Abstract
Background
Cutaneous malignant melanoma (MM) is potentially aggressive, and numerous clinically suspicious pigmented skin lesions are excised, causing unnecessary mutilation for patients at high healthcare costs, but without histopathological evidence of MM. The high number of excisions may be lowered by using more accurate diagnostics. Tape stripping (TS) of clinically suspicious lesions is a non‐invasive diagnostic test of MM that can potentially lower the number needed to biopsy/excise.
Materials and methods
The aim is to determine the diagnostic accuracy of TS in detecting MM in clinically suspicious pigmented skin lesions. This systematic review following PRISMA guidelines searched PubMed, Web of Science, and Embase (September 2022) using melanoma combined with tape stripping, adhesive patch(es), pigmented lesion assay, or epidermal genetic information retrieval.
Results
Ten studies were included. Sensitivity ranged from 68.8% (95% confidence interval [CI] 51.5, 82.1) to 100% (95% CI 91.0, 100). Specificity ranged from 69.1% (95% CI 63.8, 74.0) to 100% (95% CI 78.5, 100). A pooled analysis of five studies testing the RNA markers LINC00518 and PRAME found a sensitivity of 86.9% (95% CI 81.7, 90.8) and a specificity of 82.4% (95% CI 80.8, 83.9).
Conclusion
Overall quality of studies was low, and the reliability of sensitivity and specificity is questionable. However, TS may supplement well‐established diagnostic methods as pooled analysis of five studies indicates a moderate sensitivity. Future studies are needed to obtain more reliable data as independent studies with no conflict of interest.
Keywords: adhesive patch, diagnostic accuracy, epidermal genetic information retrieval, malignant melanoma, nevus, pigmented lesion assay, skin cancer, tape stripping
1. INTRODUCTION
Cutaneous malignant melanoma (MM) is one of the most aggressive skin cancers. In 2020, more than 300,000 people were diagnosed with melanoma globally, and more than 50 000 patients died of MM globally. 1 Early detection of melanoma makes a vast difference in overall survival. 2 Therefore, a quick and accurate diagnosis and fast, efficient treatment of MM are imperative for overall survival.
A suspicious lesion is diagnosed by excision and subsequent histopathology examination; this constitutes the reference standard for MM diagnosis. 3 In a study by Malvehy et al., 2014, diagnostic efficacy was studied in MM. 4 The observed sensitivity was 70.6% and specificity was 81.4% in a group of dermatologists, including visual‐ and dermoscopic assessment. A high number of clinically suspicious pigmented skin lesions are therefore excised annually, where histopathological examination demonstrates no signs of MM. There is no medical need to remove these lesions, and the unnecessary removal burdens dermatologists, surgeons, lab workers and pathologists.
The tape stripping (TS) method has been used in other skin diseases such as atopic dermatitis and psoriasis. 5 TS of pigmented lesions is a novel non‐invasive diagnostic test for MM. Several potential biomarkers exist, such as RNA, cells and lipids. MM expresses a different RNA profile on the surface than normal skin and nevi, which can be used diagnostically. 6 An adhesive patch is placed on the pigmented lesion. After delineation of the lesion by a surgical marker pen or a standard dark colour pen, the patch is immediately removed, and cells from the stratum corneum are left on the patch to be analysed. TS can be repeated with a new patch on the same lesion multiple times. If the method detects, for example, specific RNA markers at a certain threshold, the TS test is positive (TS+). Likewise, if the method does not detect RNA markers at a certain threshold, the TS test is negative (TS‐). Some RNA markers are downregulated in MM, and the TS method will detect the lack of these specific RNA markers and the test will be positive (TS+). A disadvantage of TS is that the adhesive patch does not necessarily work on mucous membranes, palms, soles, and nails or if there is bleeding or serous exudation. 7 A commercially available test that can examine pigmented lesions is referred to as pigmented lesion assay (PLA).
Exfoliative cytology is a non‐invasive method that uses skin cells from the stratum corneum to identify disease by examining the structure and characteristics of these cells. 8 Typically, the examiner scrapes the skin cells using a scalpel or curette and then smears the cells on a glass slide. Another way of collecting the skin cells is to use double‐stick tape; one side sticks to the glass slide and the other sticks to the skin. After the cells are smeared or taped onto the glass slide, the cells can be stained with various stain types, for example, toluidine blue and then examined under a microscope.
Numbers needed to biopsy/excise (NNB) is a metric used to assess the accuracy and cost‐effectiveness of MM diagnostics. A systematic review from 2019 included 46 articles and found an NNB ranging from 2.2 to 287. The weighted mean NNB was 15.6. 9 The NNB partly depends on the prevalence of MM in the patient population and on the performers’ ability to distinguish MM from other skin conditions. Theoretically, when TS is introduced in MM diagnostics, this may lower the NNB.
2. MATERIAL AND METHODS
This study aims to determine the diagnostic accuracy of TS for detecting MM in clinically suspicious pigmented lesions. A meta‐analysis was made to examine the test's accuracy by comparing the existing studies. The primary outcomes are sensitivity and specificity for the index test TS. This systematic review based on PRISMA guidelines is registered through PROSPERO (CRD42022312716).
The inclusion criteria of this study were the index test “TS” and analysis of the tissue on tape. The reference standard was skin biopsy and histopathological examination. The target disease was MM. Studies were on patients with pigmented lesions or lesions clinically suspicious of MM and included details on true positives, true negatives, false positives, and false negatives. Articles were in Danish, English or German. The exclusion criteria of this study were animal and ex vivo studies and articles in other languages.
On 14 September 2022, literature searches were performed on PubMed, Web of Science and Embase to retrieve studies on TS and MM. Reference lists were manually searched for additional studies to include. The search was performed according to PRISMA guidelines. 10 The MeSH and Emtree terms “melanoma” and the word “melanoma” were combined with the terms: TS, adhesive patch(es), PLA or epidermal genetic information retrieval (EGIR). Two authors (Ida Marie Nedergaard Thomsen and Mette Mogensen) screened the articles and agreed on which articles should be included in this review.
The following data from the included studies were extracted: information on lesions, index test, reference standard, true positive, true negative, false positive, false negative, population characteristics, study details and setting. Two authors (Ida Marie Nedergaard Thomsen and Ida M. Heerfordt) independently extracted data from the included studies, registering all data in Microsoft Excel ver. 16.66.1 (22101101).
Only data on the true negative, true positive, false negative and false positive were used in the review to guarantee sensitivity, specificity and 95% confidence interval (95% CI) were identically calculated. Sensitivity, specificity and 95% CI were calculated in the statistical software “R Studio” (ver. 1.4.11.06). The calculations from R studio are shown in Supplementary 1.
QUADAS‐2 tool was used to judge the Risk of Bias and Applicability. Two authors (Ida Marie Nedergaard Thomsen and Ida M. Heerfordt) independently performed the judgement and if disagreement between reviewers, they would discuss the different evaluations. A summary was made to visualise the Risk of Bias and Applicability judgement. 11
3. RESULTS
Figure 1 shows a PRISMA flow diagram of the search results. Fifty‐two articles met the inclusion/exclusion criteria. Fourteen hits were meeting, or conference abstracts and no articles were found. Sixteen hits were scoping reviews, and TS of MM was only mentioned in short. 5 , 7 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 Five articles had other outcomes where sensitivity and specificity were not examined. 26 , 27 , 28 , 29 , 30 Three articles were “response letters” to other articles with no relevant data. 31 , 32 , 33 One article used formalin‐fixed paraffin‐embedded tissue block samples instead of in vivo skin on patients, 34 and one article examined melasma instead of MM. 35 The last two articles consisted of an article which analysed another article's data, 36 and an article which was “A Health Technology Assessment”. 37 In total, 28 articles were excluded, and ten articles were included in this review.
FIGURE 1.
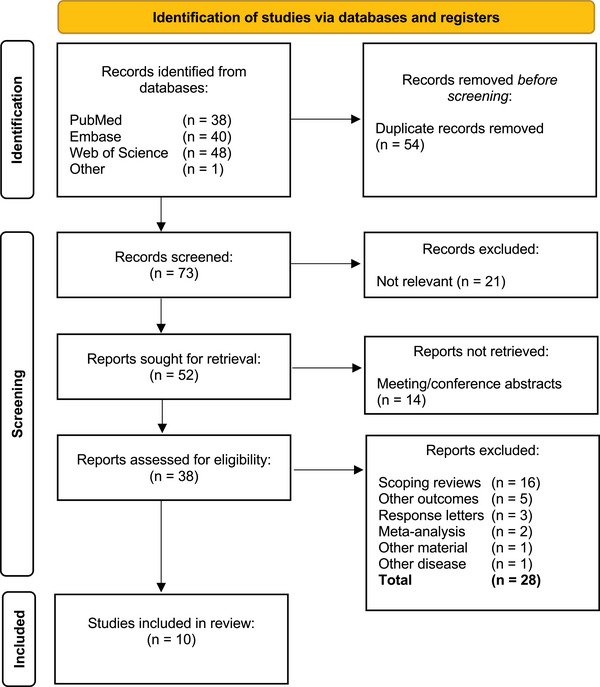
PRISMA Flow diagram.
The extracted data were combined into five categories, as shown in Table 1. One study separated their data and had two datasets: TS+ and TS‐. 38 These two datasets were combined in the review to calculate sensitivity and specificity. Four studies showed results from when they made the index test and when they validated the index test. 6 , 39 , 40 , 41 The results used in this review are the results from the test validations.
TABLE 1.
Extracted data and results.
| Skelsey et al., | Shah et al., | Hornberger et al., | Ferris et al., | Ferris et al., | Gerami et al., | Yao et al., | Gerami et al., | Wachsman et al., | Berardi et al., | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2021 | 2019 | 2018 | 2018 | 2017 a | 2017 | 2016 | 2014 | 2011 | 1992 | |||
| Index test | Genes tested | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 17 | 0 | |
| Name of genes | LINC00518 + PRAME | LINC00518 + PRAME | LINC00518 + PRAME | LINC00518 + PRAME | LINC00518 + PRAME | LINC00518 + PRAME | LINC00518 + PRAME | LINC00518 + CMIP | LINC00518, CMIP, PRAME + 14 genes | ‐ | ||
| Name of method | PLA | PLA | PLA | PLA | PLA | PLA | PLA | Adhesive patch method | EGIR | TSTB | ||
| Location of analysis | USA b | USA b | USA b | USA b | USA b | USA b | USA b | USA b | USA b | Italy | ||
| Patch pr. lesion | 4 | 4 | N/A | 4 | 4 | 4 | 4 | 4 | 4 | 1 | ||
| Reference standard and lesions | Total lesions | TS‐ 1233 | TS+ 313 | 20 | 319 | 381 | 60 a | 398 | 73 | 64 | 128 | 150 |
| Assummed non‐melanoma c | 1223 | ‐ | 14 | 288 d | 329 e | ‐ | ‐ | 42 f | ‐ | ‐ | ‐ | |
| Total biopsies | 10 | 313 | 6 | 42 | 55 | 60 a | 398 | 73 | 64 | 128 | 150 | |
| Histopathological melanoma | 10 | 59 | 6 | 15 | 20 | 8 a | 87 | 31 | 42 | 39 | 32 | |
| Histopathological nevus | N/A | 213 | N/A | 14 | 28 | 44 a | 253 | N/A | 21 | 89 | 98 | |
| Other histopathological diagnosis | N/A | 41 (keratoses and scars after biopsy) | N/A | 2 | 4 seborrheic or actinic keratoses | 8 (5 lentigines, 3 keratoses) a | 58 (seborrheic keratosis, lentigo simplex BBC and fibrosis) | N/A | 1 | 0 | 20 (1 lentigo simplex, 7 seborrhoeic keratosis, 5 BCC, 7 other) | |
| Location on body | N/A | N/A | N/A | N/A | N/A | N/A | Info | N/A | Info | Info b | N/A | |
| Accuracy | True positive | 59 | 6 | 14 | 19 | 8 | 79 | 22 | 41 | 39 | 22 | |
| True negative | 1223 | 14 | 279 | 329 | 46 | 215 | 37 | 16 | 76 | 115 g | ||
| False negative | 10 | 0 | 1 | 1 | 0 | 8 | 9 | 1 | 0 | 10 h | ||
| False positive | 254 | 0 | 25 | 32 | 6 | 96 | 5 | 6 | 13 | 3 | ||
| Sensitivity (95% CI) | 85.5 (75.3; 91.9) | 100 (61.0; 100) | 93.3 (70.1; 98.8) | 95.0 (76.4; 99.1) | 100 (67.6; 100) | 90.8 (82.9; 95.3) | 71.0 (53.4; 83.9) | 97.6 (87.6; 99.6) | 100 (91.0; 100) | 68.8 (51.5; 82.1) | ||
| Specificity (95% CI) | 82.8 (80.8; 84.6) | 100 (78.5; 100) | 91.8 (88.2; 94.4) | 91.1 (87.7; 93.6) | 88.5 (77.1; 94.6) | 69.1 (63.8; 74.0) | 88.1 (75.0; 94.8) | 72.7 (51.8; 86.8) | 85.4 (76.6; 91.3) | 97.5 (92.9; 99.2) | ||
| Study details and Setting | Type of study | Cohort study | CSS | Case report | Registry study (CSS) | Registry study (CSS) | CSS | CSS | CSS | CSS | CSS | CSS |
| Eligibility criteria | N/A | N/A | N/A | N/A | N/A | N/A | Info | N/A | Info | Info | N/A | |
| Follow‐up time (if no biopsy) | 36 months | ‐ | 3‐6 months | 3‐6 months | 3‐6 months | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | |
| Samples collected | Aug 2017 ‐ Aug 2018 | July 2018 ‐ June 2019 | N/A | N/A | May 2016 ‐ June 2017 PLA(+) | N/A | N/A | N/A | 2004 ‐ 2010 DermTech | N/A | Sep 1990 ‐ Sep 1991 | |
| Conflic of interest | Yes | Yes | No | Yes i | Yes i | Yes | Yes i | Yes i | Yes i | Yes i | N/A | |
| Samples collected | 5 clinical sites (USA) | 53 practices (USA) | Pigmented lesion clinic (USA) | 2 derm. sites (USA) | 4 derm. sites (USA) | N/A a | 28 derm. sites (USA, Europe and Australia) | N/A | Derm. sites (USA 18, Europe 2 and Australia 1) | USA | Italy (Dept. of Dermatology) | |
| Population characteristics | Total patients | 1233 | N/A | 1 | N/A | 381 | N/A | 398 j | N/A | 64 | N/A | 142 |
| Median/mean age (year) | N/A | Median: 48 | 64 | N/A | N/A | N/A | Median: 49 | N/A | Mean: 48.4% k | Mean: 52.9 l | Mean: 29 | |
| Age range (year) | N/A | N/A | ‐ | N/A | N/A | N/A | 19‐97 | N/A | 18‐83 | 19‐95 l | (2‐91) | |
| Male/Female | N/A | 39.2%/60.8% | 1/0 | N/A | N/A | N/A | 218/179 | N/A | 42/22 | 42%/58% l | 49/93 | |
Lesions from the same pool of patients as Gerami et al., 2017.
Test analysed at the same American company.
Assummed non‐melanoma if no biopsy was performed at follow‐up.
11 lesions had a histopathological diagnosis "non‐melanoma".
Three lesions had a histopathological diagnosis "non‐melanoma".
42 lesions had a histopathological diagnosis "non‐melanoma".
27 non‐significant, 78 true‐negative.
Seven non‐significant, three true‐negative.
Funded/partly funded by the same American company.
One patient with no male/female information.
Calculated using multiple means.
Info both validation set and training set.
Abbreviations: BCC, basal cell carcinoma; CSS, cross‐sectional study; derm, dermatology; EGIR, epidermal genetic information retrieval; N/A, not available; PLA, pigmented lesion assay; TSTB, tape stripping toluidine blue.
All ten studies included data on the true negative, true positive, false negative, and false positive. 6 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 Sensitivity ranged from the lowest 68.8% (95% CI 51.5, 82.1) 46 to the highest 100% (95% CI 91.0, 100). 6 Two other studies also had a sensitivity of 100% but with a wider 95% CI. 42 , 45 Specificity ranged from the lowest 69.1% (95% CI 63.8, 74.0) 40 to the highest 100% (95% CI 78.5, 100). 42 All included studies tested the lesions with tape strips, 6 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 and the results are shown in Table 1. Nine studies examined RNA markers, 6 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 and one examined the cells using exfoliative cytology. 46 One study tested for 17 MM RNA markers to determine if the suspected lesions were melanoma. 6 Another study tested the lesions for two RNA markers: overexpression of LINC00518 and downregulation of CMIP. 39 A third study made a test that tested for overexpression of two RNA markers: LINC00518 and PRAME. 41 The same two RNA markers were used in the remaining six studies. 38 , 40 , 42 , 43 , 44 , 45 One study called their test EGIR). 6 and another study called their test “Adhesive patch method”. 39 Seven studies all called their test Pigmented lesion Assay “PLA”. 38 , 40 , 41 , 42 , 43 , 44 , 45 In all nine RNA marker studies, the tests were analysed at the same American company, and all used four patches per lesion for the analysis. 6 , 38 , 39 , 40 , 41 , 42 , 44 , 45 The exfoliative cytology study called their test “TS toluidine blue”. They used one tape strip per lesion and analysed the tests in their own lab. 46
Skelsey et al. included the most (1546 lesions), 38 and Shah et al. included the least (20 lesions). 42 Gerami et al. performed biopsies on 398 lesions and was the study with the most biopsied lesions. 40 All studies had information on how many melanomas were biopsied. 6 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 Eight studies demonstrated data on how many nevi and other skin lesions were biopsied, 6 , 38 , 39 , 40 , 43 , 44 , 45 , 46 including actinic keratoses, seborrheic keratoses, fibrosis, basal cell carcinoma and lentigo simplex. Only three studies had information on the tested lesions' location on the body. 6 , 39 , 40
All ten studies examined the accuracy of TS. Nine articles did a cross‐sectional study, 6 , 38 , 39 , 40 , 41 , 43 , 44 , 45 , 46 and one article did a case report. 42 Three studies had information on inclusion and exclusion criteria 6 , 39 , 40 and are shown in Supplementary 2.
Nine studies had data on the country of sample retrieval. In one study, the samples were from Italy. 46 Five studies had samples from The United States. 6 , 38 , 42 , 43 , 44 Three studies had samples from Australia, The United States, and Europe. 39 , 40 , 45 Eight studies had information on the place of the test: a dermatology site. 38 , 39 , 40 , 43 , 44 , 45 , 46 Only one study wrote that dermatologists chose the lesions. 40 No studies had data on who did the TS, for example, doctors or trained staff. Four articles had information on when samples were collected. 38 , 39 , 44 , 46 The collection time for the four studies did not overlap. Three studies did not biopsy all lesions tested. Instead, TS‐lesion patients had follow‐up appointments. 38 , 42 , 43 , 44 Three studies had a follow‐up period of 3–6 months, 42 , 43 , 44 and one study had a follow‐up period of 36 months. 38
Eight studies have a potential conflict of interest (COI) as they are all associated with the same American company. 6 , 38 , 39 , 40 , 41 , 43 , 44 , 45
Six studies had information on the patient's age and sex. 6 , 38 , 39 , 40 , 42 , 46 One study only had data on TS+ patients. 38 In three of these six studies, most patients were female, 6 , 38 , 46 and in the three other studies, the majority were male. 39 , 40 , 42 The mean age of the included patients ranged from 29 46 to 52.9 years. 6 The case report included one patient, who was 64 years old. 42 In two studies, the median ages were 48 38 and 49 years, 40 respectively. In one study, patients under 18 years were included. 46
A meta‐analysis of two forest plots shows the sensitivity and specificity of all the included studies and pooled analysis of five included studies (Figure 2). The pooled analysis includes studies that used the RNA markers LINC00518 and PRAME. 38 , 40 , 41 , 42 , 44 Two studies were not included in the analysis because there was a risk of the same lesions being included twice. 43 , 45 The pooled analysis found a sensitivity of 86.9% (95% CI 81.7, 90.8) and specificity of 82.4% (95% CI 80.8, 83.9).
FIGURE 2.
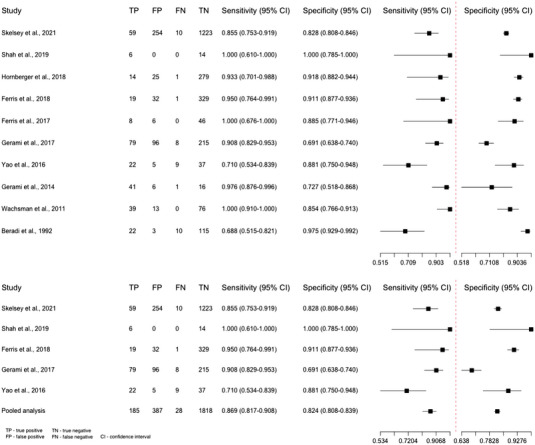
Forest plots of tape stripping—all included studies and a pooled analysis.
The Risk of Bias and Applicability results are demonstrated in Figure 3. If lesions were not randomly selected, the standard reference was not the same for every lesion and all patients were not included in the analysis, the studies scored high.
FIGURE 3.
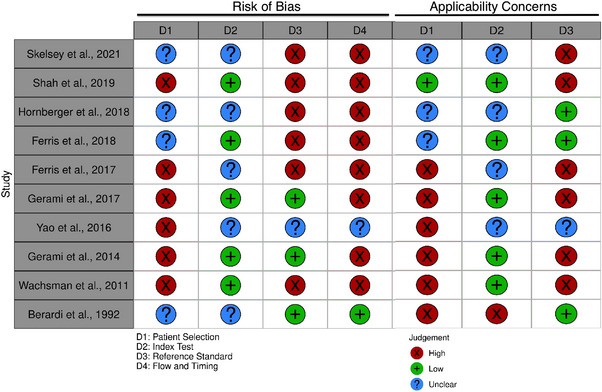
Risk of bias and applicability concerns summary.
4. DISCUSSION
We examined the diagnostic accuracy of TS to detect MM. The metanalysis suggests moderate sensitivity and specificity and the Risk of Bias and Applicability results display low quality of the included studies, especially on patient selection, reference standard and “flow and timing”.
TS must have high sensitivity to ensure the test diagnoses all MM. At the same time, it is crucial that TS also obtains high specificity to avoid unnecessary biopsies and excisions. The pooled analysis of the “PLA” test had a sensitivity of 86.9% (95% CI 81.7, 90.8) and a specificity of 82.4 % (95% CI 80.8, 83.9). In contrast, the “EGIR” test had a sensitivity of 100 (95% CI 91.0, 100) and a specificity of 85.4% (95% CI 76.6, 91.3). 6 As the PLA test is commercially available, it is essential to encourage independent studies exploring which RNA markers have the highest diagnostic accuracy for the diagnosis of MM.
Only one study of TS and exfoliative cytology is included in the review. Even though the method can distinguish MM from nevi, the demonstrated RNA analysis is more accurate.
The studies lacked information, especially on population characteristics and settings. An example was the number of patients; four papers did not write how many patients were included in their studies. 6 , 41 , 43 , 45 The RNA markers may vary from patient to patient; therefore, some patients’ melanoma and skin types may be better to test with TS than others. Of the six papers that did have information on the number of included patients, 38 , 39 , 40 , 42 , 44 , 46 one paper only had partial information on the number of patients included. Four studies had the same number of lesions as patients. 38 , 39 , 40 , 44 One study had one patient and 20 lesions 42 and one study had 142 patients and 150 lesions, which suggests some patients had more than one lesion tape‐stripped. 46
Shah et al. included the lowest number of patients, as it was a case report with one patient 42 and Skelsey et al. included the highest number of patients, with at least 1233 patients. 38
Six studies had a reference standard (histopathological examination) for every included lesion. 6 , 39 , 40 , 41 , 45 , 46 A limitation of the four most recent studies is the lack of histopathological examination for TS‐lesions. Instead, they did a follow‐up after 3–36 months and assumed that TS‐lesions not biopsied at follow‐up were true negatives. 38 , 42 , 43 , 44 When studies do not have the same standard reference, the studies get a high risk of bias.
Details on who performed the TS were missing in all the included studies, and only one study specifically stated the lesions were selected by a dermatologist. 40
There is a potential risk of overlap of patients. One study 45 used lesions from the same pool of patients as another study. 40 Two studies are registry studies. 43 , 44 Both studies had one lesion that tested TS‐ and was biopsied the same day. This was not the typical procedure for their studies when a lesion tested negative. It could be the same lesion and perhaps an overlap in patients in the studies. There is an additional risk of patient overlap as two studies are registry studies, and six studies do not have information on when patients were included.
As the quality of the included studies is low, the reliability of sensitivity and specificity is questionable. Some of the studies show high sensitivity. 6 , 39 , 42 , 45 However, the pooled analysis indicates moderate sensitivity. New studies with no COI and transparent population selection, characteristics and settings are required. All included lesions should undergo a biopsy to find a more reliable sensitivity and specificity in future studies.
Most of the studies included are conducted in the United States, with only a few patients from Europe and Australia. Australia and New Zealand (NZ) in 2020 had the most diagnosed MM pr. 100 000 citizens worldwide, followed by western Europe. 47 A new prospective cohort study from Denmark is being conducted, but more studies are needed in Europe and Australia/NZ to assess regional differences and ensure the test works on European and Australian/NZ skin. 48
No randomised control trials have yet been made to find the difference between TS and no TS and how it affects the patient's prognosis, which would be very relevant.
Included papers present TS as a supplement to the already established evaluation of clinically suspicious lesions (visual inspection, dermoscopy and clinical photography). 42 The test should be used for the “clinically ambiguous lesions” and not “definitive melanomas”. 38 The proportion of diagnosed TS+ lesions should therefore be in situ or pT1a MM. One of the included papers studied how dermatologists' mean biopsy sensitivity and specificity improved when TS was incorporated into their decision on when or when not to biopsy. Mean biopsy sensitivity improved from 95.0% to 98.6% (p = 0.01) and specificity increased from 32.1% to 56.9% (p < 0.001). 45 As the histopathological examination is still the golden standard, TS will not likely replace MM's standard diagnostics. Another non‐invasive method to combine with TS would be reflectance confocal microscopy which offers bedside in vivo microscopy of suspicious lesions with moderate to high diagnostic accuracy and a reduction of numbers needed to excise by 43% shown in a randomized diagnostic trial of more than 3000 patients. 49
5. CONCLUSION
As the overall quality of the included studies is low, the reliability of sensitivity and specificity is questionable. Some of the studies show high sensitivity and specificity. The pooled analysis indicates moderate sensitivity and specificity. The pooled analysis examines RNA markers LINC00518 and PRAME and found a sensitivity of 86.9% (95% CI 81.7, 90.8) and specificity of 82.4% (95% CI 80.8, 83.9). TS with RNA markers is more accurate than exfoliative cytology. Lastly, TS should be done by a person with knowledge of skin cancers to only test lesions where MM is suspected.
CONFLICT OF INTEREST
None declared.
FUNDING INFORMATION
The authors received no financial support for the research, authorship, and/or publication of this article.
Supporting information
Supporting information
Thomsen IMN, Heerfordt IM, Karmisholt KE, Mogensen M. Detection of cutaneous malignant melanoma by tape stripping of pigmented skin lesions – A systematic review. Skin Res Technol. 2023;29:e13286. 10.1111/srt.13286
Systematic review registration: PROSPERO CRD42022312716
DATA AVAILABILITY STATEMENT
All the data for this study will be made available upon reasonable request to the corresponding author.
REFERENCES
- 1. Sung H, Jacques F, Rebecca LS, et al. Global cancer statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209‐249. [DOI] [PubMed] [Google Scholar]
- 2. Papageorgiou C, Zoe A, Sofia‐Magdalini M, et al. Melanoma: staging and follow‐up. Dermatol Pract Concept. 2021;11(1):2021162S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kittler, H Evolution of the clinical, dermoscopic and pathologic diagnosis of melanoma. Dermatol Pract Concept. 2021;11(1):2021163S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Malvehy J, Hauschild A, Curiel‐Lewandrowski C, et al. Clinical performance of the Nevisense system in cutaneous melanoma detection: an international, multicentre, prospective and blinded clinical trial on efficacy and safety. Br J Dermatol. 2014;171(5):1099‐1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barber C, Boiko S. Tape stripping: investigational, diagnostic and therapeutic uses in dermatology. Clin Dermatol. 2022;40(4):355‐362. [DOI] [PubMed] [Google Scholar]
- 6. Wachsman W, Morhenn V, Palmer T, et al. Noninvasive genomic detection of melanoma. Br J Dermatol. 2011;164(4):797‐806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heibel HD, Hooey, L , Cockerell, CJ. A review of noninvasive techniques for skin cancer detection in dermatology. Am J Clin Dermatol. 2020;21(4):513‐524. [DOI] [PubMed] [Google Scholar]
- 8. Ferrante di Ruffano L, Jacqueline D, Naomi C, et al. Exfoliative cytology for diagnosing basal cell carcinoma and other skin cancers in adults. Cochrane Database Syst Rev. 2018;12(12):Cd013187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nelson KC, Susan MS, Kathylynn S, Suephy CC, Clara C‐L, Evaluation of the number‐needed‐to‐biopsy metric for the diagnosis of cutaneous melanoma: a systematic review and meta‐analysis. JAMA Dermatol. 2019;155(10):1167‐1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Page MJ, Joanne EM, Patrick MB, The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372(n/a):n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McGuinness LA, Higgins, JPT. Risk‐of‐bias VISualization (robvis): an R package and Shiny web app for visualizing risk‐of‐bias assessments. Res Synth Methods. 2020;(n/a). [DOI] [PubMed] [Google Scholar]
- 12. Babino G, Francesca S, Aimilios L, Caterina L, Elvira M, Giuseppe A. Tape stripping: a very short‐term follow‐up procedure for suspicious black lesions. J Am Acad Dermatol. 2015;72(6):e151‐e152. [DOI] [PubMed] [Google Scholar]
- 13. Blumenberg, M. Skinomics: past, present and future for diagnostic microarray studies in dermatology. Expert Rev Mol Diagn. 2013;13(8):885‐894. [DOI] [PubMed] [Google Scholar]
- 14. Dorrell DN, Strowd, LC. Skin cancer detection technology. Dermatol Clin. 2019;37(4):527‐536. [DOI] [PubMed] [Google Scholar]
- 15. Fink, C , Haenssle, HA. Strategies for the noninvasive diagnosis of melanoma. Hautarzt. 2016;67(7):519‐528. [DOI] [PubMed] [Google Scholar]
- 16. Fried L, Andrea T, Shirin B, Tracey NL, David P, Jennifer AS. Technological advances for the detection of melanoma: advances in molecular techniques. J Am Acad Dermatol. 2020;83(4):996‐1004. [DOI] [PubMed] [Google Scholar]
- 17. Jung JM, Ji YC, Woo JL, Sung EC, Mi WL, Chong HW. Emerging minimally invasive technologies for the detection of skin cancer. J Pers Med. 2021;11(10):951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee N, Scope, A , Rabinovitz, H. Assessing skin cancer using epidermal genetic information retrieved by adhesive patch skin surface sampling. Dermatol Clin. 2017;35(4):521. [DOI] [PubMed] [Google Scholar]
- 19. Paliwal S, Byeong HH, Kenneth YT, Samir M. Diagnostic opportunities based on skin biomarkers. Eur J Pharm Sci. 2013;50(5):546‐556. [DOI] [PubMed] [Google Scholar]
- 20. Patel JK, Sailesh K, Oliver AP, Sadegh A, George E, Brian B. Newer technologies/techniques and tools in the diagnosis of melanoma. Eur J Dermatol. 2008;18(6):617‐631. [DOI] [PubMed] [Google Scholar]
- 21. Pathania YS, Zoe A, Gabriel S, Anant P, Stephan G, Mohamad G. Non‐invasive diagnostic techniques in pigmentary skin disorders and skin cancer. J Cosmet Dermatol. 2021;721(2):444‐450. [DOI] [PubMed] [Google Scholar]
- 22. Rivers JK, Copley MR, Svoboda R, Rigel DS. Non‐invasive gene expression testing to rule out melanoma. Skin Therapy Lett. 2018;23(5):1‐4. [PubMed] [Google Scholar]
- 23. Varedi A, Laura JG, Caroline CK, et al. Use of new molecular tests for melanoma by pigmented‐lesion experts. J Am Acad Dermatol. 2020;82(1):245‐247. [DOI] [PubMed] [Google Scholar]
- 24. Wassef C, Rao, BK. Uses of non‐invasive imaging in the diagnosis of skin cancer: an overview of the currently available modalities. Int J Dermatol. 2013;52(12):1481‐1489. [DOI] [PubMed] [Google Scholar]
- 25. Skudalski L, Reid W, Philip EK, Jane MG‐K. Melanoma: How and when to consider clinical diagnostic technologies. J Am Acad Dermatol. 2022;86(3):503‐512. [DOI] [PubMed] [Google Scholar]
- 26. Brouha B, Laura KF, Maral KS, et al. Real‐world utility of a non‐invasive gene expression test to rule out primary cutaneous melanoma: a large US registry study. J Drugs Dermatol. 2020;19(3):257‐262. [PubMed] [Google Scholar]
- 27. Ferris LK, Ronald LM, Pedram G, et al. Noninvasive analysis of high‐risk driver mutations and gene expression profiles in primary cutaneous melanoma. J Invest Dermatol. 2019;139(5):1127‐1134. [DOI] [PubMed] [Google Scholar]
- 28. Ferris LK, Darrell SR, Daniel MS, et al. Impact on clinical practice of a non‐invasive gene expression melanoma rule‐out test: 12‐month follow‐up of negative test results and utility data from a large us registry study. Dermatol Online J. 2019;25(5):13030. [PubMed] [Google Scholar]
- 29. Yao ZX, Ronald M, Talisha A, Burkhard J. An adhesive patch‐based skin biopsy device for molecular diagnostics and skin microbiome studies. J Drugs Dermatol. 2017;16(10):979‐986. [PubMed] [Google Scholar]
- 30. Kim S, Paris F, Franklin A, et al. Effect of plasticizers on drug‐in‐adhesive patches containing 5‐fluorouracil. Int J Pharm. 2022;611:121316. [DOI] [PubMed] [Google Scholar]
- 31. Beatson M, Weinstock, MA. Further consideration of the pigmented lesion assay. JAMA Dermatol. 2019;155(3):393. [DOI] [PubMed] [Google Scholar]
- 32. Siegel DM, Hornberger, J. Further consideration of the pigmented lesion assay – reply. JAMA Dermatol. 2019;155(3):393‐394. [DOI] [PubMed] [Google Scholar]
- 33. Rigel DS, John WW, Maral KS, Gary P, Michael DH, Burkhard J. Response to Marchetti et al. J Invest Dermatol. 2022;142(1):232‐234. [DOI] [PubMed] [Google Scholar]
- 34. Jansen B, Doyle H, Ronald M, Maesa H, Zuxu Y. Gene expression analysis differentiates melanomas from spitz nevi. J Drugs Dermatol. 2018;17(5):574‐576. [PubMed] [Google Scholar]
- 35. Xu J, Haojie L, Haixin L, et al. Tape stripping and lipidomics reveal skin surface lipid abnormity in female melasma. Pigment Cell Melanoma Res. 2021;34(6):1105‐1111 [DOI] [PubMed] [Google Scholar]
- 36. Marchetti MA, Japbani KN, Silvia EM, Stephen WD. Real‐world application of a noninvasive two‐gene expression test for melanoma diagnosis. J Invest Dermatol. 2021;141(9):2303‐2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. OntarioHealth , Pigmented lesion assay for suspected melanoma lesions: a health technology assessment. Ont Health Technol Assess Ser. 2021;21(5):1‐81. [PMC free article] [PubMed] [Google Scholar]
- 38. Skelsey M, Brouha B, Rock J, et al. Non‐invasive detection of genomic atypia increases real‐world NPV and PPV of the melanoma diagnostic pathway and reduces biopsy burden. SKIN J Cutaneous Med. 2021;5(5):512–523. [Google Scholar]
- 39. Gerami P, John PA, Tara JP, Howard SR. Development of a novel noninvasive adhesive patch test for the evaluation of pigmented lesions of the skin. J Am Acad Dermatol. 2014;71(2):237‐244. [DOI] [PubMed] [Google Scholar]
- 40. Gerami P, Zuxu Y, David P, et al. Development and validation of a noninvasive 2‐gene molecular assay for cutaneous melanoma. J Am Acad Dermatol. 2017;76(1):114‐120.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yao ZX, Talisha A, Margaret O, Carol S, Darryl G, Burkhard J. Analytical characteristics of a noninvasive gene expression assay for pigmented skin lesions. Assay Drug Dev Technol. 2016;14(6):355‐363. [DOI] [PubMed] [Google Scholar]
- 42. Shah A, John H, Scott RF, et al. Use of the Pigmented Lesion Assay to rapidly screen a patient with numerous clinically atypical pigmented lesions. JAAD Case Rep. 2019;5(12):1048‐1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hornberger J, Siegel, DM . Economic analysis of a noninvasive molecular pathologic assay for pigmented skin lesions. JAMA Dermatol. 2018;154(9):1025‐1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ferris LK, Pedram G, Maral KS, et al. Real‐world performance and utility of a noninvasive gene expression assay to evaluate melanoma risk in pigmented lesions. Melanoma Res. 2018;28(5):478‐482. [DOI] [PubMed] [Google Scholar]
- 45. Ferris LK, Burkhard J, Jonhan H, et al. Utility of a noninvasive 2‐gene molecular assay for cutaneous melanoma and effect on the decision to biopsy. JAMA Dermatol. 2017;153(7):675‐680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Berardi P, Arcangeli F. The tape stripping toluidine blue (TSTB) method in the diagnosis of malignant melanoma: an investigator‐blind study. Melanoma Res. 1992;2(2):93‐100. [DOI] [PubMed] [Google Scholar]
- 47. Globocan . Melanoma of skin: age standardized (World) incidens rate, melanoma of skin. 2020. Available from: https://gco.iarc.fr/today/data/factsheets/cancers/16‐Melanoma‐of‐skin‐fact‐sheet.pdf
- 48. Heerfordt IM, Jeppe DA, Peter AP, et al. Detection of cutaneous malignant melanoma using RNA sampled by tape strips: a study protocol. PLoS One. 2022;17(9):e0274413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pellacani G, Francesca F, Silvana C, et al. Effect of reflectance confocal microscopy for suspect lesions on diagnostic accuracy in melanoma: a randomized clinical trial. JAMA Dermatol. 2022;158(7):754‐761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Data Availability Statement
All the data for this study will be made available upon reasonable request to the corresponding author.


