Abstract
Bacterial htrA genes are typically activated as part of the periplasmic stress response and are dependent on the extracytoplasmic sigma factor rpoE. A putative promoter region, P1, of the ςE-type heat-inducible promoters has previously been identified upstream of the htrA gene of Bartonella henselae. Further analysis of the htrA mRNA by primer extension demonstrated that transcription initiates from P1 and a second region downstream of P1. This second promoter region, termed P2, had no sequence identity to ςE-type heat-inducible promoters. Promoter regions were cloned individually and in tandem into pANT3 upstream of a promoterless version of the green fluorescent protein (GFP) gene (gfpmut3) and transformed into B. henselae by electroporation. The contiguous promoter region containing both P1 and P2 were necessary for the optimal transcriptional activation of the htrA gene. Promoter activity at 37°C was distinctively higher than at 27°C. However, thermal induction at 47°C did not increase expression of gfpmut3. Invasion of human microvascular endothelial cells (HMEC-1) by B. henselae resulted in the formation of well-defined vacuoles containing clusters of bacteria exhibiting marked expression of gfpmut3 transcribed from the P1-P2 region. In addition, a moderate yet significant increase in the ratio of bacterial GFP to DNA was detected for intracellular bacteria compared to extracellular bacteria, indicating upregulation of htrA upon invasion of HMEC-1. The activation of specific genes in the intracellular environment may help us better understand the novel pathogenic mechanisms used by this bacterium.
Bartonella henselae is a gram-negative fastidious, facultative intracellular bacillus that has been identified as an emergent pathogen capable of causing a variety of disease syndromes including cat scratch disease, bacillary angiomatosis, and bacillary peliosis hepatis (5). Cat scratch disease is usually benign and self-limiting and is characterized by lymphadenopathy related to a cat scratch or cat exposure (33). Potentially serious multiorgan complications resulting from bacteremia or systemic disease are not uncommon. These complications include encephalopathy and Parinaud's oculoglandular syndrome among many other presentations (8). Conversely, B. henselae infections in immunocompromised individuals tend to be systemic and include fever with bacteremia and bacillary angiomatosis, which is characterized by abnormal proliferation of vascular endothelial cells.
Prokaryotic organisms are equipped with stress response mechanisms which enable them to survive under extreme environmental adversities. While many papers have been published regarding the biology of stress response proteins in microorganisms of medical significance, the stress responses in recently described emergent pathogens remain less characterized. Heat shock proteins (HSPs) are a highly conserved group of proteins. HtrA (high-temperature requirement A), GroEL, GroES, and DnaK are among the HSPs induced in bacteria when confronted with stressful insults such as temperature increase. HtrA is probably the least studied and characterized bacterial HSP. HtrA, also termed DegP or Do protease (40), is an envelope-associated serine protease first described in Escherichia coli (28), which is involved in the degradation of periplasmic misfolded proteins in certain gram-negative bacteria (41).
We have previously described a B. henselae clone immunoreactive with hyperimmune rabbit serum to B. henselae which expressed a 66-kDa antigen in E. coli with homology to other htrA genes (3). Cloning and sequencing of the putative htrA gene of B. henselae showed an open reading frame of 1,509 nucleotides which codes for 503 amino acids (3). A significant level of amino acid sequence conservation between the stress response protein encoded by the htrA gene of B. henselae and those of Brucella abortus (59%), E. coli (37%), and Salmonella enterica serovar Typhimurium (36%) was observed (3). The deduced protein exhibited features shared by other bacterial HtrA stress response proteins, including a catalytic domain and a serine protease active site, GNSGGP (34). The identity of the B. henselae htrA gene was further confirmed by the ability of the cloned gene to complement an htrA E. coli mutant (S. Resto-Ruiz and B. E. Anderson, Abstr. Eur. Working Group Rickettsia Am. Soc. Rickettsiol. Joint Meet., abstr. 169, 1999). Sequencing of the 5′ regulatory region upstream of B. henselae htrA suggested a putative promoter (P1) with regions of notable homology to the ςE-type heat shock promoters (3).
In this report we confirm that P1 serves as a promoter region for B. henselae htrA and describe a second functional promoter region, P2, found within an adjacent region located upstream of the htrA gene of B. henselae. Furthermore, optimal transcriptional activation from this regulatory region required the presence of P1 and P2 in tandem. Upregulation upon thermal induction and a moderate intracellular activation in human microvascular endothelial cells were detected for the tandem promoters. The findings reported by this study will advance our knowledge of the regulation of the htrA gene and describes methodology that will assist further study of intracellular activation of B. henselae genes.
MATERIALS AND METHODS
Bacteria and plasmids.
E. coli strains JM109 and DH12S (Table 1) were grown overnight in Luria-Bertani broth (Difco, Detroit, Mich.) at 37°C with shaking at 200 rpm. Strains harboring selected plasmids derived from pANT3 were grown using medium supplemented with 50 μg of kanamycin per ml (k50). Competent E. coli cells were prepared for standard transformation as previously described (24). The promoterless vector pANT3 was used to assess promoter activity in B. henselae. A modified version of the gene encoding green fluorescent protein (GFP) from the jellyfish Aequorea victoria that has been optimized for flow cytometric analysis (gfpmut3) is located in this plasmid (26).
TABLE 1.
Description of vectors, resulting constructs, and E. coli strains
| Plasmid or strain | Application or genotype | Source or reference |
|---|---|---|
| Plasmids | ||
| pBlue1-1 | pBluescript/htrA regulatory region | 3 |
| pANT3 | Promoterless gfpmut3; used to evaluate htrA promoter activity in B. henselae | 26 |
| pSIR11 | pANT3/B. henselae htrA P1 | This work |
| pSIR12 | pANT3/B. henselae htrA P2 | This work |
| pSIR13 | pANT3/B. henselae htrA P1-P2 | This work |
| pSIR14 | pANT3/B. henselae htrA P1R | This work |
| pSIR15 | pANT3/B. henselae htrA P2R | This work |
| pSIR16 | pANT3/B. henselae htrA P1R-P2R | This work |
| E. coli strains | ||
| JM109 | F′ traD36 lacIq Δ(lacZ)M15 proA+ B+/e14−(McrA−) Δ(lac-proAB) thi gyrA96 (Nalr) endA1 hsdR17(rK− mK+ mK+) relA1 supE44 recA1 | |
| DH12S | mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 deoR Δ(ara, leu)7697 araD139 galU galK nupG rpsL/ F′ proAB+ lacIqZΔM15 |
All B. henselae strains were grown on heart infusion agar (HIA) (Difco) supplemented with 5% rabbit blood; 200 μg of streptomycin per ml (S200; Sigma Chemical Co., St. Louis, Mo.) and/or k50 (Boehringer Mannheim Corp., Indianapolis, Ind.) were added as required. At all times, B. henselae plates were incubated at 37°C with 5% CO2 and humidity to saturation. Parallel plates were grown at 27°C or shifted to 47°C for promoter activity experiments. The following strains were used in this work: Houston-1 ATCC 49882 (recovered from a human immunodeficiency virus-infected patient) (35) and 882str (streptomycin-resistant laboratory-adapted strain of ATCC 49882) (26) (gift from Anthea Lee, Stanford University, Stanford, Calif.).
PCR amplification and cloning of promoter regions.
Single or combined promoters in the forward or reverse orientations were amplified by PCR using the primer pairs in Table 2 and B. henselae Houston-1 genomic DNA as template. PCRs were performed in a MiniCycler (MJ Research, Watertown, Mass.) following the description of EasyStart PCR Mix-in-a-Tube (Molecular Bio-Products, San Diego, Calif.), using Taq polymerase (Gibco/BRL, Rockville, Md.). The initial cycle denaturing step of 94°C for 4 min was followed by 3 cycles of 94°C for 1 min, 42°C for 1 min, and 67°C for 2 min and 30 cycles of 94°C for 1 min, 50°C for 2 min, and 70°C for 7 min). The resulting PCR products were subsequently cloned into pANT3. Purified pANT3 was digested with BamHI and XbaI according to optimal digestion conditions recommended by the supplier (New England Biolabs, Beverly, Mass.). PCR products including B. henselae htrA promoters P1, P2, P1R (R denotes reverse orientation), P2R, P1-P2, or P1R-P2R amplified with added BamHI and XbaI restriction sites, were digested with those enzymes, loaded in a 0.7% agarose gel, and purified by GENECLEAN (Bio 101, Vista, Calif.). Restriction endonuclease digested pANT3 and PCR-amplified promoter regions were ligated overnight at 14°C using T4 DNA ligase (New England Biolabs). Ligation products were washed with a Microcon 100 device and used to transform either E. coli strain JM109 or DH12S by the method of Hanahan (24). Finally, cells were plated in Luria-Bertani agar supplemented with k50. Plasmid DNA was examined by a minilysate procedure as previously described (38) and confirmed by sequencing to ensure the presence and correct orientation of cloned promoter regions in pANT3. The T7 quick denature plasmid sequencing kit (Amersham, Life Science, Piscataway, N.J.), which is based on the dideoxy chain termination sequencing system developed by Sanger (39), was used with primer PANT3 (5′ TGC AAT GCG TCT AGG ATC GAG ACA AAG 3′).
TABLE 2.
Primer pairs used for the PCR amplification of htrAa promoter regions and the kanamycin resistance gene
| Region amplified | Primer | Sequence |
|---|---|---|
| P1 PCR | P1 | 5′ AAAAGGATCCTACTTTGAATGCTAG 3′ |
| P1A | 5′ AAAATCTAGATTTTTTCCTGTAACA 3′ | |
| P2 PCR | P2 | 5′ AAAAGGATCCTCATCAGGATGTATT 3′ |
| P2A | 5′ AAAATCTAGAAACAAACCTTACCGT 3′ | |
| P1Rb PCR | P1R | 5′ AAAATCTAGATACTTTGAATGCTAG 3′ |
| P1AR | 5′ AAAAGGATCCTTTTTTCCTGTAACA 3′ | |
| P2Rb PCR | P2R | 5′ AAAATCTAGATCATCAGGATGTATT 3′ |
| P2AR | 5′ AAAAGGATCCAACAAACCTTACCGT 3′ | |
| Kanamycin resistance gene | TnKn903-1 | 5′ CCGATGCGCCAGAGTTGTTTCTGAA 3′ |
| TnKn903-2 | 5′ ACCTATTAATTTCCCCTCGTCAAAA 3′ |
Primer pairs P1-P2A and P1R-P2AR were used to amplify the tandem promoter regions P1-P2 and P1R-P2R, respectively.
R, used to clone promoter regions in the reverse orientation in pANT3.
Bacterial RNA isolation.
Total RNA was isolated from B. henselae using Tri Reagent (Molecular Research Center, Inc., Cincinnati, Ohio) after heat shock at 42°C for 15 min. Bacterial cultures were harvested from 3-day-old HIA plates by scraping the plates with a sterile cotton swab into heart infusion broth and collecting the cells by centrifugation for 5 min at 4°C in a microcentrifuge tube at 12,000 × g. One milliliter of Tri Reagent was used to resuspend approximately 107 bacterial cells. Homogenates were transferred to RNase-free microcentrifuge tubes preloaded with Phase Lock Gel (5′→3′ Prime, Inc., Boulder, Colo.) and stored at room temperature for 5 min. Next, 200 μl of chloroform per ml of Tri Reagent used was added, and the mixture was mixed well for 15 s. Suspensions were held at room temperature for 5 to 15 min and then spun at 12,000 × g for 15 min at 4°C. The aqueous phase was transferred to a clean tube, and 500 μl of isopropanol was added to precipitate the RNA. After vortexing, tubes were incubated at room temperature for 15 min, and this was followed by centrifugation at maximum speed for 15 min at 4°C. Supernatant was discarded and the pellet was mixed and vortexed with 1 ml of 75% ethanol. At this point samples were either stored at −80°C in the 75% ethanol or treated for immediate use as follows. If the pellet floated after vortexing, the sample was spun at a maximum of 12,000 × g for 5 min at 4°C; otherwise, the wash was done at 7,500 × g. Pellets were air dried at room temperature for 10 to 20 min, with caution taken not to over dry. Excess ethanol on the inside walls of the tube was removed using a sterile cotton swab. RNA was resuspended in 20 to 30 μl of sterile RNase-free water containing 1 μl of RNasin (40 U/μl). Concentrations and purity were determined by optical density at 260 or 280 nm. Samples were also run in formaldehyde agarose gels to visualize the relative quality of RNA by examining the relative intactness of 5S, 16S, and 23S rRNA as compared to known RNA markers.
Primer extension.
Putative transcription start sites were mapped by primer extension. Primers PrEX1 (5′ AAC AAA CCT TAC CGT TTT CTG TCT AAA 3′) and PrEX2 (5′ GAA AAA CTT ACT GCG GCT AAT GTT 3′), reverse and complementary to the sense strand located approximately 100 bases downstream of the putative transcription initiation sites, were end labeled. One microliter of nuclease-free water containing 66 ng of primer, 3 μl of nuclease-free water, 1 μl of forward exchange 10× buffer, 3 μl of [γ-32P]ATP (3,000 Ci/mmol), and 1 μl of T4 polynucleotide kinase (8 to 10 U/μl) was incubated at 37°C for 10 min. To inactivate the T4 polynucleotide kinase, the reaction mixture was heated at 90°C for 2 min and chilled on ice before quick spinning, followed by the addition of 90 μl of nuclease-free water. Labeled primers were stored at −20°C.
The avian myeloblastosis virus (AMV) reverse transcriptase primer extension system (Promega Corp., Madison, Wis.) was used for 5′ end mapping. A mixture containing 1 μl of end-labeled primer, 150 μg of total RNA in 5 μl of nuclease-free water, and 5 μl of AMV primer extension 2× buffer was heated for 20 min at 58°C. Annealing was done by allowing the tubes to cool at room temperature for 10 min. Subsequently, 9 μl of a master reverse transcriptase extension mixture composed of 5 μl of AMV primer extension 2× buffer, 1.4 μl of sodium pyrophosphate (40 mM), 1 U of AMV reverse transcriptase, and 2.6 μl of nuclease-free water was added to the tube containing the annealed reaction mixture. Primer extension was performed at 42°C for 30 min. The reaction was stopped by the addition of 20 μl of loading dye. Approximately 15 μl of the final products was loaded into a 38-by-50-cm, 8% denaturing polyacrylamide gel containing 8 M urea. Control reaction mixtures, including a primer specific for the B. henselae groESL promoter, were also loaded. Sequencing reactions of pBlue1-1 (Table 1) served as the reference to map the start site of transcription. A constant voltage of 1,700 V was applied to the Bio-Rad Sequi-Gen nucleic acid sequencing cell in 1× Tris-borate-EDTA buffer. After electrophoresis, the gel was fixed in 20% methanol with 10% acetic acid, and dried for 1 h at 80°C in a vacuum gel dryer. The gel was exposed to X-ray film with an intensifying screen for 1 week before developing.
Electroporation of B. henselae.
Electrocompetent B. henselae cells were prepared from two strains, wild-type (wt) Houston-1 ATCC 49882 and streptomycin-resistant 882str. The contents of two 3-day-old plates were aseptically harvested with a sterile cotton swab into 1 ml of heart infusion broth (Difco) at 22°C. Competent cells were prepared by serial washes with 10% glycerol–water (22). Cell suspensions were collected by centrifugation at 3,000 × g for 5 min at 4°C. Pellets were washed by resuspending in 1 ml of ice-cold 10% glycerol–water, a total of three times. After the final wash, cells were counted using a Petroff-Hauser counter and resuspended in a final volume of 100 μl at an approximate density of 109 cells/ml. Plasmid was prepared from E. coli DH12S or JM109 strains containing pANT3 with putative promoter sequences using the Quantum Prep Plasmid Midi Prep kit (Bio-Rad, Hercules, Calif.). Varied amounts of plasmid (0.1, 0.3, and 0.5 μg) derived from DH12S or JM109 in 4 μl were added to vials containing 40 μl of competent wt or 882str B. henselae cells. The mixture was allowed to stabilize on ice for 15 min and transferred to precooled 0.1-cm-diameter cuvettes (Bio-Rad). B. henselae cells were electroshocked using the Bio-Rad Pulse Controller II for 4.6 ms, at field strengths equivalent to 12.5 or 15 kV/cm and constant capacitance of 25 μF. Each electroporated sample was immediately resuspended in 1 ml of room temperature SB broth (RPMI 1640 with glutamine [Mediatech Inc., Herndon, Va.], 1% HEPES buffer, 1% sodium pyruvate [Sigma], 5% heat inactivated fetal calf serum [HyClone, Logan, Utah], and 5% rabbit blood lysate prepared as previously described [22]). Subsequent to recovery for 7 h at 37°C with 5% CO2, wt Houston-1 and 882str were plated into HIA-k50 and HIA-k50-s200, respectively. Dilutions of the recovered cells, wt and 882str, were plated in parallel control medium HIA or HIA-s200, respectively, and incubated as previously stated. Total DNA from transformant colonies was prepared as previously described (2). Approximately 100 ng of the total DNA was used as template to identify true transformants containing pANT3 derivatives by PCR using primers TN903Kn-1 and TN903Kn-2 (Table 2). To further corroborate the identity of B. henselae as the transformed organism, a coding region of the htrA gene specific to B. henselae and corresponding to 414 bp was amplified using primers CAT1 and CAT2 (4).
Promoter activity.
The gfpmut3 system in B. henselae was analyzed by fluorescent microscopy and flow cytometry as follows. A 177-bp tandem regulatory sequence (P1-P2) upstream of B. henselae htrA (3) was ligated into the BamHI-XbaI site of pANT3 and used to transform competent E. coli DH12S or JM109 cells. Plasmid midi preps were used to prepare plasmid for insert orientation confirmation by dideoxy sequencing and to electroporate 882str cells as described above. In addition, individual promoters (P1 or P2) were cloned as were their reverse counterparts (P1R or P2R). Reverse tandem promoter sequences were also cloned (P1R-P2R). B. henselae transformants were screened and analyzed by fluorescence microscopy using the Eclipse E400 (Nikon, Melville, N.Y.) with a fluorescein isothiocyanate filter. Fluorescence emission was assessed by flow cytometric analysis using a FACSCalibur (Becton Dickinson Immunocytometry Systems, San Jose, Calif.) equipped with an argon laser. Promoter activity was analyzed by fluorescence intensity as a function of GFP expression when excited at a wavelength of 488 nm. Analysis was performed for bacteria simultaneously grown at 27 or 37°C and for bacteria grown at 37°C and heat shocked at 47°C for 5, 10, or 15 min.
Human microvascular endothelial cell culture.
HMEC-1 is a human microvascular endothelial cell line of dermal origin immortalized by transfection with plasmid PBR-322 containing sequences from the simian virus 40 Large T antigen (1) (Materials Transfer Agreement, Centers for Disease Control and Prevention, Atlanta, GA). Stock cultures of HMEC-1 were seeded in 75-cm2 flasks containing 20 ml of MCDB131 medium with 1 μg of hydrocortisone per ml, 10 ng of epithelial growth factor per ml, 10% fetal bovine serum, and 1× penicillin-streptomycin (Mediatech, Inc.). All incubations involving HMEC-1 culture and cocultivation with B. henselae were done at 37°C, 5% CO2, and humidity to saturation. Cultures were refreshed with 2 ml of medium every 2 days until confluency was reached. The medium was removed from cell monolayers, replaced with 20 ml of supplemented MCDB131 without antibiotics, and allowed to adapt for 1 day. Cell monolayers were washed three times with medium without antibiotics with gentle rocking, rinsed with 5 ml of HEPES buffer and treated with trypsin-EDTA (Clonetics, San Diego, Calif.) to detach cells. After the cells showed circular shape, the flask was tapped gently to loosen cells, and after 30 s the previous step was repeated and 5 ml of Trypsin-neutralizing solution (Clonetics) was added. The total volume was removed to a clean tube. HMEC-1 were spun at 3,500 rpm (Beckman GS-6R, rotor GH-3.8) for 10 min. The concentration was adjusted to 105 cells/ml using MCDB131. Sterile microscope slides were placed in sterile petri dishes (one slide/dish) with 3 ml of MCDB131 medium without antibiotics. One ml of cells was aseptically added to the petri dishes and incubated for 1 to 2 days until approximately 50% confluency was achieved on the slide.
Intracellular induction of htrA promoters.
Three-day-old B. henselae 882str cells harboring pSIR13 (P1-P2 htrA promoter sequence), pSIR16 (P1R-P2R), or pANT3 without insert (negative control) were harvested from HIA plates using a sterile cotton swab and resuspended in MCDB131 without antibiotics at a concentration of ∼107 bacteria/ml. One ml of the corresponding bacterial suspension was diluted 1:2 in MCDB131, and the final volume of 2 ml was added to the HMEC-1 grown on slides to give an approximate multiplicity of infection of 100. Slides containing HMEC-1 and bacteria were incubated for 1 or 2 days. Slides were removed from petri dishes, fixed in 4% paraformaldehyde, and washed in phosphate-buffered saline (PBS) several times. All slides were covered with coverslips in mounting medium of antifade–4,6 diamidino-2 phenylindole (DAPI) (Vector, Burlingame, Calif.). The fluorescent stain DAPI was used to stain eukaryotic and bacterial DNA. Immunofluorescence was observed with a Leitz Orthoplan 2 microscope and images were captured by a charge-coupled device (CCD) camera with the Smart Capture program (Vysis, Downers Grove, Ill.). Bacterial pixel densities were analyzed and expressed as number of pixels per area of study. The ratio of expressed GFP to the DNA concentration in a given area was determined both intracellularly and extracellularly. Vacuole formation was used as an indicator of intracellular expression.
Statistical analysis.
Statistical significance was determined by the Student's t test. Values of P of <0.05 were considered significant. Analysis was performed using Microsoft Excel 97 (Microsoft Corp., Redmond, Wash.). Experiments were performed three or more times. Results shown as bar graphs represent the mean value ± the standard deviation of the mean.
RESULTS
Mapping of the transcription initiation site.
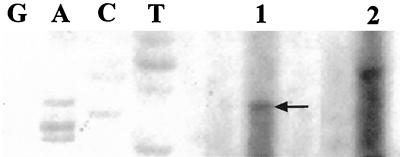
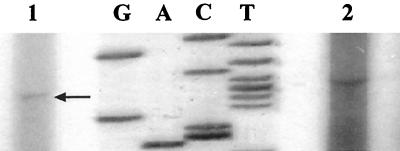
The transcription start site was mapped by primer extension using total RNA from B. henselae. The resulting cDNAs were loaded on an 8% acrylamide gel along with pBlue1-1 (pBluescript/htrA regulatory region [Fig. 1]) sequencing reaction mixtures to determine the complementary site of transcription initiation. Extension products included a band at the cytosine residue corresponding to the guanine in position 61 (Fig. 2, lane 1), which is 178 nucleotides upstream of the start of translation and six bases downstream of promoter P1 (Fig. 1). However, despite the fact that this band was identified in three independent experiments, it was not observed to be a sharp band indicative of transcription initiation from a strong promoter. In addition, the distance between htrA promoter P1 and the start codon was relatively long (184 bases). To determine the presence of any promoter regions in addition to P1, a second primer (PrEx2), beginning 207 bases downstream of P1, and extending beyond the start of translation, was used in primer extension reactions. A second band corresponding to an additional transcription start site, different in location from the one previously described, was mapped to the adenine in position 165 (Fig. 3, lane 1). This finding suggested the presence of a second promoter within the 5′ upstream regulatory region of the htrA gene (Fig. 1). Parallel primer extension reactions were done using total RNA isolated from E. coli harboring the construct pBlue1-1 (pBlue/htrA). Primer extensions reactions with both PrEx1 and PrEx2 resulted in cDNA products from E. coli clones which mapped to the same sites as those resulting from B. henselae cDNAs (data not shown). These results confirm the detected transcription start sites for B. henselae htrA regardless of the source of RNA utilized. Finally, a control extension reaction consisting of a primer specific for B. henselae groESL was performed in all experiments to assess the quality of B. henselae RNA (Fig. 2, lane 2; Fig. 3, lane 2).
FIG. 1.
The 5′ regulatory region of B. henselae htrA. The region with extensive homology to ςE-type stress response promoters is indicated (P1) and boxed, with the corresponding direction of transcription (dotted line with arrow) and site of transcription initiation (guanine at position 61) (boldface type) as shown. A second site of transcription initiation from promoter region P2 is indicated in boldface type (adenine at position 165) and putative −35 and −10 promoter regions are overlined. The methionine codon representing the start of translation is boxed and the putative ribosome binding site is underlined and labeled (RBS). Sequences complementary to the oligonucleotides used for primer extension are underlined and labeled (PrEx1 and PrEx2). Sequence numbering is identical to that of the sequence with GenBank accession no. L20127, a description of which has been previously published (3).
FIG. 2.
Mapping of the transcription start site of P1. Total RNA and a 5′ end-labeled primer PrEx1 were used in primer extension reactions. The resulting cDNAs and sequencing reactions (GACT) from pBlue1-1, which harbors the htrA regulatory region, were loaded on an 8% acrylamide gel. The start of transcription was mapped to G61 and the complementary cytosine residue on the other strand is indicated (arrow). Lanes: 1, B. henselae htrA; 2, B. henselae groEL control.
FIG. 3.
Mapping of the transcription start site of P2. Total RNA and a 5′ end-labeled primer PrEx2 were used in primer extension reactions. The resulting cDNAs and sequencing reactions from pBlue1-1, harboring the htrA regulatory region, were loaded on an 8% acrylamide gel. The start of transcription was mapped to A165, and the complementary thymine residue on the other strand is indicated (arrow). Lanes: 1, B. henselae htrA; 2, B. henselae groEL control.
Electroporation of B. henselae.
Electroporation of B. henselae cells was performed with various pANT3 derivatives harboring the htrA promoters using 0.3 μg of plasmid. Regardless of the B. henselae host strain (Houston-1 or 882str), electroporation with lower plasmid DNA concentrations (0.1 μg) was unproductive. An additional factor shown to influence electrotransformation was electric field strength. In general, the most significant results were observed with 882str, 0.3 μg of pANT3 derived from DH12S, and a field strength of 12.5 kV/cm with an overall efficiency of transformation of 1.06 × 105 transformants per μg of DNA. Optimal field strengths and conditions previously reported for B. bacilliformis and B. quintana are similar to those identified in this study (22, 36). A kanR-derived amplicon of 510 bp was observed in all screened clones tested by PCR (data not shown). To further corroborate the identity of B. henselae as the transformed organism, a coding region of the htrA gene specific to B. henselae and corresponding to 414 bp was amplified using primers CAT1 and CAT2 (not shown). Control electrotransformations without plasmid DNA did not yield transformants.
Promoter activity. (i) Steady-state activity (flow cytometry and fluorescent microscopy).
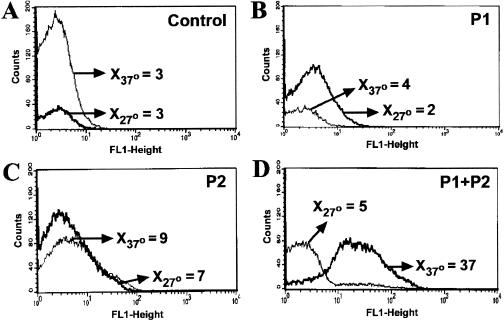
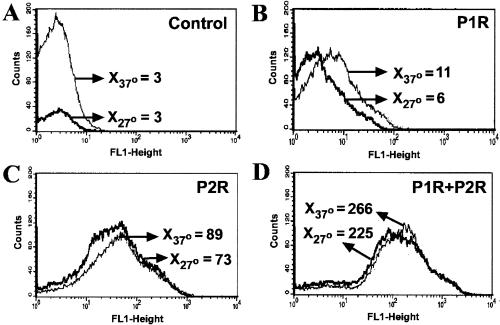
Promoter regions P1 and P2, including the site of transcription initiation mapped by primer extension and short flanking regions, were cloned into pANT3 and transformed into B. henselae 882str. In addition, the tandem P1-P2 region which contains each promoter region and the short region in between was also cloned. Expression of GFP in B. henselae transformants analyzed by flow cytometry varied with the different promoter constructs. In contrast to the pANT3 negative control (Fig. 4A), GFP-expressing bacteria harboring P1-P2 in pSIR13 produced a discrete fluorescence distribution at 37°C with a mean fluorescence channel between 101 and 102 on a logarithmic scale (Fig. 4D). GFP expression was also evident by fluorescent microscopy as shown by green emission throughout the entire bacteria (see Fig. 6A). However, strong expression of GFP in reporter plasmids harboring promoter regions P1 and P2 alone (pSIR11 and pSIR12) were not demonstrable by flow cytometry (Fig. 4B and C) and were only marginally fluorescent by fluorescence microscopy (data not shown). Promoters from the B. henselae htrA 5′ region were also cloned in the reverse orientation relative to gfpmut3 of pANT3. Although these regions were initially cloned into pANT3 to serve as controls, they were also shown to initiate transcription of the reporter gene. In particular P2R (Fig. 5C) and both promoters in tandem cloned in the reverse orientation were shown to result in strong GFP fluorescence (Fig. 5D). Thus, the region 5′ of B. henselae htrA is capable of initiating transcription in both orientations, suggesting the presence of a second unknown gene on the opposite strand from htrA and diverging away from a common regulatory region.
FIG. 4.
Thermal induction of htrA promoter regions P1, P2, or P1-P2 in B. henselae analyzed by flow cytometry. B. henselae 882str cells harboring pSIR11, pSIR12, or pSIR13 were grown at 27 or 37°C. Bacterial growth was harvested after 2 days, resuspended in PBS and analyzed in the FACSCalibur. B. henselae 882str was electrotransformed with the following plasmids: (A) pANT3, (B) pSIR11 (harboring P1), (C) pSIR12 (harboring P2), (D) pSIR13 (harboring P1+P2). Arrows indicate the mean fluorescence channel (X) at 27°C and 37°C. Data shown corresponds to one representative set of histograms.
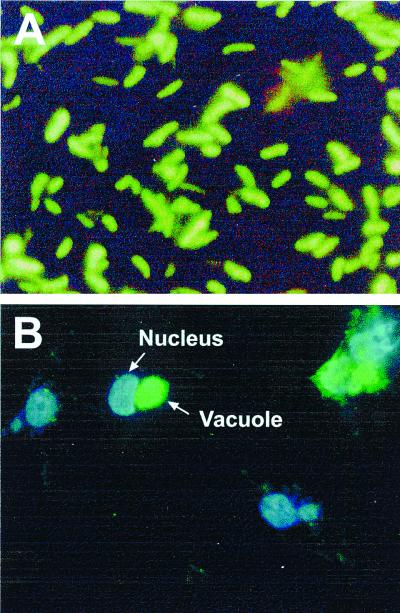
FIG. 6.
Fluorescence microscopy of GFP transcribed from tandem promoter regions P1-P2 of pSIR13. (A) Expression of GFP in B. henselae pSIR13/882str by fluorescence microscopy. Green bacilli indicate uniform GFP expression throughout the bacterial surface. Overall magnification was ×20,000. (B) Invasion of HMEC-1 by B. henselae harboring pSIR13. Slides were stained with the DNA dye DAPI (blue fluorescence). Images were captured with Vysis software. The HMEC-1 nucleus is depicted in blue. Multiple intracellular bacteria are shown in the vacuole containing GFP-expressing B. henselae. Overall magnification was ×400.
FIG. 5.
Thermal induction of htrA promoter regions in the reverse orientation in B. henselae 882str by flow cytometry. Bacteria harboring pSIR14, pSIR15, or pSIR16 were grown at 27 or 37°C. Bacterial growth was harvested after two days, resuspended in PBS, and analyzed in the FACSCalibur. B. henselae 882str was electrotransformed with the following plasmids: pANT3 (A), pSIR14 (harboring P1R) (B), pSIR15 (harboring P2R) (C), and pSIR16 (harboring P1R-P2R) (D). Arrows indicate the mean fluorescence channel (X) at 27 and 37°C. One representative set of histograms is displayed.
(ii) Temperature induction (flow cytometry).
Due to the fastidious characteristics of B. henselae, two different strategies were used to evaluate induction by temperature. First, bacteria grown at 37°C followed by heat shock at 47°C was utilized. No significant change in GFP expression was noted under these growth conditions. Second, parallel bacterial plates harboring the htrA promoter constructs were grown in duplicate at 27 and 37°C. Two-day-old bacterial cultures were used for all flow cytometric analysis. Results showed a significant increase in GFP expression at 37°C compared to that at 27°C only for tandem htrA promoters P1-P2 (Fig. 4D). The inactivity of P1 or P2 independently, compared to the expression driven by P1-P2, indicates that the entire upstream region is necessary for optimal expression and suggests interdependency of the contiguous promoter for the regulation of htrA. Unlike P1R, P2R and P1R-P2R demonstrated promoter activity by driving the expression of GFP regardless of temperature induction (Fig. 5). These reverse promoter regions may be involved in the transcriptional regulation of genes upstream and divergent from htrA in a non-heat-inducible manner.
(iii) Intracellular induction (fluorescent microscopy).
Experiments using cocultivation and invasion of HMEC-1 by B. henselae 882str harboring different plasmid constructs were performed to evaluate the intracellular induction of htrA promoters. B. henselae 882str(pSIR13), which contains both P1 and P2, was used for these experiments, and the same region in reverse (pSIR16) was used as a control. The formation of a distinct vacuole or invasome within the endothelial cell was readily observed as strong green fluorescence between 20 and 24 h after infection of HMEC-1 with B. henselae 882str (Fig. 6B). Bacterial and endothelial DNA were stained with DAPI. Therefore, vacuole and nucleus were represented by blue luminescence using the Vysis software. However, under examination for GFP-expressing bacteria, only the vacuole emitted green fluorescence (Fig. 6B). Extracellular bacteria were also detected and observed predominantly in small undefined clusters with the 40× objective. For subsequent analysis (blue and green fluorescence measurements) intracellular bacteria were defined as those within a vacuole and extracellular bacteria were defined as amorphous clusters distal to the nuclei of host cells.
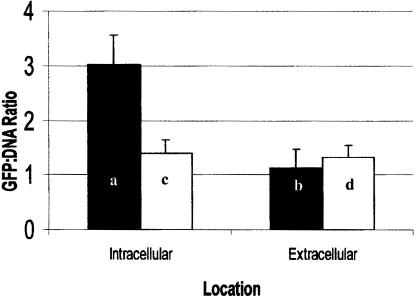
To determine if the B. henselae htrA promoter region is activated in HMEC-1, intracellular and extracellular ratios of GFP to DNA were determined for bacteria harboring P1-P2 and P1R-P2R tandem promoters. Tandem promoters P1-P2 were moderately activated (∼2.7-fold) within the intracellular environment compared to extracellular bacteria counterpart (Fig. 7). This increase was significant according to the statistical t test (P = 0.002). However, P1R-P2R tandem promoters were not significantly induced upon entry into HMEC-1 compared to the extracellular expression (P = 0.38). The intracellular difference observed between both constructs was also significant (P = 0.009). However, differences for the extracellular ratios from both constructs were not (P = 0.47). Thus, increased GFP-to-DNA ratio is not due to the difference in DNA content of intracellular versus extracellular bacteria.
FIG. 7.
Intracellular induction of B. henselae promoter regions P1-P2 (■) and P1R-P2R (□) in HMEC-1. The ratio of GFP to DNA was used to determine intracellular activation of the B. henselae htrA promoter regions. A moderate but significant (Pa/b = 0.002) 2.7-fold increase in the intracellular activity of P1-P2 was observed. Promoter constructs in the reverse orientation were not significantly induced (Pc/d = 0.38).
DISCUSSION
Despite the ubiquitous nature of B. henselae, little is known about the interaction of B. henselae with its host. This pathogen has been shown to invade a variety of animal and human cell types including human epithelial (6) and human endothelial cells (12, 14). Invasion of human endothelial cells occurs predominantly by clusters of B. henselae resulting in the uptake of bacteria into vacuoles that have been termed invasomes (14). The interaction of B. henselae with human endothelial cells also results in their activation and proliferation, ultimately leading to angiogenesis in infected individuals. The B. henselae-induced proliferation of endothelial cells is a distinctive trait shared by some but not all members of the genus. However, invasion of bacteria is not required for cell proliferation, since subcellular fractions have also been shown to mediate endothelial cell proliferation (10, 32). To date, the molecular basis for host cell entry and survival by B. henselae remains largely uncharacterized. In other bacteria, stress response proteins have been shown to be required for optimal intracellular growth and survival (20, 21). In particular, HtrA has been shown to be important for intracellular invasion and virulence (7, 15, 16, 27, 31). Hence, the focus of this study is on htrA expression in B. henselae and analysis of the corresponding 5′ regulatory region.
The majority of the studies aimed at the understanding of the transcriptional regulation of htrA, arise from E. coli. First, in E. coli the stress response is segregated between the cytoplasm and the periplasm. While the cytoplasmic compartment is regulated by ς32 (RpoH) (23), the alternative sigma factor ςE (RpoE or sigma 24), required by the htrA promoter for initiation of transcription (29), is the element controlling the extracytoplasmic or periplasmic response. This transcription factor functions as a regulator of the expression of at least 10 genes (11), which encode proteins assigned to periplasmic functions, including HtrA (17, 28, 29, 41). Second, although the E. coli htrA gene is transcribed from one promoter (29), the presence of tandem promoters has been documented for other bacterial genes (17). Multiple promoters may be required to direct the expression of a given gene(s). Previously, a B. henselae htrA promoter, P1, with homology to the ςE-type heat-inducible promoters, was described (3). Nevertheless, the presence of an additional promoter upstream of htrA was suspected due to the longer distance in the sequence between P1 and the start of translation. The additional htrA regulatory region, P2, detected by primer extension, showed identity to the htrA gene of B. abortus.
To further analyze the htrA regulatory region, we compared the activity of a promoter with that of other promoters, considered the effect of certain parameters on promoter activity, and evaluated interdependency of adjacent promoters. Two regulatory regions, P1 and P2, may drive the expression of htrA in B. henselae. To confirm the authenticity of these transcription sites and to eliminate the possibility of nonspecific stops by the reverse transcriptase resulting from secondary structure in the RNA, promoterless vectors were used to determine the activity of these promoter regions in B. henselae. A major obstacle found during the progress of this work was the limited gene transfer mechanisms allowing introduction of DNA into B. henselae. Other members of the genus Bartonella have been transformed by electroporation (22, 36); however, only conjugation, requiring cell-cell contact between donor and recipient cells, has been reported for B. henselae (13, 26).
Although restriction mechanisms are not well characterized for this organism, its resistance to accept exogenous DNA by any of the transformation methods suggests the presence of a restriction modification system. Our attempts to transform B. henselae by standard transformation involving DNA uptake by cells subsequent to a short heat shock were unproductive. Given the successful conjugation of recipient B. henselae 882str with the E. coli donor harboring the promoterless pANT3 reported by Lee (26), we attempted to introduce this plasmid into B. henselae directly by electroporation. Electroporation of 882str was more productive than Houston-1 wt. The best efficiency was observed with 882str, using approximately 0.3 μg of pANT3 derived from DH12S and a field strength of 12.5 kV/cm with an overall transformation efficiency in the order of 1.06 × 105 transformants per μg of DNA. Accordingly, these conditions and strain 882str were used for subsequent studies. Similar results were described for the electroporation of B. bacilliformis and B. quintana (22, 36). To our knowledge, this constitutes the first report describing the uptake of exogenous DNA by B. henselae using electroporation.
The electroporation of B. henselae allowed us to expand the spectrum of htrA promoter analysis. Under steady-state conditions at 37°C, the activities of singular P1 or P2 were lower independently than in tandem, using gfpmut3 in B. henselae (Fig. 4). The increased activity demonstrated by P1-P2 suggests interdependency of these contiguous promoters in the regulation of B. henselae htrA. Regardless of the evident activity of P1-P2 at 37°C, no obvious increase in activity was observed when shifted from 37 to 47°C (not shown). This prompted the comparative analysis of induction between 27 and 37°C. Thermal induction was observed in bacteria grown at 37°C compared to those grown at 27°C. However, only bacteria harboring the tandem construct P1-P2 were upregulated at 37°C (Fig. 4). These contrasting temperatures may represent biological relevance for B. henselae since the vector (cat flea) and natural vertebrate host (cat) provide a temperature adaptation similar to those used during the experiments. The discrete histogram distributions at 27 versus 37°C suggest that the B. henselae htrA promoter region P1-P2 works synergistically and exhibits heat-inducible properties typical of the ςE-type heat-inducible promoters. Although other bacterial htrA regulatory regions show identity with ςE-type heat inducible promoters, tandem promoters are a unique feature displayed by the B. henselae htrA.
Internal controls were designed by cloning the htrA promoters in the reverse orientation (P1R, P2R, and P1R-P2R) into the promoterless vector pANT3. Surprisingly, the promoter activity assays showed that the reverse orientations were active, therefore potentially behaving as bidirectional promoters. The level of activity of P1R in this system was low (Fig. 5). However, an interesting feature was the ability of P2R, surpassed by that of P1R-P2R, to drive the expression of GFP regardless of the temperature used (Fig. 5). It is possible to speculate that either one or two of the reverse htrA promoter regions could be involved in the regulation of other genes found upstream and divergent from htrA. The lack of thermal induction in P1R, P2R, or P1R-P2R suggests that the reverse constructs may include promoter regions responsible for transcription of genes that are not heat inducible. The B. henselae genome sequence project showed the presence of an open reading frame corresponding to rpsK and rplO genes upstream of htrA (S. Andersson, personal communication). These genes code for two distinct, small and large, ribosomal proteins. Expression of these ribosomal proteins may correlate with the bacterial growth stage used during these experiments. Variability in the promoter activities of the reverse tandem construct P1R-P2R, analyzed by flow cytometry versus fluorescent microscopy, suggest that the putative genes transcribed divergently from htrA may be differentially expressed in response to different environmental conditions. Under conditions conducive to rapid growth in blood agar, it is likely that expression of these ribosomal proteins is increased in the bacteria. However, promoter activity of P1R-P2R decreased in B. henselae upon invasion of eukaryotic cells suggesting rpsK and rplO may be downregulated in the stressful intracellular environment. Regions with sequence similarity to −35 and −10 promoter regions can be found on the divergent strand. Overlapping regulatory sequences are not unusual in bacteriophages and bacteria with small chromosomes due to the limited space available for noncoding regulatory regions. However, conclusive results regarding the bidirectionality of P1 or P2 require additional studies.
The results observed in the invasion assays performed in this study were consistent with the formation of a well-defined structure within HMEC-1 upon interaction and invasion by B. henselae. GFP was markedly expressed by htrA tandem promoters P1+P2 within vacuoles in HMEC-1 (Fig. 6B). The activity of tandem promoters P1-P2 appeared to be upregulated upon invasion of HMEC-1 compared to the extracellular GFP expression. The ratio of GFP fluorescence to DNA represented by DAPI staining was determined for bacteria inside and outside the cell. A moderate 2.7-fold increase in the ratio of GFP to DNA was observed in intracellular bacteria harboring the recombinant plasmid pSIR13. To determine if the ratio was artificially increased due to retardation of bacterial DNA replication in the intracellular environment, the reverse tandem promoters in pSIR16 were introduced into 882str, used to infect HMEC-1, and utilized as controls. The ratio of intracellular GFP to DNA corresponding to bacteria harboring P1R-P2R was similar to the extracellular ratios of P1-P2 and P1R-P2R, suggesting that changes in the intracellular ratio of P1-P2 are not the result of decreased bacterial DNA replication. It is not clear what distinct factors in the intracellular environment modulate the expression of these inducible promoters. Considering the well-documented activity of other members of this class of stress response proteases (25, 30, 37), it is likely that the protease activity of B. henselae HtrA contributes to the intracellular survival of this bacterium. The removal of damaged proteins by HtrA prior to accumulation of toxic levels has been described (25, 30, 37). Upregulation of HSPs upon invasion of host cells has been previously documented. The B. henselae htrA results are consistent with the intracellular activation reported for the Salmonella htrA promoter in epithelial cells (18). HSPs other than HtrA have also been implicated in intracellular induction. Studies involving Legionella pneumophila Hsp60 report the increased expression of Hsp60 after bacterial association with host cells (19). Not only was Hsp60 synthesis upregulated upon invasion of monocytes by virulent L. pneumophila, but Hsp60 expression in bacterial cell surface was enhanced, as was Hsp60 secretion into the phagosome (19). In addition, recombinant Hsp60-coated beads were internalized by HeLa cells and the formation of a compact endosome was also described. Thus, stress response proteins may have ubiquitous functions of significant benefit to bacterial survival.
The B. henselae HtrA protein and htrA mutants have potential utility as diagnostic antigen and vaccine candidates, respectively. A recombinant well-defined skin test antigen would alleviate the safety concerns associated with the human-derived antigen previously used for diagnosis. Perhaps the area of greatest public health relevance is the use of htrA mutants as vaccine strains and in the development of vaccine vectors (7, 9, 18, 27, 31). Various reports describe htrA mutants of S. enterica serovar Typhimurium (7), B. abortus (9), Yersinia enterocolitica (27), and S. enterica serovar Typhi (31) as less virulent than their wt counterparts. Recently, studies involving S. enterica serovar Typhi strain CVD 908-htrA as a live attenuated vaccine advanced to a phase 2 clinical trial (42). Cats are the reservoir for B. henselae. The development of an attenuated B. henselae htrA mutant could result in a useful candidate vaccine for cat immunization to prevent transmission to humans.
This study expands our understanding of the basic transcriptional regulation of B. henselae htrA. The attenuation described for other bacterial htrA mutants (9) is interesting and suggests that a B. henselae htrA mutant may also be less virulent than its wt counterpart. Furthermore, the advances described in the phase 2 clinical trial involving S. enterica serovar Typhi htrA live attenuated vaccine for humans supports the potential of this application. The tools for genetic manipulation discussed in this work can be applied towards the development of a B. henselae htrA mutant. We anticipate these findings will provide groundwork for the development of such a B. henselae htrA mutant which may be suitable for cat immunization as a live attenuated vaccine.
ACKNOWLEDGMENTS
Sandra I. Resto-Ruiz was supported by NIH training grant T32DA07245-10. This work was also supported by NIH grant R29AI38178.
We thank Anthea Lee and Stanley Falkow of Stanford University for providing pANT3, DH12S, and 882str. HMEC-1 was obtained by materials transfer agreement with the Centers for Disease Control and Prevention. We thank the developers of HMEC-1, Edwin Ades of CDC and Thomas Lawley of Emory University, for making this cell line available.
REFERENCES
- 1.Ades E W, Candal F J, Swerlick R A, George V G, Summers S, Bosse D C, Lawley T J. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J Investig Dermatol. 1992;99:683–690. doi: 10.1111/1523-1747.ep12613748. [DOI] [PubMed] [Google Scholar]
- 2.Anderson B, Goldsmith C, Johnson A, Padmalayam I, Baumstark B. Bacteriophage-like particle of Rochalimaea henselae. Mol Microbiol. 1994;13:67–73. doi: 10.1111/j.1365-2958.1994.tb00402.x. [DOI] [PubMed] [Google Scholar]
- 3.Anderson B, Jones D, Burgess A. Cloning, expression and sequence analysis of the Bartonella henselae gene encoding the HtrA stress-response protein. Gene. 1996;178:35–38. doi: 10.1016/0378-1119(96)00330-7. [DOI] [PubMed] [Google Scholar]
- 4.Anderson B, Sims K, Regnery R, Robinson L, Schmidt M J, Goral S, Hager C, Edwards K. Detection of Rochalimaea henselae DNA in specimens from cat scratch disease patients by PCR. J Clin Microbiol. 1994;32:942–948. doi: 10.1128/jcm.32.4.942-948.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson B E, Neuman M A. Bartonella spp. as emerging human pathogens. Clin Microbiol Rev. 1997;10:203–219. doi: 10.1128/cmr.10.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batterman H J, Peek J A, Loutit J S, Falkow S, Tompkins L S. Bartonella henselae and Bartonella quintana adherence to and entry into cultured human epithelial cells. Infect Immun. 1995;63:4553–4556. doi: 10.1128/iai.63.11.4553-4556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumler A J, Kusters J G, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carithers H A. Cat-scratch disease. An overview based on a study of 1,200 patients. Am J Dis Child. 1985;139:1124–1133. doi: 10.1001/archpedi.1985.02140130062031. [DOI] [PubMed] [Google Scholar]
- 9.Chatfield S N, Strahan K, Pickard D, Charles I G, Hormaeche C E, Dougan G. Evaluation of Salmonella typhimurium strains harbouring defined mutations in htrA and aroA in the murine salmonellosis model. Microb Pathog. 1992;12:145–151. doi: 10.1016/0882-4010(92)90117-7. [DOI] [PubMed] [Google Scholar]
- 10.Conley T, Slater L, Hamilton K. Rochalimaea species stimulate human endothelial cell proliferation and migration in vitro. J Lab Clin Med. 1994;124:521–528. [PubMed] [Google Scholar]
- 11.De Las Penas A, Connolly L, Gross C A. ςE is an essential sigma factor in Escherichia coli. J Bacteriol. 1997;179:6862–6864. doi: 10.1128/jb.179.21.6862-6864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dehio C. Interactions of Bartonella henselae with vascular endothelial cells. Curr Opin Microbiol. 1999;2:78–82. doi: 10.1016/s1369-5274(99)80013-7. [DOI] [PubMed] [Google Scholar]
- 13.Dehio C, Meyer M. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J Bacteriol. 1997;179:538–540. doi: 10.1128/jb.179.2.538-540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dehio C, Meyer M, Berger J, Schwarz H, Lanz C. Interaction of Bartonella henselae with endothelial cells results in bacterial aggregation on the cell surface and the subsequent engulfment and internalisation of the bacterial aggregate by a unique structure, the invasome. J Cell Sci. 1997;110:2141–2154. doi: 10.1242/jcs.110.18.2141. [DOI] [PubMed] [Google Scholar]
- 15.Elzer P H, Hagius S D, Robertson G T, Phillips R W, Walker J V, Fatemi M B, Enright F M, Roop R M., II Behaviour of a high-temperature-requirement A (HtrA) deletion mutant of Brucella abortus in goats. Res Vet Sci. 1996;60:48–50. doi: 10.1016/s0034-5288(96)90130-7. [DOI] [PubMed] [Google Scholar]
- 16.Elzer P H, Phillips R W, Robertson G T, Roop R M., II The HtrA stress response protease contributes to resistance of Brucella abortus to killing by murine phagocytes. Infect Immun. 1996;64:4838–4841. doi: 10.1128/iai.64.11.4838-4841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson J W, Gross C A. Identification of the sigma E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- 18.Everest P, Frankel G, Li J, Lund P, Chatfield S, Dougan G. Expression of LacZ from the htrA, nirB and groE promoters in a Salmonella vaccine strain: influence of growth in mammalian cells. FEMS Microbiol Lett. 1995;126:97–101. doi: 10.1111/j.1574-6968.1995.tb07398.x. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez R C, Logan S M, Lee S H, Hoffman P S. Elevated levels of Legionella pneumophila stress protein Hsp60 early in infection of human monocytes and L929 cells correlate with virulence. Infect Immun. 1996;64:1968–1976. doi: 10.1128/iai.64.6.1968-1976.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garduño R A, Faulkner G, Trevors M A, Vats N, Hoffman P S. Immunolocalization of Hsp60 in Legionella pneumophila. J Bacteriol. 1998;180:505–513. doi: 10.1128/jb.180.3.505-513.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garduño R A, Garduño E, Hoffman P S. Surface-associated Hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect Immun. 1998;66:4602–4610. doi: 10.1128/iai.66.10.4602-4610.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grasseschi H A, Minnick M F. Transformation of Bartonella bacilliformis by electroporation. Can J Microbiol. 1994;40:782–786. doi: 10.1139/m94-123. [DOI] [PubMed] [Google Scholar]
- 23.Grossman A D, Erickson J W, Gross C A. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell. 1984;38:383–390. doi: 10.1016/0092-8674(84)90493-8. [DOI] [PubMed] [Google Scholar]
- 24.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 25.Kim K I, Park S C, Kang S H, Cheong G W, Chung C H. Selective degradation of unfolded proteins by the self-compartmentalizing HtrA protease, a periplasmic heat shock protein Escherichia coli. J Mol Biol. 1999;294:1363–1374. doi: 10.1006/jmbi.1999.3320. [DOI] [PubMed] [Google Scholar]
- 26.Lee A K, Falkow S. Constitutive and inducible green fluorescent protein expression in Bartonella henselae. Infect Immun. 1998;66:3964–3967. doi: 10.1128/iai.66.8.3964-3967.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S R, Dorrell N, Everest P H, Dougan G, Wren B W. Construction and characterization of a Yersinia enterocolitica O:8 high-temperature requirement (htrA) isogenic mutant. Infect Immun. 1996;64:2088–2094. doi: 10.1128/iai.64.6.2088-2094.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipinska B, Fayet O, Baird L, Georgopoulos C. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J Bacteriol. 1989;171:1574–1584. doi: 10.1128/jb.171.3.1574-1584.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipinska B, Sharma S, Georgopoulos C. Sequence analysis and regulation of the htrA gene of Escherichia coli: a sigma 32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 1988;16:10053–10067. doi: 10.1093/nar/16.21.10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipinska B, Zylicz M, Georgopoulos C. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J Bacteriol. 1990;172:1791–1797. doi: 10.1128/jb.172.4.1791-1797.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowe D C, Savidge T C, Pickard D, Eckmann L, Kagnoff M F, Dougan G, Chatfield S N. Characterization of candidate live oral Salmonella typhi vaccine strains harboring defined mutations in aroA, aroC, and htrA. Infect Immun. 1999;67:700–707. doi: 10.1128/iai.67.2.700-707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maeno N, Oda H, Yoshiie K, Wahid M R, Fujimura T, Matayoshi S. Live Bartonella henselae enhances endothelial cell proliferation without direct contact. Microb Pathog. 1999;27:419–427. doi: 10.1006/mpat.1999.0315. [DOI] [PubMed] [Google Scholar]
- 33.Margileth A M. Cat scratch disease. Adv Pediatr Infect Dis. 1993;8:1–21. [PubMed] [Google Scholar]
- 34.Pallen M J, Wren B W. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- 35.Regnery R L, Anderson B E, Clarridge J E D, Rodriguez-Barradas M C, Jones D C, Carr J H. Characterization of a novel Rochalimaea species, R. henselae sp. nov., isolated from blood of a febrile, human immunodeficiency virus-positive patient. J Clin Microbiol. 1992;30:265–274. doi: 10.1128/jcm.30.2.265-274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reschke D K, Frazier M E, Mallavia L P. Transformation of Rochalimaea quintana, a member of the family Rickettsiaceae. J Bacteriol. 1990;172:5130–5134. doi: 10.1128/jb.172.9.5130-5134.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roop R M, II, Fletcher T W, Sriranganathan N M, Boyle S M, Schurig G G. Identification of an immunoreactive Brucella abortus HtrA stress response protein homolog. Infect Immun. 1994;62:1000–1007. doi: 10.1128/iai.62.3.1000-1007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seol J H, Woo S K, Jung E M, Yoo S J, Lee C S, Kim K J, Tanaka K, Ichihara A, Ha D B, Chung C H. Protease Do is essential for survival of Escherichia coli at high temperatures: its identity with the htrA gene product. Biochem Biophys Res Commun. 1991;176:730–736. doi: 10.1016/s0006-291x(05)80245-1. [DOI] [PubMed] [Google Scholar]
- 41.Strauch K L, Johnson K, Beckwith J. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol. 1989;171:2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tacket C O, Sztein M B, Wasserman S S, Losonsky G, Kotloff K L, Wyant T L, Nataro J P, Edelman R, Perry J, Bedford P, Brown D, Chatfield S, Dougan G, Levine M M. Phase 2 clinical trial of attenuated Salmonella enterica serovar Typhi oral live vector vaccine CVD 908-htrA in U.S. volunteers. Infect Immun. 2000;68:1196–1201. doi: 10.1128/iai.68.3.1196-1201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]