Abstract
Adherence of enterohemorrhagic Escherichia coli (EHEC) to intestinal epithelium is essential for initiation of the infection. To identify genes involved in adherence, an EHEC O157:H7 strain (O157Sakai) was mutagenized by mini-Tn5Km2, where Km refers to kanamycin resistance, and 4,677 insertion mutants were screened for their ability to form microcolonies (MC) on Caco-2 cells. The less adherent mutants were divided into three groups: those with no adherent ability (designated as class 1 mutants, n = 10), those less adherent than the wild type (class 2 mutants, n = 16), and those unable to form MC but which adhered in a diffuse manner (class 3 mutants, n = 1). The sites of insertion in class 1 mutants were all found within genes of the locus for enterocyte effacement (LEE) thought to be required for type III protein secretion. Indeed, the class 1 mutants failed to secrete type III secreted proteins such as EspA and Tir into the culture medium. The insertions in class 2 mutants were outside the LEE, and all the mutants except one were able to secrete type III proteins into the culture medium. The class 3 mutant had the insertion in the tir gene in the LEE and was deficient in Tir and intimin expression, suggesting that in the absence of intimin-Tir, O157Sakai can still adhere to Caco-2 cells but in a diffused manner. This was confirmed by construction of a nonpolar eae (encoding intimin) mutant. Examination of the eae mutant together with O157Sakai and one of the class 1 mutants for the ability to form MC revealed that EHEC initially adhered diffusely at 1.5 h after infection. Following washing out of the nonadherent bacteria, while wild-type EHEC bacteria developed MC for another 2 to 3 h on Caco-2 cells, the eae mutant diffusely adhered throughout the infection without forming MC. MC with O157Sakai but not the diffusely adherent eae mutant could evoke F-actin condensation beneath the bacterium. Our results suggest that EHEC encodes additional adherence-associated loci and that the type III secreted proteins are involved in the initial diffuse adherence, while the intimin-Tir interaction is required for the subsequent development of MC.
Enterohemorrhagic Escherichia coli (EHEC) is responsible for a range of illnesses, including nonbloody diarrhea, hemorrhagic colitis, and hemolytic uremic syndrome. The pathogenesis of EHEC (represented by O157:H7) has been indicated to be associated with several characteristics, including the production of Shiga-like toxins, the capacity to express an attaching and effacing intestinal lesion, and the presence of EspP encoded by a gene borne by a 90-kb plasmid (2, 15, 34, 41). Among these characteristics, bacterial attachment to the intestinal epithelium constitutes an early, essential step in the illness.
The adherence of EHEC strains (a subset of Shiga-like toxin-producing E. coli called STEC) to epithelial cells has been variously reported; in some studies, EHEC strains exhibited a diffuse pattern of adherence on epithelial cells (37), while in other studies they formed microcolonies (19, 28). Some of the adherent mechanisms in EHEC are common to enteropathogenic E. coli (EPEC). Adherence of EPEC is thought to consist of three stages: nonintimate adherence, signal transduction, and intimate adherence (9, 34). EHEC also has factors involved in the last two stages. In some late stages of infection, it has been reported that O157:H7 EHEC strains intimately attach to and develop microcolonies (MC) on epithelial cells and induce F-actin condensation underneath MC that requires expression of intimin and type III secreted proteins, including EspA, EspB, EspD, and Tir/EspE (4, 14, 17, 19, 22, 28). However, a prominent difference between EHEC and EPEC in the attachment to epithelial cells has been indicated to lie in the initial stage. The initial attachment of EPEC to epithelial cells is thought to be mediated by bundle-forming pili (BFP), which facilitates autoaggregation of bacteria, thus leading to the formation of MC on epithelial cells (34). EHEC strains do not have BFP; instead, the bacteria seem to utilize other putative adherence factors. Using an O26 STEC strain, EspA protein, one of the type III-mediated secretion proteins encoded by genes in the locus of enterocyte effacement (LEE), has been suggested to be required at the initial stage of bacterial adherence to the host cells (11). To date, several putative bacterial factors have been indicated to be involved in the adherence of EHEC to epithelial cells. Although the precise role of each is not fully understood, the factors include (i) the EspA filament, the product of the espA gene in the LEE; (ii) intimin (the only established adhesin) and Tir (a receptor for intimin that is translocated into host cells via a type III secretion system), the products of the eae and tir genes in the LEE; (iii) the product of the pas gene in the LEE; (iv) a product(s) of pO157; (v) a 94-kDa putative outer membrane protein; (vi) O157 lipopolysaccharide (LPS); (vii) Efa1; and (viii) Iha (7, 11, 14, 20, 21, 23, 34, 35, 46).
The entire genome of one O157:H7 strain, RIMD 0509952 (referred to as O157Sakai in this study), (48), has been sequenced (K. Makino and H. Shinagawa, personal communication). Accordingly, the chromosome of O157Sakai is estimated to be ∼5.6 Mb, with approximately 1,550 kb of strain-specific DNA, which does not exist in the E. coli K-12 chromosome (36). Analysis of the strain-specific DNA sequences of O157Sakai suggests several pathogenicity islands, including the LEE, a gene cluster partly homologous to the Salmonella enterica serovar Typhimurium pathogenicity island I (SPI-I), and phage lambda-like DNA elements encoding VT-1 and VT-2 (25; T. Tobe and C. Sasakawa, unpublished data). So far, only a limited number of genes in the LEE and other loci have been shown to be involved in the adherence of EHEC O157:H7 to host cells (34), and much about the adherence, especially the initial attachment, is only poorly understood. We, therefore, undertook to identify the adherence factors involved in the initial attachment of O157Sakai using random transposon mutagenesis and investigated the process of the development of MC on Caco-2 cells.
MATERIALS AND METHODS
Bacterial strains, media, and tissue culture.
The O157:H7 strain RIMD 0509952 (O157Sakai in this paper) was originally isolated from a patient in the biggest of several outbreaks to occur in 1996 in the city of Sakai, Japan (24, 48). EPEC strain (O111: NM) B171-8 was described previously (40).
EHEC and EPEC were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 0.45% glycerol (DMEM-glycerol) without glucose through all experiments, since this medium enhanced the adherence efficiency of O157Sakai more than standard DMEM containing glucose or L broth (H. Abe and C. Sasakawa, unpublished data). Caco-2 cells were maintained in DMEM supplemented with gentamicin (100 μg/ml) and kanamycin (50 μg/ml), l-glutamine (2 mM), 100 μM MEM nonessential amino acid solutions (Gibco BRL), and 10% fetal calf serum in a humidified atmosphere of 5% CO2 at 37°C.
Construction of mini-Tn5Km2 mutants.
To obtain a selectable marker for conjugation, plasmid pSC101 (45), carrying the tetracycline resistance gene, was introduced into O157Sakai by electroporation with a Gene Pulser electroporater (Bio-Rad Laboratories, Hercules, Calif.) in a 10% glycerol solution at 2.5 kV/25 μF. The resulting strain O157Sakai-pSC101, named O157T, was verified to have retained the phenotype of the parent in terms of adherence properties, including the levels of intimin expression and secretion of EspA, EspB, and Tir into culture medium, from immunoblottings with specific antibodies for intimin, EspA, EspB, and Tir proteins (data not shown). The mini-Tn5Km2-bearing plasmid pUT mini-Tn5Km2 (6), where Km refers to kanamycin resistance, was introduced into O157T from an E. coli K-12 derivative, SM17 λpir (5), by conjugation, and the transposon was inserted randomly into the chromosome. The insertion mutants thus obtained were purified on agar plates, and the individual clones were kept in 50% glycerol in L broth at −80°C.
Screening the mini-Tn5Km2 mutants library for adherence ability.
The insertion mutants were grown in DMEM-glycerol overnight at 37°C and were inoculated at a multiplicity of infection of approximately 50:1 onto semiconfluent Caco-2 cell monolayers grown on 96-well microtiter plates filled with DMEM with 10% fetal calf serum, which was replaced with fresh DMEM-glycerol. Bacteria and cells were incubated for 2 h, washed five times with 1 ml of sterile phosphate-buffered saline (PBS), and incubated with fresh DMEM-glycerol for another 2.5 h. These incubation times were determined to be the minimum for visualization of MC formed by O157Sakai in the large screening. The monolayers were fixed and stained with Giemsa solution for microscopic evaluation. Bacterial clusters on Caco-2 cells consisting of eight or more bacteria were considered microcolonies. To quantitate the adherence capacity of the mutants, bacteria freshly grown in DMEM-glycerol at 37°C for 2 h were used to infect Caco-2 cells on 24-well plastic plates for 1.5 h. Then these infected monolayers were washed five times with sterile PBS (1 ml) and incubated with fresh DMEM-glycerol for another 2.5 h. The numbers of MC were scored as the sum of 20 microscopic fields. This protocol was used for all adherence and fluorescence actin staining (FAS) assays unless otherwise described.
Southern blot analysis.
Total DNA was extracted from each strain by sodium dodecyl sulfate (SDS) lysis, pronase treatment, phenol-chloroform extraction, and ethanol precipitation (26). Total DNA was digested for about 2 h with the restriction endonuclease EcoRI, EcoRV, or PstI, none of which digest a DNA-specific probe for mini-Tn5Km2; was separated by electrophoresis in 0.7% agarose; and was transferred to Hybond-N+ positively charged nylon membranes (Amersham, Little Chalfont, England). A 1.7-kb BamHI fragment that contains the kanamycin resistance gene of mini-Tn5Km2 was labeled using the psoralen-biotin nonisotopic labeling kit (Ambion, Austin, Texas) and served as the DNA probe for hybridization by the method of Southern hybridization (Hybond-N, version 2.0; Amersham) under stringent conditions (7% SDS–1 mM EDTA–0.5 mM NaHPO4 [pH 7.2], 65°C).
Cloning mini-Tn5Km2 fragments.
The EcoRI, EcoRV, or PstI digest of total DNA from each strain was ligated to EcoRI-, EcoRV-, or PstI-digested pBluescript II KS(+) (43) and was used to transform E. coli DH5α by selecting for kanamycin-resistant transformants. Clones of the EcoRI, EcoRV, or PstI fragment containing the mini-Tn5Km2 insert were obtained. The region flanking mini-Tn5Km2 was sequenced by primers to the T3 or T7 promoter on pBluescript II KS(+).
Protein secretion and expression.
Bacteria were grown to mid-logarithmic phase in DMEM-glycerol at 37°C for detecting proteins secreted into the culture medium and expressed in bacteria. The bacterial density of each culture was adjusted by optical density at 580 nm in all sample preparations. One milliliter of culture supernatant was filtered by a 0.45-μm-pore-size filter and was concentrated by trichloroacetic acid (6% [vol/vol]) precipitation, and the pellets were analyzed by SDS-12% polyacrylamide gel electrophoresis (PAGE). Immunoblotting with EspA-, EspB-, or Tir-specific antiserum was performed as described previously (47). Antiserum against each of EspA, EspB, and Tir was prepared by immunizing rabbits with a synthetic oligopeptide which corresponded to the amino acid sequence of the C-terminal part of each protein. The amounts of intimin in whole bacterial lysates prepared from equal amounts of cultures were determined by immunoblotting with an intimin-specific antiserum. Antiserum against intimin was prepared by immunizing rabbits with purified His-tagged intimin, which was expressed from plasmid pEB312 provided by L. J. Gansheroff of Uniformed Services University of the Health Sciences (29, 47).
Construction of a nonpolar eae mutant.
Bacteria were cultured at 30°C for all constructions. Two sets of oligonucleotides, (i) primer 1 (5′-GCGCGGGATCCCATCGTTTCGTC-3′) and primer 2 (5′-TGGCTCCCGGGACTCGCCGCTTGTTGTG-3′) and (ii) primer 3 (5′-GATGTCCCGGGGTAAGTCTCAAACGCAAGC-3′) and primer 4 (5′-GTTGTGAGCTCAGTTGTTGCTTGATGTG-3′), were used to amplify DNA fragments of 1,027 and 944 bp corresponding, respectively, to the 5′ and 3′ ends of the eae gene and additional flanking regions from chromosomal DNA of strain O157Sakai by PCR using Takara ExTaq (Takara Shuzo Co., Ltd., Otsu, Japan). The corresponding oligonucleotides contained restriction sites for the endonucleases BamHI, SmaI, and SacI (shown in bold in the primer sequence). These enzymes were used for ligation of both amplification products and the aphA-3 cassette (31) and for subsequent cloning of the resulting fragment into the oriRR6kγ lacZα sacB plasmid pMW91 (32). The resulting recombinant suicide plasmid was introduced by conjugation into O157Sakai or O157T and integrated into the chromosome. Further cultivation in the presence of 5% sucrose with 5 μg of kanamycin/ml led to a second round of recombination and the excision of the plasmid with the wild-type eae gene. Bacteria were confirmed to harbor a mutated eae allele by PCR using the combination primer 1-primer 4 and two primers outside primers 1 and 4, respectively. For complementation, the complete eae gene of strain O157Sakai was amplified using primer 1 and primer 4 and was cloned into the pSC101 derivative pMW118 vector. The eae mutants, O157SakaiΔeae and the Δeae strain, were confirmed to secrete EspA and Tir into the culture medium at a level similar to that of the wild type and were able to form MC on Caco-2 cells at 4 h postinfection when the cloned eae gene was introduced on a plasmid. Furthermore, it was confirmed that there was no alteration in the length of the LPS of O157SakaiΔeae and the Δeae strain by the agglutination assay with antiserum specific for O157 and in the electrophoretic profiles of LPS in an SDS-polyacrylamide gel treated with silver staining as previously described (16). The resultant two nonpolar eae mutants derived from O157Sakai and O157T showed the same phenotypes in all experiments in this study.
FAS.
The ability of O157Sakai derivatives to induce intimate attachment was assessed using the FAS test (19).
RESULTS
Isolation of adherent defective mutants of O157Sakai.
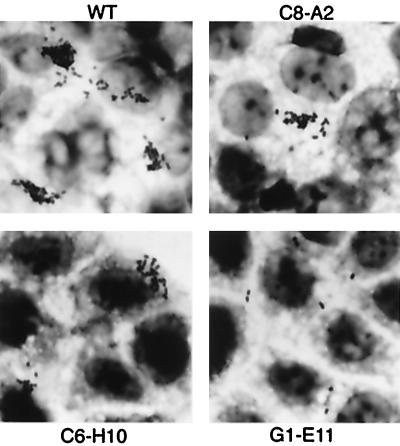
To identify factors involved in the initial adherence of EHEC to epithelial cells, we randomly mutagenized O157T with mini-Tn5Km2 and collected a total of 4,677 independent insertion mutants. Each of the mutants was used to infect Caco-2 cells for 1.5 h, and after the nonadherent bacteria had been washed out, the infected Caco-2 cells were further incubated in fresh DMEM for another 2.5 h to allow the adherent bacteria to develop MC visible by Giemsa staining (see Materials and Methods). By the methods described, we identified 713 mutants that were potentially nonadherent or less adherent, 548 mutants that showed increased adherence, and 228 mutants that grew slowly. In this study, only potentially nonadherent or less adherent mutants were further screened. From the 713 mutants, 29 mutants which reproducibly exhibited the less adherent phenotype were further characterized for the pattern of adherence, growth in DMEM-glycerol, and FAS-positive phenotype (an ability to condense F-actin in host cells beneath bacterial attachment). Twelve of the 29 insertion mutants, named A3-C10, A3-E9, A5-E4, A6-E7, B4-B2, B6-H7, B8-E7, B8-F12, B9-F9, C8-C2, D5-B12, and F4-B2, lost adherent capacity almost completely (less than 0.5% of wild-type adherence capacity). Since two mutants, F4-B2 and C8-C2, showed poor growth in DMEM at 37°C when growth rates were measured spectrophotometrically at 580 nm, they were eliminated from the collection, and the remaining 10 mutants were tentatively classified as class 1 strains (Table 1). The other 17 mutants were also significantly less adherent to Caco-2 cells, to an extent ranging from 1 to 40% of the wild-type level. Thus, all these variants except one were tentatively grouped as class 2 mutants. The one mutant exception, named G1-E11, did not show an MC pattern of adherence; instead, it diffusely adhered to the host cells at 4 h postinfection (Fig. 1) and was tentatively grouped as a class 3 mutant.
TABLE 1.
Adherence and other characteristics of strains
| Strains | Adherencea | EspA secretionb | Intiminb | FASc | Growthd | MCe |
|---|---|---|---|---|---|---|
| WTf | ++++ | + | + | + | N | + |
| Class 1 | ||||||
| A3-C10 | − | − | + | ND | N | − |
| A3-E9 | − | − | + | ND | N | − |
| A5-E4 | − | − | + | ND | N | − |
| A6-E7 | − | − | + | ND | N | − |
| B4-B2 | − | − | + | ND | N | − |
| B6-H7 | − | − | + | ND | N | − |
| B8-E7 | − | − | + | ND | N | − |
| B8-F12 | − | − | + | ND | N | − |
| B9-F9 | − | − | + | ND | N | − |
| D5-B12 | − | − | + | ND | N | − |
| Class 2 | ||||||
| A3-H2 | + | +/− | + | + | N | + |
| C6-E9 | ++ | + | + | + | N | + |
| C6-F9 | + | + | + | + | N | + |
| C6-H10 | ++ | + | + | + | N | + |
| C7-A10 | + | + | + | + | N | + |
| C8-A2 | + | + | + | + | N | + |
| F4-H4 | + | + | + | + | N | + |
| F5-F4 | + | + | + | + | N | + |
| F6-B9 | ++ | + | + | + | N | + |
| F6-D11 | ++ | + | + | + | N | + |
| G2-B12 | ++ | + | + | + | N | + |
| G4-B6 | + | + | + | + | N | + |
| G4-B12 | + | + | + | + | N | + |
| G4-C10 | ++ | + | + | + | N | + |
| G4-E10 | + | + | + | + | N | + |
| H7-C4 | + | + | + | + | N | + |
| Class 3 | ||||||
| G1-E11 | + | + | − | − | N | − |
Adherence was divided by the mean adherence (100%) of the O157Sakai-T parental strain. ++++, >80%; +++, 80 to 50%; ++, 50 to 25%; +, 25 to 1%; −, <1%. Data are the means of triplicate determinations, which differed by <15%, from representative experiments.
EspA in culture supernatant and intimin in whole bacterial cell were detected by anti-EspA and anti-intimin antibody, respectively. +, detected; −, not detected; +/−, detected at a reduced level.
FAS assays of infected Caco-2 cells were performed by using rhodamine-phalloidin. ND, not determined because nonadherent to Caco-2 cells.
Growth rates of each mutant and the parental wild type were essentially the same. N, normal.
Ability to form microcolonies; +, positive; −, negative.
WT, wild type.
FIG. 1.
Adherence phenotypes of class 2 mutants and class 3 mutants are shown. The C8-A2 and C6-H10 mutants are representatives of class 2 mutants. The G1-E11 mutant shows a diffused adherence phenotype. The bacteria grown in DMEM-glycerol for 2 h were used to infect Caco-2 cell monolayers. The infected monolayers were incubated for 1.5 h and washed five times with PBS. After another 2.5 h of incubation, the monolayers were again washed three times, fixed with methanol, and stained with Giemsa solution to visualize the adherent bacterial colonies. WT, wild type.
Sites of mini-Tn5Km2.
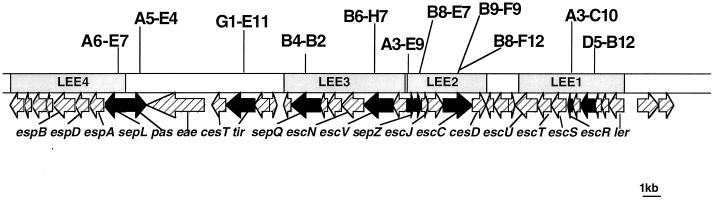
To confirm that class 1, 2, and 3 mutants possessed a single copy of mini-Tn5Km2, we performed Southern blotting of total DNA from each of the mutants digested with EcoRI, EcoRV, or PstI using a mini-Tn5Km2-specific DNA probe (see Materials and Methods). The results indicated that all of the class 1, 2, and 3 mutants had a single copy of mini-Tn5Km2 in their chromosomes (data not shown). Thus, we directly estimated the site of the transposon by sequencing the DNA flanking the mini-Tn5Km2 as described in Materials and Methods. The sites of mini-Tn5Km2 on the chromosome were estimated based on sequence homology between the flanking DNA and sequences in a DNA database of O157Sakai or E. coli K-12 genome sequences (K. Makino and H. Shinagawa, unpublished data; Escherichia coli www home page [http://mol.genes.nig.ac.jp/ecoli/] or E. coli database collection [ECDC] [http://susi.bio.uni-giessen.de/ecdc/ecdc.html]). Accordingly, all of the class 1 mutants were found to possess mini-Tn5Km2 in either the sepL, pas, sepQ, escV, sepZ, L0038, escC, escR, or L0051 gene at the LEE (Fig. 2). Although not all of the LEE gene products have been well characterized, based on the amino acid sequence similarity of SepL, Pas, SepQ, EscV, SepZ, L0038, EscC, EscR, and L0051 to the corresponding EPEC proteins (13, 34, 38), the genes were assumed to be involved in type III secretion.
FIG. 2.
Genetic organization of the EHEC O157:H7 LEE adapted from Perna et al. (38). Insertion sites of all class 1 mutants and the class 3 mutant (G1-E11) are shown. Genes with a mini-Tn5Km2 insertion are shown by black arrows. Operons LEE1 to LEE4 were those of the EPEC LEE (30).
The sites of insertion in the class 2 mutants were estimated by similar methods, and the mini-Tn5Km2 insertions were allocated outside the LEE. Each of the sites of A3-H2, C8-A2, F4-H4, F6-B9, F6-D11, G4-B6, G4-E10, G4-B12, H7-C4, and F5-F4 were mapped in regions highly homologous (>95%) to the DNA sequence of the E. coli K-12 genome, while the insertion sites of C6-E9, C6-F9, C6-H10, G2-B12, G4-C10, and C7-A10 were placed in sequences specific to the O157Sakai genome (Table 2). The class 3 mutant (G1-E11) had the mini-Tn5Km2 in the tir gene in the LEE (Fig. 2).
TABLE 2.
Products encoded by the gene witha or in the vicinity of mini-Tn5Km2
| Strain | Gene product | Site (min) on E. coli K-12 genomeb | Accession no. or reference |
|---|---|---|---|
| A3-H2 | Hypothetical 17.8-kDa protein (yfjG) | 59.2 | P52121 (1) |
| C6-E9 | Putative membrane protein | – | AE000234 |
| C6-F9 | Putative membrane protein | – | AE000234 |
| C6-H10 | Putative membrane protein | – | AE000234 |
| C7-A10 | Putative membrane protein | – | AE000234 |
| C8-A2 | Hypothetical 8-membrane-spanning 37.6-kDa protein (yiaH) | 80.2 | P37669 |
| F4-H4 | Putative NAGC-like transcriptional regulator (yphHI) | 57.8 | AE000341 |
| ABC transporter-periplasmic binding protein YPHF precursor | P77269 | ||
| Hypothetical 127.3-kDa protein | P76585 | ||
| Serine hydroxy transferase (glyA) | 38 | ||
| F5-F4 | Putative potassium channel (kch) | 28.3 | P31069 (32) |
| F6-B9 | Hypothetical 10-membrane-spanning 58.4-kDa protein | 27.2 | P40877 |
| F6-D11 | Hypothetical 40.2-kDa protein | 64.7 | Q46803 |
| Hypothetical sigma 54-dependent transcriptional regulator | Q46802 | ||
| G2-B12 | Putative adherence factor (efa1) | – | AF159462 (34) |
| G4-B6 | Arsenite (efflux) pump membrane protein (arsF) | 78.59 | 7 |
| Origopeptidase A (prlC) | 3 | ||
| G4-B12 | Hypothetical 27.7-kDa protein | 14.6 | P77627 |
| G4-C10 | (orf3 of EPEC pathogenicity island LIM) | – | 46 |
| G4-E10 | Lysine-specific permease (lysP) | 48.5 | 43 |
| Hypothetical transcriptional regulator | P32484 | ||
| H7-C4 | Hypothetical 10-membrane-spanning 43.6-kDa protein | 87.5 | P32135 |
Products encoded by the gene with mini-Tn5Km2 are in bold type.
The insertion sites placed in the DNA sequences specific to the O157Sakai genome sequence are indicated by a dash (–). The sites of C6-E9, C6-F9, C6-H10, and C7-A10 were in the same ∼3-kb regions encoding an open reading frame partially homologous to the b1372 gene, which encodes a putative membrane protein of 1,122 amino acids of E. coli K-12. F6-D11 was placed in a region containing genes encoding a hypothetical sigma 54-dependent transcriptional regulator and a hypothetical 40.2-kDa protein homologous to ornithine carbamoyltransferase. G4-C10 was placed in orf3 in a region homologous to the EPEC pathogenicity island named LIM, encoding a chaperon-like protein involved in the stable expression of BfpA and intimin (47).
Activity of type III secretion in class 1 mutants.
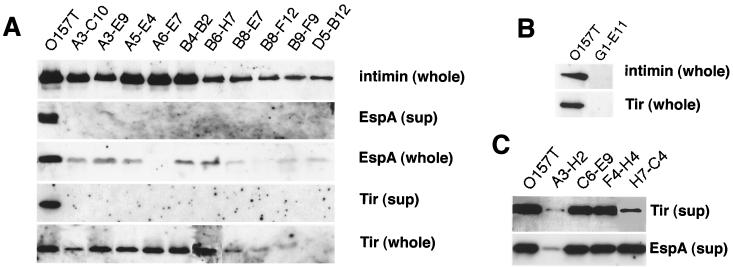
The genetic analysis of class 1 mutants suggested that their type III secretion system was inactivated. Hence, class 1 mutants were investigated for the ability to secrete EspA and Tir into the culture medium (see Materials and Methods). Immunoblot analysis with anti-EspA or anti-Tir antibody revealed that neither EspA nor Tir was secreted into the culture supernatants of class 1 mutants (Fig. 3A). In fact, analysis of the profiles of protein secreted into the culture supernatants in SDS-PAGE silver staining revealed that although 24-, 37-, 38-, and 72-kDa proteins corresponding to EspA, EspB, EspD, and Tir, respectively, were detected from the culture medium of O157T, they were not detected in those of class 1 mutants (data not shown). The whole cell levels of intimin and Tir of class 1 mutants such as A3-E9 (sepZ::mini-Tn5Km2), A5E4 (escD::mini-Tn5Km2), A6-E7 (sepL::mini-Tn5Km2), and B4-B2 (sepQ::mini-Tn5Km2) as examined by immunoblotting were similar to those of the wild type, while the remaining class 1 members showed a slight decrease in intimin and Tir expression (Fig. 3A). Although the reason for the effect of mini-Tn5Km2 insertions in genes outside the tir-cesT-eae locus on intimin expression was unclear, we concluded that the nonadherent phenotype of class 1 mutants was due to defects in type III secretion.
FIG. 3.
Levels of secretion and expression of EspA and intimin of class 1 to 3 mutants. Trichloroacetic acid-precipitated culture supernatants (sup) and bacterial cell lysates (whole) derived from equal amounts of wild-type strain O157T, class 1 mutants (A), class 3 mutants (B), and class 2 mutants (C) were resolved by SDS-12% PAGE, transferred to nitrocellulose membrane, and probed with polyclonal rabbit antisera specific to each protein indicated at the right of the photos.
Activity of type III secretion in class 2 mutants.
Although the adherence efficiency of class 2 mutants was decreased, the MC that formed at 4 h after infection were almost as large as those for the wild type (Fig. 1 and Table 1). The activity of type III secretion in class 2 mutants, as determined from the levels of secreted EspA and Tir using the immunoblots, indicated that class 2 mutants contained at least three subclasses of mutants: the first subclass (A3-H2) had decreased levels of type III secreted proteins, the second subclass had decreased levels of Tir but not of EspA (H7-C4), and the third subclass, which consists of the remaining 14 mutants, showed no change in the levels of type III secreted proteins (Table 1 and Fig. 3C). Thus, these results strongly suggested that some chromosome-encoded factors other than the proteins encoded by the genes in the LEE are involved in the adherence of EHEC to epithelial cells.
Two distinctive stages in the adherence of EHEC to epithelial cells.
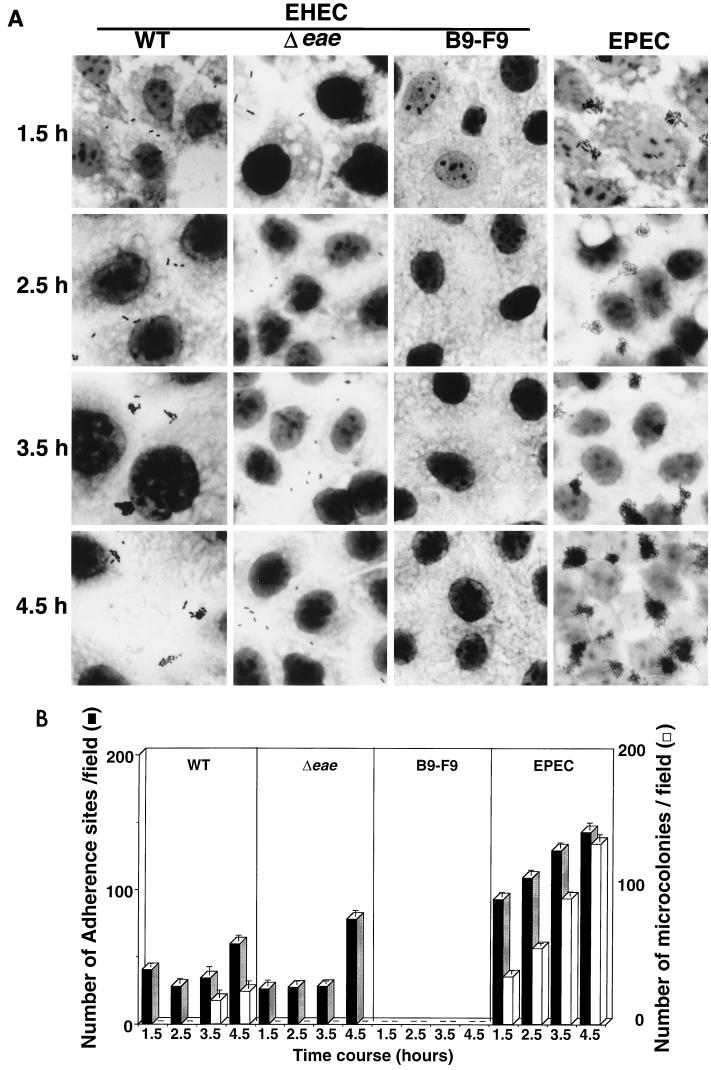
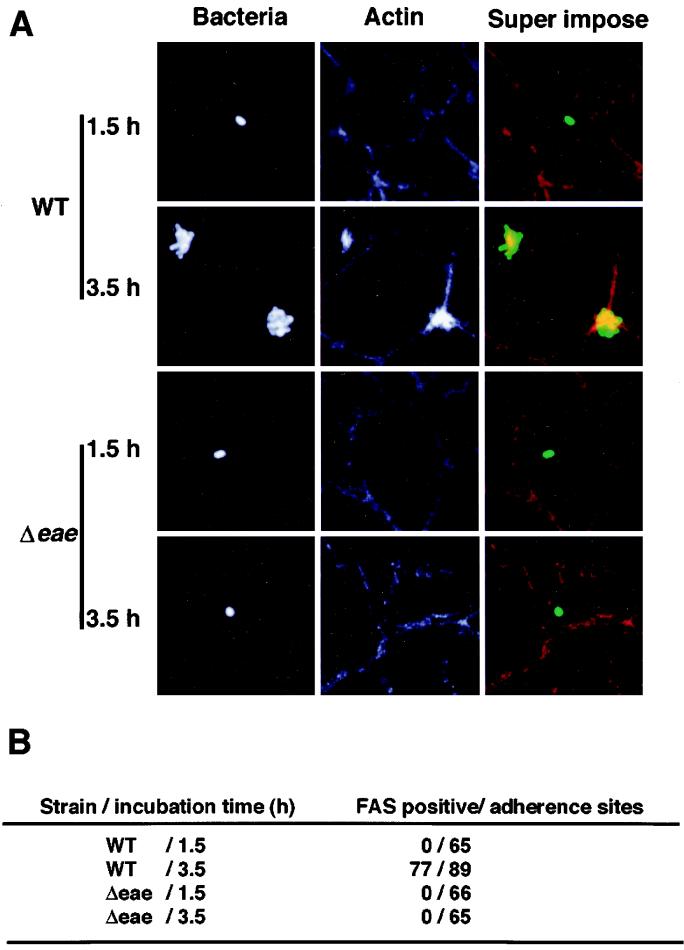
The class 3 mutant G1-E11 possessed the insertion mutation in the tir gene, thus causing a polar effect on downstream eae expression. G1-E11 displayed a diffuse pattern of adherence on Caco-2 cells even 4 h after infection and also displayed an aberrant adhesion phenotype compared with that of the wild type (Table 1). To further characterize the aberrant adhesion by G1-E11, we constructed a nonpolar eae mutant with O157T by replacing the portion containing the eae gene with an aphA-3 cassette, and we designated the nonpolar mutant the Δeae mutant. Because the Δeae mutant exhibited a diffuse pattern of adherence on Caco-2 cells similar to that exhibited by G1-E11, whereas the mutant was able to form MC on Caco-2 cells at 4 h postinfection when a cloned eae gene was introduced as a plasmid (data not shown), the Δeae mutant together with O157T, B9-F9 (one of the class 1 mutants), and an EPEC strain were investigated for their ability to adhere to Caco-2 cells. As shown in Fig. 4A, the Δeae mutant failed to form MC even at 4.5 h postinfection; instead, the bacteria adhered diffusely throughout the infection period. O157T exhibited a diffuse pattern of adherence at 1.5 h postinfection; however, the bacteria developed MC on further incubation for 2 h (at 3.5 h postinfection), suggesting that the development of MC by EHEC is a two-step process, the first step being intimin-independent diffuse adherence and the second step being intimin-dependent MC development (Fig. 4A). The quantitative assay for the numbers of adherent bacteria and visible MC supported the above notion; O157T and the Δeae mutant initially attached diffusely at 1.5 h postinfection, and after extensive washing out of the nonadherent bacteria, about 50% of the adherent bacterial population of O157T but not of the Δeae mutant formed MC (bacterial clusters of over eight bacteria) for another 2 h (Fig. 4B). To investigate the nature of the bacterial interaction with Caco-2 cells in MC development, O157T (wild type) and the Δeae mutant were examined for the ability to induce a FAS-positive phenotype at 1.5 and 3.5 h postinfection. Wild-type strain O157T did not elicit a FAS-positive phenotype at 1.5 h postinfection, but 2 h later the bacteria developing MC showed a FAS-positive phenotype at the point of bacterial attachment to Caco-2 cells (Fig. 5). In contrast, the Δeae mutant attached to Caco-2 cells at 3.5 h postinfection did not show a FAS-positive phenotype (Fig. 5), suggesting that the intimin-Tir interaction is involved in MC formation. To confirm this, O157T and the Δeae mutant in Caco-2 cells at 1.5 h were examined by scanning electron microscopy. Although pedestal-like structures were not recognized at the point of bacterial attachment of O157T or the Δeae mutant to Caco-2 cells at 1.5 h, such structures were observed underneath O157T but not the Δeae mutant at 4 h postinfection (data not shown). Under the same conditions, B9-F9 (a class 1 mutant) showed a very poor adherence capacity (less than one bacterium per field) at 1.5 h postinfection (Fig. 4B). The above results together with the changes in the adherent pattern of O157T on Caco-2 cells suggest that the diffuse pattern of adherence of EHEC to epithelial cells at the initial stage is mediated by the proteins secreted via the type III system, while at the later stage, development of MC is mediated by the intimin-Tir interaction.
FIG. 4.
Adherence behavior of O157Sakai (WT), the intimin mutant (Δeae), type III secretion mutant (B9-F9), and EPEC B171-8 (EPEC). B9-F9 is representative of class 1 mutants defective in the type III secretion system. (A) The bacteria were grown at 37°C for 2 h in DMEM-glycerol and then used to infect Caco-2 cell monolayers. Then these infected monolayers were incubated for 1.5 h and washed five times with PBS. After another 0, 1, 2, or 3 h of incubation at 37°C in DMEM-glycerol, the monolayers were again washed three times with PBS, fixed with methanol, and stained with Giemsa solution to visualize the adherent bacterial colonies. Strains and incubation times are shown above and on the left side of photos, respectively. (B) The black bars represent the total number of sites containing adherent EHEC, having either a single bacterium or a cluster of multiple bacteria. The white bars represent the number of clusters containing at least eight bacteria. The data shown are the means and standard errors of the means for 20 microscopic fields. The representative results were obtained from three independent experiments.
FIG. 5.
Photomicrographs of the intimin mutant and parental wild-type strain in a rhodamine-phalloidin assay for FAS in Caco-2 cells. (A) Caco-2 cells infected by bacteria as described in the Fig. 4 legend are shown as follows: in fluorescent views after treatment with anti-O157 LPS rabbit antibody followed by anti-rabbit goat antibody conjugated with fluorescein isothiocyanate (Bacteria), in fluorescent views of actin stained by rhodamine-phalloidin (Actin), and in a superimposed view of bacteria (green) and actin (red) (Super impose). Strains and incubation times are indicated at the left of photos. (B) Quantitative FAS assay of the O157Sakai wild type (WT) and the intimin mutant (Δeae), derived from the experiments whose results are shown in panel A.
DISCUSSION
Using mini-Tn5Km2 insertion mutagenesis, we isolated three classes of O157Sakai mutants defective in adherence to Caco-2 cells and investigated the altered phenotypes, including the site of mini-Tn5Km2 insertion. Class 1 mutants had lost almost all ability to adhere to Caco-2 cells, while class 2 mutants showed less of a capacity than the wild type. The class 3 mutant had little capacity to adhere to Caco-2 cells and had an aberrant, diffuse adherence phenotype. Class 1 and class 3 mutants were found to possess the transposon insertions in various genes at the LEE, while class 2 mutants possessed the mini-Tn5Km2 outside the LEE.
The defective adherence phenotype of class 1 mutants was accounted for by the site of the insertions in the LEE, since in all of these mutants the mini-Tn5Km2 was inserted in either the sepL, pas, sepQ, escV, sepZ, L0038, escC, escR, or L0051 gene, assumed to encode a protein involved in type III secretion (13, 14, 38). Immunoblot analysis of EspA and Tir secretion into the culture medium from class 1 mutants suggested that the type III secretion system was almost completely inactivated. Class 2 mutants showed a decrease in adherent capacity ranging from 1 to 40% of the wild-type level. A3-H2 showed the greatest decrease among class 2 mutants (1% of the wild-type level), with a marked defect in the activity of type III secretion, followed by H7-C4 (4% of the wild-type level), which was capable of secreting Tir but not EspA. Whereas the rest of the class 2 mutants showed some loss in adherence capacity, they were still capable of secreting EspA and Tir into the culture medium. Although the numbers of MC produced by the class 2 mutants were smaller than that produced by the wild type, their sizes at 4 h postinfection were similar to those of O157T. Indeed, since the numbers of initially diffuse adherent bacteria of class 2 mutants at 1.5 h postinfection were also decreased, the defects seem to lie in the initial diffuse adherence to Caco-2 cells.
In this study, we isolated a less adherent mutant (G1-E11) and classed it as a type 3 strain, since it possessed mini-Tn5Km2 in the tir gene at the LEE and showed aberrant diffuse adherence on Caco-2 cells throughout the infection, as previously described (7). The altered adherence phenotype was thus distinctive from that of class 2 mutants but rather similar to that of O157T at 1.5 h postinfection, suggesting that the initial diffuse adherence of EHEC to epithelial cells does not require the intimin-Tir interaction. To confirm the aberrant adherence phenotype of G1-E11 and to investigate the process for the development of MC by O157T, we constructed a nonpolar eae mutant (Δeae mutant) which was unable to produce intimin, and the Δeae mutant together with O157T (wild type) were examined for the ability to adhere to cells. The wild-type strain exhibited a diffuse pattern of adherence at 1.5 h postinfection and had formed MC (clusters of over eight bacteria) by 3.5 h postinfection, while the Δeae mutant adhered diffusely throughout the infection. Thus, the adherence of EHEC appears to proceed in at least two distinctive stages, diffuse adherence and MC development. The first stage of adherence in EHEC is clearly different from that in EPEC, since at 1.5 h postinfection EPEC began to form MC, often referred to as “localized adherence,” a process which is thought to be promoted by bundle pili (Fig. 4 and reference 34). Thus, the Δeae mutant is defective not in the initial stage but in the second stage. In this sense, class 1 mutants (defective in type III secretion) would have defects in the initial stage. Considering the process of bacterial adherence displayed by O157T (wild-type EHEC) compared with that of class 1 and class 3 mutants, including the Δeae mutant, the initial stage (diffuse adherence) and the subsequent, second stage (MC development) are type III dependent and intimin-Tir dependent, respectively.
Although various putative factors have been proposed to affect the adherence of EHEC to epithelial cells (see the introduction), which factor is involved in the initial stage is still unclear. EspA would be the best candidate for a protein mediating initial adherence. Recently, Ebel et al. (11) concluded that filamentous EspA-containing surface appendages mediate the initial contact between STEC and host cells based on the following observations: (i) EspA was detected as surface appendage structures attached to the host cells, (ii) the appendages were especially prominent on bacteria that had not yet induced the formation of actin pedestals, and (iii) a deletion of the espA gene completely abolished the capacity to bind to host cells. Thus, the properties of EspA may account for the aberrant, diffuse adherence phenotype displayed by G1-E11 (class 3 mutant) and also the defective adherence phenotypes of class 1 mutants (type III secretion mutants).
Although the mechanism underlying the formation of MC by EHEC is still to be elucidated, the process has been indicated to be closely associated with a FAS-positive phenotype, a typical host cellular response to the intimate bacterial attachment involved in actin condensation beneath the localized bacterial attachment (14, 34). In this study, we confirmed that the FAS-positive phenotype in O157Sakai adhering to Caco-2 cells was induced as the MC developed and that the accumulation of F-actin was evoked underneath the bacteria in MC. Similarly, the FAS-positive phenotype in class 2 mutants was evoked when bacteria developed MC. However, neither G1-E11 nor the Δeae mutant was able to form MC and induce the FAS-positive phenotype. When a cloned eae gene was introduced as a plasmid into the Δeae mutant, the bacteria began to develop MC and a FAS-positive phenotype at 3.5 h postinfection. Furthermore, a previous study indicated that adherence by EHEC to epithelial cells induced the formation of pedestal-like structures underneath the bacterium with actin polymerization (22, 42). Examination of MC developed by O157T at 4 h postinfection by scanning electron microscopy revealed that bacteria developing MC were placed over the protruded structures on the Caco-2 cells (data not shown). Therefore, it is most likely that the bacterial ability to induce MC formation requires the intimin-Tir interaction. For the MC formation of O157Sakai on epithelial cells, there would be two plausible mechanisms; one is the consequence of bacterial cell division, and the other one is recruitment of fresh bacteria. In the former case, an initial single adherent bacterium contributes to the formation of MC, while in the latter case, the receptor (Tir) clustering around the initial attached bacterium provides an additional receptor for fresh bacteria. Although we must be careful in drawing the conclusion, our data supports the former possibility. After the first 1.5 h of infection of Caco-2 cells by O157Sakai, we washed thoroughly the epithelial cells and incubated them for another 2.5 h to visualize the MC. The number of bacteria composing MC was almost parallel to the multiplied bacterial number during 2.5 h (the doubling time is approximately 40 min). Thus, the progeny of a bacterium initially attached to Caco-2 cells would remain tethered to the same site through the intimin-Tir interaction. If the hypothesis is correct, it may also account for the time delay observed in MC formation by EHEC in comparison with that by EPEC (see Fig. 4), since the BFP-entangled, multiplied EPEC has been indicated to be recruited to the epithelium as a unit (34).
It is still unclear how, exactly, the initially diffuse adherent EHEC strains developed MC in these experimental conditions, since by 1.5 h postinfection they had been extensively washed once with medium, and this was followed by incubation in fresh tissue culture medium for another 2.5 h (see Materials and Methods). During the initial adherence stage, some of the adhered bacteria would have the chance to induce the intimin-Tir interaction through type III secretion. In this sense, the ability of G1-E11 or the Δeae mutant to display the diffuse adherence phenotype on Caco-2 cells after the washing would be peculiar. In the absence of the intimin-Tir interaction, EHEC may utilize some unknown adherence factors, as is implied by the notion that class 2 mutants are involved in sustaining the bacterial association with Caco-2 cells during extensive washing.
In our study, all class 1 mutants had the transposon insertions in the LEE. As our study does not identify novel factors that play an indispensable role in adherence, it may support the importance of several LEE-encoded proteins, such as intimin, Tir, and possibly EspA, previously implicated in adherence and/or colonization (7, 10, 20, 23, 27, 28, 29). However, such other genetic loci as iha (7, 11, 14, 20, 21, 23, 34, 35, 46) would also be involved in promoting bacterial adherence, including activation of the type III secretion system in EHEC. Elliott et al. have recently reported that when the cloned LEE of EHEC, but not EPEC, was introduced into an E. coli K-12 nonadherent strain, the bacterium was unable to mediate adherence, including an attaching and effacing lesion (reference 12, or see the introduction), thus raising the possibility that genetic determinants other than LEE genes are also involved in the attachment of EHEC to epithelial cells. Although it is not clear at present how each of the mutated loci in class 2 mutants affects the activity of type III secretion or bacterial adherence, the results suggest the existence of adherence-associated loci additional to that encoded by the LEE. As mentioned above, A3-H2 and H7-C4 were defective in type III secretion. Furthermore, G2-E12 had the mini-Tn5Km2 insertions in regions homologous to that associated with bacterial adherence, such as the efa1 gene, encoding a putative adherence factor (35). G4-C10 had the mini-Tn5Km2 insertion in open reading frame 3 (orf3) in a region homologous to an EPEC pathogenicity island known as the LIM (locus for improving microcolony formation), which consists of four open reading frames and in which trcA encodes a chaperone-like protein required for stable expression of BfpA and intimin (47). Thus, further characterization of class 2 mutants may allow us to identify novel adherence-associated factors of EHEC.
In summary, our results suggest that EHEC adherence requires multiple factors encoded by the LEE and additional chromosomal loci and that the process is comprised of at least two distinctive stages, the initial diffuse stage and the MC-developing intimate adherence stage.
ACKNOWLEDGMENTS
This research was supported by the Research for the Future Program of the Japan Society for the Promotion of Science and by grant 11770135 from the Ministry of Education, Science and Culture of the Japanese Government.
We thank Chizu Sasako, Kenji Yamada, and Asaomi Kuwae for technical assistance and Alison O'Brien and Lisa Gansheroff for providing pEB312 plasmid.
REFERENCES
- 1.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 2.Boyce T G, Swerdlow D L, Griffin P M. Current concepts: Escherichia coli O157:H7 and the hemolytic-uremic syndrome. N Engl J Med. 1995;333:364–368. doi: 10.1056/NEJM199508103330608. [DOI] [PubMed] [Google Scholar]
- 3.Conlin C A, Trun N J, Silhavy T J, Miller C G. Escherichia coli prlC encodes an endopeptidase and is homologous to the Salmonella typhimurium opdA gene. J Bacteriol. 1992;174:5881–5887. doi: 10.1128/jb.174.18.5881-5887.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deibel C, Kramer S, Chakraborty T, Ebel F. EspE, a novel secreted protein of attaching and effacing bacteria, is directly translocated into infected host cells, where it appears as a tyrosine-phosphorylated 90 kDa protein. Mol Microbiol. 1998;28:463–474. doi: 10.1046/j.1365-2958.1998.00798.x. [DOI] [PubMed] [Google Scholar]
- 5.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 6.de Lorenzo V, Herrero M, Jacubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devinney R, Stein M, Reinscheid D, Abe A, Rushkowski S, Finley B B. Enterohemorrhagic Escherichia coli O157:H7 produce Tir, which is translocated to the host cell membrane but is not tyrosine phosphorylated. Infect Immun. 1999;67:2389–2398. doi: 10.1128/iai.67.5.2389-2398.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diorio C, Cai J, Marmor J, Shinder R, DuBow M S. An Escherichia coli chromosomal ars operon homolog is functional in arsenic detoxification and is conserved in gram-negative bacteria. J Bacteriol. 1995;177:2050–2056. doi: 10.1128/jb.177.8.2050-2056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donnenberg M S, Kaper J B. Enteropathogenic Escherichia coli. Infect Immun. 1992;60:3953–3961. doi: 10.1128/iai.60.10.3953-3961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnenberg M S, Tzipori S, McKee M L, O'Brien A D, Alroy J, Kaper J B. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J Clin Investig. 1993;92:1418–1424. doi: 10.1172/JCI116718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebel F, Podzadel T, Rohde M, Kresse A U, Krämer S, Deibel C, Guzmán C A, Chakraborty T. Initial binding of Shiga toxin-producing Escherichia coli to host cells and subsequent induction of actin rearrangement depend on filamentous EspA-containing surface appendages. Mol Microbiol. 1998;30:147–161. doi: 10.1046/j.1365-2958.1998.01046.x. [DOI] [PubMed] [Google Scholar]
- 12.Elliott S J, Yu J, Kaper J B. The cloned locus of enterocyte effacement from enterohemorrhagic Escherichia coli O157:H7 is unable to confer the attaching and effacing phenotype upon E. coli K-12. Infect Immun. 1999;67:4260–4263. doi: 10.1128/iai.67.8.4260-4263.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott S J, Wainwright L A, McDaniel T, MacNamara B, Donnenberg M, Kaper J B. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol. 1988;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 14.Frankel G, Philips A D, Rosenshine I, Dougan G, Kaper J B, Knuton S. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversion elements. Mol Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 15.Griffin P M, Tauxe R V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–97. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 16.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenny B, DeVinney R, Stein M, Reinscheid D J, Frey E A, Finlay B B. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 18.Knutton S, Rosenshine I, Pallen M J, Nisan I, Neves B C, Bain C, Wolff C, Dougan G, Frankel G. A novel EspA associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knutton S, Baldwin T, Williams P H, McNeish A S. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–1298. doi: 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kresse A U, Schulze K, Deibel C, Ebel F, Rohde M, Chakraborty T, Guzmán C A. Pas, a novel protein required for protein secretion and attaching and effacing activities of enterohemorrhagic Escherichia coli. J Bacteriol. 1998;180:4370–4379. doi: 10.1128/jb.180.17.4370-4379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kresse A U, Rohde M, Guzmán C A. The EspD protein of enterohemorrhagic Escherichia coli is required for the formation of bacterial surface appendages and is incorporated in the cytoplasmic membrane of target cells. Infect Immun. 1999;67:4834–4842. doi: 10.1128/iai.67.9.4834-4842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Magoun L, Luperchio S, Schauer D B, Leong J M. The Tir-binding region of enterohaemorrhagic Escherichia coli intimin is sufficient to trigger actin condensation after bacterial-induced host cell signalling. Mol Microbiol. 1999;34:67–81. doi: 10.1046/j.1365-2958.1999.01574.x. [DOI] [PubMed] [Google Scholar]
- 23.Louie M, de Azavedo J C, Handelsman M Y, Clark C G, Ally B, Dytoc M, Sherman P, Brunton J. Expression and characterization of the eaeA gene product of Escherichia coli serotype O157:H7. Infect Immun. 1993;61:4085–4092. doi: 10.1128/iai.61.10.4085-4092.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makino K, Ishii K, Yasunaga T, Hattori M, Yokoyama K, Yutsudo C H, Kubota Y, Yamaichi Y, Iida T, Yamamoto K, Honda T, Han C G, Ohtsubo E, Kasamatsu M, Hayashi T, Kuhara S, Shinagawa H. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 1998;28:1–9. doi: 10.1093/dnares/5.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Makino K, Yokoyama K, Kubota Y, Yutsudo C H, Kimura S, Kurokawa K, Ishii K, Hattori M, Tatsuno I, Abe H, Iida T, Yamamoto K, Onishi M, Hayashi T, Yasunaga T, Honda T, Sasakawa C, Shinagawa H. Complete nucleotide sequence of the prophage VT-2-Sakai carrying the verotoxin 2 genes of the enterohemorrhagic Escherichia coli O157:H7 derived from the Sakai outbreak. Genes Genet Syst. 1999;74:227–239. doi: 10.1266/ggs.74.227. [DOI] [PubMed] [Google Scholar]
- 26.Marmur J. A procedure for isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1996;3:208–218. [Google Scholar]
- 27.McKee M L, Melton-Celsa A R, Moxley R A, Francis D H, O'Brien A D. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect Immun. 1995;63:3739–3744. doi: 10.1128/iai.63.9.3739-3744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKee M L, O'Brien A D. Investigation of enterohemorrhagic Escherichia coli O157:H7 adherence characteristics and invasion potential reveals a new attachment pattern shared by intestinal E. coli. Infect Immun. 1995;63:2070–2074. doi: 10.1128/iai.63.5.2070-2074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKee M L, O'Brien A D. Truncated enterohemorrhagic Escherichia coli (EHEC) O157:H7 intimin (EaeA) fusion proteins promote adherence of EHEC strains to HEp-2 cells. Infect Immun. 1996;64:2225–2233. doi: 10.1128/iai.64.6.2225-2233.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellies J L, Elliott S J, Sperandio V, Donnenberg M S, Kaper J B. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 31.Menard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metcalf W W, Jiang W, Daniels L L, Kim S K, Haldimann A, Wanner B L. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 33.Milkman R, Bridges M M. Molecular evolution of the Escherichia coli chromosome. IV. Sequence comparisons. Genetics. 1993;133:455–468. doi: 10.1093/genetics/133.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholls L, Grant T H, Robins-Browne R M. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol Microbiol. 2000;35:275–288. doi: 10.1046/j.1365-2958.2000.01690.x. [DOI] [PubMed] [Google Scholar]
- 36.Ohnishi M, Tanaka C, Kuhara S, Ishii K, Hattori M, Kurokawa K, Yasunaga T, Makino K, Shinagawa H, Murata T, Nakayama K, Terawaki Y, Hayashi T. Chromosome of the enterohemorrhagic Escherichia coli O157:H7; comparative analysis with K-12 MG1655 revealed the acquisition of a large amount of foreign DNAs. DNA Res. 1999;31:361–368. doi: 10.1093/dnares/6.6.361. [DOI] [PubMed] [Google Scholar]
- 37.Paton A W, Voss E, Manning P A, Paton J C. Shiga toxin-producing Escherichia coli isolates from cases of human disease show enhanced adherence to intestinal epithelial (Henle 407) cells. Infect Immun. 1997;65:3799–3805. doi: 10.1128/iai.65.9.3799-3805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perna N T, Mayhew G F, Pósfai G, Elliott S, Donnenberg M S, Kaper J B, Blattner F R. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plamann M D, Stauffer L T, Urbanowski M L, Stauffer G V. Complete nucleotide sequence of the E. coli glyA gene. Nucleic Acids Res. 1983;11:2065–2075. doi: 10.1093/nar/11.7.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puente J L, Bieber D, Ramer S W, Murray W, Schoolnik G K. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol Microbiol. 1996;20:87–100. doi: 10.1111/j.1365-2958.1996.tb02491.x. [DOI] [PubMed] [Google Scholar]
- 41.Riley L W, Remis R S, Helgerson S D, McGee H B, Wells J G, Davis B R, Hebert R J, Olcott E S, Johnson L M, Hargrett N T, Blake P A, Cohen M L. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 42.Rosenshine I, Ruschkowski S, Stein M, Reinscheid D J, Mills S D, Finlay B B. A pathogenic bacterium triggers epithelial signals to form a functional bacterial receptor that mediates actin pseudopod formation. EMBO J. 1996;15:2613–2624. [PMC free article] [PubMed] [Google Scholar]
- 43.Short J M, Fernandez J M, Sorge J A, Huse W D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988;16:7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steffes C, Ellis J, Wu J, Rosen B P. The lysP gene encodes the lysine-specific permease. J Bacteriol. 1992;174:3242–3249. doi: 10.1128/jb.174.10.3242-3249.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoker N G, Fairweather N F, Spratt B G. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982;18:335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- 46.Tarr P I, Bilge S S, Vary J C, Jr, Jelacic S, Habeeb R L, Ward T R, Baylor M R, Besser T E. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect Immun. 2000;68:1400–1407. doi: 10.1128/iai.68.3.1400-1407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tobe T, Tatsuno I, Katayama E, Wu C Y, Schoolnik G K, Sasakawa C. A novel chromosomal locus of enteropathogenic Escherichia coli (EPEC), which encodes a bfpT-regulated chaperone-like protein, TrcA, involved in microcolony formation by EPEC. Mol Microbiol. 1999;33:741–752. doi: 10.1046/j.1365-2958.1999.01522.x. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe H, Wada A, Inagaki Y, Ito K, Tamura K. Outbreaks of enterohaemorrhagic Escherichia coli O157:H7 infection by two different genotype strains in Japan. Lancet. 1996;348:831–832. doi: 10.1016/s0140-6736(05)65257-9. [DOI] [PubMed] [Google Scholar]