Abstract
A comprehensive characterization of regulatory elements in the chicken genome across tissues will have substantial impacts on both fundamental and applied research. Here, we systematically identified and characterized regulatory elements in the chicken genome by integrating 377 genome-wide sequencing datasets from 23 adult tissues. In total, we annotated 1.57 million regulatory elements, representing 15 distinct chromatin states, and predicted about 1.2 million enhancer-gene pairs and 7662 super-enhancers. This functional annotation of the chicken genome should have wide utility on identifying regulatory elements accounting for gene regulation underlying domestication, selection, and complex trait regulation, which we explored. In short, this comprehensive atlas of regulatory elements provides the scientific community with a valuable resource for chicken genetics and genomics.
Decoding the chicken regulatory machinery helps to inform selection for simple and complex traits.
INTRODUCTION
The chicken was the first farm animal to have its genome sequenced and is generally considered to be one of the most important domesticated species. Globally, chicken production produces a third of all animal protein, and as one of the most intensively studied bird species, chicken fills an important gap in our understanding of the evolutionary relationship between mammals and birds (1). Chickens are also an excellent and widely used model species in evolutionary and developmental biology, immunology, and epigenetic regulation of gene expression (2).
Functional annotation of farm animal genomes is crucial for understanding the molecular mechanisms of economically important complex traits, including growth, reproduction, and disease resistance. Protein-coding sequences in humans only account for 1 to 3% of the whole genome, and nearly 90% of all phenotype-associated single nucleotide polymorphisms (SNPs) identified by genome-wide association studies (GWAS) lied within noncoding regions (3), suggesting that the activity of noncoding regions is the main driver of phenotypic variation. Therefore, the comprehensive annotation of functional regulatory elements, and especially noncoding regions, will help to address basic biological questions (4), interpret the genetic architecture underpinning disease risks and phenotypic variation (5), and improve the prediction accuracy of polygenic traits (6). The Encyclopedia of DNA Elements (ENCODE) projects and Epigenome Roadmap projects clearly demonstrated the importance of regulatory element annotation in humans and model species (7). Like the ENCODE projects, the Functional Annotation of Animal Genomes (FAANG) Consortium has made substantial contributions to the improvement of the annotation of farm animal genomes (8–11). However, current annotations of regulatory elements are still limited to only a few tissues, particularly in chickens (12), and a more complete catalog of regulatory elements is urgently needed. Furthermore, our study in pigs showed GWAS SNPs were significantly enriched in tissue-specific regulatory elements (8). Therefore, a comprehensive atlas of regulatory elements in the chicken genome would be expected to improve the identification of potential causative variants for economically important traits, with implications for breeding programs, and furthering the use of chickens in life science research.
In the current study, we systemically generated, collected, and analyzed a total of 377 epigenomic, transcriptomic, and chromatin conformation datasets from 23 major tissues in chickens (fig. S1). By combining these datasets, we built an atlas of regulatory elements across 23 tissues and explored their tissue-specific functionalities. We also predicted interactions between enhancers and their target genes, as well as super-enhancers. Furthermore, we conducted an exploratory integrative analysis of regulatory elements with selection signatures and GWAS results. In summary, we provided a comprehensive catalog of functional elements in chicken genome and demonstrated a potential role of regulatory elements in interpreting domestication and complex trait regulation in chickens.
RESULTS AND DISCUSSION
Data summary
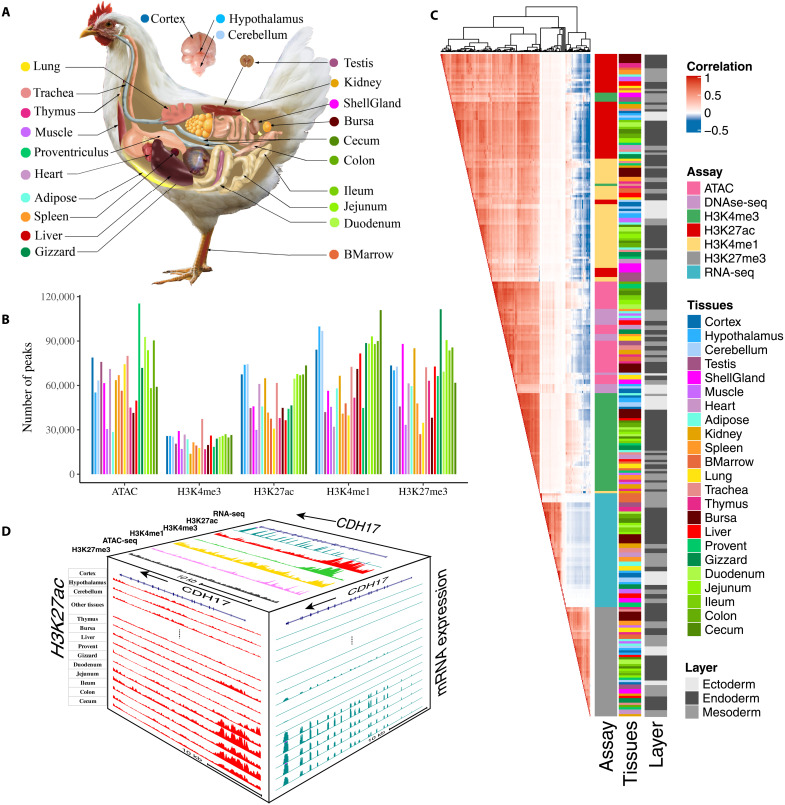
In total, we uniformly analyzed 377 genome-wide sequencing datasets from 23 major tissues in adult chicken (Fig. 1A), including chromatin immunoprecipitation sequencing (ChIP-seq) for four histone modifications (H3K4me3, H3K4me1, H3K27ac, and H3K27me3), CTCF, Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq), deoxyribonuclease sequencing (DNase-seq), reduced representation bisulfite sequencing (RRBS), RNA sequencing (RNA-seq), and Hi-C (table S1 and fig. S1). We generated 19.8 billon raw reads and retained 12.9 billon mapped reads, keeping on average 68.29% of reads after alignment and filtering (table S2). Further analysis obtained an average of 53,464, 65,843, 69,157, 23,410, and 65,811 peaks for H3K27ac, H3K27me3, H3K4me1, H3K4me3, and ATAC, or DNase-seq, respectively, with an average coverage of 4.11, 11.52, 3.84, 2.32, and 4.71% of the entire genome, respectively (Fig. 1B, table S2, and fig. S2).
Fig. 1. The summary of chicken epigenomic atlas.
(A) The 23 tissues that were profiled for building the epigenomic atlas in chickens. BMarrow, bone marrow; ShellGland: shell gland; Cortex: corticoidea dorsolateralis. (B) The average number of peaks detected for each of five epigenetic marks across tissues. (C) The relationship among assay, tissue, germ layers based on Pearson’s correlations of normalized signal in 1-kb windows stepped across the whole genome. (D) The distribution of assay signals around CDH17 gene across tissues (chr2:125,878,923-125,908,898, galGal6). Top face is jejunum. The vertical scale of UCSC tracks shows the normalized signal from 0 to 500 for RNA-seq, 0 to 150 for H3K27ac and H3K4me3, and 0 to 100 for other marks and ATAC-seq.
To evaluate relationship across different assays and tissues, we conducted hierarchical clustering of samples based on the signal intensity of epigenetic marks and gene expression profiles. The clusters clearly recapitulated the sequencing assays, followed by tissue types, and lastly biological replicates (Fig. 1C), which was consistent with results of principal components analysis (PCA) (fig. S3). The six sequence assays formed three major clusters: (i) active regulatory marks (i.e., H3K4me3, H3K27ac, H3K4me1, and ATAC), (ii) Polycomb repression (H3K27me3), and (iii) gene expression (RNA-seq). We also found that the four active regulatory marks were positively correlated with each other but were negatively correlated with H3K27me3. The signal intensity of RNA-seq (mainly within gene bodies) showed a weakly positive correlation with active regulatory marks and a negative correlation with H3K27me3. Within each assay, tissues with similar biological functions, such as different brain regions, intestine-relevant, and immune-relevant tissues, clustered together. Tissues derived from the same germ layer were also more likely to cluster together (Fig. 1C).
To explore the relationship between regulatory elements and gene expression, we first explored the distribution of signals from each mark near six different types of genes [e.g., protein-coding, long noncoding RNA (lncRNA), pseudogenes, microRNA (miRNA), small nucleolar RNA (snoRNA), and small nuclear RNA (snRNA) genes] and found that transcription start sites (TSS) of protein-coding genes showed a stronger enrichment of three active marks (i.e., ATAC, H3K4me3, and H3K27ac) but not H3K4me1, compared to the other gene types (fig. S4). We took Cadherin 17 (CDH17) as an example (Fig. 1D). The protein encoded by CDH17 is an intestinal proton-dependent peptide transporter (13) and also plays important roles in gastrointestinal cancer development (14). CDH17 was specifically and highly expressed in intestinal tissues and had a specific enrichment for H3K27ac around its TSS in intestinal tissues compared to other tissues (Fig. 1D). In addition, the TSS of CDH17 was accessible and enriched for three other active regulatory marks (e.g., H3K27ac, H3K4me3, and H3K4me1) but not for Polycomb repression (H3K27me3). These findings demonstrated coordinated gene regulation among different epigenetic marks in a gene expressed in a tissue-specific manner in the chicken.
Prediction and characterization of chromatin states
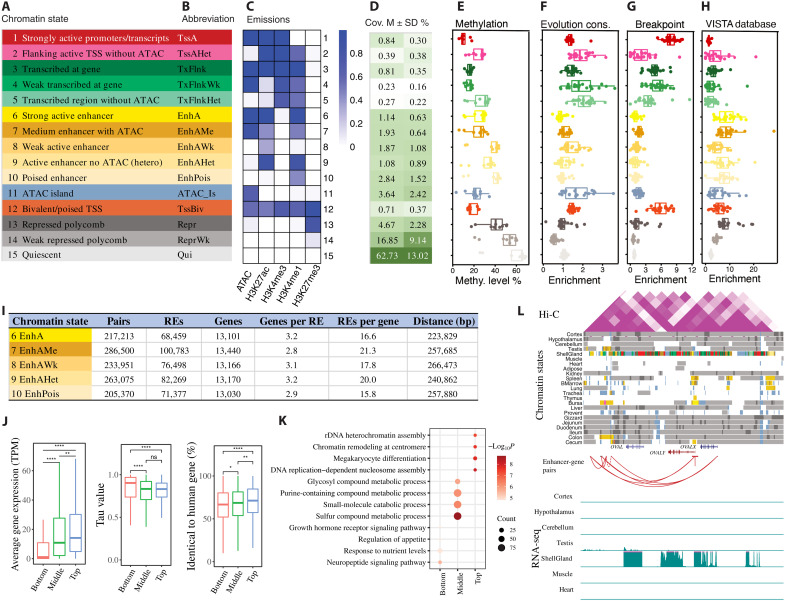
By combining all five epigenetic marks across 23 tissues via ChromHMM (15), we predicted 15 distinct chromatin states, as described in our previous pig study (8) (Fig. 2, A to C). These chromatin states mainly represented promoters (TssA, TssAHet, and TssBiv, covering 1.94% of the entire genome), TSS-proximal transcribed regions (TxFlnk, TxFlnkWk, and TxFlnkHet, covering 1.31% of the genome), enhancers (EnhA, EnhAMe, EnhAWk, EnhAHet, and EnhPois, covering 8.86% of the genome), accessible islands (ATAC island, 3.64%), repressed regions (Repr and ReprWk, covering 21.52% of the genome), and quiescent regions (Qui, 62.73%) (Fig. 2D and table S3). In total, we predicted 1,573,399 regulatory elements (excluding Qui) across 23 tissues. These included 102,907 promoters, 146,045 TSS-proximal transcribed regions, 765,400 enhancers, 351,928 ATAC islands, and 201,377 repressed regions (fig. S5). As expected, promoters had a higher enrichment in TSS, 5′ untranslated regions, and CpG islands than enhancers, and both active promoters and enhancers were significantly enriched in the TSS and gene body of expressed genes [transcripts per million (TPM) ≥ 0.1] compared to repressed genes (fig. S5E). Among these chromatin states, the activities of enhancers were the most dynamic, whereas the activities of promoters were the most conserved among tissues (fig. S6). At the DNA sequence level, promoters were also more conserved than enhancers and Polycomb repressed regions (measured by phyloP score, based on 77 vertebrates) (Fig. 2F).
Fig. 2. Discovery and characterization of chromatin states and enhancer-gene interactions in chicken.
The definition (A) and abbreviation (B) for 15 predicted chromatin states. (C) Emission probabilities of five epigenetic marks for each chromatin state. (D) Genomic coverage for each chromatin state. M, mean. (E) Methylation level in each state. Whiskers show 1.5× interquartile range. Each dot represents one tissue. (F) Fold enrichments of chromatin states for conserved region based on phyloP score. Whiskers show 1.5× interquartile range. Each dot represents one of 23 different tissues. (G) Fold enrichments of chromatin states in chicken specific evolution breakpoint. Whiskers show 1.5× interquartile range. Each dot represents one of 23 different tissues. (H) Chromatin state enrichment of enhancer datasets from VISTA database. Whiskers show 1.5× interquartile range. Each dot represents one of 23 different tissues. The y axes label on (E) to (H) has the same label as on (A). (I) The summary of enhancer-gene pairs across all tissues. RE, regulatory element. (J) The gene expression, tissue-specific value (Tau), and conservation score for genes linked to different numbers of enhancers (bottom = 1, 10 ≤ middle ≤ 20, top ≥ 50 enhancers). *P < 0.05, **P < 0.01, and ****P < 0.0001. ns, not significant. (K) The top four GO terms for the three gene groups. (L) Chromatin state landscape and mRNA expression at OVAL locus (chr2:67,664,385-67,749,248, galGal6) across 23 tissues. Vertical scale of UCSC tracks shows normalized signal from 0 to 100 for RNA-seq.
Differences in the regulatory landscape within and near evolutionary breakpoint regions (EBRs) have been found to be associated with changes in gene expression relating to species-specific traits (16). Here, to determine what types of regulatory elements were associated with chicken-specific EBRs [breakpoints with extended 1000 base pairs (bp) on each side], we found that they were significantly enriched for promoters and particularly for active promoters (TssA) (Fig. 2G). This promoter set included seven genes (i.e., ANKRD10, UVRAG, APBB3, XYLT2, XPO1, CALM2, and RCHY1) that were involved in multiple diseases relevant to brain development, immune response, and intestine function (table S4). These results suggest that chicken-specific EBRs could be associated with chicken-specific gene expression profiles. Nonetheless, further research is needed to verify this hypothesis.
Then, we further explored the relationship between DNA methylation and regulatory elements. We found that promoters had the lowest methylation level, followed by ATAC islands, enhancers, and Polycomb repressed regions (Fig. 2E), which was consistent with how these regulatory elements regulate gene expression and the general negative correlation between methylation level and gene expression. We also observed that enhancers with ATAC signal (i.e., EnhA and EnhAMe) had a lower methylation level than enhancers without ATAC, which was in line with our previous findings in pigs (8).
To validate enhancers predicted in our study in silico, we queried the VISTA database (17), which contains information on functionally validated enhancers in humans and mice. As expected, we found that strong (EnhA) and medium (EnhAme) enhancers demonstrated higher enrichments in the VISTA enhancers than the other types of enhancers (Fig. 2H). In addition, the enrichment pattern exhibited strong tissue specificity (fig. S7). These findings not only provided additional evidence for the functionality of enhancers identified in our study but also suggested potential conservation of some enhancers among humans, mice, and birds.
Linking enhancers to specific target genes is essential for understanding how gene expression is regulated. To predict the target gene(s) for each enhancer, we correlated the signal intensity of H3K27ac and the expression level of genes within the same predicted CTCF loop region (see Materials and Methods). In total, we obtained ~1.2 million enhancer-gene pairs with a median distance of 249,346 bp across all tissues. On average, each gene was linked to 18 enhancers, whereas each enhancer targeted three genes (Fig. 2I). The significance (i.e., q value) of enhancer-gene pairs was significantly negatively correlated with the distance between each enhancer and its target genes (fig. S8). We then separated genes into three groups based on the number of enhancers to which they were linked. We found that genes with more enhancers had higher gene expression levels, weaker tissue-specific expression patterns, and more sequence-level conservation compared to genes linked to fewer enhancers (Fig. 2J). Gene Ontology (GO) enrichment analysis showed that genes with only one enhancer were more likely to have a tissue-specific function (e.g., neuropeptide signaling pathway for brain), genes with the medium number of enhancers were related with metabolic processes, and genes with the highest number of enhancers were involved in basic biological functions (e.g., ribosomal DNA heterochromatin assembly) (Fig. 2K and table S5). The ovalbumin-related protein (OVAL) gene locus, as an example, illustrated how regulatory elements control gene expression across tissues (Fig. 2L). This locus contained several OVAL genes that were highly and specifically expressed in shell gland, and nearby regulatory elements had active chromatin states. On the basis of enhancer-gene pair predictions, several enhancers, located up to 50 kb away from TSS, may regulate the expression of OVAL genes (Fig. 2L). Similar patterns were also observed for TMPRSS15, VMO1, and MEIOC (figs. S9 and S10).
Tissue-specific chromatin states and their functional annotation
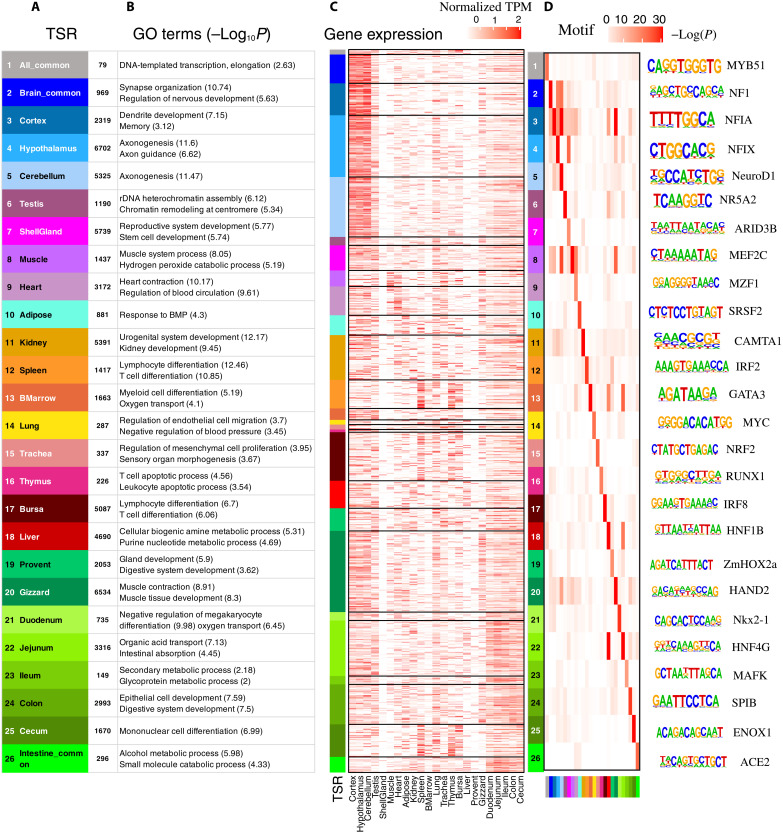
Tissue-specific regulatory elements play important roles in determining tissue identity. Here, we identified a total of 790,279 tissue-specific regulatory elements (excluding Qui), representing 50.2% of all regulatory elements identified in this study (table. S6). As expected, a great number of enhancers exhibited tissue-specific activity compared to promoters (table S6). Furthermore, we defined an additional three types of EnhAs based on their tissue specificities, including all-common EnhAs (shared among all tissues), gut-common EnhAs (shared among all gut-associated tissues), and brain-common EnhAs (shared among three brain regions). We identified a total of 64,657 tissue-specific EnhAs among 23 tissues, ranging from 149 in ileum to 6702 in hypothalamus (Fig. 3A). The GO analysis of their target genes revealed distinct biological functions, clearly reflecting the known tissue biology (Fig. 3B). For instance, genes targeted by the all-common EnhAs were involved in basic biological functions (e.g., DNA template transcription), and those targeted by brain-common EnhAs were involved in the regulation of nervous system development. Regarding the gut-associated tissues, genes targeted by EnhAs specific to each tissue had distinct functions (e.g., proventriculus for digestive system development, gizzard for muscle contraction, intestine for nutrition absorption and immune responses, and cecum for mononuclear cell differentiation). In the reproductive system, genes targeted by shell gland–specific EnhAs participated in stem cell development, while genes targeted by oocyte- and testis-specific EnhAs were involved in chromatin remodeling at the centromere, which may be related to spermospore meiosis. In addition, after lifting the regulators to human genome, these conserved enhancers also showed tissue-specific biological function and were associated with tissue-relevant phenotypes in humans and mice (fig. S11A). Furthermore, we observed that putative target genes of tissue-specific EnhAs were highly expressed in the corresponding tissues (Fig. 4C). These collective results suggest that the activities of these tissue-specific enhancers regulate the expression of their target genes, contributing to the unique biological function of each tissue.
Fig. 3. Tissue-specific strong enhancers (EnhA) and their potential functions in 23 tissues.
(A) Twenty-six modules of tissue-specific strong enhancers (EnhA) and their number in each module. (B) The GO functional enrichment for target genes of tissue-specific strong enhancers in each module. The columns represent 26 modules of strong enhancers. The rows represent GO terms in each module. (C) The expression (TPM) of EnhAs’ putative target genes within the top three GO terms of each module. The rows represent the target genes within the top GO terms of each module, and the columns represent each tissue. (D) The enrichment of transcription factor motifs in each module. The columns represent 26 modules of EnhAs. The rows represent motifs in each module.
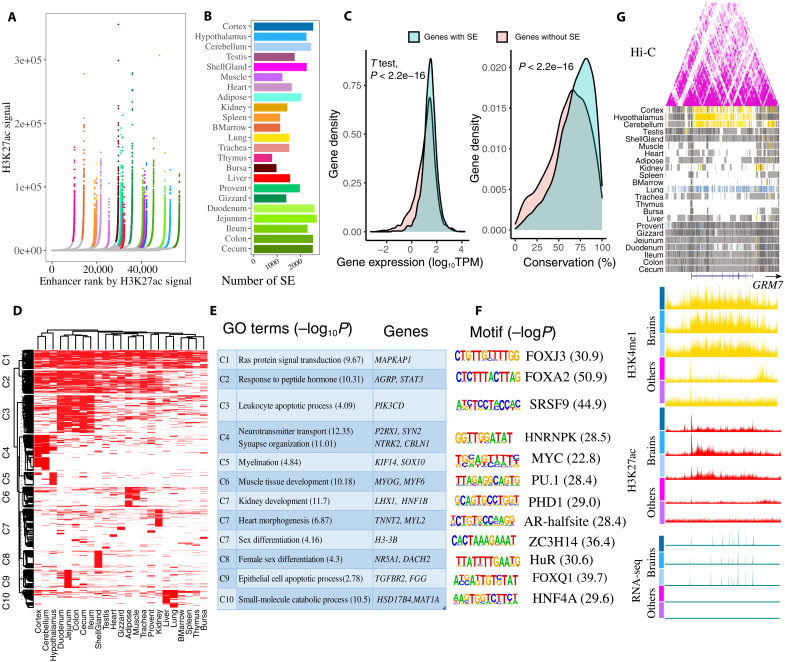
Fig. 4. Super-enhancers and their potential functions.
(A) Super-enhancers (SEs) identified by ROSE based on H3K27ac signal in each tissue. Super-enhancers are colored based on tissue and show exceptionally high signal. Tissue color is the same with (B). (B) The number of super-enhancers in each tissue. (C) The expression and conservation difference between genes with (8338) and without (8441) super-enhancers. The gene conservation score is sequence identity (%) from chicken gene to orthologous human gene. (D) Clustering of 7662 nonredundant super-enhancers based on their activity in each tissue. (E) GO function and representative genes in each super-enhancer cluster. (F) Motifs enriched in each super-enhancer cluster. (G) A brain-specific super-enhancer present at the GRM7 locus (chr12:19,167,915-19,582,314). The following sections are Hi-C, chromatin state, H3K4me1, H3K27ac, and RNA-seq. Vertical scale of UCSC tracks shows normalized signal from 0 to 100 for RNA-seq, 0 to 150 for H3K27ac, and 0 to 50 for H3K4me1.
To explore how transcriptional factors (TFs) were involved in tissue-specific regulation, we conducted motif enrichment analysis for these tissue-specific EnhAs. Multiple notable motifs were enriched in tissue-specific EnhAs, such as NF1 and NEUROD1 in the brain, MEF2C in the muscle, and HNF1B and HNF4G in the liver and intestinal tissues (Fig. 4D and figs. S12A and S13A). These findings were in line with previous results in humans (18) and pigs (8), suggesting the conservation of TF regulation of gene expression across species. Specifically, we found that the binding motif of NR5A2, which is important for horse steroidogenesis (19), was significantly enriched in testis-specific EnhAs, and the NR5A2 gene was highly expressed in chicken testis (fig. S13B). The binding motif of ARID3B, an important regulator of ovarian cancer in mammals (20), was significantly enriched in shell gland–specific EnhAs, indicating that it may also play a role in reproduction in chickens. Bursa is one of major immune organs producing B cells in chickens (21). In line with the function of bursa, the binding motif of an important TF in B cell development, IRF8, was enriched in bursa-specific EnhAs (fig. S13C). In addition, we found that expression levels of the inferred TFs were higher in the corresponding tissues than in other tissues (fig. S12B), indicating that these tissue-specific enhancers are hotspots for TF activity and play important roles in the tissue-specific regulation of gene expression.
Identification and characterization of super-enhancers
Super-enhancers, which are clusters of enhancers in close genomic proximity (within 12.5 kb) that bind various transcription factors, are proposed to play key roles in the control of cell identity, and their dysfunction may cause diseases (22, 23). Here, we identified an average number of 1823 super-enhancers across tissues, ranging from 775 in thymus to 2676 in jejunum, by integrating H3K27ac signals and enhancers (all five types of enhancers). These super-enhancers had an average size of 42,534 bp, represented 40% of all enhancers and covered 7.3% of the chicken genome (Fig. 4, A and B, and fig. S14). Each super-enhancer consisted of about 30 enhancers on average with an average distance of 664 bp between adjacent enhancers (fig. S14A). Among all five types of enhancers, the super-enhancers had many more strong-enhancers (EnhA) and fewer poised-enhancers (EnhPois) compared to the other three types of enhancers (fig. S14, B and C). Further analysis revealed that the genes regulated by super-enhancers showed significantly higher expression levels and were more conserved at the sequence level (Fig. 4C) than the genes without super-enhancers, which was consistent with previous findings in humans (24).
After merging super-enhancers from 23 tissues, we obtained a total of nonredundant 7662 super-enhancers. These super-enhancers showed strong tissue-specific activity, as only 31 of 7662 super-enhancers were identified in all tissues (fig. S14D). We then clustered all super-enhancers based on their activity in all 23 tissues and revealed distinct tissue sharing patterns for each cluster of super-enhancers (Fig. 4D). Further GO enrichment analysis of putative target genes of these different super-enhancer clusters suggested distinct biological functions for each cluster (Fig. 4E and table S7). For instance, the function of genes targeted by the super-enhancers in C1 (cluster 1) (widely active in most of tissues) were involved in basic biological functions (e.g., Ras protein signal transduction). On the other hand, the super-enhancers in C3 were specifically active in gut and immune tissues, and their target genes participated in immune function (e.g., leukocyte apoptotic process). The super-enhancers in C4 and C5 were specifically active in brain tissues, and their target genes participated in nerve development and synapse organization. The super-enhancers in C6 were specifically active in muscle and were relevant to muscle development, and C8 super-enhancers, specifically active in shell gland, were related to female sex differentiation. The super-enhancers in C10, specific to liver, participated in small-molecule catabolic processes, whereas super-enhancers in C7, specific to kidney, heart, and testis, participated in kidney development, heart morphogenesis, and sex differentiation. Some well-known genes associated with super-enhancers of each cluster are presented in Fig. 4E. Furthermore, we observed that genes with tissue-specific super-enhancers were specifically and highly expressed in the corresponding tissues (fig. S14D).
To explore potential TFs that bind super-enhancers, we also performed motif enrichment analysis for each super-enhancer cluster. We observed that distinct TFs motifs were enriched in each cluster, reflecting corresponding tissue-specific functions (Fig. 4F). Notably, HNF4A plays important roles in liver (25), and its motif was significantly enriched in C10 (liver-specific super-enhancers). The HNRNPK motif was enriched in C4, which was related to multiple brain diseases in humans (26). The motif of ZC3H14 was enriched in testis-specific super-enhancers, which was relevant with testis size in mice (27). The motif of PHD1 was enriched in kidney-specific super-enhancers, which plays a role in acute kidney injury (28). The motif of FOXQ1 enriched in C9, which plays roles in gastrointestinal function (29). Here, we took GRM7 as an example (Fig. 4G). GRM7 was linked to brain-specific super-enhancers with strong brain-specific H3K4me1 and H3K27ac signal, and it also showed strong brain-specific expression. GRM7, a neuro-related gene, is involved in schizophrenia in humans (30). These results collectively suggest that super-enhancers play a key role in tissue identity and function in chickens.
Exploring the utility of enhanced genome annotation to inform chicken biology
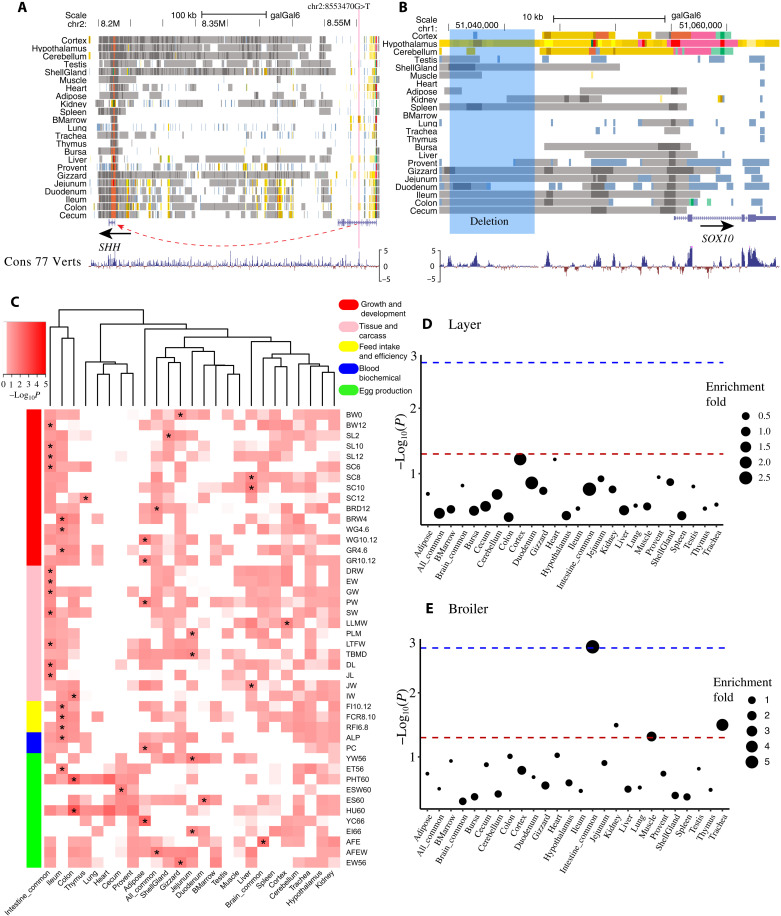
For monogenic traits, it is more feasible to identify the underlying mechanism when amino acid changes or structural variants are involved. However, there are clear examples where polymorphisms in regulators have been found such as the role of the Shh regulatory element on polydactyly in both humans and mice (31, 32), suggesting that the identification of regulator elements, especially enhancers, may provide more insights. To explore the potential role of regulatory elements on the genetic control of monogenic traits in chicken, we examined eight noncoding causal or likely causal variant(s) of seven chicken monogenic traits in the Online Mendelian Inheritance in Animals (OMIA) database (https://omia.org/home/) and found that six (75%) of them are located in active regulatory elements in at least one tissue (table S8). For instance, the potential causal SNP (rs80659072) of polydactyly (33) is located in an active enhancer of SHH in bone morrow, which has a high DNA sequence constraint score (PhyloP > 4) (Fig. 5A and fig. S15A). Another noted example is a 7.6k deletion (g.51035106_51042744delins) at 12 K upstream of SOX10 that causes dark brown/yellow plumage in chickens (34, 35). This deletion exclusively overlaps with a brain-specific enhancer region, particularly in hypothalamus (Fig. 5B and fig. S15B). Furthermore, a single causal variant (rs316090093) of the Silky/Silkie feathering in chickens (36) resides in a strong promoter (TssA) of PDSS2 with a clear ATAC-seq foot-printing across multiple chicken tissues (fig. S15C).
Fig. 5. The regulatory elements involved in chicken monogenic traits, complex traits, and selection signatures.
(A) The predicted chromatin states around a potential causal SNP (chr2:8553470G>T, rs80659072) of polydactyly (chr2:8,546,000-8,561,000) in chickens. The red line indicates the position of the SNP. (B) The predicted chromatin states around a 7.6k deletion (g.51035106_51042744delins, highlighted in blue) at the upstream of SOX10 which causes dark brown/yellow plumage in chickens (chr1:51,034,163-51,064,718). The highlighted region means the deletion region. (C) GWAS enrichment in tissue-specific enhancers (EnhA). “*” indicates that P value is the smallest in this trait, and P < 0.05. (D and E) The enrichment of tissue-specific strong enhancer (EnhA) in selection signatures of layer and broiler, respectively. The dashed lines indicate P < 0.05 (red) and FDR < 0.05 (blue).
There is a clear role of regulatory elements in complex traits; however, understanding the underlying genetic architecture of complex traits is important but challenging, as the majority of trait-associated loci have small genetic effects and exhibit linkage disequilibrium (LD) that include nearby variants (37). We hypothesized that functional annotation of regulatory elements across tissues may provide new multidimensional functional information to interpret the GWAS results of complex traits and help pinpoint potential causal variants of complex traits in chickens. Through examining GWAS summary statistics from 44 complex traits of economic importance in chickens, including growth and egg production traits (table S9) (38, 39), we observed that active regulatory elements had a higher enrichment for GWAS signals compared to quiescent regions, and TssA and TssBiv had the highest enrichments (fig. S16A), which was consistent with previous findings in pigs (8) and humans (40). To explore whether tissue-specific regulatory elements in the trait-relevant tissues had more regulatory effects on GWAS SNPs than in the other tissues, we performed GWAS signal enrichment analysis using tissue-specific EnhAs across all 44 complex traits (Fig. 5C). We found that most growth-related traits (e.g., growth and carcass weight) showed the highest enrichment in intestine-shared regulatory elements, whereas feed intake and efficiency were enriched in ileum-specific EnhAs. These findings raise new and interesting hypotheses: Genes that are actively regulated across gut tissues were essential in affecting growth, whereas genes that are specifically regulated in ileum were associated with feed efficiency. Of particular note, weight gain and growth rate from 4 to 6 weeks were significantly enriched in ileum-specific EnhAs, whereas these traits from 10 to 12 weeks were significantly enriched in adipose-specific EnhAs. This finding was in line with the principle of animal growth, in which the protein-to-fat ratio is high at early developmental stages (4 to 6 weeks) but significantly decreases during late developmental stages (10 to 12 weeks) (41, 42). Most egg production traits were significantly enriched in intestine-specific EnhAs, except for age at first egg (enriched in brain-shared EnhAs), which was consistent with a previous report that the hypothalamus-pituitary-gonad axis plays important roles in age at first egg (43).
Last, we conducted an exploratory analysis to investigate whether domestication and selection were associated with regulatory elements. By examining selective sweeps in chicken domestication (red jungle fowls versus domesticated chickens) from a previous study (table S10) (44), we found that hypothalamus and shell gland–specific enhancers were significantly depleted [false discovery rate (FDR) < 0.05] in both datasets of selective sweeps by using two different outgroups (G. g. murghi and G. g. jabouillei.) (table S12). This result provides the notion that domestication likely induced changes in brain and reproductive system via tissue-specific regulation through enhancers. Furthermore, we examined selection sweeps between modern broilers and layers (table S11 and fig. S16, B and C), and we found that intestine-shared enhancers were significantly enriched (FDR = 0.0496) for selection sweeps in broiler (Fig. 5E), representing two genes: MYO10 and CDH18 (table S12). Together, our results show that this atlas of regulatory elements provides candidates further characterization that may lead to a greater understanding of the molecular mechanisms underlying monogenic and complex traits as well as adaptive evolution and selection in chickens.
In summary, we generated and characterized a comprehensive landscape of regulatory elements in 23 adult chicken tissues. To our knowledge, this is the most comprehensive catalog of regulatory elements in any farm animals, including a total of 1,573,399 regulatory elements (102,907 promoters, 146,045 TSS-proximal transcribed regions, 765,400 enhancers, 351,928 ATAC islands, and 201,377 repressed regions). Furthermore, we identified a total of 790,279 tissue-specific regulatory elements, 7662 nonredundant super enhancers, and more than 1.2 million enhancer-gene pairs across 23 tissues. Using this rich resource, we find that GWAS signals associated with growth traits were significantly enriched in intestine-specific enhancers. We revealed that tissue-specific enhancers may be involved in different developmental stages of growth. Ileum-specific enhancers contributed to the early stage of growth, whereas adipose-specific enhancers were related to the late stage of growth. However, functional genome data from different development stages are required to further verify these hypotheses. As the FAANG project keeps going, more epigenomic data in diverse biological contexts, such as additional developmental stages, breeds, and single cells, will add more information to the chicken functional genome annotation and enhance the understanding of regulation biology in chicken. Overall, this extensive annotation of regulatory elements in the chicken genome will help researchers to decode the molecular mechanisms underlying complex traits and adaptive evolution, as well as provide a powerful resource in realizing the genetic improvement of economically important traits in poultry.
MATERIALS AND METHODS
Animals and tissues
The animal experiment was conducted according to the Animal Care and Use protocol (#18464), which was approved by the Institutional Animal Care and Use Committee, University of California, Davis. All the birds were euthanized with CO2 under U.S. Department of Agriculture (USDA) inspection. A total of four birds were selected from the F1 cross of two highly inbred lines: line 6 and line 7, maintained in the USDA, ARS, Avian Disease and Oncology Laboratory. A total of 21 tissues were collected from two male birds (20 weeks of age) as in our previous report (12, 45), including cortex, hypothalamus, cerebellum, testis, muscle, heart, adipose, kidney, spleen, bone marrow, lung, trachea, thymus, liver, proventriculus, gizzard, duodenum, jejunum, ileum, colon, and cecum. Shell glands were collected from two female birds at 20 weeks of age. Bursa tissues were collected from four birds of two highly inbred lines (two for each line) at 3 weeks of age, Fayoumi (M15.2) and Leghorn (GHs6), maintained at Iowa State University (Ames, IA, USA), which have been described previously (46). All tissues have been flash frozen in liquid nitrogen and stored at −80°C before further using.
Library construction and sequencing
RNA-seq, RRBS, ATAC-seq, and ChIP-seq (H3K4me3, H3K4me1, H3K27ac, and H3K27me3) experiments were performed on flash-frozen tissue samples. For RNA-seq experiments, total RNA was extracted using TRIzol (Thermo Fisher Scientific, #15596026) and treated with DNase I (Thermo Fisher Scientific, #EN0521). RNA integrity number (RIN) was determined using an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), and RNA samples with RIN > 8 were selected for constructing directional RNA-seq libraries using an NEBNext Ultra RNA Library Prep Kit of Illumina (New England BioLabs, #E7530L), followed by sequencing on an Illumina Hiseq 4000 platform (Illumina, San Diego, CA, USA) with paired-end 150-bp reads. RRBS datasets were generate by an Illumina HiSeq 4000 platform with 150-bp paired-end reads after library construction by Novogene (Sacramento, CA, USA). For ATAC-seq experiments, libraries were generated using a modified protocol (https://figshare.com/articles/dataset/Final_ATAC_protocol_docx/13891268) according to OmniATAC and cryopreserved nuclei protocols (47, 48). For ChIP-seq experiments, samples were processed as described previously (8). Libraries of both ATAC-seq and ChIP-seq were sequenced on an Illumina HiSeq 4000 platform with 50-bp paired-end and single-end reads, respectively.
Raw sequence data processing
In total, 217 new sequence datasets, including 140 ChIP-seq (H3K4me3, H3K4me1, H3K27ac, H3K27me3, and input control), 28 ATAC-seq, and 27 RNA-seq from 14 tissues, 18 RRBS from 9 tissues, and 4 ATAC-seq from bursa, were generated (table S1). We also uniformly analyzed another 160 existing chicken datasets, including 116 ChIP-seq (H3K4me3, H3K4me1, H3K27ac, H3K27me3, CTCF, and input control), 15 DNase-seq, 2 ATAC-seq, and 20 RNA-seq from eight tissues (i.e., adipose, cerebellum, brain cortex, hypothalamus, liver, lung, muscle, and spleen) in the FAANG pilot project (PRJEB14330) (12) and four bursa samples from PRJEB38600 and PRJEB3859 (46), three RNA-seq samples of bone marrow from PRJNA279487 (49), and four liver Hi-C datasets from PRJEB27364 (10). The processing of ChIP-seq, ATAC-seq, DNase-seq, and RNA-seq data followed the UC Davis FAANG Functional Annotation Pipeline (https://github.com/kernco/functional-annotation) (12). Briefly, the galGal6 genome assembly and Ensembl genome annotation (v100) have been used as reference. Trim Galore! (50) (v.0.6.5) was used to trim raw sequencing reads. STAR (51) (v.2.5.4a) was used for the alignment of RNA-seq reads to the reference genome, while BWA (52) (v0.7.17) was used for the alignment of sequence reads from other assays. The reads with mapping quality (MAPQ) scores less than 30 have been removed using Samtools (53) (v.1.9). For RNA-seq, the htseq-count (54) (v.0.13.5) was used to extract the read counts of genes, and then the trimmed mean of M values (TMM) and TPM were calculated as the normalized gene expression with EdgeR (v3.32.0) and StringTie2 (v.1.3.3), respectively (55). For ChIP-seq, duplicated reads were removed by Picard (v.2.18.7), and then peaks were called by MACS2 (56) (v.2.1.1). Various quality metrics of ChIP-seq (e.g., Jensen-Shannon divergence; table S2) were calculated following the method described by Kern et al. (12). Bismark (57) (v.0.22.1) pipeline was used to process RRBS data, and Juicer pipeline (58) was applied for analyzing Hi-C data, including the detection of topologically associating domains (TADs).
Sample clustering
The global relationship among all samples from different assays, tissues, and germ layers was exploited by deepTools (59) (v.3.5.0). Briefly, the mark signals (bigWig) of each sample were created by bamCompare, Z-score was normalized by scipy.stats.zscore function in Scipy (60) (v.1.8.0), and then the Z-score–normalized signals of all samples within 1-kb windows were calculated by multiBigwigSummary. The relationships among samples were measured by the Pearson’s correlation of Z-score–normalized read signals by plotCorrelation. The PCA analysis of samples was conducted using plotPCA (59) (fig. S3). The Z-score–normalized signals of protein_coding gene, lncRNA, pseudogene, miRNA, snRNA, and snoRNA were calculated by computeMatrix scale-regions function in deepTools (59) (v.3.5.0) with setting of -a 2500 -b 2500 (fig. S4).
Annotation of chromatin states
To predict chromatin states, ChromHMM (15) (v.1.20) was used to combine all the datasets of ChIP-seq (H3K4me3, H3K4me1, H3K27ac, H3K27me3, and input control), ATAC-seq, and DNase-seq from two biological replicates of 23 tissues. The 15-state model was selected because it had the maximum number of chromatin states with distinct epigenetic mark combinations. These 15 chromatin states were named according to the combinations of epigenome modifications and enrichments around TSS of genes (9, 18), including TssA, TssAHet, TxFlnk, TxFlnkWk, TxFlnkHet, EnhA, EnhAMe, EnhAWk, EnhAHet, EnhPois, ATAC_Is, TssBiv, Repr, ReprWk, and Qui.
Enrichment of chromatin states in genomic features
The enrichment of chromatin states in distinct genomic features (e.g., exon, TSS, and CpG islands) was assessed by (C/A)/(B/D) (15), where A is the number of bases in a state, B is the number of bases in a genomic feature, C is the number of bases in the overlapped region of the state and the genomic feature, and D is the number of bases in the entire genome. The genomic features also included sequence conserved regions, EBRs, and VISTA enhancers. For sequence conserved regions, we first download the chicken base-wise conservation scores (phyloP) based on 77 vertebrate genomes from UCSC (https://hgdownload.soe.ucsc.edu/goldenPath/galGal6/phyloP77way/). The conserved sites were extracted on the basis of conservation scores of more than 5 and then were merged into a conserved region by bedtools merge (61) with parameter a of -d 10 -c 5 -o mean. Given that the majority of regulatory regions are located in the noncoding region, the conserved region within the coding region was excluded in the current study. For chicken-specific chromosome evolutionary breakpoints, we compared genomes of 38 species (33 mammals, the chicken, and the alligator), identified breakpoints by the method described in recent work (16), and then extended each breakpoint 1000 bp on each side. For experimentally validated VISTA enhancers, they were obtained from the VISTA database (https://enhancer.lbl.gov/), and the positive human enhancers from all tissue (n = 998), forebrain (n = 324), and heart (n = 141) were selected. The significance of enrichment was calculated using the Fisher’s exact test.
Methylation level of chromatin states
The methylation level of each CpG site in each tissue was extracted by Bismark (v.0.22.1) (57). The average methylation level of each chromatin state in each tissue was then calculated using the bedtools map function in BEDTools (v.2.29.2) (61).
Chromatin states variability
To investigate the changes of chromatin states across tissues, the nonredundant regulatory elements across all 23 tissues were first generated (fig. S5) using the merge function in BEDtools (v.2.29.2) (61). The chromatin state variability and switching between tissues were then calculated using the methods described previously (fig. S6) (8).
Enhancer-gene pair prediction
The enhancer-gene interaction was predicted by the method described previously (8). First, CTCF-mediated loops from CTCF ChIP-seq data of eight tissues were identified by FIMO (62), following the pipeline described by Oti et al. (63). The CTCF-mediated loops were then merged to generate the non-overlapping CTCF loops using merge function in BEDtools (v.2.29.2) (61). The enhancer-gene pairs were predicated on the basis of the Spearman’s correlation of signal density of H3K27ac and gene expression (TMM) within each loop region across all 46 samples. After Benjamini-Hochberg correction for multiple testing, enhancer-gene pairs with FDR < 0.05 were considered as confident interactions. The enhancer-gene pairs were predicted for all five different kinds of enhancers annotated above. On the basis of the number of strong enhancers (EnhA) linking to a gene, genes were clustered into the following categories: (i) genes with only one EnhA (bottom, n = 1049), (ii) genes with more than 10 but less than 20 (middle, n = 3489), (iii) genes with more than 50 (top, n = 726). The tissue-specific expression of a gene was measured using the Tau method (64). The sequence conservation scores of orthologous genes between human and chicken were downloaded from Ensembl (v 100). The GO enrichment analysis of these three groups of genes was performed by clusterProfiler (v.4.4.1) (65).
Detection of tissue-specific regulatory elements
The tissue-specific regulatory elements (TSRs) for all chromatin states (except for Qui) were identified using the previous pipeline (8). Briefly, if a RRAT presents in a tissue, then it is given as “1” for this tissue, otherwise “0.” A total of 26 categories of tissue-specific regulatory elements were detected, including all-common (presented in all tissues), gut-common (presented in all five intestinal tissues), brain-common (presented in all three brain tissues), and single tissue-specific. The GO enrichment analyses of target genes of TSRs were performed using clusterProfiler (v.4.4.1) (65). The HOMER (66) (v.4.11) was used to identify motifs significantly (FDR < 0.05) enriched with tissue-specific EnhAs. The top three enriched motifs with tissue-relevant function were selected as the candidate motifs in each tissue. The expression level of corresponding TFs was obtained from RNA-seq of the same samples.
Identification of super-enhancers
The five kinds of enhancers (i.e., EnhA, EnhAMe, EnhAWk, EnhAHet, and EnhPois) in each tissue have been combined in a gff file. ROSE v1.3.1 (67, 68) was then used to identify super-enhancers with default parameters based on the H3K27ac signals of each sample in each tissue. Super-enhancers were then combined among samples within the same tissue. A total of 7662 nonredundant of super-enhancers were detected after combining and merging (at least an overlap of 10,000 bp) them across all tissues. Tissue-specific super-enhancers were then detected using the same approach as above for tissue-specific regulatory elements. The clustering of super-enhancers was conducted using k-means (n = 10) in ComplexHeatmap (v.2.9.3) (69). The GO enrichment analysis of genes with super-enhancers was performed using clusterProfiler (v.4.4.1) (65). The significant motifs of super-enhancers were identified by HOMER (66) (v.4.11) with FDR < 0.05.
Causal variants of monogenic traits in chromatin states
We downloaded 56 causal or potential causal mutations of chicken Mendelian traits/disorders in the OMIA database (https://omia.org/home/). After filtering the mutations without chromosome position information or located in coding region, we only analyzed eight noncoding causal variants (table S8). We identified whether a causal mutation is overlapped with regulatory elements using the intersect function in BEDTools (v.2.29.2) (61).
GWAS signal enrichment of chromatin states
The GWAS summary statistics of 44 complex traits was obtained from two distinct chicken populations. The first one is an intercross population derived from two divergent chicken lines (a native Chinese breed and a broiler line) (38), including 33 complex traits with different sample size (table S9). These traits included body weight, growth rate, organ weight, feed intake, efficiency, and so on (table S9). All samples were resequenced on MGISEQ-2000 platform to approximately 0.8× depth. The BaseVar algorithm was used to identify polymorphic sites and infer allele frequencies and STITCH to impute genotypes (7.9M SNPs) (70). The linear mixed model, implemented in the fastGWA package (71), was used to perform the GWAS based on common SNPs with minor allele frequency (MAF) > 0.05. The second population with 1512 F2 hens, derived from a White Leghorn and Dongxiang reciprocal cross (39), included 11 egg production traits. The birds were genotyped using the chicken 600K SNP genotyping array and then imputed to sequence level (4.8M SNPs) using Beagle (version 5.2). GWAS for these traits was performed with the same approach as above. The GWAS signal enrichment of 44 complex traits for each chromatin state across 23 tissues was conducted using a genotype cyclical permutation test (10,000 times) (72).
Selection signature enrichment analysis of chromatin states
The selection signatures between domesticated chickens and wild ones detected by locus-specific branch length (LSBL) using two outgroups (G. g. murghi or G. g. jabouillei) were obtained from a previous study (44). The selection signatures between layers and broilers were detected using the same method based on a public dataset, including whole-genome sequence data from 35 red jungle fowl (RJF), 40 broilers, and 50 layers (73). The trimmomatic-0.39 with parameters “LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36” was used to filter out low-quality reads and bases. Clean reads were mapped to the chicken reference genome (Galgal6) using BWA-MEM with default settings except for the “-t 8 -M” options (52). The mapped bam files were sorted using SortSam, and the duplicates of genome were marked using MarkDuplicates in Picard Tools (v.1.56) (http://picard.sourceforge.net). Reads mapped around InDels were realigned using RealignerTargetCreator and IndelRealigner tools from the Genome Analysis Toolkit (GenomeAnalysisTK-3.7.0, GATK). The UnifiedGenotyper function in GATK -3.7.0 with default parameter was used to call variants, and then VariantFiltration with options “QUAL < 40.0 MQ < 25.0 MQ0 >= 4 && ((MQ0/(1.0*DP)) > 0.1)” was used to filter out low-quality variants (44). The variants with more than two alleles were discarded. The LSBL statistics was used to estimate differentiation of each SNP in either Broiler or Layer with RJF as outgroup. The sliding windows analysis with 20 adjacent SNPs was applied, and the top 0.1% windows with the largest LSBL statistics were retrieved, which were used to perform the enrichment analysis of TSR. LSBL was calculated on the basis of the FST value among the three groups using the formula: LSBL = [FST(AB) + FST(AC) − FST(BC)]/2 (74). The fold enrichment of selection signatures for chromatin states was performed by the Genomic Association Test (GAT) (75). We used GAT to estimate the significance (P value) based on 10,000 simulations of the regions in all chicken chromosomes. Multiple testing (FDR) were performed by P adjusted based on the “BH” method in R. Genes associated with the enrichment were extracted from enhancer-gene pairs. The LD (r2)between target SNPs with surrounding variants was calculated using plink (v1.9) (76) with options: --ld-window-kb 1000 --ld-window 99999 --ld-snp targets.snp --r2 --ld-window-r2 0.
Acknowledgments
Funding: This study was supported by Agriculture and Food Research Initiative Competitive grants nos. 2020-67015-31175 and 2015-67015-22940 (H.Z.) from the USDA National Institute of Food and Agriculture, Multistate Research Project NRSP8 and NC1170 (H.Z.), and the California Agricultural Experimental Station (H.Z.) and partial support from National Natural Science Foundation of China (NSFC 31961133003 and 31902143).
Author contributions: H.Z., L.F., and Z.P. conceived and designed the study. Yi.W. and H.H.C. were responsible for sample collection. Yi.W., Z.P., L.A., Y.B., and M.M.H. performed ChIP-seq, ATAC-seq, and RNA-seq. N.T. and K.W. contributed to RRBS data collection. Z.P., S.G., J.D., S.W., and M.S. conducted bioinformatic analysis. Yu.W., X.Z., C.Z., C.S., N.Y., and X.H. were responsible for chicken GWAS data collection and analysis. M.W. and Z.P. were responsible for chicken selection signature analysis. Z.P., Yi.W., L.F., M.W., and H.Z. wrote the initial draft of the manuscript. M.M.H., N.Y., and X.H. revised the manuscript. All coauthors contributed to the final manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All the high-throughput sequencing data from 15 tissues generated in this study were deposited in European Nucleotide Archive (ENA) with accession number PRJEB53920 (www.ebi.ac.uk/ena/browser/view/PRJEB37735). High-throughput sequencing data of eight tissues in the FANNG pilot project are available the Gene Expression Omnibus (GEO) under accession number GSE158430 (www.ebi.ac.uk/ena/browser/view/PRJEB14330). All raw data are also available through the FAANG portal (https://data.faang.org/dataset). All processed data are publicly available at https://farm.cse.ucdavis.edu/~zhypan/Chicken_FAANG/ and https://doi.org/10.6084/m9.figshare.13480425. Chromatin states of chicken are available through the UCSC Genome Browser: http://genome.ucsc.edu/s/zhypan/galGal6_FAANG_V1. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The pipeline for RNA-seq, ATAC-seq, DNase-seq, and ChIP-seq processing is available at GitHub (https://github.com/kernco/functional-annotation) and Zenodo (https://doi.org/10.5281/zenodo.4540293) (77). The RRBS pipeline and other processing codes are publicly available at GitHub (https://github.com/zhypan/Functional-Annotation-of-Pig and https://github.com/zhypan/FAANG_chicken) and Zenodo (https://doi.org/10.5281/zenodo.5338812 and https://doi.org/10.5281/zenodo.6609263) (78, 79).
Supplementary Materials
This PDF file includes:
Figs. S1 to S16
Legends for tables S1 to S12
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S12
REFERENCES AND NOTES
- 1.D. W. Burt, Chicken genome: Current status and future opportunities. Genome Res. 15, 1692–1698 (2005). [DOI] [PubMed] [Google Scholar]
- 2.T. H. Beacon, J. R. Davie, The chicken model organism for epigenomic research. Genome 64, 476–489 (2021). [DOI] [PubMed] [Google Scholar]
- 3.H. Giral, U. Landmesser, A. Kratzer, Into the wild: GWAS exploration of non-coding RNAs. Front. Cardiovasc. Med. 5, 181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.C. M. Carnielli, F. V. Winck, A. F. Paes Leme, Functional annotation and biological interpretation of proteomics data. Biochim. Biophys. Acta 1854, 46–54 (2015). [DOI] [PubMed] [Google Scholar]
- 5.E. Cano-Gamez, G. Trynka, From GWAS to function: Using functional genomics to identify the mechanisms underlying complex diseases. Front. Genet. 11, 424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.C. Márquez-Luna, S. Gazal, P.-R. Loh, S. S. Kim, N. Furlotte, A. Auton; 23andMe Research Team, A. L. Price, Incorporating functional priors improves polygenic prediction accuracy in UK Biobank and 23andMe data sets. Nat. Commun. 12, 6052 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D. U. Gorkin, I. Barozzi, Y. Zhao, Y. Zhang, H. Huang, A. Y. Lee, B. Li, J. Chiou, A. Wildberg, B. Ding, B. Zhang, M. Wang, J. S. Strattan, J. M. Davidson, Y. Qiu, V. Afzal, J. A. Akiyama, I. Plajzer-Frick, C. S. Novak, M. Kato, T. H. Garvin, Q. T. Pham, A. N. Harrington, B. J. Mannion, E. A. Lee, Y. Fukuda-Yuzawa, Y. He, S. Preissl, S. Chee, J. Y. Han, B. A. Williams, D. Trout, H. Amrhein, H. Yang, J. M. Cherry, W. Wang, K. Gaulton, J. R. Ecker, Y. Shen, D. E. Dickel, A. Visel, L. A. Pennacchio, B. Ren, An atlas of dynamic chromatin landscapes in mouse fetal development. Nature 583, 744–751 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Z. Pan, Y. Yao, H. Yin, Z. Cai, Y. Wang, L. Bai, C. Kern, M. Halstead, G. Chanthavixay, N. Trakooljul, K. Wimmers, G. Sahana, G. Su, M. S. Lund, M. Fredholm, P. Karlskov-Mortensen, C. W. Ernst, P. Ross, C. K. Tuggle, L. Fang, H. Zhou, Pig genome functional annotation enhances the biological interpretation of complex traits and human disease. Nat. Commun. 12, 5848 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.L. Fang, S. Liu, M. Liu, X. Kang, S. Lin, B. Li, E. E. Connor, R. L. Baldwin VI, A. Tenesa, L. Ma, G. E. Liu, C. J. Li, Functional annotation of the cattle genome through systematic discovery and characterization of chromatin states and butyrate-induced variations. BMC Biol. 17, 68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.S. Foissac, S. Djebali, K. Munyard, N. Vialaneix, A. Rau, K. Muret, D. Esquerré, M. Zytnicki, T. Derrien, P. Bardou, F. Blanc, C. Cabau, E. Crisci, S. Dhorne-Pollet, F. Drouet, T. Faraut, I. Gonzalez, A. Goubil, S. Lacroix-Lamandé, F. Laurent, S. Marthey, M. Marti-Marimon, R. Momal-Leisenring, F. Mompart, P. Quéré, D. Robelin, M. S. Cristobal, G. Tosser-Klopp, S. Vincent-Naulleau, S. Fabre, M. H. Pinard-van der Laan, C. Klopp, M. Tixier-Boichard, H. Acloque, S. Lagarrigue, E. Giuffra, Multi-species annotation of transcriptome and chromatin structure in domesticated animals. BMC Biol. 17, 108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.S. Liu, Y. Yu, S. Zhang, J. B. Cole, A. Tenesa, T. Wang, T. G. McDaneld, L. Ma, G. E. Liu, L. Fang, Epigenomics and genotype-phenotype association analyses reveal conserved genetic architecture of complex traits in cattle and human. BMC Biol. 18, 80 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.C. Kern, Y. Wang, X. Xu, Z. Pan, M. Halstead, G. Chanthavixay, P. Saelao, S. Waters, R. Xiang, A. Chamberlain, I. Korf, M. E. Delany, H. H. Cheng, J. F. Medrano, A. L. van Eenennaam, C. K. Tuggle, C. Ernst, P. Flicek, G. Quon, P. Ross, H. Zhou, Functional annotations of three domestic animal genomes provide vital resources for comparative and agricultural research. Nat. Commun. 12, 1821 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.N. Couto, Z. M. al-Majdoub, S. Gibson, P. J. Davies, B. Achour, M. D. Harwood, G. Carlson, J. Barber, A. Rostami-Hodjegan, G. Warhurst, Quantitative proteomics of clinically relevant drug-metabolizing enzymes and drug transporters and their intercorrelations in the human small intestine. Drug Metab. Dispos. 48, 245–254 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.N. C. Panarelli, R. K. Yantiss, M. M. Yeh, Y. Liu, Y.-T. Chen, Tissue-specific cadherin CDH17 is a useful marker of gastrointestinal adenocarcinomas with higher sensitivity than CDX2. Am. J. Clin. Pathol. 138, 211–222 (2012). [DOI] [PubMed] [Google Scholar]
- 15.J. Ernst, M. Kellis, ChromHMM: Automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.M. Farré, J. Kim, A. A. Proskuryakova, Y. Zhang, A. I. Kulemzina, Q. Li, Y. Zhou, Y. Xiong, J. L. Johnson, P. Perelman, W. E. Johnson, W. C. Warren, A. V. Kukekova, G. Zhang, S. J. O'Brien, O. A. Ryder, A. S. Graphodatsky, J. Ma, H. A. Lewin, D. M. Larkin, Evolution of gene regulation in ruminants differs between evolutionary breakpoint regions and homologous synteny blocks. Genome Res. 29, 576–589 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A. Visel, S. Minovitsky, I. Dubchak, L. A. Pennacchio, VISTA Enhancer Browser—A database of tissue-specific human enhancers. Nucleic Acids Res. 35, D88–D92 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roadmap Epigenomics Consortium, A. Kundaje, W. Meuleman, J. Ernst, M. Bilenky, A. Yen, A. Heravi-Moussavi, P. Kheradpour, Z. Zhang, J. Wang, M. J. Ziller, V. Amin, J. W. Whitaker, M. D. Schultz, L. D. Ward, A. Sarkar, G. Quon, R. S. Sandstrom, M. L. Eaton, Y.-C. Wu, A. R. Pfenning, X. Wang, M. Claussnitzer, Y. Liu, C. Coarfa, R. A. Harris, N. Shoresh, C. B. Epstein, E. Gjoneska, D. Leung, W. Xie, R. D. Hawkins, R. Lister, C. Hong, P. Gascard, A. J. Mungall, R. Moore, E. Chuah, A. Tam, T. K. Canfield, R. S. Hansen, R. Kaul, P. J. Sabo, M. S. Bansal, A. Carles, J. R. Dixon, K.-H. Farh, S. Feizi, R. Karlic, A.-R. Kim, A. Kulkarni, D. Li, R. Lowdon, G. N. Elliott, T. R. Mercer, S. J. Neph, V. Onuchic, P. Polak, N. Rajagopal, P. Ray, R. C. Sallari, K. T. Siebenthall, N. A. Sinnott-Armstrong, M. Stevens, R. E. Thurman, J. Wu, B. Zhang, X. Zhou, A. E. Beaudet, L. A. Boyer, P. L. De Jager, P. J. Farnham, S. J. Fisher, D. Haussler, S. J. M. Jones, W. Li, M. A. Marra, M. T. McManus, S. Sunyaev, J. A. Thomson, T. D. Tlsty, L.-H. Tsai, W. Wang, R. A. Waterland, M. Q. Zhang, L. H. Chadwick, B. E. Bernstein, J. F. Costello, J. R. Ecker, M. Hirst, A. Meissner, A. Milosavljevic, B. Ren, J. A. Stamatoyannopoulos, T. Wang, M. Kellis, Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R. Valdez, C. A. Cavinder, D. D. Varner, T. H. Welsh Jr., M. M. Vogelsang, N. H. Ing, Dexamethasone downregulates expression of several genes encoding orphan nuclear receptors that are important to steroidogenesis in stallion testes. J. Biochem. Mol. Toxicol. 33, e22309 (2019). [DOI] [PubMed] [Google Scholar]
- 20.A. Bobbs, K. Gellerman, W. M. Hallas, S. Joseph, C. Yang, J. Kurkewich, K. D. Cowden Dahl, ARID3B directly regulates ovarian cancer promoting genes. PLOS ONE 10, e0131961 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.H. Wang, S. Jain, P. Li, J. X. Lin, J. Oh, C. Qi, Y. Gao, J. Sun, T. Sakai, Z. Naghashfar, S. Abbasi, A. L. Kovalchuk, S. Bolland, S. L. Nutt, W. J. Leonard, H. C. Morse III, Transcription factors IRF8 and PU. 1 are required for follicular B cell development and BCL6-driven germinal center responses. Proc. Natl. Acad. Sci. U.S.A. 116, 9511–9520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D. Hnisz, B. J. Abraham, T. I. Lee, A. Lau, V. Saint-André, A. A. Sigova, H. A. Hoke, R. A. Young, Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.X. Wang, M. J. Cairns, J. Yan, Super-enhancers in transcriptional regulation and genome organization. Nucleic Acids Res. 47, 11481–11496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.T. Van Groningen, J. Koster, L. J. Valentijn, D. A. Zwijnenburg, N. Akogul, N. E. Hasselt, M. Broekmans, F. Haneveld, N. E. Nowakowska, J. Bras, C. J. M. van Noesel, A. Jongejan, A. H. van Kampen, L. Koster, F. Baas, L. van Dijk-Kerkhoven, M. Huizer-Smit, M. C. Lecca, A. Chan, A. Lakeman, P. Molenaar, R. Volckmann, E. M. Westerhout, M. Hamdi, P. G. van Sluis, M. E. Ebus, J. J. Molenaar, G. A. Tytgat, B. A. Westerman, J. van Nes, R. Versteeg, Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet. 49, 1261–1266 (2017). [DOI] [PubMed] [Google Scholar]
- 25.A. L. Hunter, T. M. Poolman, D. Kim, F. J. Gonzalez, D. A. Bechtold, A. S. I. Loudon, M. Iqbal, D. W. Ray, HNF4A modulates glucocorticoid action in the liver. Cell Rep. 39, 110697 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R. Valverde, L. Edwards, L. Regan, Structure and function of KH domains. FEBS J. 275, 2712–2726 (2008). [DOI] [PubMed] [Google Scholar]
- 27.J. Rha, S. K. Jones, J. Fidler, A. Banerjee, S. W. Leung, K. J. Morris, J. C. Wong, G. A. S. Inglis, L. Shapiro, Q. Deng, A. A. Cutler, A. M. Hanif, M. T. Pardue, A. Schaffer, N. T. Seyfried, K. H. Moberg, G. J. Bassell, A. Escayg, P. S. García, A. H. Corbett, The RNA-binding protein, ZC3H14, is required for proper poly (A) tail length control, expression of synaptic proteins, and brain function in mice. Hum. Mol. Genet. 26, 3663–3681 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.J. Schödel, B. Klanke, A. Weidemann, B. Buchholz, W. Bernhardt, M. Bertog, K. Amann, C. Korbmacher, M. Wiesener, C. Warnecke, A. Kurtz, K. U. Eckardt, C. Willam, HIF-prolyl hydroxylases in the rat kidney: Physiologic expression patterns and regulation in acute kidney injury. Am. J. Pathol. 174, 1663–1674 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.M. Yang, Q. Liu, M. Dai, R. Peng, X. Li, W. Zuo, J. Gou, F. Zhou, S. Yu, H. Liu, M. Huang, FOXQ1-mediated SIRT1 upregulation enhances stemness and radio-resistance of colorectal cancer cells and restores intestinal microbiota function by promoting β-catenin nuclear translocation. J. Exp. Clin. Cancer Res. 41, 70 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.T. Ohtsuki, M. Koga, H. Ishiguro, Y. Horiuchi, M. Arai, K. Niizato, M. Itokawa, T. Inada, N. Iwata, S. Iritani, N. Ozaki, H. Kunugi, H. Ujike, Y. Watanabe, T. Someya, T. Arinami, A polymorphism of the metabotropic glutamate receptor mGluR7 (GRM7) gene is associated with schizophrenia. Schizophr. Res. 101, 9–16 (2008). [DOI] [PubMed] [Google Scholar]
- 31.L. A. Lettice, S. J. Heaney, L. A. Purdie, L. Li, P. de Beer, B. A. Oostra, D. Goode, G. Elgar, R. E. Hill, E. de Graaff, A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12, 1725–1735 (2003). [DOI] [PubMed] [Google Scholar]
- 32.J. Sharpe, L. Lettice, J. Hecksher-Sørensen, M. Fox, R. Hill, R. Krumlauf, Identification of sonic hedgehog as a candidate gene responsible for the polydactylous mouse mutant Sasquatch. Curr. Biol. 9, 97–100 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Q. Chu, Z. Yan, J. Zhang, T. Usman, Y. Zhang, H. Liu, H. Wang, A. Geng, H. Liu, Association of SNP rs80659072 in the ZRS with polydactyly in Beijing You chickens. PLOS ONE 12, e0185953 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.T. Zhu, M. Liu, S. Peng, X. Zhang, Y. Chen, X. Lv, W. Yang, K. Li, J. Zhang, H. Wang, H. Li, Z. Ning, L. Wang, L. Qu, A deletion upstream of SOX10 causes light yellow plumage colour in chicken. Genes 13, 327 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.U. Gunnarsson, S. Kerje, B. Bed’hom, A. S. Sahlqvist, O. Ekwall, M. Tixier-Boichard, O. Kämpe, L. Andersson, The Dark brown plumage color in chickens is caused by an 8.3-kb deletion upstream of SOX10. Pigment Cell Melanoma Res. 24, 268–274 (2011). [DOI] [PubMed] [Google Scholar]
- 36.C. Feng, Y. Gao, B. Dorshorst, C. Song, X. Gu, Q. Li, J. Li, T. Liu, C. J. Rubin, Y. Zhao, Y. Wang, J. Fei, H. Li, K. Chen, H. Qu, D. Shu, C. Ashwell, Y. da, L. Andersson, X. Hu, N. Li, A cis-regulatory mutation of PDSS2 causes silky-feather in chickens. PLOS Genet. 10, e1004576 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.C. A. Boix, B. T. James, Y. P. Park, W. Meuleman, M. Kellis, Regulatory genomic circuitry of human disease loci by integrative epigenomics. Nature 590, 300–307 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Y. Wang, X. Cao, C. Luo, Z. Sheng, C. Zhang, C. Bian, C. Feng, J. Li, F. Gao, Y. Zhao, Z. Jiang, H. Qu, D. Shu, Ö. Carlborg, X. Hu, N. Li, Multiple ancestral haplotypes harboring regulatory mutations cumulatively contribute to a QTL affecting chicken growth traits. Commun. Biol. 3, 472 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J. Yuan, K. Wang, G. Yi, M. Ma, T. Dou, C. Sun, L. J. Qu, M. Shen, L. Qu, N. Yang, Genome-wide association studies for feed intake and efficiency in two laying periods of chickens. Genet. Sel. Evol. 47, 82 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.H. K. Finucane, B. Bulik-Sullivan, A. Gusev, G. Trynka, Y. Reshef, P.-R. Loh, V. Anttila, H. Xu, C. Zang, K. Farh, S. Ripke, F. R. Day; ReproGen Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; RACI Consortium, S. Purcell, E. Stahl, S. Lindstrom, J. R. B. Perry, Y. Okada, S. Raychaudhuri, M. J. Daly, N. Patterson, B. M. Neale, A. L. Price, Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D. Murawska, The effect of age on growth performance and carcass quality parameters in different poultry species. Poult. Sci., 33–50 (2017). [Google Scholar]
- 42.G. Wang, W. K. Kim, M. A. Cline, E. R. Gilbert, Factors affecting adipose tissue development in chickens: A review. Poult. Sci. 96, 3687–3699 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Y. G. Tan, X. L. Xu, H. Y. Cao, W. Zhou, Z. Z. Yin, Effect of age at first egg on reproduction performance and characterization of the hypothalamo-pituitary-gonadal axis in chickens. Poult. Sci. 100, 101325 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.M. S. Wang, M. Thakur, M. S. Peng, Y. Jiang, L. A. F. Frantz, M. Li, J. J. Zhang, S. Wang, J. Peters, N. O. Otecko, C. Suwannapoom, X. Guo, Z. Q. Zheng, A. Esmailizadeh, N. Y. Hirimuthugoda, H. Ashari, S. Suladari, M. S. A. Zein, S. Kusza, S. Sohrabi, H. Kharrati-Koopaee, Q. K. Shen, L. Zeng, M. M. Yang, Y. J. Wu, X. Y. Yang, X. M. Lu, X. Z. Jia, Q. H. Nie, S. J. Lamont, E. Lasagna, S. Ceccobelli, H. G. T. N. Gunwardana, T. M. Senasige, S. H. Feng, J. F. Si, H. Zhang, J. Q. Jin, M. L. Li, Y. H. Liu, H. M. Chen, C. Ma, S. S. Dai, A. K. F. H. Bhuiyan, M. S. Khan, G. L. L. P. Silva, T. T. le, O. A. Mwai, M. N. M. Ibrahim, M. Supple, B. Shapiro, O. Hanotte, G. Zhang, G. Larson, J. L. Han, D. D. Wu, Y. P. Zhang, 863 genomes reveal the origin and domestication of chicken. Cell Res. 30, 693–701 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.M. Tixier-Boichard, S. Fabre, S. Dhorne-Pollet, A. Goubil, H. Acloque, S. Vincent-Naulleau, P. Ross, Y. Wang, G. Chanthavixay, H. Cheng, C. Ernst, V. Leesburg, E. Giuffra, H. Zhou; Collaborative Working Group , Tissue resources for the functional annotation of animal genomes. Front. Genet. 12, 666265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.G. Chanthavixay, C. Kern, Y. Wang, P. Saelao, S. J. Lamont, R. A. Gallardo, G. Rincon, H. Zhou, Integrated transcriptome and histone modification analysis reveals NDV infection under heat stress affects bursa development and proliferation in susceptible chicken line. Front. Genet. 11, 567812 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.M. R. Corces, A. E. Trevino, E. G. Hamilton, P. G. Greenside, N. A. Sinnott-Armstrong, S. Vesuna, A. T. Satpathy, A. J. Rubin, K. S. Montine, B. Wu, A. Kathiria, S. W. Cho, M. R. Mumbach, A. C. Carter, M. Kasowski, L. A. Orloff, V. I. Risca, A. Kundaje, P. A. Khavari, T. J. Montine, W. J. Greenleaf, H. Y. Chang, An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.M. Halstead, C. Kern, P. Saelao, G. Chanthavixay, Y. Wang, M. E. Delany, H. Zhou, P. J. Ross, Systematic alteration of ATAC-seq for profiling open chromatin in cryopreserved nuclei preparations from livestock tissues. Sci. Rep. 10, 5230 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.H. Sun, P. Liu, L. K. Nolan, S. J. Lamont, Avian pathogenic Escherichia coli (APEC) infection alters bone marrow transcriptome in chickens. BMC Genomics 16, 690 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.F. Krueger, Trim Galore!: A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files (2015); https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
- 51.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.H. Li, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 (2013).
- 53.H. Li, B. Handsaker, A. Wysoker, T. Fennell, J. Ruan, N. Homer, G. Marth, G. Abecasis, R. Durbin; 1000 Genome Project Data Processing Subgroup , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.S. Anders, P. T. Pyl, W. Huber, HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.S. Kovaka, A. V. Zimin, G. M. Pertea, R. Razaghi, S. L. Salzberg, M. Pertea, Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20, 278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Y. Zhang, T. Liu, C. A. Meyer, J. Eeckhoute, D. S. Johnson, B. E. Bernstein, C. Nusbaum, R. M. Myers, M. Brown, W. Li, X. S. Liu, Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.F. Krueger, S. R. Andrews, Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.N. C. Durand, M. S. Shamim, I. Machol, S. S. P. Rao, M. H. Huntley, E. S. Lander, E. L. Aiden, Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.F. Ramírez, D. P. Ryan, B. Grüning, V. Bhardwaj, F. Kilpert, A. S. Richter, S. Heyne, F. Dündar, T. Manke, deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.P. Virtanen, R. Gommers, T. E. Oliphant, M. Haberland, T. Reddy, D. Cournapeau, E. Burovski, P. Peterson, W. Weckesser, J. Bright, S. J. van der Walt, M. Brett, J. Wilson, K. J. Millman, N. Mayorov, A. R. J. Nelson, E. Jones, R. Kern, E. Larson, C. J. Carey, İ. Polat, Y. Feng, E. W. Moore, J. V. Plas, D. Laxalde, J. Perktold, R. Cimrman, I. Henriksen, E. A. Quintero, C. R. Harris, A. M. Archibald, A. H. Ribeiro, F. Pedregosa, P. van Mulbregt; SciPy 1.0 Contributors , SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.A. R. Quinlan, BEDTools: The Swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 11.12.1–11.12.34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.C. E. Grant, T. L. Bailey, W. S. Noble, FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.M. Oti, J. Falck, M. A. Huynen, H. Zhou, CTCF-mediated chromatin loops enclose inducible gene regulatory domains. BMC Genomics 17, 252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.I. Yanai, H. Benjamin, M. Shmoish, V. Chalifa-Caspi, M. Shklar, R. Ophir, A. Bar-Even, S. Horn-Saban, M. Safran, E. Domany, D. Lancet, O. Shmueli, Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21, 650–659 (2005). [DOI] [PubMed] [Google Scholar]
- 65.T. Wu, E. Hu, S. Xu, M. Chen, P. Guo, Z. Dai, T. Feng, L. Zhou, W. Tang, L. Zhan, X. Fu, S. Liu, X. Bo, G. Yu, clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.S. Heinz, C. Benner, N. Spann, E. Bertolino, Y. C. Lin, P. Laslo, J. X. Cheng, C. Murre, H. Singh, C. K. Glass, Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.W. A. Whyte, D. A. Orlando, D. Hnisz, B. J. Abraham, C. Y. Lin, M. H. Kagey, P. B. Rahl, T. I. Lee, R. A. Young, Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.J. Lovén, H. A. Hoke, C. Y. Lin, A. Lau, D. A. Orlando, C. R. Vakoc, J. E. Bradner, T. I. Lee, R. A. Young, Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Z. Gu, R. Eils, M. Schlesner, Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 70.R. Yang, X. Guo, D. Zhu, C. Tan, C. Bian, J. Ren, Z. Huang, Y. Zhao, G. Cai, D. Liu, Z. Wu, Y. Wang, N. Li, X. Hu, Accelerated deciphering of the genetic architecture of agricultural economic traits in pigs using a low-coverage whole-genome sequencing strategy. Gigascience 10, giab048 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.L. Jiang, Z. Zheng, T. Qi, K. E. Kemper, N. R. Wray, P. M. Visscher, J. Yang, A resource-efficient tool for mixed model association analysis of large-scale data. Nat. Genet. 51, 1749–1755 (2019). [DOI] [PubMed] [Google Scholar]
- 72.L. Fang, W. Cai, S. Liu, O. Canela-Xandri, Y. Gao, J. Jiang, K. Rawlik, B. Li, S. G. Schroeder, B. D. Rosen, C. J. Li, T. S. Sonstegard, L. J. Alexander, C. P. van Tassell, P. M. VanRaden, J. B. Cole, Y. Yu, S. Zhang, A. Tenesa, L. Ma, G. E. Liu, Comprehensive analyses of 723 transcriptomes enhance genetic and biological interpretations for complex traits in cattle. Genome Res. 30, 790–801 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.S. Qanbari, C. J. Rubin, K. Maqbool, S. Weigend, A. Weigend, J. Geibel, S. Kerje, C. Wurmser, A. T. Peterson, I. L. Brisbin, R. Preisinger, R. Fries, H. Simianer, L. Andersson, Genetics of adaptation in modern chicken. PLOS Genet. 15, e1007989 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.M. D. Shriver, G. C. Kennedy, E. J. Parra, H. A. Lawson, V. Sonpar, J. Huang, J. M. Akey, K. W. Jones, The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Hum. Genomics 1, 274–286 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.A. Heger, C. Webber, M. Goodson, C. P. Ponting, G. Lunter, GAT: A simulation framework for testing the association of genomic intervals. Bioinformatics 29, 2046–2048 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.C. C. Chang, C. C. Chow, L. C. A. M. Tellier, S. Vattikuti, S. M. Purcell, J. J. Lee, Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.C. Kern, Functional annotations of three domestic animal genomes provide vital resources for comparative and agricultural research (2021); https://github.com/kernco/functional-annotation, 10.5281/zenodo.4540293. [DOI] [PMC free article] [PubMed]
- 78.Z. Pan, Pig genome functional annotation enhances the biological interpretation of complex traits and human disease (2021); https://github.com/zhypan/Functional-Annotation-of-Pig, 10.5281/zenodo.5338812. [DOI] [PMC free article] [PubMed]
- 79.Z. Pan, An atlas of regulatory elements in 23 chicken tissues informs complex trait variation and domestication selection (2022); https://github.com/zhypan/FAANG_chicken, 10.5281/zenodo.6609263. [DOI]
- 80.B. Dorshorst, R. Okimoto, C. Ashwell, Genomic regions associated with dermal hyperpigmentation, polydactyly and other morphological traits in the Silkie chicken. J. Hered. 101, 339–350 (2010). [DOI] [PubMed] [Google Scholar]
- 81.J. Li, M. O. Lee, J. Chen, B. W. Davis, B. J. Dorshorst, P. B. Siegel, M. Inaba, T. X. Jiang, C. M. Chuong, L. Andersson, Cis-acting mutation affecting GJA5 transcription is underlying the Melanotic within-feather pigmentation pattern in chickens. Proc. Natl. Acad. Sci. U.S.A. 118, e2109363118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.G. W. Zhang, Y. Liao, W. X. Zhang, Y. Wu, A. Liu, A new dominant haplotype of MC1R gene in Chinese black plumage chicken. Anim. Genet. 48, 624 (2017). [DOI] [PubMed] [Google Scholar]
- 83.J. Li, M.-O. Lee, B. W. Davis, P. Wu, S.-M. Hsieh Li, C.-M. Chuong, L. Andersson, The crest phenotype in domestic chicken is caused by a 197 bp duplication in the intron of HOXC10. G3 11, jkaa048 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.J. Li, M. O. Lee, B. W. Davis, S. Lamichhaney, B. J. Dorshorst, P. B. Siegel, L. Andersson, Mutations upstream of the TBX5 and PITX1 transcription factor genes are associated with feathered legs in the domestic chicken. Mol. Biol. Evol. 37, 2477–2486 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.C. Bortoluzzi, H. J. Megens, M. Bosse, M. F. L. Derks, B. Dibbits, K. Laport, S. Weigend, M. A. M. Groenen, R. P. M. A. Crooijmans, Parallel genetic origin of foot feathering in birds. Mol. Biol. Evol. 37, 2465–2476 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S16
Legends for tables S1 to S12
References
Tables S1 to S12