurally related, fragmentation patterns generated by tandem mass spectrometry (LC–MS/MS) were analysed using the Global Natural Product Social Molecular Networking (GNPS)30. GNPS identified relationships between these masses (Extended Data Fig. 2g) and connections to metabolites not initially detected (exact M: 1,391.60 and 706.32) (Extended Data Fig. 2h). By assuming losses of single amino acids, a partial amino acid sequence from the DynB leader peptide was identified. With this confirmation, we concluded that the known post-translational modifications of the darobactins—a carbon-carbon connection and ether connection—could not explain the masses of these products.
Cryo-EM microED technique yields novel antibiotic structure
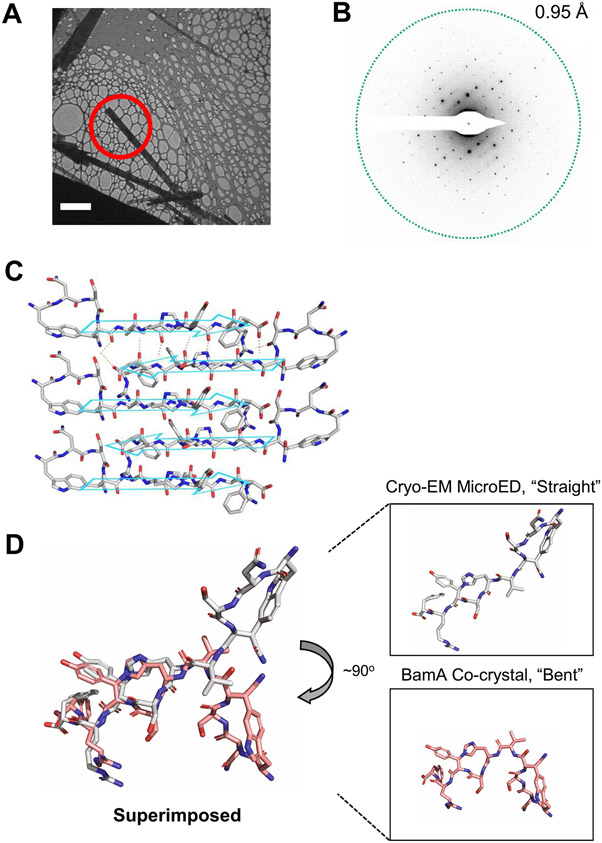
To determine the structure of these compounds, we employed cryo-electron microscopy (cryo-EM) micro-electron diffraction (microED), a technique recently adapted to small molecules. This approach does not require large crystals (Extended Data Fig. 3a) and has been shown to be applicable to seemingly amorphous powders31. To obtain crystals, 1 mg of the dominant metabolite was split among 1,536 crystallization conditions in microtitre plates and remotely monitored for chiral crystals by second-order nonlinear imaging (Fig. 3d) at the Hauptman-Woodward Institute32. Within 24 h, nanocrystals amenable to cryo-EM microED were detected and subjected to electron diffraction.
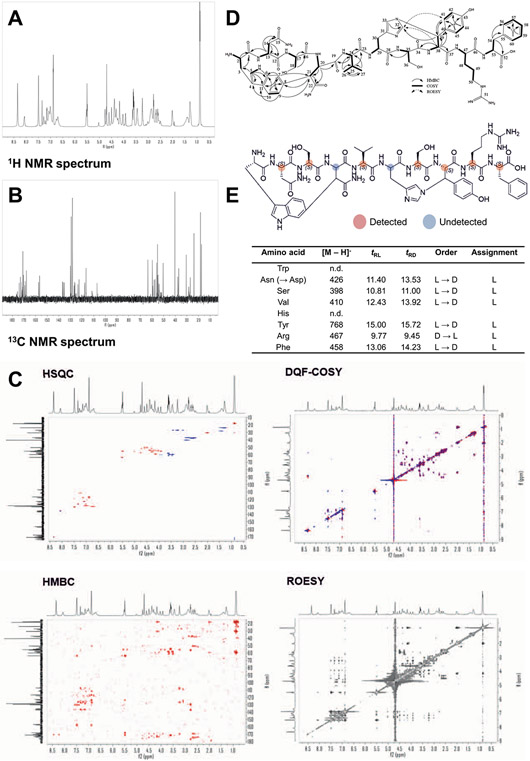
Crystals were transferred to a gold grid and visualized using bright-field imaging on a transmission cryo-electron microscope (TEM) (Extended Data Fig. 3a). These were rotated as they were struck by an electron beam, and the resulting diffraction data were captured by a high-speed camera (Extended Data Fig. 3b). This resulted in a 1.05-Å-resolution dataset with a high degree of completeness (Extended Data Fig. 3c and Supplementary Table 4). The novel compound, here named dynobactin A, is apparently the first natural-product antibiotic of unknown structure solved de novo with this approach (Fig. 3e, PDB 7T3H). Dynobactin A is a deca-peptide of sequence W1N2S3N4V5H6S7Y8R9F10. Two closed rings are present, with a carbon-carbon bond formed between the W1 C6 and the β-carbon of N4 (dashed green box, Fig. 3e) and an unusual nitrogen-carbon linkage between the H6 imidazole Nε2 and the β-carbon of Y8 (dashed orange box, Fig. 3e). These connections create unfused 4- and 3-constituent rings respectively, resulting in a flexible peptide (Extended Data Fig. 3d), contrasting with the fused rings of darobactins. Further supporting structural characterizations by Marfey’s analysis and full assignment by NMR were also performed (Extended Data Fig. 4 and Supplementary Table 5). The solubility of dynobactin A was tested side-by-side with darobactin A. Dynobactin A showed water solubility in excess of 200 mg ml−1, whereas darobactin A began to gel and show signs of insolubility at 10 mg ml−1 in water. The flexibility and additional chargeable site probably contribute to this order of magnitude difference in solubility for dynobactin.
Ring 1 of dynobactin A is a 4-constituent ring (W1-to-N4) bearing similarities to the 3-constitutent xenorceptide ring 1 (W1-to-N3)33. Ring 2 (H6-to-Y8) contains an unprecedented linkage, the closest analogous feature being a histidine Nε2 linked to an alanine β-carbon reported for a marine natural product34. The second ring of dynobactin A is distinct from both cyclophane closures in the darobactins and is apparently the first example of an N-C cyclization formed by an rSAM enzyme35,36. The lack of exogenous oxygen in the structure may suggest different mechanisms underpinning biosynthesis between DarE and DynA.
Dynobactin A targets BamA
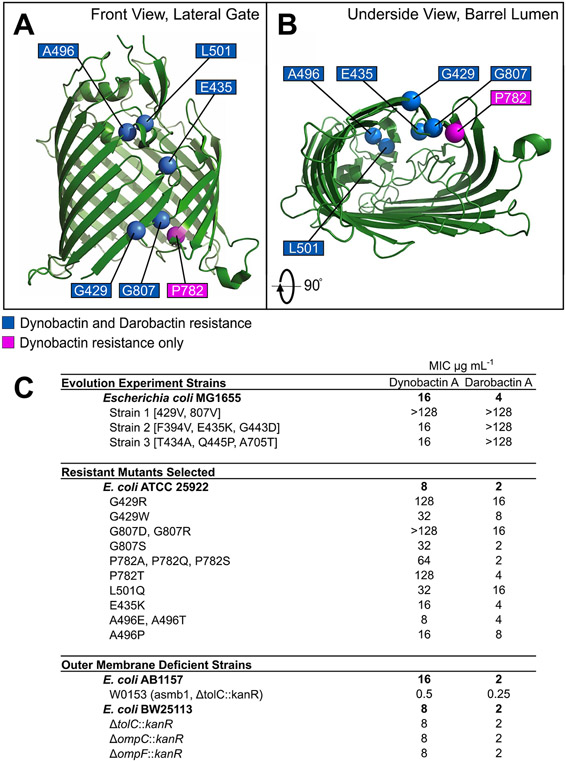
A high-density E. coli culture was plated onto Muller-Hinton II agar containing dynobactin at 2x and 4x minimum inhibitory concentrations (MIC), and resistant colonies were obtained at a frequency of 2 × 10−8 and 3 × 10−9, respectively. The bamA gene of 96 stable mutant colonies was PCR-amplified, and in all cases mutations were observed within bamA (Extended Data Fig. 5). Several mutations conferring strong resistance to dynobactin were identified at the same residues reported for darobactin: BamA G429 and G807. We also identified a new mutation outside the β1 and β16 strands, L501Q, conferring resistance to both darobactin (8x MIC) and dynobactin (4x MIC).
Unique mutations only conferring dynobactin resistance were also present at site P782 (8x MIC). To confirm whether a BamA mutation was solely responsible for resistance, we constructed an E. coli strain carrying BamA mutation P782T by lambda red recombination37,38, whereafter the resistance phenotype matched. We also tested three mutants with high resistance to darobactin obtained in a previously reported evolution experiment14. Two of the three mutants (strain 2 (F394V, E435K, G443D) and strain 3 ((T434 A, Q445P, A705T)) showed no dynobactin MIC shift when compared with the parental strain; however, strain 1 (G429V, G807V) showed high resistance to dynobactin (Table 1). Taken together, these findings indicate that BamA is the dynobactin target, and that not all resistance mutations confer cross-resistance.
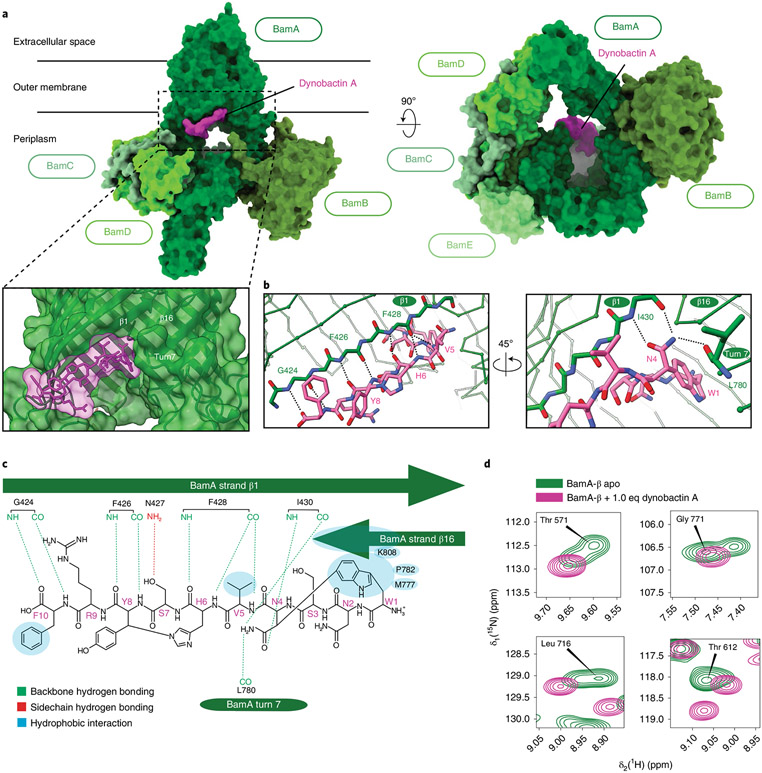
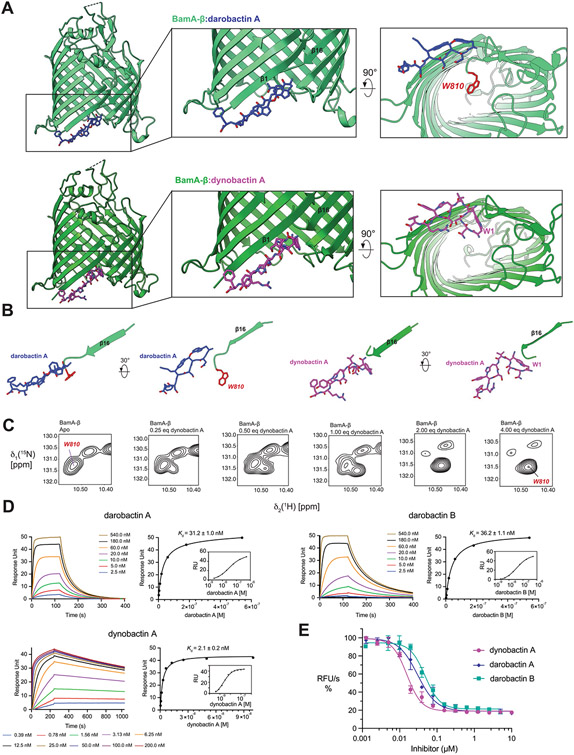
To resolve the mechanism of action, we determined the structure of the BAM complex with bound dynobactin A by cryo-EM (Fig. 4a, and Supplementary Fig. 1 and Table 6). The reconstruction at 3.6 Å resolution revealed that dynobactin binds in the lateral gate region of BamA formed by strands β1 and β16. To examine this interaction, a crystal structure of the BamA β-barrel domain (BamA-β) in detergent micelles with bound dynobactin A at 2.5 Å resolution was observed (Fig. 4b, Extended Data Figs. 6 and 7a-d, and Supplementary Table 7). Interestingly, the dynobactin binding site only partially overlaps with darobactin15. Residues N4–F10 occupy roughly the same binding site as darobactin; however, N-terminal residues W1N2S3 protrude into the lumen of BamA.
Fig. 4 ∣. Dynobactin A binds BamA lateral gate.
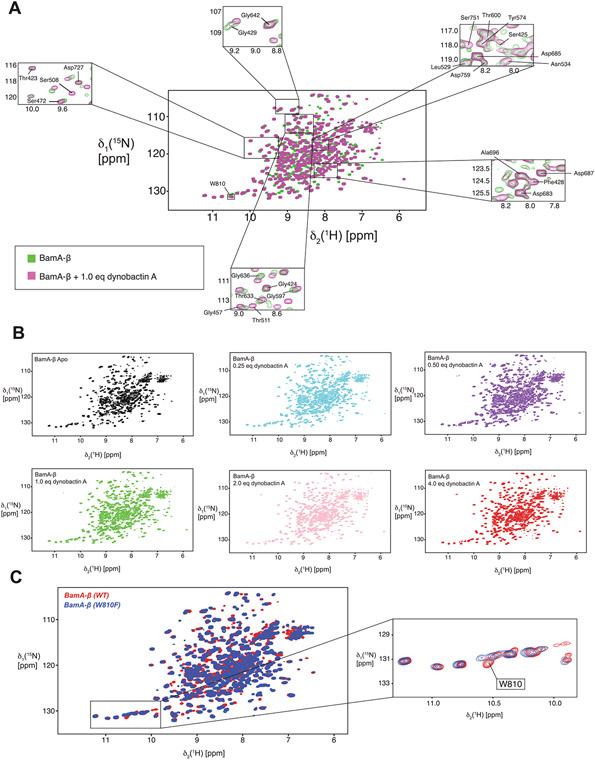
a, Three-dimensional structure of the BAM complex in DDM detergent micelles with bound dynobactin A, resolved by cryo-EM to a resolution of 3.6 Å. The proteins are shown as surfaces with colours as annotated. b, Close-up view of the dynobactin A binding site in a 2.5 Å crystal structure of dynobactin A bound to BamA-β. Amino acids involved in dynobactin A binding are shown in stick representation and polar contacts are shown as dashed lines: blue, nitrogen; red, oxygen; green and magenta (BamA and dynobactin, respectively), carbon. c, Schematic representation of the interactions between dynobactin A and BamA. d, Selected regions from a 2D [15N,1H]-TROSY spectrum of BamA-β in LDAO detergent micelles in the absence (green) and presence (magenta) of dynobactin A. The four selected amino acids show doublet peaks resolved into a singlet upon dynobactin A addition. Full spectra are shown in Extended Data Fig. 8.
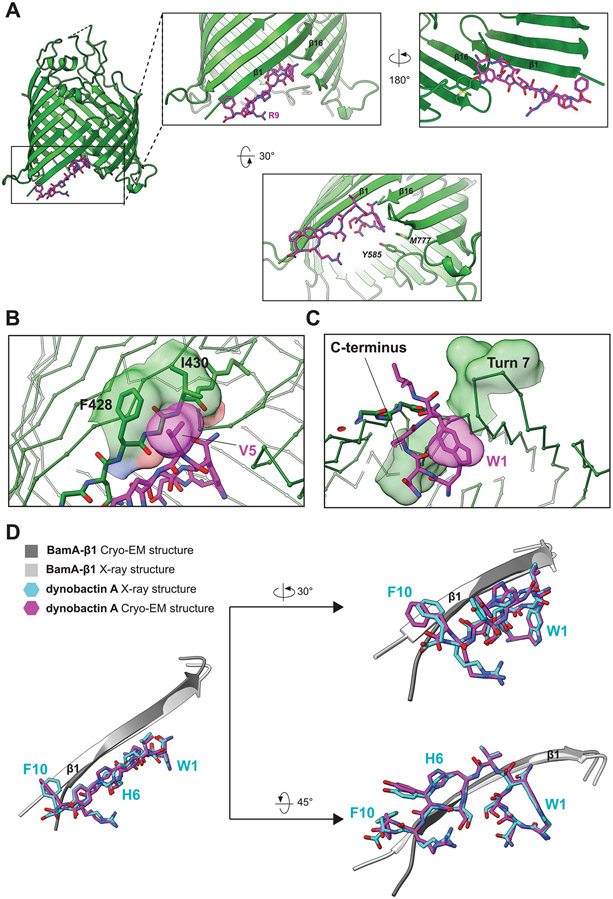
The binding interaction is formed by multiple contacts including a set of backbone-mediated hydrogen bonds (Fig. 4b and Extended Data Fig. 7a). Residues F426 and F428 form backbone-backbone hydrogen bonds to dynobactin at Y8 and H6, respectively. The BamA N427 side chain targets the S7 hydroxyl via its amine proton; notably, this same side chain interacts via its carbonyl oxygen with N2 side chain amine in darobactin A15. The dynobactin C-terminal phenylalanine residue binds in the recognition pocket, paralleling darobactin15. The adjacent residue R9 extends toward the polar head groups of membrane lipids. Further interactions were observed to Y585 and M777 located in turns 4 and 7 of BamA, respectively (Extended Data Fig. 7a). A non-canonical hydrogen bond was observed between F428 and the amide proton of H6, resulting in bifurcated hydrogen bonding between the carbonyl group of F428, simultaneously to H6 and V5 of dynobactin A (Fig. 4b,c). Additionally, the V5 side chain makes a hydrophobic contact with BamA side chains F428 and I430 (Extended Data Fig. 7b). Moreover, I430 demonstrated two hydrogen bond backbone interactions via its amide proton and carbonyl oxygen to the N4 side chain (Fig. 4b,c). Uniquely, dynobactin residue W1 features a large hydrophobic interaction with the BamA C-terminus (Extended Data Fig. 7c). Taken together, these structural data demonstrate that the BamA-dynobactin interaction is mediated through hydrogen bonds and hydrophobic interactions.
Additionally, dynobactin A binding to BamA-β was assessed using solution NMR spectroscopy and was shown to induce substantial chemical shift perturbations (Extended Data Fig. 8a). We observed that addition of one equivalent of dynobactin A to BamA-β resulted in selection of a singlet peak out of the doublets, selecting an ensemble state resembling the outward closed and gate closed conformations, similar to darobactin A39, as exemplified by Thr571, Gly771, Leu716 and Thr612 (Fig. 4d).
Notably, dynobactin W1 occupies the same spatial position occupied by W810 of BamA-β in the complex with darobactin A15 (PDB:7NRF) (Extended Data Fig. 6a,b). The N-terminal extension W1N2S3, unique to dynobactin, thus displaces the BamA C-terminus further into the barrel lumen, resulting in a flexibly disordered W810 (Extended Data Fig. 6b). This was corroborated by monitoring titration of BamA-β and dynobactin A by two-dimensional (2D) [15N,1H]-TROSY spectra. W810 strongly responded to the addition of dynobactin, its signal switching from a doublet in the apo form to a new position not populated in the apo form (Extended Data Figs. 6c and 8); this peak was assigned after confirmation by mutagenesis.
To quantify the strength of target–ligand associations, surface plasmon resonance was used to measure dissociation constants of darobactin A, darobactin B and dynobactin A. Measurements indicated that dynobactin A interacted an order of magnitude more strongly with BamA, with a disassociation rate constant 30–70 times slower than both darobactins tested (Supplementary Table 8). In contrast to the darobactins which bind mono-exponentially, dynobactin associated with BamA in a bimodal manner, consistent with a second binding step whereby a rearrangement increases affinity.
Efficacy of dynobactin in vitro and in vivo
To assess activity of these compounds in vitro, we performed a functional assay where the BAM complex mediates folding of the protein OmpT. Notably, this assay was performed here in outer membrane vesicles (OMVs), leveraging the native outer membrane40. This assay demonstrated that dynobactin A is a more potent inhibitor than both darobactin A and B (Extended Data Fig. 6e and Supplementary Fig. 2). Quantitative assessment of the data yielded IC50 values of 30 ± 6 nM, 48 ± 8 nM and 16 ± 2 nM for darobactin A, darobactin B and dynobactin A, respectively (Supplementary Fig. 2). To test whether the values of log IC50 differed significantly between pairs of datasets, the extra sum-of-squares F-test implemented in GraphPad Prism was used. In both cases, the hypothesis that the log IC50 was the same for both datasets was rejected at the 95% confidence level (P = 0.05), indicating a significant difference in the log IC50 between dynobactin A and darobactin A, and between dynobactin A and darobactin B (Supplementary Fig. 2). Remarkably, dynobactin A is thus the most potent of the three antibiotics in vitro. Dynobactin A is approximately 4-fold less potent against whole cells as compared with darobactin A (Fig. 5a, Table 1, and Extended Data Figs. 5 and 9). We therefore sought to determine whether target access was responsible. We first tested a strain with a defective outer membrane. E. coli W0153 contains a mutant lpxC allele, diminishing lipopolysaccharide production and outer membrane integrity41. W0153 additionally carries a tolC deletion, disabling several multi-drug resistance efflux pumps42. Combined, these facilitate access to the periplasm, the location of the BamA β1 strand. The MICs against E. coli W0153 for both dynobactin A and darobactin A improved (0.5 and 0.25 μg ml−1, respectively) (Extended Data Fig. 5). When considering the molarity of these drugs, the difference in potency between the two seems negligible (<2-fold). The MICs of both dynobactin A and darobactin A did not change significantly in a ΔtolC:kanR knockout43 or in knockouts of major porins ompC and ompf44,45. These results indicate that the outer membrane serves as a partial barrier for both compounds, and more so for dynobactin.
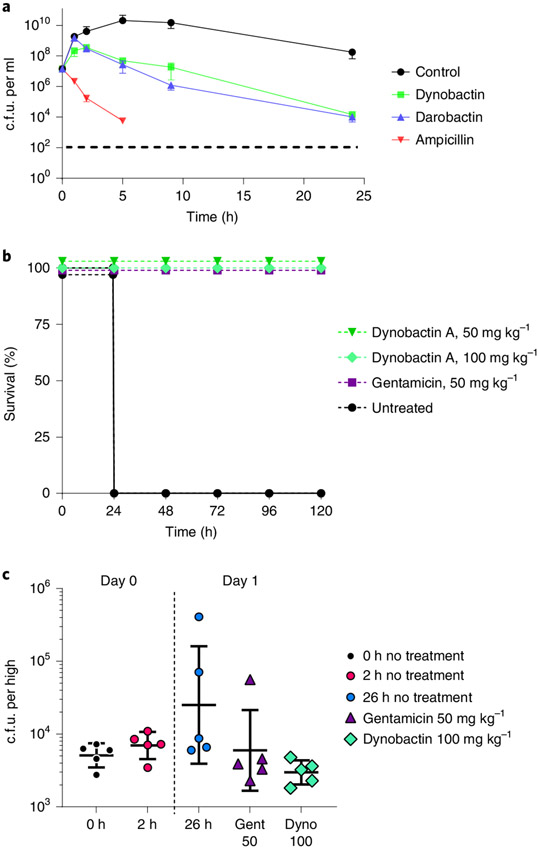
Fig. 5 ∣. Efficacy of dynobactin A.
a, Time-dependent killing of E. coli ATCC 25922 by dynobactin, darobactin and ampicillin. Antibiotics were added at 4x their respective MICs. Time points are graphed as the mean c.f.u. ± s.d. The experiment was performed in biological triplicate. Ampicillin-killed E. coli fell below the limit of detection, denoted by a dashed line. b, Mouse septicaemia model, wherein mice were inoculated with a lethal dose of multidrug-resistant E. coli AR350, followed by administration of a single intraperitoneal dose of antibiotics: dynobactin 50 mg kg−1, dynobactin 100 mg kg−1 and gentamicin 50 mg kg−1 at 1 h post infection; an untreated mouse group was included. Four mice were tested per group. c, In a neutropenic thigh model of E. coli AR350 infection, gentamicin (purple triangle) or dynobactin (green diamond) were delivered to groups of mice (n = 5) by intraperitoneal injection at 2 h post infection (red circle). Infection was monitored over 26 h. At 26 h post infection (blue circle), thighs were homogenized, serially diluted and plated in triplicate for c.f.u. Error bars represent c.f.u. mean ± s.d. for each group.
A mouse septicaemia model and thigh infection model utilizing a polymyxin-resistant E. coli AR350 mcr-1 were used to assess efficacy of dynobactin A as described previously14. In the septicaemia model, lethality was observed after 24 h of infection (>90% mice). Mice that received dynobactin A were completely protected and displayed no signs of toxicity (Fig. 5b). In a thigh model, mice were killed after 24 h to determine the E. coli burden. Dynobactin at 100 mg kg−1 was efficacious, decreasing E. coli burden and showing comparable results to 50 mg kg−1 gentamicin (Fig. 5c). Taken together, results indicate that dynobactins have development potential as therapeutics against Gram-negative pathogens.
Discussion
The permeability barrier of Gram-negative species poses a unique challenge to drug discovery5,45. Notably, there are two conserved, essential surface proteins in Gram-negative bacteria—BamA of the BAM complex insertase and LptD, a lipopolysaccharide translocator46. A large, 14-constituent murepavadin polycation was found to bind to LptD47,48. A derivative of murepavadin linked to polymyxin binds lipopolysaccharide and BamA49. Polycations have a liability—nephrotoxicity—that has so far prevented development of murepavadin and its derivatives. BamA is especially challenging, being a classically ‘undruggable’ target. BamA is a non-enzymatic β-barrel protein lacking a defined catalytic center. A synthetic compound library screen identified MRL-494, which binds BamA with a modest MIC, but has off-target membrane activity50. Notably, MRL-494 activity was unaltered by mutations affecting outer membrane permeability, suggesting action at the surface of BamA. Genentech took a different approach, raising a BamA-targeting monoclonal antibody51; however, its activity necessitates truncated lipopolysaccharide to reach its target. We recently reported a natural product—darobactin A—produced by Photorhabdus, targeting BamA14. Darobactin is a 7mer peptide with two fused rings, which creates a preformed β-strand in the peptide backbone. This leads to a tight β-β interaction with the BamA lateral gate, the site of peptide entry15. The lateral gate appears to be the Achilles’ heel of BamA.
In search of novel antibiotics acting against Gram-negative bacteria, we undertook a computational approach, looking for compounds only distantly related to darobactins. Natural products form orthologous groups, where enzymes are related but have diverged far enough that their products are no longer analogues52. For example, a search for BGCs that share a Ca2+-binding domain with that of daptomycin led to the discovery of chemically unrelated malacidins17. Our search indicated a large family of RiPP operons sharing partial homology with rSAM enzyme DarE, but carrying distinct propeptides; we named these dynobactins. Phylogenetic tree construction and examination of SPASM motifs guided our discovery of this novel RiPP class. In addition to the dynobactins, other unexplored clades neighbouring the darobactins and dynobactins (DarW and DynW) present further avenues for enquiry. One of the dynobactin propeptides is found in Photorhabdus, a group of bacteria likely to make non-toxic compounds as a result of their symbiosis with nematodes. To facilitate identification of a dynobactin RiPP, we employed differential screening, resolving the bottleneck of non-specifically acting compounds, revealing a compound that inhibited the growth of E. coli but not S. aureus. Cryo-EM microED rapidly determined its structure, revealing a 10mer peptide with distinct cyclophane rings. Dynobactin A has two unfused rings, one bearing unprecedented N-C cyclization for rSAM-SPASM enzymes, histidine Nε2 to a tyrosine β-carbon. Analysis of resistant mutants suggested that the target is BamA. Cryo-EM of BAM-dynobactin and X-ray analysis of the BamA-dynobactin complex displayed lateral gate binding, an unexpected protrusion and additional interactions from within the β-barrel lumen. Apparently, the dynobactin backbone first forms β-β interactions with the lateral gate, enabling additional hydrophobic interactions, resulting in specific and tight binding. Functional in vitro assays show stronger BamA binding by dynobactin than darobactin, further supported by surface plasmon resonance measurements. The activity of dynobactin A increases 32-fold against an E. coli mutant with a defective outer membrane, but is unaffected by a ΔtolC knockout in multi-drug resistance efflux, suggesting slower permeation of dynobactin A to the interface of the outer membrane and the periplasm as compared with darobactin A. This explains its lower whole-cell activity. How dynobactin A reaches its target is unclear; the cut-off for permeating the outer membrane is 600 Da53,54, while dynobactin is 1,304 Da. We speculate that dynobactin may enter along the BamA/membrane interface, explaining penetration and why several resistant mutations lie outside the lateral gate.
Why two different BamA inhibitors are produced by P. australis is an intriguing question. Not all BamA mutations provide resistance to both compounds; this would suggest that expression of the two antibiotics decreases effective resistance frequency. Notably, mutations in BamA can decrease fitness in vivo14. Mutation at BamA G807 confers resistance to both darobactin and dynobactin, and was previously reported to increase susceptibility of E. coli to vancomycin and rifampicin55. This suggests that an aberrant outer membrane results from these mutations. Previously, all tested darobactin-resistant mutants resulted in loss of virulence;14 many of these same mutations confer dynobactin resistance. This means that in vivo resistance frequency to these compounds may be very low.
Dynobactin A showed no cytotoxicity in concentrations up to 1 mg ml−1, and had efficacy in mouse septicaemia and thigh infection models with E. coli. Based on our computational analyses, there are approximately 16 unique dynobactins coded by the BGCs found in the NCBI database56, providing a diverse starting set for drug development. Unlike darobactins, dynobactin A has excellent water solubility in excess of 200 mg ml−1—an attractive property. Apart from natural diversity, RiPP operons offer straightforward approaches to medicinal chemistry optimization by cloning/amino acid replacement. A recent report describes that a simple Phe/Trp substitution of the last amino acid of darobactins expands their spectrum and potency57. A similar approach will be available to modify dynobactins. The discovery of dynobactins shows that computational analysis is a fruitful approach to finding new antibiotic leads and furthers our understanding of targeting BamA. Darobactins and dynobactins are probably only the first two examples of natural-product antibiotics acting against the ‘undruggable’ surface proteins of Gram-negative bacteria.
Methods
Identification of putative RiPP clusters.
NCBI BLAST was queried with the sequence of the darobactin biosynthetic enzyme DarE (WP_152962147.1). Results from these queries were manually screened or mined by using the RiPPER search tool to identify putative propeptides in the genomic neighbourhood of BLAST hits (https://github.com/streptomyces/ripper). The outputs of these screens were included in our iterative search strategy: rSAM enzymes scored with a high-likelihood-to-neighbour RiPPs were used as queries in BLAST and this output was examined using RiPPER. The putative RiPP-tailoring enzymes were organized by creating a multi-sequence alignment utilizing ClustaΩ21. This online tool additionally generated a phylogenetic tree, which was visualized using the UGENE platform58.
Screening conditions.
Strains were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, https://www.dsmz.de/), Belgian Co-ordinated Collections of Micro-organisms (BCCM/LMG, https://bccm.belspo.be/about-us/bccm-lmg), kindly provided by Dr Nick Waterfield at the University of Warwick, or kindly provided by Dr Aunchalee Thanwisai from Naresuan University. Fermentations of these strains were grown in tryptic soy broth. Briefly, supernatant was collected from cells grown in 10 ml tryptic soy broth in 50 ml falcon tubes after growth for 8 d at 28 °C with 200 r.p.m. shaking. Aliquots were concentrated by centrifugal evaporation or lyophilisation, reconstituted at 20x and then assayed on lawns of test organisms: E. coli, P. aeruginosa and S. aureus. Lawns of bacteria were prepared by diluting overnight cultures of the respective bacteria in Mueller Hinton II (MHII) broth, adjusting the final concentration to optical density (OD)600 = 0.03, and flooding MHII agar plates with this dilute liquid culture. The excess culture solution was removed and the plates allowed to dry in a sterile biosafety cabinet. Concentrated Photorhabdus and Xenorhabdus culture supernatants were then spotted on these lawns, allowed to dry, incubated at 37 °C overnight and examined for inhibition zones.
Strain fermentation and purification of dynobactin A.
Photorhabdus australis was grown as 5 ml overnight culture in starter culture tubes incubated at 28 °C with 200 r.p.m. shaking, inoculated 1:100 into a 2 l Erlenmeyer flask with 500 ml tryptic soy broth and incubated for 8–10 d. Culture supernatant (10 l) was collected by centrifugation at 10,000 g for 10 min, and incubated overnight with agitation with 2 kg polymeric resin XAD16N (20–60 mesh, Sigma-Aldrich). Resin was washed using 20 l of doubly deionized water (ddH2O), and dynobactin was eluted from XAD16N resin utilizing 4 l of acidified methanol (0.1% (v/v) formic acid). The eluate was concentrated using a rotary evaporator, exchanged into water containing 0.1% formic acid and applied to a cation exchange chromatography column. The eluate was loaded onto the cation exchange resin (200 ml of SP Sepharose Fast Flow) and flushed with 0.1% formic acid (v/v) in ddH2O at pH 3. The bound dynobactin A was then eluted using a step gradient of 50 mM ammonium acetate: pH 5 and then pH 7. The resin was then washed with 0.5 M NaOH and 0.5 M NaCl. Each gradient step consisted of 10 column volumes of the respective buffers. The pH 7 eluate, showing biological activity, was concentrated by rotary evaporator or lyophilised, and further purified by reverse-phase high-performance liquid chromatography (RP-HPLC) using a C18 column (Agilent 1200, XBridge Prep C18, 5 μm OBD, 19 × 250 mm column). RP-HPLC conditions were as follows: solvent A, ddH2O with 0.1% (v/v) formic acid; solvent B, acetonitrile with 0.1% (v/v) formic acid. A flow rate of 15 ml min−1 was used over a gradient of 2–20% solvent B, incrementing 1% per minute. A photodiode array enables UV monitoring (210 to 400 nm) for peak picking. Dynobactin A could be collected with purity in excess of 98% by UV analysis.
Mass spectrometry analyses.
To determine the exact mass of dynobactin A, an Agilent 1260 Infinity liquid chromatography system coupled to a 6530 quadrupole time-of-flight mass spectrometer equipped with electrospray ionization (Agilent Technologies) was leveraged for LC–MS/MS analyses of dynobactin A. A Waters XBridge C18 column (100 mm × 2.1 mm, 3.5 μm) was utilized for the separation, with a flow rate of 200 μl min−1 with solvent A (0.1% (v/v) formic acid in water) and solvent B (0.1% (v/v) formic acid in acetonitrile). The initial concentration of 2% solvent B was maintained for 2 min, followed by a linear gradient to 30% over 30 min. MS parameters were as follows: gas temperature, 300 °C; gas flow, 7 l min−1; nebulizer, 35 psi; fragmentor voltage, 175 V; skimmer voltage, 65 V. Acquisition was set in positive mode at ‘Auto MS/MS’ with the following parameters: mass range for MS 111–3,000 m/z with 2 spectra per second; mass range for MSMS, 50–3,000 m/z with 4 spectra per second; collision energy, ‘use formula’ mode, slope = 1 and offset = 10; max precursor per cycle, 10; active exclusion after 3 spectra with released after 0.5 min. MassHunter qualitative analysis software (v10.1, Agilent Technologies) was used for qualitative analysis to acquire dynobactin A exact mass and fragmentation. An accurate mass spectrometry analysis identified a compound with an exact mass of 1,305.5799 [M + H]+, which is consistent with the molecular formula of C60H77N18O16 (calculated [M + H]+ = 1,305.5765, Δ2.6 ppm).
Molecular networking analyses.
Using the GNPS website (http://gnps.ucsd.edu) online workflow (https://ccms-ucsd.github.io/GNPSDocumentation/), mass spectrometry data from biologically active P. australis ion exchange fractions were first filtered by removing MS/MS fragment ions within ±17 Da of the precursor m/z. These MS/MS spectra were then window filtered by selecting only the top 6 fragment ions within a set threshold of ±50 Da. A precursor ion mass tolerance was set to 0.1 Da and an MS/MS fragment ion tolerance set to 0.02 Da. From this selection, a network was generated, having its edges filtered for a minimum cosine score of 0.4 and greater than 3 matched peaks. Further, edges between two nodes were kept dependent on each respective node appearing in a corresponding node’s top 10 most similar nodes. Additionally, the maximum molecular family size was set to 100, pruning the lowest scoring edges until a molecular family was within this set range. The network was then queried against GNPS’s spectral libraries. The spectra were then filtered in, following the parameters of the input data. All matches kept between network spectra and library spectra were required to have a score above 0.7 and at least 6 matched peaks.
Dynobactin A structure elucidation by Cryo-EM microED.
To optimize crystallization conditions, 500 μl 2.5 mg ml−1 dynobactin A dissolved in water was screened at the high-throughput crystallization center of the Hauptman-Woodward Institute, where crystals were detected by SONICC59. Two conditions were selected that provided promising needle- and rod-shaped crystals suitable for microED: 20% ethanol, 0.1 M Tris, pH 8.5 and 1 M MgSO4, 0.1 M Tris, pH 8.5. Crystals were fragmented and dispersed by vortexing with a mixture of 0.1 and 0.5-mm-diameter glass beads (Biospec Products). To transfer the crystals from Eppendorf tubes into TEM grids (lacey carbon Au 300 mesh, Ted Pella), we adapted a pressure-assisted backside blotting method60 before plunge freezing the grids in liquid ethane and transferring into liquid nitrogen. The grids were placed in the cartridge and cryo-transferred to the TEM for investigation. All images and diffraction data were collected at cryogenic temperature on a Thermo Fisher Talos Arctica TEM operating at an acceleration voltage of 200 keV. Crystals were identified both in low-magnification imaging and diffraction modes. Diffraction images from selected crystals were recorded as a movie with a 4 K × 4 K CMOS (Thermo Fisher Ceta) camera as the crystals were continuously rotated under the electron beam. Data collection was accomplished using a constant tilt rate of ~0.6° s−1 over an angular wedge of ~65°. Individual crystals were isolated using a selected area aperture to reduce the background noise, and their eucentric heights were adjusted to stay in the aperture over the entire tilt range. Because of their long flat morphology, these crystals were distributed on the grid with a preferred orientation, which complicated the generation of datasets with a high degree of completeness. Thus, it was necessary to accumulate diffraction data from multiple crystals to increase completeness. Each crystal was identified in bright-field imaging mode and continuous-rotation electron diffraction data from 19 crystals were collected accordingly. The diffraction movies were saved as SER files and converted into SMV format using a local script. The collected data from various crystals were indexed and integrated using XDS61. Integrated diffraction intensities from each crystal were selectively merged and scaled in XSCALE. Input files with the appropriate formats were prepared with XPREP, and the structure was solved ab initio by direct methods in SHELXT62 and refined with SHELXL62 integrated within Olex 2, leading to the final structure at the cut-off of 1.05 Å resolution. SHELXPRO was used for generating the PDB deposition, 7T3H.
Marfey’s analysis.
For Marfey’s analysis, dynobactin (1 mg) was hydrolysed in 0.5 ml 6 M HCl at 100 °C for 1 h. The hydrolysate was then dried and divided into two portions. To each hydrolysate, 100 μl 1 N NaHCO3 and 100 μl of either N-α-(5-fluoro-2,4-dinitrophenyl)-l-leucinamide (l-FDLA) or d-FDLA (1% (w/v) in acetone) were added and the mixtures were heated at 40 °C for 1 h. Afterwards, 20 μl 2 M HCl was added to neutralize the mixtures and a 20 μl aliquot of each reaction mixture was dissolved in 20 μl CH3CN. The resulting mixture was analysed by LC–MS63.
NMR structural studies of dynobactin A.
The NMR spectra were recorded on a Bruker Biospin Advance II 900 NMR spectrometer (900 MHz for 1H and 225 MHz for 13C) at Korea Basic Science Institute (KBSI) in Ochang, Korea. NMR spectra were obtained using 12 mg dynobactin dissolved in D2O or DMSO-d6 and chemical shifts were referenced to residual solvent signal. This analysis was performed with Bruker Topspin software 4.1.3.
MIC determination.
MICs were determined by microtitre plate growth assay in triplicate. Strains of tested aerobic bacteria were grown as overnight cultures in MHII broth, diluted 1:100 in MHII broth, incubated at 37 °C with shaking at 200 r.p.m. and allowed to grow for 2 h to reach exponential phase. These exponential phase cultures were further diluted to OD600 = 0.001 in MHII broth and 90 μl was added to round-bottom 96-well plates. To microtitre plate wells, 10 μl of purified compounds were added from a 2-fold serial dilution created in sterile ddH2O, and activity was assessed after overnight incubation at 37 °C. MIC was determined as the concentration at which no cell growth was detected.
Strains of tested anaerobic bacteria were grown under anaerobic conditions (Coy vinyl anaerobic chamber, 37 °C, 5% H2, 10% CO2, 85% N2) for 3 d in brain-heart infusion (BHI) broth supplemented with 0.5% yeast extract, 0.1% l-cysteine hydrochloride and 15 μg ml−1 hemin. Stationary phase cultures were diluted 1:100 in the modified BHI broth (BHI-YCH), and 90 μl was seeded into each well of a 96-well plate. Purified compound (10 μl) from a serial dilution in sterile ddH2O was added to a final volume of 100 μl. Final MIC was determined as above.
Cytotoxicity.
Effects of dynobactin on mammalian cells were studied using a microtitre plate Alamar blue assay (MABA/resazurin) in triplicate. Pharynx squamous carcinoma cell line FaDu (ATCC HTB-43), liver hepatocyte carcinoma cell line HepG2 (ATCC HB-8065) and human embryonic kidney cell line HEK293 with red-fluorescent-protein tag (HEK293-RFP; GenTarget SC007) were grown in Eagle’s medium supplemented with 10% fetal bovine serum (FBS) in a 96-well flat-bottom, tissue culture-treated plate (Corning) and incubated at 37 °C with 5% CO2. The medium was aspirated and fresh FBS-supplemented Eagle’s medium containing purified compound was added. A final concentration of 0.15 mM resazurin (Acros Organics) was added to the plate after 72 h incubation with purified compounds, and incubated for 3 h. Absorbance of each well was read using a BioTek Synergy H1 plate reader, observing at 544 nm and 590 nm.
Time-dependent killing.
An overnight culture of E. coli ATCC 25922 was grown in MHII broth and subcultured in 50 ml (1:1,000) into a baffled 250 ml Erlenmeyer flask, with 200 r.p.m. shaking at 37 °C for 2 h. Culture was then aliquoted to 15 ml starter culture tubes at 2 ml each, and drug was delivered in triplicate at 4x their respective MICs (Dynobactin 32 μg ml−1, Darobactin 8 μg ml−1, Amp 16 μg ml−1). From each tube, 30 μl was sampled at each time point, serially diluted (10-fold) in phosphate-buffered saline in a 96-well plate, and 10 μl of each dilution was spread onto MHII agar for counting of colony-forming units (c.f.u.).
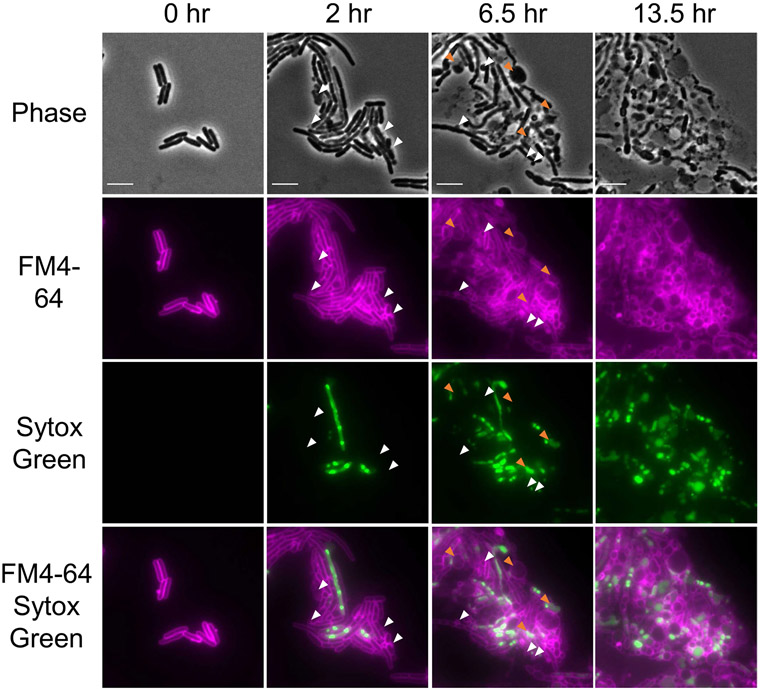
This was also visualized with fluorescence microscopy as previously described14. In brief, E. coli MG1655 was subcultured from an overnight culture into MHIIB (1:10,000) and grown for 2 h at 37 °C. Cells were concentrated and spotted on agarose pads (1.5%) containing dynobactin A (128 μg ml−1), FM4–64 (10 μg ml−1) and Sytox Green (0.5 μM) dyes from Molecular Probes. Killing was monitored using a ZEISS LSM 710 confocal microscope equipped with a ×63 oil immersion objective lens. The two signals from FM4–64 and Sytox Green were collected after excitation at 488 nm, alongside a differential interference contrast image. Acquisition and recording of differential interference contrast, FM4–64 and Sytox Green signals were performed every 15 min under a temperature of 37 °C, all within a thermostatic chamber. NIS-Elements 5.20.02 was used to acquire fluorescence microscopy images.
Resistance studies.
To generate resistant mutants, an overnight culture of E. coli ATCC 25922 was grown in MHII broth and subcultured in 200 ml (1:100) in a 2 l Erlenmeyer flask, with 200 r.p.m. shaking at 37 °C for roughly 2 h. The OD600 of the cells were observed, and cells were concentrated to an equivalent OD600 = 5.0, 0.5 and 0.05 and plated on MHII plates containing different concentrations of dynobactin or darobactin. Meanwhile, a 10-fold serial dilution series of the cells was plated to enumerate c.f.u., and these plates were incubated over several days at 37 °C to identify resistant colonies. Colonies that grew in the presence of drug were isolated and transferred to fresh plates containing drugs. The BamA gene of resistant colonies was amplified using PCR primers (5’-GCTGGGATGACAGCGGAAC-3’, 5’-TTCACAGCAGTCTGGATACGAG-3’) with an initial 2 min denaturing step of 98 °C, followed by 30 cycles of 98 °C (10 s), 60 °C (30 s) and 72 °C (1.5 min), with a final extension step of 72 °C for 2 min. Amplicons were then Sanger sequenced at Psomagen, using two PCR primers (5’-ACTATCTGGATCGCGGTTATGC-3’ and 5’-CTTCCCGCATCAGGCCAG-3’).
Solution NMR spectroscopy.
A construct of BamA(421–810) (C690S, C700S) with N-terminal Tobacco Etch Virus cleavable His6-tag was expressed and purified as described previously64. Briefly, the protein was expressed in perdeuterated M9 medium with 1.0 g l−1 15N-ammonium chloride. BamA-β was isolated as inclusion bodies from the cell lysates and purified via immobilized-metal affinity chromatography (IMAC). The IMAC fractions containing unfolded BamA-β were precipitated and the protein was refolded in 50 mM Tris-HCl, 300 mM NaCl, 500 mM l-arginine monohydrochloride, pH 8.0 and 1% (w/v) lauryldimethylamine-N-oxide (LDAO). After dialysis, the sample was subjected to ion exchange chromatography and size exclusion chromatography, with a final buffer of 20 mM HEPES, pH 7.5, 150 mM NaCl and 0.1% (w/v) LDAO. The protein was concentrated to a final concentration of 250 μM for NMR experiments. 2D [15N,1H]-TROSY-HSQC experiments were measured on a 700 MHz Bruker spectrometer equipped with a cryogenic probe. Transients (128) were accumulated at a sample temperature of 37 °C, with 1,024 and 256 complex points in the 1H and 15N dimensions, respectively. The data were processed using Topspin 3.6.2 and analysed using NMR Sparky65. Titration of dynobactin A with the BamA-barrel was performed in five steps accounting for 0.25, 0.5, 1.0, 2.0 and 4.0 equivalents of dynobactin A in molar equivalents with respect to the apo BamA-β concentration. Sequence-specific resonance assignments of BamA-β were established previously66.
Target:ligand crystallization and structure determination.
BamA-β (10 mg ml−1 in 20 mM Tris pH 7.5, 150 mM NaCl, 0.35% C8E4, 0.05% LDAO) with a 2-fold excess of dynobactin A was crystallized in a sitting-drop vapour diffusion experimental setup. Crystals were grown in 0.12 M lithium sulfate, 0.02 M Tris pH 7.5, 0.1 M sodium citrate pH 5.0 and 20% (v/v) PEG30, and directly flash frozen in liquid nitrogen. Data were collected at the SLS beamline X06DA (Swiss Light Source, Paul Scherrer Institute, Switzerland) at 100 K, indexed and integrated with XDS61, and scaled using Aimless67. The structure was solved by molecular replacement using the crystal structure of 7NRF.pdb15 as search model using the programme Phaser68. Model building was performed with Coot69, ligand restraints were generated with PRODRG70, and the model was refined in PHENIX71. MolProbity72 was used to evaluate the final model. The programme ChimeraX73,74 was used to visualize the atomic model. Data and refinement statistics are summarized in Supplementary Table 6. The atomic coordinates have been deposited in the RCSB Protein Data Bank and are available under the accession code 7R1V.
Target:ligand cryo-EM sample preparation and data collection.
A 4 μl aliquot of full-length BAM complex in 20 mM Tris pH 8.0, 150 mM NaCl and 0.05% (w/v) n-dodecyl β-d-maltopyranoside (DDM), at a concentration of 7 mg ml−1 with a 10-fold molar excess of dynobactin A was applied to a Quantifoil R2/1 holey carbon copper grid (Quantifoil Micro Tools) vitrified by plunging into liquid ethane using a Vitrobot (FEI, Vitrobot III). One dataset was collected using a Titan Krios electron microscope (FEI) operated at 300 kV, a GIF Quantum LS imaging filter (Gatan) and a K2 Summit (Gatan) operating in counting mode using SerialEM75. Images were acquired at 0.8–3.0 μm defocus and a nominal magnification of ×165,000, corresponding to a pixel size of 0.82 Å (Supplementary Table 5). Movies were collected with a total dose of approximately 48 e− Å−2 per 9 s exposure and fractionated over 40 frames using beam-image shift to record 4 holes at 8 images per hole.
Target:ligand cryo-EM data processing and analysis.
Micrographs (4,499) were corrected for beam-induced drift using patch motion and the contrast transfer function (CTF) parameters for each micrograph were determined using Patch CTF in cryoSPARC76. Particles were picked with the blob picker function in cryoSPARC and subjected to reference-free 2D classification. Heterogeneous refinement was started with identical volumes based on 7NRI.pdb. The class with the most structural features was used for non-uniform refinement76, followed by local refinement using an automatically determined global mask, which resulted in a reconstruction at 3.9 Å resolution. Using global and local CTF refinement followed by additional rounds of non-uniform and local refinements, a final map at a global resolution of 3.6 Å was obtained. Modelling started by fitting the BamABCDE-darobactin complex 7NRI.pdb15 into the EM density map in UCSF Chimera77. Model building was performed in Coot69 and real-space refinement was carried out in PHENIX71,78. Validation was done using the cryo-EM validation tools in PHENIX71,78. The map resolution range was determined from local resolution calculation in cryoSPARC, and the model resolution range was determined by calculating a map at 6 Å resolution in Chimera from the final model. This map was converted into a mask by thresholding and was applied to the local resolution map to obtain the histogram of local resolution in the model region.
BAM-mediated OmpT folding assay.
The inhibitory effects of dynobactin A, and darobactins A and B on BAM-mediated folding were measured in a fluorescence-based assay where the activity of BAM-folded OmpT was quantified by cleavage of the self-quenching fluorogenic reporter peptide Abz-Ala-Arg-Arg-Ala-Tyr(NO2)-NH2 (GenScript). The BAM complex was presented in OMVs, and the functional assay was otherwise performed as previously described40,79,80. In brief, BAM-OMVs pre-incubated with colistin were incubated with the respective compound of interest (darobactin A, B or dynobactin A) at variable concentrations between 10 to 0.001 μM for 10 min at 37 °C. Folding reactions were initiated with the addition of a 100 μM reporter peptide plus a periplasmic chaperone SurA–OmpT mixture comprised respectively of 50 and 2 μM. Fluorescence emission was monitored at regular intervals of 40 s for approximately 2 h at 37 °C, with excitation and emission wavelengths of 320 nm and 410 nm, and 25 nm and 20 nm bandwidths, respectively. Initial reaction rates were determined, normalized and plotted against the inhibitor concentration. Data were collected on a TECAN Spark plate reader with SPARKCONTROL v.2.3 software. Inhibition dose-response curves (relative fluorescence units vs log ([inhibitor] (M))) were fitted to the ‘log(inhibitor) vs response – variable slope (four parameters)’ model in Graphpad Prism (version 9.2.0). The 95% confidence intervals on log IC50 and IC50 were calculated using the extra sum-of-squares F-test implemented in the software.
Surface plasmon resonance.
Surface plasmon resonance measurements were conducted on a Biacore T100 (Cytiva). BamA-β with an N-terminal Avi-tag (GLNDIFEAQKIEWHE) was biotinylated via BirA and captured on a streptavidin-coated sensor chip SA (Cytiva), resulting in capture levels of 3,500 resonance units (RU) for darobactin A and B and 1,900 RU for dynobactin after ten blank injections. All runs were conducted with PBS buffer (10 mM phosphate, 150 mM NaCl, 0.1% (w/v) LDAO, pH 7.4). The different ligands were injected in increasing concentrations in triplicate for a multi-cycle experiment with the following parameters: for darobactin A and B (2.5, 5, 10, 20, 60, 180, 540 nM), contact time 120 s, dissociation time 900 s, flow rate 30 μl min−1; for dynobactin A (0.39, 0.78, 1.56, 3.13, 6.25, 12.5, 25, 50, 100, 200 nM), contact time 240 s, dissociation time 1,200 s, flow rate 30 μl min−1. For dynobactin A, an additional 30 s regeneration step using 0.25% (w/v) sodium dodecyl sulfate was added after each cycle. Reference and blank-subtracted data were analysed with the Biacore T200 Evaluation Software 3.0 using the steady-state affinity model.
Animal studies.
Animal studies were approved by the Northeastern Institutional Animal Care and Use Committee, and performed at Northeastern University in accordance with institutional animal care and use policies, with 12 h day/night cycle at room temperature (~22 °C) and humidity (~40%). Experiments were neither randomized nor blinded, as it was not deemed necessary. Female CD-1 mice (20–25 g, experimentally naïve, 6 weeks old) from Charles River were used for all studies.
Septicaemia model.
Dynobactin A was assessed for the ability to treat multidrug-resistant E. coli AR350 (CDC) in a mouse model of septicaemia. BHI medium containing 5% mucin was used as a vehicle to deliver to each mouse 106 E. coli via a 0.5 ml intraperitoneal injection. This dosage of E. coli achieves a >90% mortality rate within 24 h of infection. At 1 h post infection, mice were given intraperitoneal injections of drug dissolved in sterile ddH2O, resulting in doses of 50 mg kg−1 dynobactin A, 100 mg kg−1 dynobactin A, or 50 mg kg−1 gentamicin as a positive control. A control group received the vehicle without the drug. Mice were monitored over the course of 7 d.
Thigh infection model.
A neutopenic thigh infection model was used to assess systemic efficacy of dynobactin A. Briefly, cyclophosphamide (150 mg kg−1) was administered to mice 4 d in advance of infection, and a smaller dose the day before infection (100 mg kg−1). An overnight culture of the pathogen was diluted to roughly 106 cells per ml (1:500 dilution), and 100 μl was delivered to the mice by intramuscular injection into the thigh. Actual infectious load was determined by c.f.u. plate counting. One group of mice was killed at 2 h post infection, at the time of drug delivery. Dynobactin A (100 mg kg−1) and a 50 mg kg−1 gentamicin positive control were delivered as single IP injections, each condition being tested in 5 mice. At 24 h post infection, mice were humanely euthanized and thighs were aseptically removed, weighed and homogenized for assessment of bacterial burden remaining at the site of infection. Homogenates were serially diluted and plated onto MHII agar to quantify c.f.u.
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
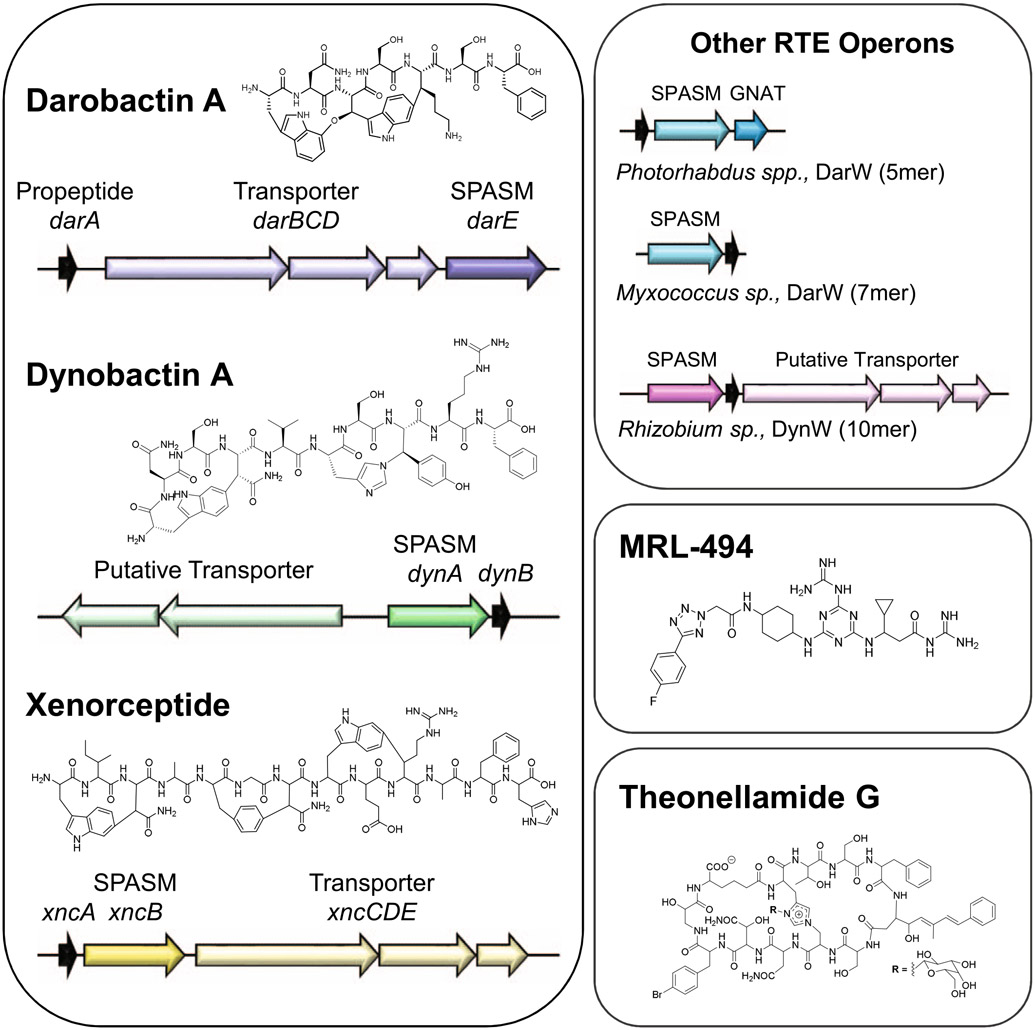
Extended Data Fig. 1 ∣. Compound and Operon structures.
Shown are darobactin and putatively-related cyclophane RiPP products (dynobactin A, xenorceptide), BamA-targeting synthetic molecule (MRL-494), and theonellamide G which contains an unusual histidine Nε2 to alanine β-carbon linkage, similar to dynobactin histidine Nε2 to tyrosine β-carbon linkage.
Extended Data Fig. 2 ∣. Rapid identification of target metabolites.
Total ion chromatogram (TIC) and Escherichia coli MG1655 activity assay. Panels A-C correspond to C18 (A), PFP (B), and Phenyl (C) columns applied to a 0.1% (v/v) formic acid in water:acetonitrile gradient condition. Panels (D), (E), and (F) correspond to the same columns applied to a 0.1% (v/v) formic acid in water:methanol condition. Timeslice fractions which inhibit E. coli growth are indicated by highlighted boxes containing a ‘+’ symbol. (G) Summary of networked masses in antimicrobial ion exchange fraction. (H) Structure of biologically-active metabolites purified from Photorhabdus australis with GNPS network relationship to the dominant metabolite (dynobactin A).
Extended Data Fig. 3 ∣. CryoEM microED structure determination.
MicroED data collection and analysis of 19 independent crystals of dynobactin A yielded structure, further details are elaborated within Methods. (A) Bright-field TEM image of dynobactin crystals (Scale bar: 5 μm). (B) Electron diffraction pattern with resolution ring at 0.95 Å. (C) 2D crystal packing arrangements of dynobactin. The intramolecular hydrogen bonds are shown as dashed lines for the top two molecules. The crystallographic b axis is parallel to the vertical direction of the figure. (D) Dynobactin A shows flexible conformations. Left shows superimposition dynobactin A microED structure (straight) and the dynobactin A structure observed in co-crystal with target BamA (bent). Right panels depict individual structures side-by-side. Separation of the two macrocycle rings in dynobactin A allows for free rotation about 4 bonds, creating an approximate 90° kink in the dynobactin A structure.
Extended Data Fig. 4 ∣. Dynobactin Secondary Structural Confirmations.
Full NMR assignment available in Supplementary Table 4. (A) 1H NMR spectrum (900 MHz, D2O). (B) 13C NMR spectrum (225 MHz, D2O). (C) 2D NMR spectra recorded in D2O (top left HSQC, top right DQF-COSY, bottom left HMBC, bottom right ROESY). (D) Key 2D NMR correlations in D2O and DMSO-d6. (E) Retention times (tR, min) of FDLA derivatives from dynobactin A Marfey’s analysis.
Extended Data Fig. 5 ∣. Target identification and resistance mutations.
BamA crystal structure (green) with labeled resistance mutation sites identified in this paper: (A) front view of lateral gate and (B) view of the barrel lumen from the periplasmic side (underside). Sites identified which gave resistance to both compounds are labeled in blue, and a mutation site which gives resistance to only dynobactin A is labeled in magenta. (C) Table listing MICs for bacteria from the previously described darobactin-resistance evolution experiment15, the isolated dynobactin-resistant mutants from this study, and other E. coli strains with outer membrane deficiencies (that is porin or efflux knockouts).
Extended Data Fig. 6 ∣. Unique features of dynobactin A binding.
(A) Comparison of the co-crystal structure of BamA-β with bound darobactin A (PDB:7NRF) and with bound dynobactin A (this work). In the close-up panels, W810 is highlighted in red. (B) Comparison of the orientation of the compound relative to strand β16 of BamA in the two structures. The bulky C-terminal extension of dynobactin A displaces the C-terminus of BamA further into the barrel lumen, with residue W810 becoming flexibly disordered (C) Selected region of a 2D [15N1H]-TROSY spectrum of BamA-β in LDAO micelles upon titration with dynobactin A. Tentative assignment for indole W810 is indicated. (D) Affinity measurements of darobactins and dynobactin A to BamA-β via Surface Plasmon Resonance, sensorgrams and the corresponding steady-state affinity plots show dynobactin A binds one order of magnitude tighter to BamA-barrel than the darobactins. (E) Efficacy of the compounds in inhibiting BAM-mediated folding in native outer membrane vesicles (OMVs) (data are presented as mean values ± SD, n = 2). Fitting of these data resulted in IC50 values of 30 ± 6 nM, 48 ± 8 nM, and 16 ± 2 nM for darobactin A, darobactin B and dynobactin A with a 95% confidence interval, respectively.
Extended Data Fig. 7 ∣. Cryo-EM and X-ray structure comparison of dynobactin A-bound BamA.
(A) X-ray crystal structure of BamA-β with bound dynobactin A at 2.5 Å resolution. Zoomed-in panels highlight specific residues involved in the interaction. (B) Hydrophobic interaction between V5 of dynobactin A to the side chains of F428 and I430 of BamA in the co-crystal structure. (C) Interaction between W1 of dynobactin A and BamA. (D) Superimposition of the X-ray and cryo-EM structures (β-strand 1 only). The comparison shows a high degree of similarity in the conformation of dynobactin A between the cryo-EM and X-ray structure.
Extended Data Fig. 8 ∣. Solution NMR spectroscopy of BamA-β interacting with dynobactin A.
(A) 2D [15N,1H]-TROSY spectra of apo BamA-β in LDAO micelles (green) overlaid with BamA-β with 1.0 eq of dynobactin A (magenta). Zoomed-in panels show selected resonances. Tentatively assigned W810 is indicated with a frame on the spectrum. (B) 2D [15N,1H]-TROSY spectra of BamA-β in a titration experiment with increasing concentration of dynobactin A, as indicated, from black to red. (C) NMR spectrum of mutant W810F to confirm the assignment of W810.
Extended Data Fig. 9 ∣. Time-lapse microscopy of E. coli undergoing dynobactin A treatment.
E. coli MG1655 cells were spotted onto a 1.5% agarose pad containing dynobactin A (8x MIC), the membrane stain FM4-64 10 μg mL−1 (false-colored in magenta), and membrane permeabilization stain Sytox Green 0.5 μM (false-colored in green). Cells were incubated at 37 °C in a thermostatic chamber and imaged every 15 minutes under the microscope. The panels and selected time points were chosen to best represent the population of E. coli MG1655 undergoing dynobactin A treatment. White arrows indicate representative examples of membrane blebbing; orange arrows indicate examples of swelling or cell lysis. Scale bars, 5 μm. This experiment is representative of two biologically-independent experiments performed, each showing similar results.
Extended Data Table 1 ∣.
Strains screened with putative RTEs
| Strains Screened | Species | rSAM-SPASM | Clade |
|---|---|---|---|
| DSM 17609 | Photorhabdus australis | WP_036772804.1 | DarE |
| W34 | WP_036771064.1 | DarW | |
| NT11.2 | WP_036772055.1 | DynA | |
| WT11.2 | WP_036768348.1 | XyeB | |
| DSM 25263 | Photorhabdus heterorhabditis | WP_054476603.1 | DarE |
| WP_149617347.1 | DynA | ||
| WP_214085659.1 | XyeB | ||
| DSM 23271 | Photorhabdus stackebrandtii | WP_166288413.1 | DarW |
| DSM 22387 | Photorhabdus tasmaniensis | WP_133813880.1 | DynA |
| DSM 17907 | Xenorhabdus kozodoii | WP_099142115.1 | DynA |
| LMG 30828 | Pectobacterium parvum | WP_181892556.1 | DynA |
| LMG 30175 | Pandoraea terrae | VVE19333.1 | DynA |
| LMG 20602 | Pandoraea capi | VVE41900.1 | DynA |
| DSM 2132 | Rhodothalassium salexigens | WP_132706374.1 | DynA |
| DSM 102598 | Devosia sp. | WP_062629801.1 | DynW |
| ATCC 43676 | Rhizobium sullae | WP_027510658.1 | DynW |
Lists bacteria screened for the presence of RiPP products. Putatively identified RTEs are listed for each strain.
Supplementary Material
Acknowledgements
Photorhabdus australis isolates were kindly shared by N. Waterfield at the University of Warwick as well as by A. Thanwisai from Naresuan University.
Crystallization screening at the National Crystallization Center at HWI was supported through NIH grant R24GM141256.
B.-K.Y. thanks L. M. Henling for fruitful discussions. The microED data were collected at the Caltech cryo-EM facility. We thank S. Chen for assistance and the Beckman Institute for their generous support of the cryo-EM facility and the Molecular Observatory at Caltech;
the Korea Basic Science Institute, Ochang, Korea, for providing NMR (900 MHz) data;
the staff of beamlines X06DA and X06SA at the Paul Scherrer Institute, Villigen, Switzerland, for support with crystallographic data collection; and the BioEM lab of the University of Basel for support with cryo-EM data acquisition. Calculations were performed at sciCORE (http://scicore.unibas.ch/) scientific computing core facility at the University of Basel.
This project was supported by the National Institutes of Health grant P01 AI118687 (K.L.), the Swiss National Science Foundation grants 177084 (T.M.) and 187170 (S.H.), and the National Center of Competence in Research AntiResist (180541).
Footnotes
Competing interests
The authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41564-022-01227-4.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41564-022-01227-4.
Data availability
Data supporting the findings of this study are available within the paper and its Supplementary Information, and have been submitted to publicly available databases. Crystal structures are available through PDB: microED dynobactin A structure (7T3H), BamA:dynobactin A X-ray co-crystal (7R1V), BAM complex:dynobactin A cryo-EM (7R1W, EMD-14242). Any other data or datasets from the current study are available upon reasonable request to the corresponding authors.
References
- 1.Brown ED & Wright GD Antibacterial drug discovery in the resistance era. Nature 529, 336–343 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Lewis K The science of antibiotic discovery. Cell 181, 29–45 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Ling LL et al. A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tacconelli E et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis 18, 318–327 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Zgurskaya HI, Rybenkov VV, Krishnamoorthy G & Leus IV Trans-envelope multidrug efflux pumps of Gram-negative bacteria and their synergism with the outer membrane barrier. Res. Microbiol 169, 351–356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richter MF et al. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature 545, 299–304 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker EN et al. Implementation of permeation rules leads to a FabI inhibitor with activity against Gram-negative pathogens. Nat. Microbiol 5, 67–75 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gavrish E et al. Lassomycin, a ribosomally synthesized cyclic peptide, kills Mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem. Biol 21, 509–518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quigley J et al. Novel antimicrobials from uncultured bacteria acting against Mycobacterium tuberculosis. mBio 10.1128/mbio.01516-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shahsavari N et al. A silent operon of Photorhabdus luminescens encodes a prodrug mimic of GTP. mBio 10.1128/mbio.00700-22(2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leimer N et al. A selective antibiotic for lyme disease. Cell 10.1016/j.cell.2021.09.011(2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crawford JM & Clardy J Bacterial symbionts and natural products. Chem. Commun 47, 7559–7566 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tobias NJ, Shi YM & Bode HB Refining the natural product repertoire in entomopathogenic bacteria. Trends Microbiol. 26, 833–840 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Imai Y et al. A new antibiotic selectively kills Gram-negative pathogens. Nature 576, 459–464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaur H et al. The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase. Nature 593, 125–129 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Rutledge PJ & Challis GL Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol 13, 509–523 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Hover BM et al. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol 3, 415–422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stokes JM et al. A deep learning approach to antibiotic discovery. Cell 180, 688–702.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. J. Mol. Biol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 20.Santos-Aberturas J et al. Uncovering the unexplored diversity of thioamidated ribosomal peptides in Actinobacteria using the RiPPER genome mining tool. Nucleic Acids Res. 47, 4624–4637 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sievers F et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grell TA, Goldman PJ & Drennan CL SPASM and twitch domains in S-adenosylmethionine (SAM) radical enzymes. J. Biol. Chem 290, 3964–3971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montalbán-López M et al. New developments in RiPP discovery, enzymology and engineering. Nat. Prod. Rep 38, 130–239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akiva E et al. The structure–function linkage database. Nucleic Acids Res. 42, D521–D530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grove TL, Lee KH, St Clair J, Krebs C & Booker SJ In vitro characterization of AtsB, a radical SAM formylglycine-generating enzyme that contains three [4Fe-4S] clusters. Biochemistry 47, 7523–7538 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldman PJ et al. X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification. Proc. Natl Acad. Sci. USA 110, 8519–8524 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q, Mo T, Ding W, Han Y & Deng Z The research on post-translational modification of RiPPs Xye catalyzed by CyFE PacB. Synth. Biol. J 10.12211/2096-8280.2021-080 (2021). [DOI] [Google Scholar]
- 28.Vesth T et al. Veillonella, firmicutes: microbes disguised as gram negatives. Stand. Genom. Sci 9, 431–448 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antunes LC et al. Phylogenomic analysis supports the ancestral presence of LPS-outer membranes in the Firmicutes. eLife 5, e14589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang M et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones CG et al. The CryoEM method MicroED as a powerful tool for small molecule structure determination. ACS Cent. Sci 4, 1587–1592 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haupert LM & Simpson GJ Screening of protein crystallization trials by second order nonlinear optical imaging of chiral crystals (SONICC). Methods 55, 379–386 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen TQN et al. Post-translational formation of strained cyclophanes in bacteria. Nat. Chem 12, 1042–1053 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Matsunaga S & Fusetani N Theonellamides AE, cytotoxic bicyclic peptides, from a marine sponge Theonella sp. J. Org. Chem 60, 1177–1181 (1995). [Google Scholar]
- 35.Nicolet Y Structure–function relationships of radical SAM enzymes. Nat. Catal 3, 337–350 (2020). [Google Scholar]
- 36.Benjdia A, Balty C & Berteau O Radical SAM enzymes in the biosynthesis of ribosomally synthesized and post-translationally modified peptides (RiPPs). Front. Chem 5, 87 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu D et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA 97, 5978–5983 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datsenko KA & Wanner BL One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaur H et al. Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach. J. Biomol. NMR 10.1007/s10858-019-00250-8(2019). [DOI] [PubMed] [Google Scholar]
- 40.Rath P et al. High-throughput screening of BAM inhibitors in native membrane environment. Nat. Commun Preprint at 10.21203/rs.3.rs-1465417/v1(2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kloser A, Laird M, Deng M & Misra R Modulations in lipid A and phospholipid biosynthesis pathways influence outer membrane protein assembly in Escherichia coli K-12. Mol. Microbiol 27, 1003–1008 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Koronakis V, Sharff A, Koronakis E, Luisi B & Hughes C Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405, 914–919 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Webber MA & Piddock LJ The importance of efflux pumps in bacterial antibiotic resistance. J. Antimicrob. Chemother 51, 9–11 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Jaffe A, Chabbert YA & Semonin O Role of porin proteins OmpF and OmpC in the permeation of beta-lactams. Antimicrob. Agents Chemother 22, 942–948 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srinivas N et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327, 1010–1013 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Silver LL A Gestalt approach to Gram-negative entry. Bioorg. Med. Chem 24, 6379–6389 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Konovalova A, Kahne DE & Silhavy TJ Outer membrane biogenesis. Annu. Rev. Microbiol 71, 539–556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melchers MJ et al. Pharmacokinetics and pharmacodynamics of murepavadin in neutropenic mouse models. Antimicrob. Agents Chemother 63, e01699–01618 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luther A et al. Chimeric peptidomimetic antibiotics against Gram-negative bacteria. Nature 576, 452–458 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Hart EM et al. A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proc. Natl Acad. Sci. USA 116, 21748–21757 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Storek KM et al. Monoclonal antibody targeting the beta-barrel assembly machine of Escherichia coli is bactericidal. Proc. Natl Acad. Sci. USA 115, 3692–3697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnston CW et al. Assembly and clustering of natural antibiotics guides target identification. Nat. Chem. Biol 12, 233–239 (2016). [DOI] [PubMed] [Google Scholar]
- 53.O’Shea R & Moser HE Physicochemical properties of antibacterial compounds: implications for drug discovery. J Med. Chem 51, 2871–2878 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Nikaido H Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev 67, 593–656 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lundquist K, Bakelar J, Noinaj N & Gumbart JC C-terminal kink formation is required for lateral gating in BamA. Proc. Natl Acad. Sci. USA 115, E7942–E7949 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sayers EW et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 49, D10–D17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Groß S et al. Improved broad-spectrum antibiotics against Gram-negative pathogens via darobactin biosynthetic pathway engineering. Chem. Sci 12, 11882–11893 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okonechnikov K, Golosova O, Fursov M & Team U Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28, 1166–1167 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Luft JR et al. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J. Struct. Biol 142, 170–179 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Zhao J et al. A simple pressure-assisted method for MicroED specimen preparation. Nat. Commun 12, 5036 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kabsch WXDS Acta Crystallogr. D 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheldrick GM SHELXT - integrated space-group and crystal-structure determination. Acta Crystallogr. A 71, 3–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fujii K, Ikai Y, Oka H & Suzuki M & Harada K A nonempirical method using LC/MS for determination of the absolute configuration of constituent amino acids in a peptide: combination of Marfeys method with mass spectrometry and its practical application. Anal. Chem 69, 5146–5151 (1997). [Google Scholar]
- 64.Bibow S, Bohm R, Modaresi SM & Hiller S Detergent titration as an efficient method for NMR resonance assignments of membrane proteins in lipid-bilayer nanodiscs. Anal. Chem 92, 7786–7793 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Lee W, Tonelli M & Markley JL NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hartmann JB, Zahn M, Burmann IM, Bibow S & Hiller S Sequence-specific solution NMR assignments of the beta-barrel insertase BamA to monitor its conformational ensemble at the atomic level. J. Am. Chem. Soc 140, 11252–11260 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Evans PR & Murshudov GN How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCoy AJ et al. Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Schuttelkopf AW & van Aalten DM PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D 60, 1355–1363 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Adams PD et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002). [DOI] [PubMed] [Google Scholar]
- 72.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goddard TD et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pettersen EF et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 76.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Pettersen EF et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 78.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thoma J et al. Protein-enriched outer membrane vesicles as a native platform for outer membrane protein studies. Commun. Biol 1, 23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hagan CL, Kim S & Kahne D Reconstitution of outer membrane protein assembly from purified components. Science 328, 890–892 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available within the paper and its Supplementary Information, and have been submitted to publicly available databases. Crystal structures are available through PDB: microED dynobactin A structure (7T3H), BamA:dynobactin A X-ray co-crystal (7R1V), BAM complex:dynobactin A cryo-EM (7R1W, EMD-14242). Any other data or datasets from the current study are available upon reasonable request to the corresponding authors.