Abstract
Following spillover, viruses must adapt to new selection pressures exerted by antiviral responses in their new hosts. In mammals, cellular defense mechanisms often include viral nucleic acid editing pathways mediated through protein families apolipoprotein-B mRNA-editing complex (APOBEC) and Adenosine Deaminase Acting on ribonucleic acid (ADAR). APOBECs induce C→U transitions in viral genomes; the APOBEC locus is highly polymorphic with variable numbers of APOBEC3 paralogs and target preferences in humans and other mammals. APOBEC3 paralogs have shaped the evolutionary history of human immunodeficiency virus, with compelling bioinformatic evidence also for its mutagenic impact on monkeypox virus and severe acute respiratory syndrome coronavirus 2. ADAR-1 induces adenose-to-inosine (A→I) substitutions in double-stranded ribonucleic acid (RNA); its role in virus adaptation is less clear, as are epigenetic modifications to viral genomes, such as methylation. Nucleic acid editing restricts evolutionary space in which viruses can explore and may restrict viral-host range.
Current Opinion in Virology 2023, 60:101326
This review comes from a themed issue on Adaptation of viruses to new host
Edited by Silke Stertz and Xander de Haan
For complete overview about the section, refer “Adaptation of viruses to new host (2023)”
https://doi.org/10.1016/j.coviro.2023.101326
1879–6257/© 2023 Published by Elsevier B.V.
Introduction
The majority of human pathogenic infectious diseases are zoonotic in origin [1]. These include several of the deadliest recently emergent pathogens, including human immunodeficiency virus (HIV), Ebola virus (EBOV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The viral and host factors leading to pathogen spillover are of great medical consequence for global public health but are largely poorly understood [2]. Owing to climate change, it is expected that opportunities for viral incursions into human populations will increase due to novel viral-host encounters in nature [3]. It was estimated that of the 214 ribonucleic acid (RNA) viruses known to be able to infect humans (data from mid-2017) [4], only 26 were human-specific. Of the remainder with potential zoonotic origins or cocirculating between hosts, over 80% showed restricted or nontransmissibility between humans. Viruses that are of the greatest consequence for public health are therefore those members of a highly select pool that are both pathogenic and able to sustain long-term transmission chains within human populations, a short list that is prominently led by SARS-CoV-2 and HIV-1. Therefore, it is imperative to better understand the determinants of post-spillover viral success.
Upon first infection in a novel host, viruses must begin adapting to the potentially hostile intracellular and extracellular environment they encounter on infection. The evolution of an initially poorly optimized virus to a highly transmissible pathogen can help identify the general fitness barriers that must be overcome by zoonotic viruses for them to succeed in their new host population. Most attention to date has been paid to changes in the viral proteome. For instance, point mutations in the SARS-CoV-2 spike protein can improve infectivity of new target cells (e.g. S:D614G) and similarly for GP:E82A in EBOV 5, 6. While perhaps less appreciated, selection for adaptive changes is not restricted to viral proteins and can act on nucleotide sequences independently of their coding.
Here, we review a significant nucleotide-selective pressure that impacts the short- and long-term adaptation of novel viruses to replication and transmission in humans — direct nucleic acid editing, principally by innate, immune-activated antiviral proteins. Direct effects on virus nucleic acids are part of a wider spectrum of pressures acting on virus composition, such as suppression of immunoreactive dinucleotides such as CpG (C followed by G) and UpA (U followed by A)(reviewed in [7]).
Main text of review
In general, the early adaptive events of most human pathogens are unknown because spillover occurred historically well before the invention and large-scale deployment of sequencing technologies. For that reason, this review will largely focus on recently emergent or reemergent zoonotic viruses, such as SARS-CoV-2 and monkeypox virus (MPXV). It is important to note that these selective pressures necessarily acted on currently circulating pathogens at the time of early adaptation even without human observation. In some instances, indirect evidence of these selective pressures can be identified bioinformatically, as will be discussed later, based on deviations from random (statistically ‘expected’) nucleotide and dinucleotide distributions.
Nucleic acid editing
Apolipoprotein-B mRNA-editing complex
The apolipoprotein-B mRNA-editing complex (APOBEC) family is an expansive group of proteins that deaminate cytidines (C) to uridines (U) in RNA and single-stranded deoxyribonucleic acid. The family has extant members throughout the tree of life and may have been present in the last eukaryotic common ancestor, although their antiviral function appears to be more recently emerged 10, 11. In humans, there are eleven APOBEC proteins: APOBEC1, APOBEC2, APOBEC3 (A3A, A3B, A3C, A3D, A3F, A3G, and A3H), APOBEC4, and activation-induced deaminase (AID). Placental mammals contain orthologs of all human APOBEC proteins with the APOBEC3 subgroup demonstrating particularly extensive diversification (reviewed in [12]). Functionally, APOBEC proteins deaminate specific nucleotides based on the surrounding base context (sequence preference) and the presence of secondary-structure elements (structural preference) ( Table 1). These sites are often referred to as deamination hotspots in the literature.
Table 1.
Deamination context preferences of human APOBEC proteins.
| Name | Sequence context | Structural context (DNA/RNA) |
|---|---|---|
| APOBEC1 | 5′-AC-3′ | n.d./n.d. |
| AID | 5′-WRC-3′ | n.d./n.d. |
| A3A | 5′-TC-3′ | Open/loop |
| A3B | 5′-TC-3′ | Open/n.d. |
| A3C | 5′-TC-3′ | Open/n.d. |
| A3D | 5′-TC-3′ | Open/n.d. |
| A3F | 5′-TC-3′ | Open/n.d. |
| A3G | 5′-CC-3′ | Open/loop |
| A3H | 5′-TC-3′ | Open/n.d. |
APOBEC2 and APOBEC4 do not have defined sequence context preferences and are excluded from this table. Structural context preferences for APOBEC3 proteins as described for DNA in McDaniel et al. [47] and for RNA in Sharma and Baysal [48]. W = A/T, R = A/G, n.d. = not determined. For structural context, ‘open’ signifies outside of a defined secondary-structure element (e.g. stem, loop, and bulge).
Adapted from Ratcliff and Simmonds [10].
Some members of the APOBEC family play roles in cellular mRNA editing to modulate gene expression; an APOBEC1-mediated C→U mutation of the ApoB mRNA produces a UAA stop codon and truncation of the encoded Apolipoprotein-B protein version of ApoB48 with a modified biological function 13, 14, 15, 16. AID is an essential component of the vertebrate adaptive immune system. Genomic DNA editing and the activation of mismatch repair mechanisms induces somatic hypermutation of immunoglobulin genes, thereby generating the wide antibody repertoire required for pathogen recognition and affinity maturation 17, 18, 19, 20, 21. In addition to these cellular roles, members of the APOBEC family have evolved a range of functions in innate immunity, particularly among members of the APOBEC3 subgroup. Their expression is interferon (IFN)-inducible with antiviral activity governed by deaminase-dependent and -independent pathways [10].
While APOBEC3 proteins are well-known for their ability to induce lethal mutagenesis in HIV, they can also hypermutate retroelements and DNA viruses [10]. Over many generations, APOBEC activity leads to the fixation of beneficial, neutral, and slightly deleterious C→T- driven substitutions, these progressively deplete genomes of preferred deamination sites (5′TC-3′ and 5′-CC-3′) and decrease their overall total G+C content. Some researchers have taken advantage of this loss of preferred dinucleotides (e.g. 5′-TC-3′ and/or 5′-GA-3′) to detect the ‘footprint’ of APOBEC activity in RNA viruses, DNA viruses, and retroelements 8, 9•, 22. For SARS-CoV-2, the accumulation of C→U substitutions has been particularly apparent due to the availability of an unprecedented number of high-quality full-genome sequences of SARS-CoV-2 strains throughout the pandemic, and an intrinsically low overall substitution rate that might otherwise mask editing-induced changes 9•, 10, 23. After over two years of evidence for APOBEC-driven hypermutation in SARS-CoV-2 being limited to bioinformatic analyses, the first experimental evidence for this phenomena was recently published by Nakata et al. (2023) [24]. In their study, overexpression of APOBEC3A, but not other APOBECs, induced low levels of C→U changes in viral RNA, although it is unclear that the proportion of these reached consensus. Importantly, the C→U mutations observed in that study match the structural and sequence context preferences for APOBEC3A in human mRNA transcripts described in Sharma et al. (2015) [25].
The site preference of APOBEC-associated editing necessarily restricts the evolutionary space available for human-infecting viruses to sample, with mutations that introduce additional preferred deamination sites likely to be purged by APOBEC activity. Indeed, sites whose deamination fails to be fixed may represent strongly targeted locations where viruses are unable to tolerate C→U transitions because of phenotypic effects from associated amino acid substitutions, or interruption of RNA secondary structures encoded within the genome. If these are in locations encoding proteins or RNA secondary structure, they may provide a starting point for antiviral drug development.
A challenge in interpreting the relevance of nucleic acid editing to pathogen spillover is the limited knowledge of the antiviral APOBEC activity in nonhumans. Lentiviruses encode viral infectivity factor (Vif) to antagonize APOBEC restriction; these proteins are found in retroviruses that infect humans [26], nonhuman primates [27], felines [28], and even-toed ungulates 29, 30, 31, strongly suggestive of a conserved evolutionary requirement to evade APOBEC hypermutation in these diverse mammalian species. Indeed, lentiviruses infecting odd-toed ungulates do not contain Vif and avoid APOBEC restriction through a novel mechanism [32]. For these nonhuman APOBECs, site preferences are largely undescribed. In rodents, several studies point to an APOBEC3 sequence context preference of 5′-WYC-3′, although the evidence is mixed for specific -1 and -2 preferences 33, 34.
During the 2022/23 outbreak of clade IIb of the large dsDNA MPXV, the observed overabundance of TC→TT and GA→AA substitutions relative to historical samples has been suggested to be due to the action of human APOBEC3 [35]. Reconstructing the evolutionary history of the clade-IIb and clade-IIa phylogenies revealed an APOBEC-driven signal on the former only; for clade IIb, 91% of mutations were C→T or G→A versus only 13% for clade IIa. Further, 93% of the observed APOBEC-like mutations for clade IIb were in APOBEC3′s preferred sequence context. The clade-specific evolutionary history is strongly suggestive of active APOBEC-driven hypermutation in the host or reservoir species for MXPV clade IIb over the last five years. The ongoing and progressive accumulation of TC→TT changes detected in currently circulating lineages of MPXV during the 2022 human outbreak lends further support to a direct role of human-to-human transmission on the MPXV sequence change [36]. Overall, this mutational spectra provide evidence of sustained, undocumented transmission of MPXV in human populations since approximately 2017. We direct the readers to O’Toole et al. (2023) [35] for an excellent discussion of the potential evolutionary consequences of APOBEC-driven hypermutation on MPXV.
The apparent lack of an APOBEC signal in clade IIa and clade I supports the traditional reservoir host (currently assumed to be African rodents) not having the capacity for APOBEC-driven hypermutation of viruses or MPXV being able to antagonize the pathway in that host (but not in humans). This raises the question as to whether APOBEC activity can act as a barrier to cross-species transmission. For primate and feline lentiviruses, APOBECs are known to impact host species range (due to an inability for the respective lentivirus Vif proteins to antagonize novel host species’ APOBEC proteins) 27, 28•. Whether APOBECs play a more general role in determining host range for viruses remains unclear and needs further investigation (i.e. does knocking out APOBEC expression in mammalian cell lines make them more permissive to in vitro infection by nonhuman infective viruses?).
Adenosine Deaminase Acting on ribonucleic acid
Adenosine Deaminase Acting on ribonucleic acid (ADAR) is a family of RNA editing enzymes that catalyze adenose-to-inosine (A→I) editing in dsRNA [37]. Inosine is read as cytosine by ribosomes and viral RNA-dependent RNA polymerases, leading to protein diversity if induced on transcripts and A→G transitions if induced on viral genomic RNA. Humans have three ADAR proteins (ADAR-1, ADAR-2, and ADAR-3), of which ADAR-1 is IFN-inducible and the focus of this section.
ADAR-1 activity can be either proviral or antiviral, dependent on the infecting virus and host cell and whether viral or host RNAs are being edited [37]. While the effects of cellular mRNA editing may be induced by any virus (including dsDNA viruses) that provokes a cellular IFN response, editing of viral RNA sequences by ADAR is limited to those virus groups that replicate through a dsRNA phase, and where the RNA replication intermediate is accessible within the cytoplasm where ADAR is expressed. In general, dsRNA is thought to be an activator of multiple intracellular innate immune pathways [38], and many RNA viruses occlude the dsRNA through a variety of methods. In contrast to APOBEC, sites of ADAR-1-induced A→I editing are not known to occur preferentially in specific favored sequence contexts within dsRNA substrates [39]. Therefore, the ‘evolutionary footprint’ left by ADAR-1 activity is solely the progressive loss of adenines; detecting this change bioinformatically is more difficult than for APOBEC.
ADAR-induced hypermutation of viruses or viral transcripts is ubiquitous throughout the kingdom animalia; there is even evidence of A→I editing by ADAR-1 of Ostreid Herpesvirus-1 transcripts during replication in pacific oysters [40]. As such, any differential impact of ADAR-1 in different host species will likely be due to a virus’ inability to evade or antagonize the ADAR-1 pathway, either directly or through downregulation of the cellular IFN antiviral response (impacting both ADAR-1 and APOBEC3 in humans). The ability of emergent SARS-CoV-2 variants of concern to antagonize the IFN pathway in humans to a greater degree than the ancestral strain, demonstrates this is a characteristic that can be gained as viruses adapt [41].
Ribonucleic acid modifications
Biochemical modifications to mRNAs beyond changes to their nucleotide sequences in the cellular epitranscriptome have been hypothesized to be functionally relevant for cellular gene expression since the 1970s. The effects and functional importance of comparable RNA modifications in the genomes of RNA viruses on their cellular interactions and replication, has recently become an area of active research [42], facilitated by the advent of sequencing technologies that can report nucleic acid methylation status, such as Oxford Nanopore [43]. In human cells, viral transcripts may be subject to the whole host of modifications encountered on cellular RNA sequences, including methylation of adenosines (m6A, m1A) or cytosines (m5C), ribose methylation in tRNAs (2′-O-methylation), and conversion of uridine to pseudouridine 42, 44, 45. These modifications largely occur in specific sequence contexts (e.g. DR(mA)CH for m6a, where D = A, G, or U; R = G or A; and H = A, C, or U) [42]. As such, selection can act on these modifications; substitutions leading to proviral modification will be selected for, while those leading to antiviral modifications will be selecting against. Post-transcriptional RNA modifications represent another dimension limiting the evolutionary space that a virus can explore and different modification pathways in different species may impact host range.
Despite the many potential effects of RNA editing on virus gene expression and cellular recognition, its scope to contribute to host adaptation appears limited. Owing to the high degree of conservation of RNA modification machinery among mammals [46], epigenetic modifications of RNA are unlikely to play a significant role in zoonotic virus host range, irrespective of their other potential functional effects. Further, unlike A→I or C→U editing, there currently is no evidence that post-transcriptional modifications induce substitutions that can be passed on to progeny viruses. While a young field with interesting potential to define viral RNA regulation, its impact on characterizing viral evolution is as of yet unclear.
Conclusions
This review highlights the potential roles of nucleic acid editing and epigenetic modifications on restricting or modulating virus gene expression and fitness. Overviews of these mechanisms are presented in Fig. 1. While epigenetic modification pathways are highly conserved across all animal groups and may not confer host specificity, an ability of a virus to successfully evade the antiviral effects of the more genetically and structurally variable ADAR-1 and particularly APOBEC family members may require a substantial degree of short- term adaptive changes to successful replication in a new host. The recent observations of TC→TT editing in MPXV and the excess of C→U substitutions in SARS-CoV-2 genome sequences flags up the intriguing possibility that these have arisen so prominently because the viruses are unable to counteract genome editing by APOBEC or related cellular defense editing pathways of a new host. A compelling parallel might be host specificity of HIV-1 and other lentiviruses, which depends, in part, on the development of effective evasion strategies for the potent host defense protein, tripartite motif-containing protein 5 (TRIM5α) [36]. This binds to incoming retroviral capsids and prevents initiation of reverse transcription, and as with APOBECs, an ability to evade or counteract its antiviral effects represents a major host range determinant. Although anti-APOBEC evasion pathways are poorly characterized functionally, the mutational signatures observed in MPXV and SARS-CoV-2 genomes may be a reflection of a similar failure to fully counteract APOBEC-mediated editing in the ‘wrong’ host. Furthermore, the observation that the APOBEC locus is highly variable, with major differences in the numbers and target specificities of paralogs of APOBEC3 between mammalian species and evidence for strong positive selection for sequence diversification [37], lends further support to the idea that it may be involved in a highly active arms race with viruses, with progressive functional changes in APOBEC3 to maintain antiviral activity against DNA and RNA viruses.
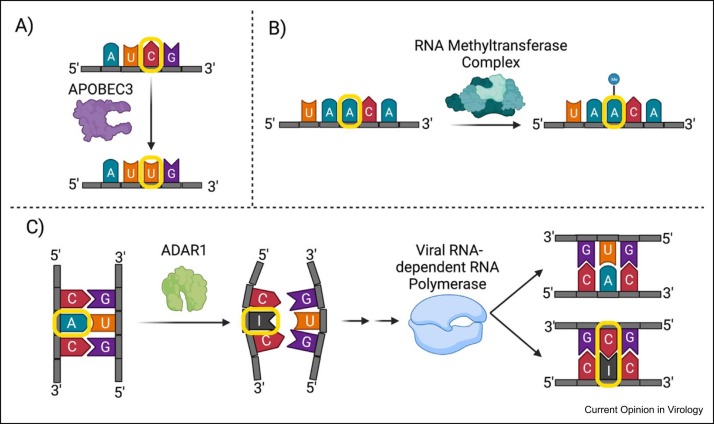
Figure 1.
Nucleotide modifications induced by APOBEC, the RNA methyltransferase complex, and ADAR. (a) Deamination of cytidine to uracil by APOBEC3 inducing a C→U mutation in the preferred sequence context of 5′-UC-3′. (b) m6a RNA methylation in an example of the DR(mA)CH sequence context. Other, nondepicted epitranscriptomic changes include cytosine and ribose methylation and conversion of uridine to pseudouridine. (c) ADAR-1-driven A→I substitutions in dsRNA leading to destabilization of the RNA secondary structure as inosine, similar to guanine, preferentially pair with cytosine. Also depicted is the strand-specific impact of A→I substitutions on viral evolution.
In the future, experimental investigation of the effects of APOBEC expression and other mediators of virus genome editing on determining host range in cell culture, and the use of deep sequencing analysis to compare site editing frequencies on virus passage in cell lines from different hosts (e.g. bat, rodent, and primates), will provide insights into its role in cellular antiviral responses and the degree to which it can shape the short- and long-term evolutionary trajectories of vertebrate viruses.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
JR was supported by the Marshall Aid Commemoration Commission and the Clarendon Fund. The authors acknowledge Cecilia Jay for assistance in preparing Figure 1 on BioRender.com.
Data Availability
No data were used for the research described in the article.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
-
•
of special interest
-
••
of outstanding interest
- 1.Taylor L.H., Latham S.M., Woolhouse M.E.J. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woolhouse M.E.J., Brierley L., McCaffery C., Lycett S. Assessing the epidemic potential of RNA and DNA viruses. Emerg Infect Dis. 2016;22:2037–2044. doi: 10.3201/eid2212.160123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson C.J., Albery G.F., Merow C., Trisos C.H., Zipfel C.M., Eskew E.A., Olival K.J., Ross N., Bansal S. Climate change increases cross-species viral transmission risk. Nature. 2022;607:555–562. doi: 10.1038/s41586-022-04788-w. [DOI] [PubMed] [Google Scholar]
- 4.Woolhouse M.E.J., Brierley L. Epidemiological characteristics of human-infective RNA viruses. Sci Data. 2018;5 doi: 10.1038/sdata.2018.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ladner J.T., Wiley M.R., Mate S., Dudas G., Prieto K., Lovett S., Nagle E.R., Beitzel B., Gilbert M.L., Fakoli L., et al. Evolution and spread of Ebola virus in Liberia, 2014–2015. Cell Host Microbe. 2015;18:659–669. doi: 10.1016/j.chom.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaunt E.R., Digard P. Compositional biases in RNA viruses: causes, consequences and applications. WIREs RNA. 2022;13 doi: 10.1002/wrna.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poulain F., Lejeune N., Willemart K., Gillet N.A. Footprint of the host restriction factors APOBEC3 on the genome of human viruses. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Simmonds P., Ansari M.A. Extensive C->U transition biases in the genomes of a wide range of mammalian RNA viruses; potential associations with transcriptional mutations, damage- or host-mediated editing of viral RNA. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009596. [DOI] [PMC free article] [PubMed] [Google Scholar]; Simmonds and Ansari (2021) use a range of bioinformatic methods to show similar signatures of C→U editing in the genomes of a range of RNA viruses to that observed in SARS-CoV-2. The association of excess C→U mutations with possession of large-scale RNA secondary structure in virus genomes throws open new questions about the physiological and evolutionary roles of RNA folding in interactions with host cell defenses.
- 10.Ratcliff J., Simmonds P. Potential APOBEC-mediated RNA editing of the genomes of SARS-CoV-2 and other coronaviruses and its impact on their longer term evolution. Virology. 2021;556:62–72. doi: 10.1016/j.virol.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conticello S.G., Thomas C.J.F., Petersen-Mahrt S.K., Neuberger M.S. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 12.Münk C., Willemsen A., Bravo I.G. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol Biol. 2012;12 doi: 10.1186/1471-2148-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S.H., Habib G., Yang C.Y., Gu Z.W., Lee B.R., Weng S.A., Silberman S.R., Cai S.J., Deslypere J.P., Rosseneu M. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987;238:363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- 14.Powell L.M., Wallis S.C., Pease R.J., Edwards Y.H., Knott T.J., Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50:831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- 15.Teng B., Burant C.F., Davidson N.O. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 16.Davidson N.O., Innerarity T.L., Scott J., Smith H., Driscoll D.M., Teng B., Chan L. Proposed nomenclature for the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme: APOBEC-1. RNA. 1995;1:3. [PMC free article] [PubMed] [Google Scholar]
- 17.Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 18.Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., Alt F.W. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 19.Dickerson S.K., Market E., Besmer E., Papavasiliou F.N. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muramatsu M., Sankaranand V.S., Anant S., Sugai M., Kinoshita K., Davidson N.O., Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 21.Pham P., Bransteitter R., Petruska J., Goodman M.F. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 22.Anwar F., Davenport M.P., Ebrahimi D. Footprint of APOBEC3 on the genome of human retroelements. J Virol. 2013;87:8195–8204. doi: 10.1128/JVI.00298-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simmonds P. Rampant C→U hypermutation in the genomes of SARS-CoV-2 and other coronaviruses: causes and consequences for their short- and long-term evolutionary trajectories. mSphere. 2020;5 doi: 10.1128/mSphere.00408-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Nakata Y., Ode H., Kubota M., Kasahara T., Matsuoka K., Sugimoto A., Imahashi M., Yokomaku Y., Iwatani Y. Cellular APOBEC3A deaminase drives mutations in the SARS-CoV-2 genome. Nucleic Acids Res. 2023;51:783–795. doi: 10.1093/nar/gkac1238. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nakata et al. (2023) demonstrate induction of C→U substitutions in SARS-CoV-2 viral RNA in an APOBEC3A overexpression model in HEK 293T cells. This is the first published experimental evidence of APOBEC driven hypermutation in SARS-CoV-2 supporting the wealth of bioinformatic evidence.
- 25.Sharma S., Patnaik S.K., Taggart R.T., Kannisto E.D., Enriquez S.M., Gollnick P., Baysal B.E. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat Commun. 2015;6 doi: 10.1038/ncomms7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desimmie B.A., Delviks-Frankenberrry K.A., Burdick R.C., Qi D., Izumi T., Pathak V.K. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J Mol Biol. 2014;426:1220–1245. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaba A., Flath B., Chelico L. Examination of the APOBEC3 barrier to cross species transmission of primate lentiviruses. Viruses. 2021;13 doi: 10.3390/v13061084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Kosugi Y., Uriu K., Suzuki N., Yamamoto K., Nagaoka S., Kimura I., Konno Y., Aso H., Willett B.J., Kobayashi T., et al. Comprehensive investigation on the interplay between feline APOBEC3Z3 proteins and feline immunodeficiency virus Vif proteins. J Virol. 2021;95 doi: 10.1128/JVI.00178-21. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kosugi et al. (2021) investigates the ability of APOBEC proteins from eighteen different feline species to antagonize eight different feline immunodeficiency virus Vif proteins. The experimental methodology provides a framework to evaluate the contribution of APOBEC to virus host range.
- 29.Bhatia S., Patil S.S., Sood R. Bovine immunodeficiency virus: a lentiviral infection. Indian J Virol. 2013;24:332–341. doi: 10.1007/s13337-013-0165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su X., Wang H., Zhou X., Li Z., Zheng B., Zhang W. Jembrana disease virus Vif antagonizes the inhibition of bovine APOBEC3 proteins through ubiquitin-mediate protein degradation. Virology. 2018;519:53–63. doi: 10.1016/j.virol.2018.03.028. [DOI] [PubMed] [Google Scholar]
- 31.Fu Y., Lu D., Su Y., Chi H., Wang J., Huang J. The Vif protein of caprine arthritis encephalitis virus inhibits interferon production. Arch Virol. 2020;165:1557–1567. doi: 10.1007/s00705-020-04637-z. [DOI] [PubMed] [Google Scholar]
- 32.Bogerd H.P., Tallmadge R.L., Oaks J.L., Carpenter S., Cullen B.R. Equine infectious anemia virus resists the antiretroviral activity of equine APOBEC3 proteins through a packaging-independent mechanism. J Virol. 2008;82:11889–11901. doi: 10.1128/JVI.01537-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacMillan A.L., Kohli R.M., Ross S.R. APOBEC3 inhibition of mouse mammary tumor virus infection: the role of cytidine deamination versus inhibition of reverse transcription. J Virol. 2013;87:4808–4817. doi: 10.1128/JVI.00112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bishop K.N., Holmes R.K., Sheehy A.M., Davidson N.O., Cho S.-J., Malim M.H. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 35••.O’Toole Á., Neher R.A., Ndodo N., Borges V., Gannon B., Gomes J.P., Groves N., King D.J., Maloney D., Lemey P., et al. Putative APOBEC3 deaminase editing in MPXV as evidence for sustained human transmission since at least 2016. BioRXiv. 2023 doi: 10.1101/2023.01.23.525187. [DOI] [PMC free article] [PubMed] [Google Scholar]; O’Toole and Rambaut (2022) compare the evolutionary history of different clades of MPXV, providing convincing evidence of clade-specific evolution driven by an APOBEC-like evolutionary pressure. Supplementary figure 3 displays an inspired method for isolating an APOBEC footprint in virus phylogenies.
- 36•.Isidro J., Borges V., Pinto M., Sobral D., Santos J.D., Nunes A., Mixão V., Ferreira R., Santos D., Duarte S., et al. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat Med. 2022;28:1569–1572. doi: 10.1038/s41591-022-01907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; Isidro et al. (2022) demonstrates ongoing CU hypermutation of MPXV during its circulation in the 2022 global outbreak. This result supports human-driven evolution of the virus and provides evidence for undetected transmission over several years.
- 37.Samuel C.E. ADARs, viruses and Innate Immunity. Curr Top Microbiol Immunol. 2012;353:163–195. doi: 10.1007/82_2011_148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hur S. Double-stranded RNA sensors and modulators in innate immunity. Annu Rev Immunol. 2019;37:349–375. doi: 10.1146/annurev-immunol-042718-041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barraud P., Allain F.H.-T. ADAR proteins: double-stranded RNA and Z-DNA binding domains. Curr Top Microbiol Immunol. 2012;353:35–60. doi: 10.1007/82_2011_145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosani U., Bortoletto E., Montagnani C., Venier P. ADAR-editing during Ostreid Herpesvirus 1 infection in Crassostrea gigas: facts and limitations. mSphere. 2022;7 doi: 10.1128/msphere.00011-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo K., Barrett B.S., Morrison J.H., Mickens K.L., Vladar E.K., Hasenkrug K.J., Poeschla E.M., Santiago M.L. Interferon resistance of emerging SARS-CoV-2 variants. Proc Natl Acad Sci. 2022;119 doi: 10.1073/pnas.2203760119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gokhale N.S., Horner S.M. RNA modifications go viral. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu L., Seki M. Recent advances in the detection of base modifications using the Nanopore sequencer. J Hum Genet. 2020;65:25–33. doi: 10.1038/s10038-019-0679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furuse Y. RNA modifications in genomic RNA of influenza A virus and the relationship between RNA modifications and viral infection. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22179127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McIntyre W., Netzband R., Bonenfant G., Biegel J.M., Miller C., Fuchs G., Henderson E., Arra M., Canki M., Fabris D., et al. Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res. 2018;46:5776–5791. doi: 10.1093/nar/gky029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 47••.McDaniel Y.Z., Wang D., Love R.P., Adolph M.B., Mohammadzadeh N., Chelico L., Mansky L.M. Deamination hotspots among APOBEC3 family members are defined by both target site sequence context and ssDNA secondary structure. Nucleic Acids Res. 2020;48:1353–1371. doi: 10.1093/nar/gkz1164. [DOI] [PMC free article] [PubMed] [Google Scholar]; McDaniel et al. (2020) systematically evaluate the structural preferences of human APOBEC3 proteins on small DNA transcripts, demonstrating DNA targets prefer to be outside of defined secondary elements. This result contrasts with similar research conducting on RNA targets.
- 48.Sharma S., Baysal B.E. Stem-loop structure preference for site-specific RNA editing by APOBEC3A and APOBEC3G. PeerJ. 2017;5 doi: 10.7717/peerj.4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data were used for the research described in the article.