Abstract
Introduction:
Placental parenchymal lesions are commonly encountered and carry significant clinical associations. However, they are frequently missed or misclassified by general practice pathologists. Interpretation of pathology slides has emerged as one of the most successful applications of machine learning (ML) in medicine with applications ranging from cancer detection and prognostication to transplant medicine. The goal of this study was to use a whole-slide learning model to identify and classify placental parenchymal lesions including villous infarctions, intervillous thrombi (IVT), and perivillous fibrin deposition (PVFD).
Methods:
We generated whole slide images from placental discs examined at our institution with infarct, IVT, PVFD, or no macroscopic lesion. Slides were analyzed as a set of overlapping patches. We extracted feature vectors from each patch using a pretrained convolutional neural network (EfficientNetV2L). We trained a model to assign attention to each vector and used the attentions as weights to produce a pooled feature vector. The pooled vector was classified as normal or 1 of 3 lesions using a fully connected network. Patch attention was plotted to highlight informative areas of the slide.
Results:
Overall balanced accuracy in a test set of held-out slides was 0.86 with receiver-operator characteristic areas under the curve of 0.917 to 0.993. Cases of PVFD were frequently miscalled as normal or infarcts, the latter possibly due to the perivillous fibrin found at the periphery of infarctions. We used attention maps to further understand some errors, including one most likely due to poor tissue fixation and processing.
Discussion:
We used a whole-slide learning paradigm to train models to recognize three of the most common placental parenchymal lesions. We used attention maps to gain insight into model function, which differed from intuitive explanations.
Keywords: placenta pathology, machine learning, infarction, intervillous thrombus, perivillous fibrin, artificial intelligence
INTRODUCTION
The placenta
The placenta is the temporary organ formed during gestation that performs nutritional, respiratory, excretory, endocrine, and immune-regulatory functions throughout fetal development.[1–3] The normal placental disc is comprised primarily of terminal villi, which contain fetal capillaries and act as the site of uptake of oxygen and nutrients and excretion of carbon dioxide and waste.[4]
Placental parenchymal lesions
Vascular and/or inflammatory pathologic conditions can affect placental function and compromise pregnancy, and identification of these lesions histologically can be important in assessment of placental function. Several lesions have been identified that form in the villous portion of the placenta. We have chosen to focuses on a subset of macroscopic lesions that cause localized injury or replacement of villi.[5,6] Specifically, we focus on villous infarctions, intervillous thrombi, and perivillous fibrin deposition.
Villous infarctions, (hereafter infarcts), are areas where there is a loss of maternal blood flow with subsequent ischemic necrosis of the villi. They are thus analogous to ischemic infarcts seen at other sites in the body including the myocardium, kidney, and central nervous system. Infarcts are seen in ~8% of placentas examined at our institution. They are associated with hypertension in pregnancy, preeclampsia, growth restriction, stillbirth, and risk of cerebral palsy.[7–11] Microscopically, infarctions are characterized by loss of definition in the villi and collapse of the intervillous space or replacement by loose blue-grey necrotic material (Figure 1).
Figure 1. Placental anatomy and parenchymal lesions:
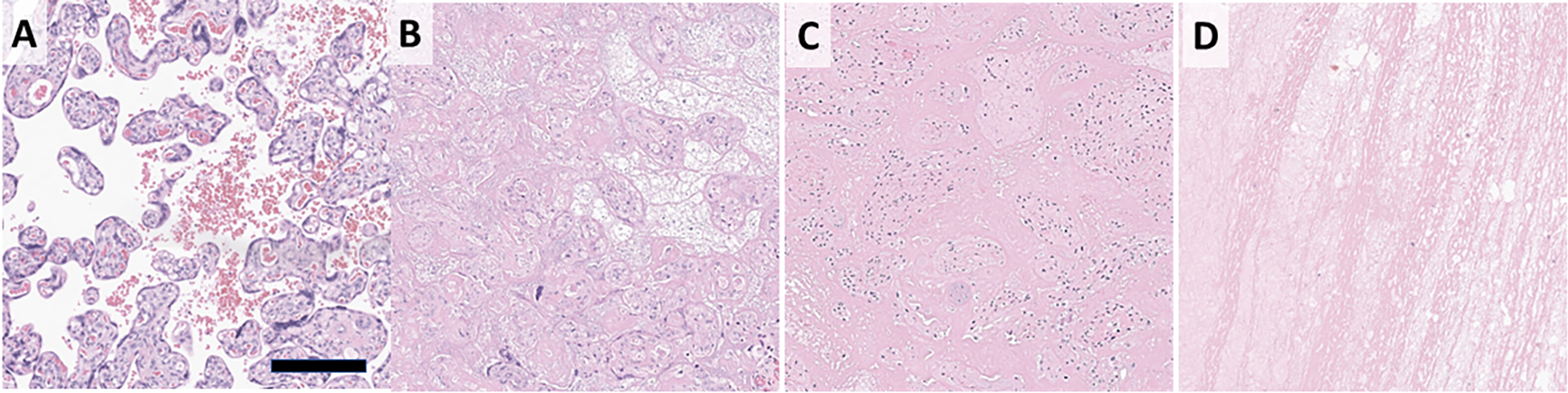
Normal placental histology showing villi are bathed in maternal blood (A). Infarctions are due to loss of maternal blood flow and characterized by indistinct villi that are either back-to-back or with loose material between (B). PVFD shows loss of syncytiotrophoblast layer from the villous surface with replacement of the intervillous space by dense pink fibrin (C). IVT displace villi and show characteristic lines of Zahn (D). Scale bar 200 μm.
Intervillous thrombi (IVT), are foci of clotted blood in the intervillous space. In contrast to other macroscopic lesions, IVT show no clear association with adverse pregnancy or neonatal outcomes.[12–14] However, they are frequently encountered it is of value to distinguish them when there is high interobserver variability or in low resource settings. Microscopic examination of IVT show areas with few to no villi containing alternating areas of fibrin and red cells – the Lines of Zahn (Figure 1).
Perivillous fibrin deposition (PVFD) can present in several forms, of which two are considered here. Massive perivillous fibrin deposition (MPVFD) involves at least 30% of the placental parenchyma with significant compromise of placental function and associated fetal growth restriction and stillbirth.[15–17] Conversely, focal PVFD forms foci up to a few cm in greatest dimension involving a small proportion of the placenta. Pathogenesis of focal PVFD is unclear but they may represent a reparative response following trophoblast injury due to turbulent flow.[18,19] Focal PVFD may also be seen as a feature of chronic inflammatory pathology.[20] Despite these striking clinical differences, MPVFD and focal PVFD appear similar microscopically, and may represent ends of a spectrum with “borderline” MPVFD between.[16] The intervillous space is filled with dense fibrin, while terminal villi lose their outer syncytiotrophoblast layer (Figure 1).
While these lesions are clinically significant, there is high interobserver variability in their diagnosis, limiting their utility in broader clinical practice. Literature is sparse, but in examples given to community pathologists, misdiagnosis rates of 68.4% for MPVFD, 57.1% for IVT, and 32.4% for infarction were reported, with most of the errors being underdiagnosis.[21] This underlines the sparseness of perinatal pathology expertise and the potential utility of diagnostic aids.
Machine learning (ML) and digital pathology
Interpretation of pathology slides has emerged as one of the most successful applications of AI/ML in medicine with applications ranging from cancer detection and prognostication to transplant medicine.[22–26] In the realm of placental pathology, accurate diagnosis of macroscopic lesions requires expert microscopic examination.[18,27] Machine learning approaches promise to reduce interobserver variation and make expert-level examination available in general practice and in low and middle income countries.[28–30]
Digital pathology challenges machine learning practitioners due to the large size of scanned whole slide images (WSI). While typical image classification software uses small images as input (e.g. 256 × 256 pixels), WSI are routinely 120,000 × 80,000 pixels.[31,32] In this task, models and humans are confronted with a similar problem – identifying which parts of the slide diagnostically relevant. To address this, we employ multiple instance learning, a domain of machine learning where models are given several related datapoints and asked to infer a single label.[33] MIL has demonstrated remarkable success in identifying cancers of unknown primary, and cardiac allograft rejection and is a component of some clinically used systems.[22,34–38]
From slide to diagnosis, whole slide learning in practice
A notional illustration of whole-slide classification is given in Figure 2. A hematoxylin and eosin (H&E) stained slide is examined. The slide contains an area of infarct (red outlined squares) with the remainder normal placental tissue (black squares). The tissue is divided into patches – smaller 256 × 256 pixel images that are typical of modern ML. The patches are fed to a fixed feature extraction subnetwork, EfficientNetV2L.[39] This subnetwork has been pre-trained to parse images into common patterns like edges, simple shapes, and textures.[39,40] This results in a feature vector, a list of numbers indicating how much the image represents each pattern. To identify salient patches, the feature vectors are fed into an attention subnetwork. In a properly functioning model, feature vectors from the infarct (red-shaded) are assigned a high attention while those of normal tissue (grayscale) are assigned a low value. An average feature vector, weighted by the attention values, is generated (weighted features). In a well-functioning model this will largely reflect the lesion. The weighted feature vector is fed into a classifier subnetwork that generates a value between 0 and 1, with values >0.5 classified having infarction. During training, the parameters of the attention subnetwork and classification subnetwork are tuned to maximize accuracy in a set of training slides. The feature extraction subnetwork is fixed. By tapping into the attention subnetwork, the attention value assigned to each feature vector and therefore each patch can be visualized as an attention map, with high attention (yellow) given to the diagnostic areas and low attention (blue) to normal placental tissue. For this work, four attention subnetworks and four classifications are generated, one each for normal, infarct, IVT, and PVFD.
Figure 2. Whole-slide learning method - diagram:
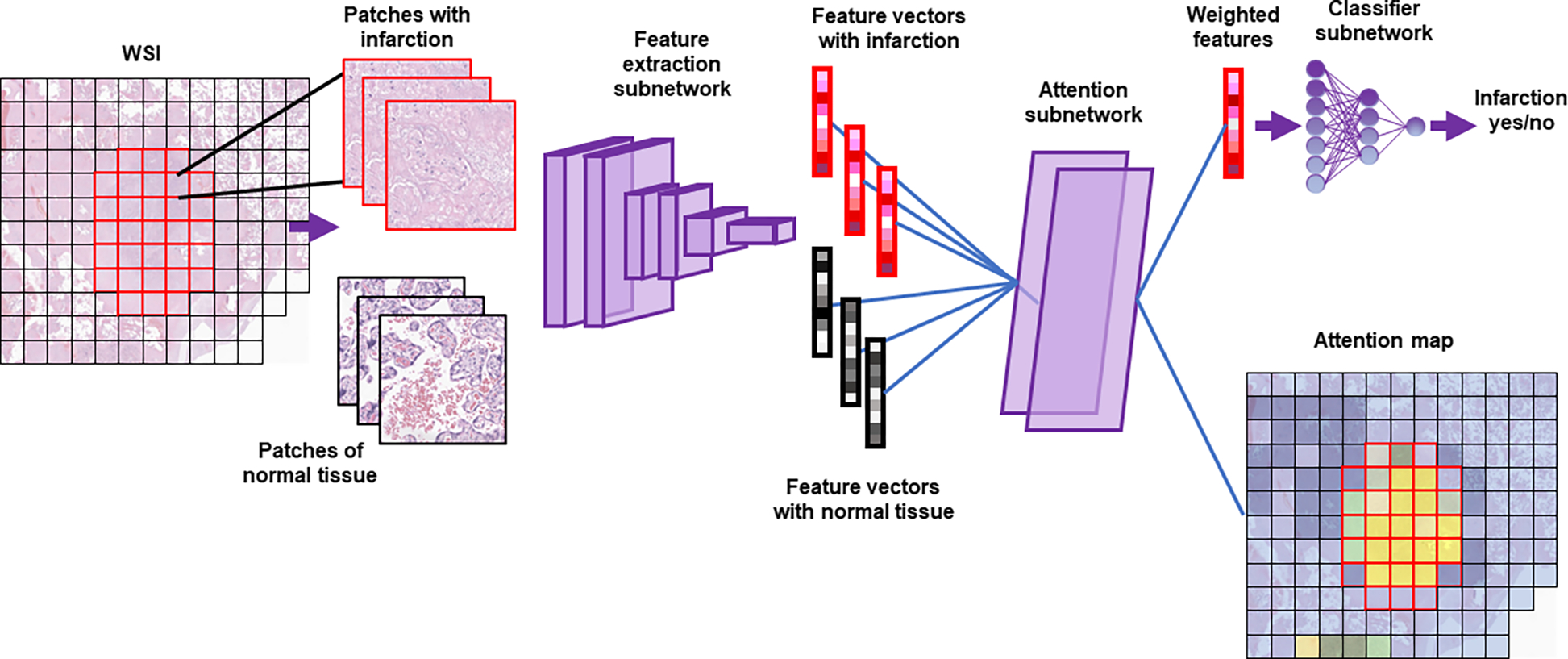
A whole slide image (WSI) is divided into a set of smaller images (patches). Images pass through a feature extraction network to become feature vectors. The attention subnetwork generates an attention value for each vector and uses that value to produce a weighted average (weighted features). The classifier subnetwork then generates a single label for the whole slide. Attention values for each patch are plotted on the attention map (blue-yellow: low-high attention). Note that a separate attention subnetwork and classifier subnetwork exist for each diagnosis, though only that for infarction is shown.
The goal of this study is to use whole slide learning to classify infarcts, IVT, and PVFD. We have selected these lesions because they are commonly encountered in pathology practice. Additionally, diagnosis of these lesions involves integrating information across large regions of the slide drawing on the strength of the whole slide learning method. Finally, to the best of our knowledge, models for classifying these lesions have not been previously reported.
METHODS
Patients, diagnoses
We identified patients that underwent delivery and placental examination at our institution between 2011 and 2022 and had slides available for scanning. Our scanning program is ongoing and the material scanned represents a mixture of prospectively scanned slides and targeted archival retrieval, e.g. for MPVFD cases. Slides were digitized on a Leica GT450 scanner with a 40x objective magnification (0.263 microns per pixel). Corresponding placental pathology reports were extracted from the institutional electronic data warehouse (EDW). Natural language processing was used to parse reports.[41] Placentas with retroplacental hemorrhage / hematoma or chorangioma or with multiple classes of lesions were excluded. Placentas were classified as having infarct, IVT, PVFD, or none of these (normal). PVFD cases included MPVFD and focal PVFD, which was defined as a macroscopic placental lesion characterized by encasement of villi in perivillous fibrin. Both lesions are distinguished from the normal degree of perivillous fibrin by the formation of macroscopic lesions.[42]
Patient and diagnosis data was stored in REDcap.[43] Slide images were reviewed by a perinatal pathologist aware of the original diagnosis using Digital Slide Archive (DSA).[44] The study was approved by our local institutional review board (STU00214052).
Feature extraction
One slide was used per patient. Non-tissue regions were identified and ignored by using Otsu’s method on low-power slide images. Machine learning was performed using TensorFlow version 2.8. Portions of the 10x image containing tissue were broken into 256 × 256-pixel patches with 64-pixel overlaps. This resulted in 5,000 – 16,000 patches per slide, depending on the amount of tissue present. Patches were submitted to the feature extraction backbone of EfficientNetV2L, trained on ImageNet.[39]
Attention, classification
Feature vectors were submitted to the network as N × D arrays, where N is the number of patches analyzed from that slides and D is the number of features in the vector from one patch, 1536 for EfficientNetV2. Feature vectors underwent further dimensionality reduction using a fully connected layer with 512 outputs and a rectified linear unit (relu) activation. Attention is generated using the dot product of two parallel 256-neuron layers with hyperbolic tangent and sigmoid activations. Four attentions are generated in parallel, one for each potential diagnosis. Attention values are used in the weights for weighted pooling of the dimensionality reduced layers, which then go through four fully connected classifier layers to produce one score for each diagnosis. The four scores undergo a softmax transformation to produce the final output – four scores that sum to 1.0 and represent the relative confidence of the model that the slide represents each diagnosis. For example, scores of [1, 0, 0, 0] would represent complete confidence that the slide is normal placenta, and not infarct, IVT, or PVFD (respectively). Scores of [0, 0.5, 0, 0.5] represent a slide that the model is equally confident could be infarction or PVFD. Note that confidence is a reflection of model output, not epistemic uncertainty or biologic reality. The model can by highly confident and dead wrong.
Training, validation, testing
The dataset consists of 790 cases, including 219 normal, 193 infarct, 186 IVT, and 92 PVFD. The dataset was split 70:15:15 into training, validation, and testing sets. The splits were stratified, such that the ratio of each diagnosis was consistent between the sets. The model was trained to minimize the categorical crossentropy between the actual and model-predicted diagnoses. The Adam optimizer, an initial learning rate of 1e-4, cosine decay with 1000 steps and restarts were used. Model performance was monitored throughout training by following the categorical crossentropy of the validation data. Up to 100 epochs were allowed, but training was stopped when the categorical crossentropy of the validation data did not improve after 15 epochs.
Attention maps
Attention values were output from the model as 1 × N arrays, with 1 attention value for each of the N examined patches. Attention values were normalized with 255 indicating the highest attention. Patches not analyzed were assigned a value of 0. TFR files contain similar 1 × N arrays of each tile’s left edge and upper edge, which are used to produce a 2 dimensional array, where each pixel corresponds to a single patch. False color and overlay of slide images was performed using MatPlotLib.
Convolutional whole slide models
We tested two alternative models for whole slide learning (Figure 3). These models use the same feature extractor network to generate feature vectors but use convolutional neural networks in subsequent steps. Convolutional neural networks have shown extreme facility in classifying images that present as 3-dimensional matrices with the shape W × H × C, where W and H are the width and height of the image and C is the number channels, often 3 (red, green blue).[45] While the main model treats each feature vector independently, the convolutional models arrange the feature vectors based on their location in the original image. The resulting data structure forms a 3-dimensional matrix with the shape of W × H × F, where W and H are the number of patches horizontally (Width) and vertically (Height) present in the image and F is the number of features in each patch. Convolutional neural networks can treat this matrix as they would an image, identifying patterns among neighboring patches. In principle, these models should be superior at tasks that require identification of patterns larger than a single patch.
Figure 3, Convolutional-based models:
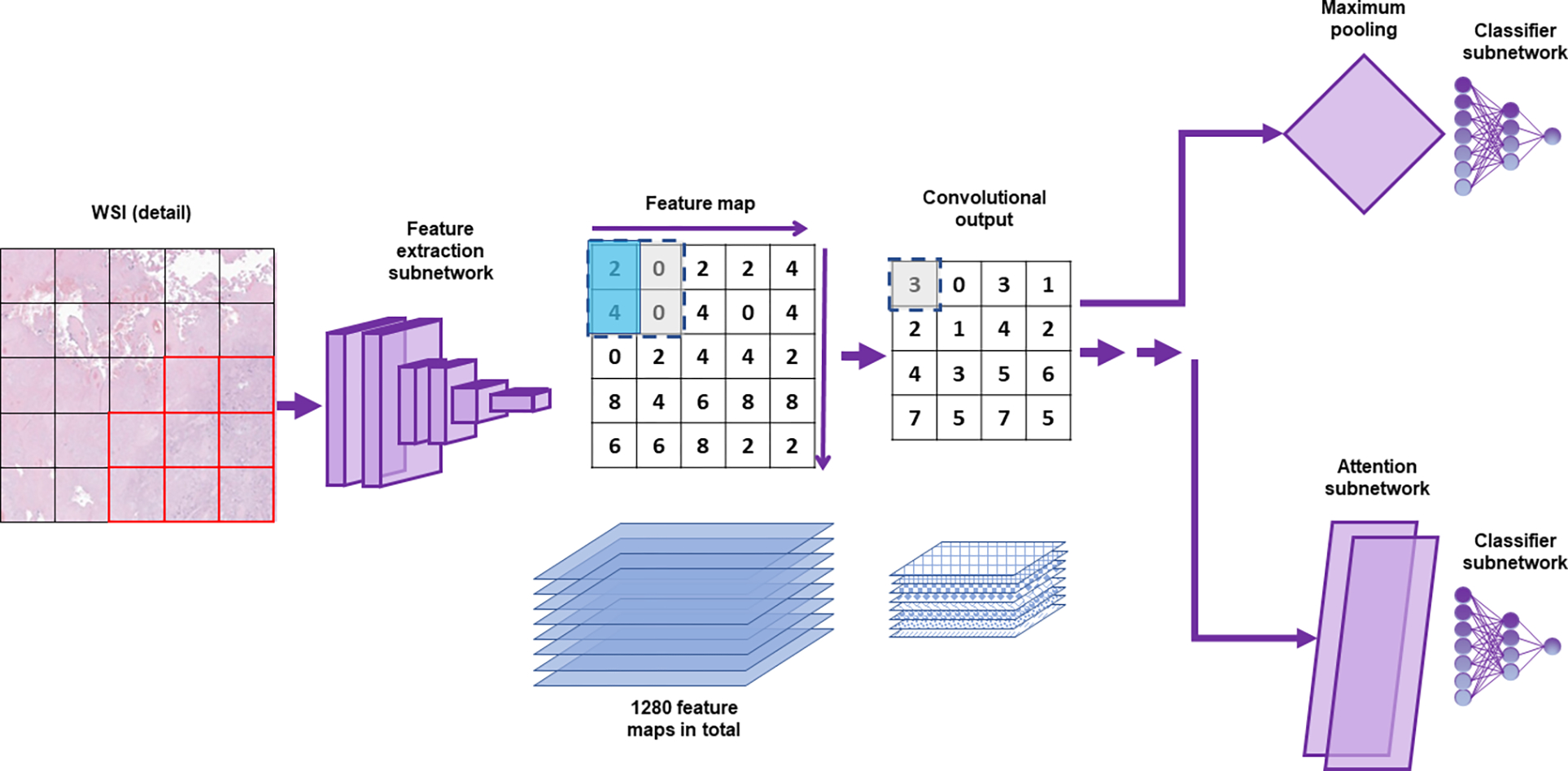
A whole slide image (WSI, detail of slide from Figure 2) is divided into a set of smaller images (patches). Images pass through a feature extraction network to become feature vectors. Unlike the dense attention model, the values across the slide from a single feature formed into 2-dimensional feature maps. There is one feature map for each of the 1280 features. A simplified 2×2 kernel is shown, which outputs the average of the values in the upper left and lower left squares (tinted aqua). The kernel is moved across and down the feature map, giving the Convolutional output shown. In the convolutional simple model, the maximum value for each feature map is used to generate a feature vector, which is then classified (upper path). For the convolutional attention model, an additional convolutional step is performed (not shown) and the feature vectors are submitted to an attention subnetwork.
We used two convolutional models, “convolutional simple” and “convolutional attention”. In the convolutional simple model, the structured matrix of feature vectors is submitted to a 2-dimensional convolutional layer with 128 filters and a kernel size of 3. Kernel size of 3 means that the feature vectors are studied in 3 × 3 groups – that is, each vector is studied in the context of its immediate neighbors. Each filter is a different pattern 3 × 3 pattern that is sought in each 3 × 3 region for each feature. For example, a filter might check whether the 2nd feature in the feature vector is above a certain threshold in the central patch, the patch up and to the left, and the patch down and to the right, forming a diagonal line. After convolution, the simple model uses 2-dimensional global max pooling to find the 3 × 3 region with the highest signal for each filter. That information is submitted to a fully connected classifier layer that outputs softmax values for each of the 4 classes.
In the convolutional attention model, the structured matrix of feature vectors is submitted first to a convolutional layer with 256 filters and a kernel size of 3, followed by a second convolutional layer with 128 filters and a kernel size of 3. The first convolutional layer is similar to the convolutional layer in the convolutional simple model, albeit with more filters. However, the second convolutional layer operates on the output from the first convolutional layer. This allows more complex patterns to be built up, for example an open diamond adjacent to a diagonal line. At this point, the output is split into 4 branches – one for each class. For each class, an attention map is generated by using a convolutional layer with a single filter and a kernel size of 1, that is, it uses only the information from a single patch at a time. A consensus feature vector is generated by calculating an average of the 2nd convolutional layer output, weighted for the attention. Each of the 4 consensus feature vectors is assigned a score, then they are brought together and go through a softmax to produce the expected output of 4 values, 1 for each class, that sum to 1.
RESULTS
We developed at dataset of 790 cases, including 219 normal, 193 infarct, 186 IVT, and 92 PVFD. Slides were split into a training set of 543 cases, 120 validation cases, and 127 test cases. The model was trained through 29 epochs before early stopping was triggered due to non-improvement in the validation categorical crossentropy (Supplementary Figure 1). The test set was evaluated, producing a balanced accuracy of 0.86 and a Matthews correlation coefficient (MCC) of 0.86. Further metrics are presented in Table 1 and Figure 4.
Table 1:
Performance of the dense attention model
| Precision | Recall | F1-score | |
|---|---|---|---|
| Normal | 0.94 | 0.96 | 0.95 |
| Infarct | 0.93 | 0.87 | 0.9 |
| IVT | 0.91 | 0.94 | 0.92 |
| PVFD | 0.67 | 0.67 | 0.67 |
| Overall | 0.9 | 0.9 | 0.90 |
Figure 4, Confusion matrix for the dense attention model:
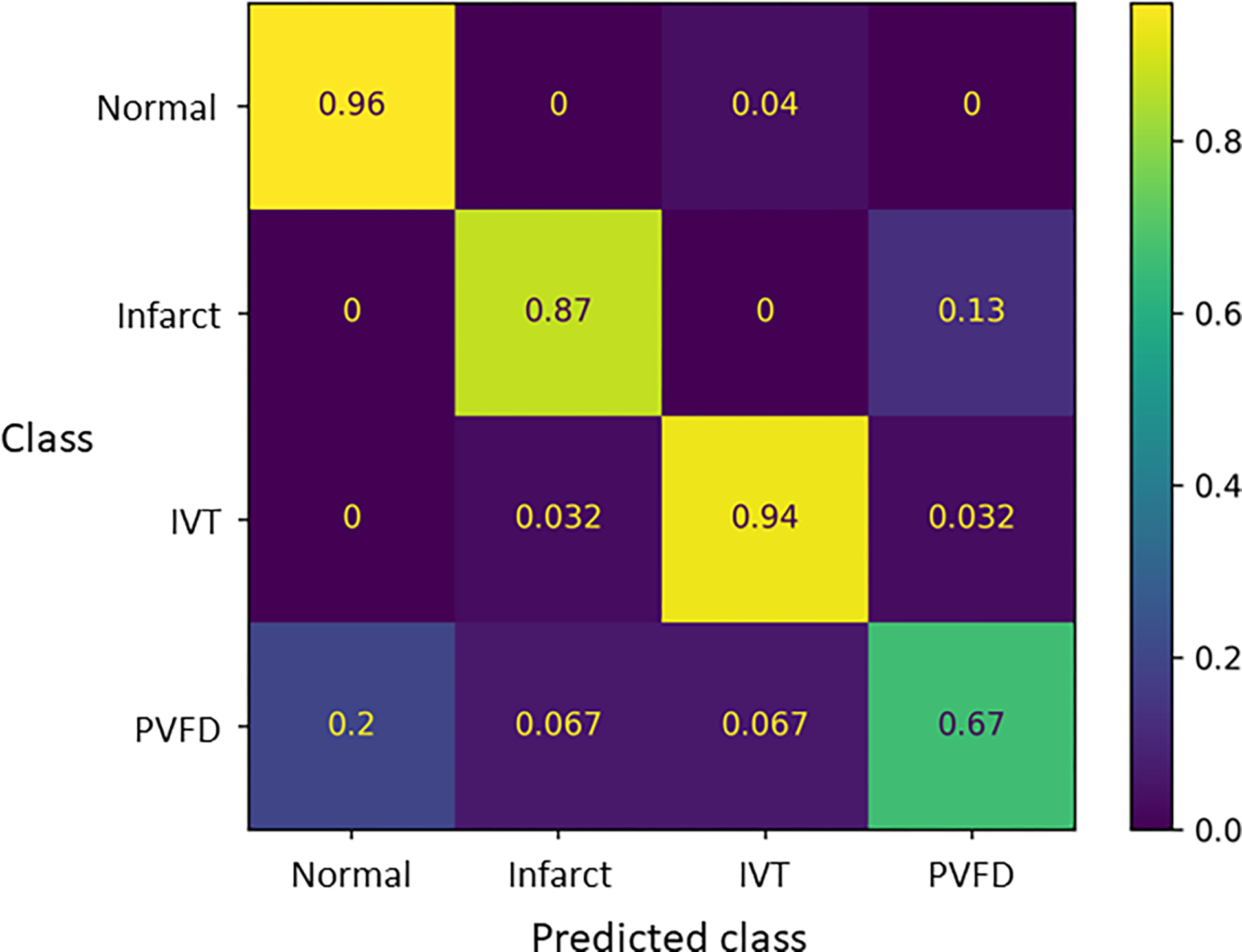
Values show the proportion of cases classified into each class for each lesion (or normal).
Model performance
We compared the performance of our network with two convolutional neural networks to capture information about patches in the context of their neighbors (see Methods). The convolutional information is then either simply averaged (“convolutional simple”) or used to generate an attention map with weights assigned to high attention groups of patches (“convolutional attention”). We generated receiver-operator characteristic curves for each classifier for each lesion (Supplementary Tables 1, 2, and 3, Supplementary Figures 2 and 3). Performance was slightly inferior, with balanced accuracies 0.79 and 0.82 and MCC of 0.78 and 0.8 for the convolutional simple and convolutional attention models, respectively.
Examination of errors
Some trends emerge in the errors. We had the largest number of normal slides, and they were identified with the highest accuracy. Concordantly, we had the smallest number of slides with PVFD and they showed the lowest sensitivity. Normal slides were rarely called as one of the lesions, while each lesions was sometimes miscalled as normal. For each class, we manually reviewed cases in the test set that were misclassified, and discuss a few illustrative examples.
One case of subchorionic PVFD was miscalled as normal, possibly due to large portions or normal. Small amounts of subchorionic fibrin are frequently encountered, and are included in the training set as “normal” if they are below the diagnostic threshold (Figure 5A). Some cases of normal were miscalled as IVT (Figure 5B). One IVT was surrounded by compressed villi, and was misclassified as an infarct (Figure 5C).
Figure 5, Morphology of misclassified cases:
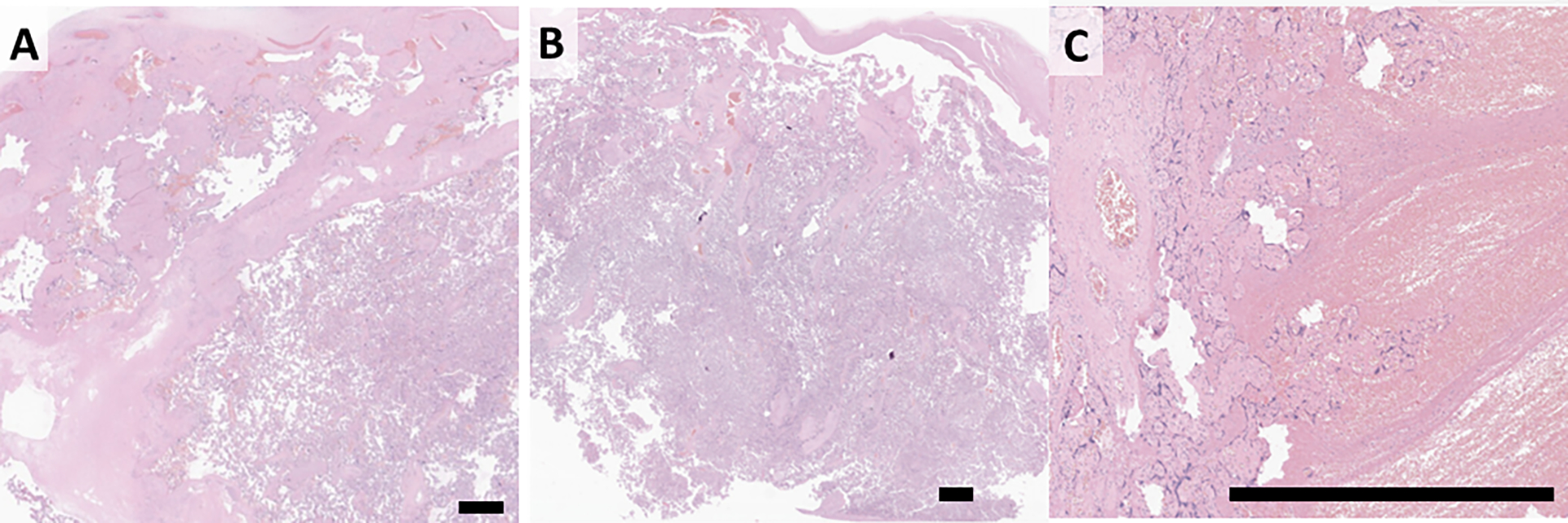
A: Focus of PVFD miscalled as normal, possibly due to large portions or normal. B: Section of normal placenta miscalled as IVT. The basis for this error is non-obvious. C: IVT miscalled as infarct, possibly related to compressed villi around the edge. Scale bar = 1 mm
DISCUSSION
Placental parenchymal lesions are commonly encountered and carry significant clinical associations. [46,47] However, they are frequently missed or misclassified by pathologists in general.[21] Artificial intelligence raises the possibility of rapid, uniform diagnosis of these findings. Our findings indicate that lesions can be distinguished from one another and from normal tissue.
Our study is noteworthy for the use of whole-slide learning. In contrast to approaches in which human annotators select a portion of the image for analysis, the model ingests the entire image to produce a unified diagnosis. This represents an increase in sophistication for the model, but also exposes challenges and errors that stem from the heterogeneity of data across a whole slide.
Even the slides of normal placenta in our data set contain multiple tissue types. In addition to the stem and terminal villi, sections of normal parenchyma often contain decidua – uterine lining transformed under the influence of progesterone, and chorionic plate – a layer of fibrous tissue supporting the largest fetal vessels and dividing the villi from the amniotic fluid. Pathology guidelines recommend sampling the interface between normal and abnormal tissues.[5] Therefore slides of parenchymal lesions generally contain stem and terminal villi as well as the lesion of interest. Lesions themselves are often heterogeneous, with both IVT and infarcts showing villi trapped in PVFD around their edges.[48]
Attention maps in explaining and improving models
When human experts and AI disagree, methods are needed to evaluate why the AI renders its diagnosis. However, AI models often operate as “black boxes” that lack the ability to explain their predictions. This hampers efforts to improve models in development and reason with their findings after deployment. [49] While several approaches have been employed, this study uses attention maps.[50–54]
We examined the attention maps for appropriately and misclassified cases to gain insight into model performance and possible improvements. In an appropriately classified case of IVT, the model showed high attention for IVT within the IVT (Figure 6). IVT frequently have a thin surrounding layer of fibrin with or without entrapped villi. Examination of the attention values for PVFD showed the model appropriately attending to areas of fibrin. This did not result in misclassification.
Figure 6, attention maps for understanding appropriate classification.
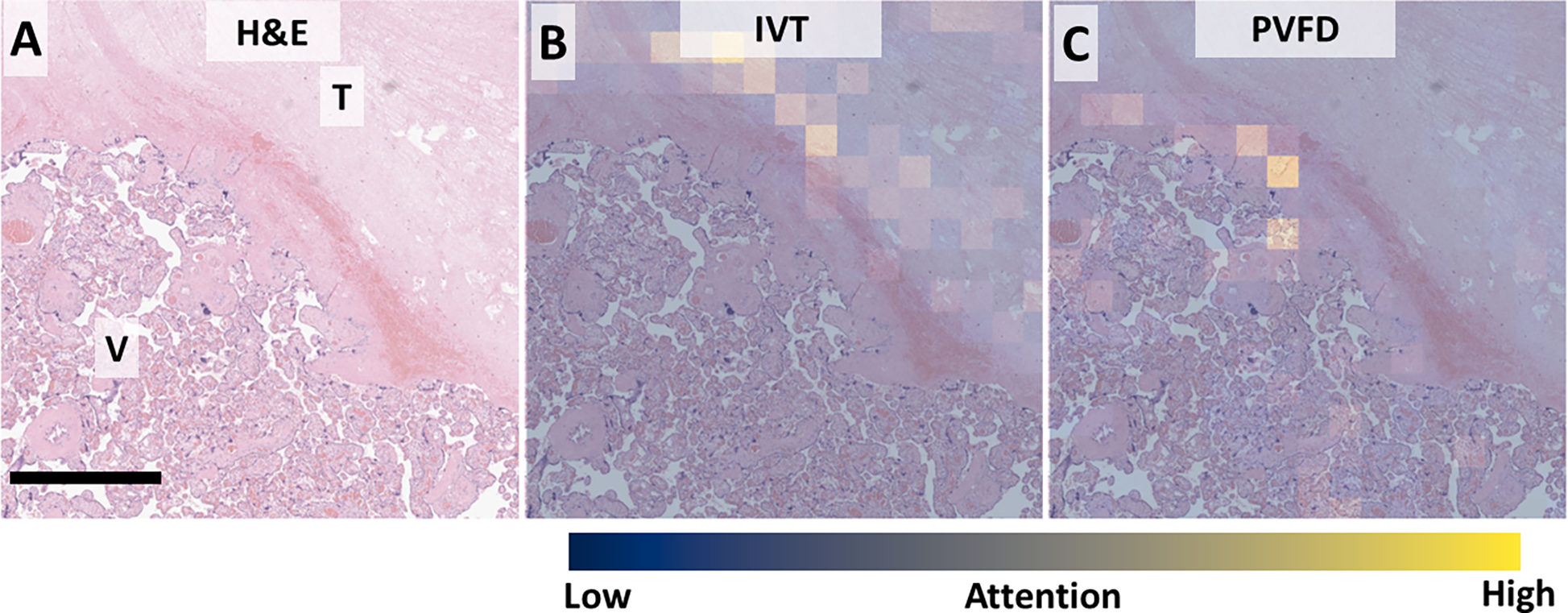
A: Low power view showing the interface between IVT (T) and normal villi (V). B: Attention for IVT is appropriately highest within the IVT and overlapping lines of Zahn. C: Attention for PVFD highlighting expected thin rim of fibrin between IVT and normal tissue. Scale bar: 250 μm. Attention: Blue – low, Yellow – high.
To examine the utility of attention maps for understanding misclassification, we examined the attention values for an IVT that was misclassified as infarct (Figure 7). In contrast to the more typical IVT seen in Figure 5, the perivillous fibrin surrounding this IVT contains large numbers of compressed villi. Despite significant attention in lines of Zahn and perivillous fibrin, a single high-attention patch seems to have overthrown the diagnosis.
Figure 7, attention maps for understanding inappropriate classification.
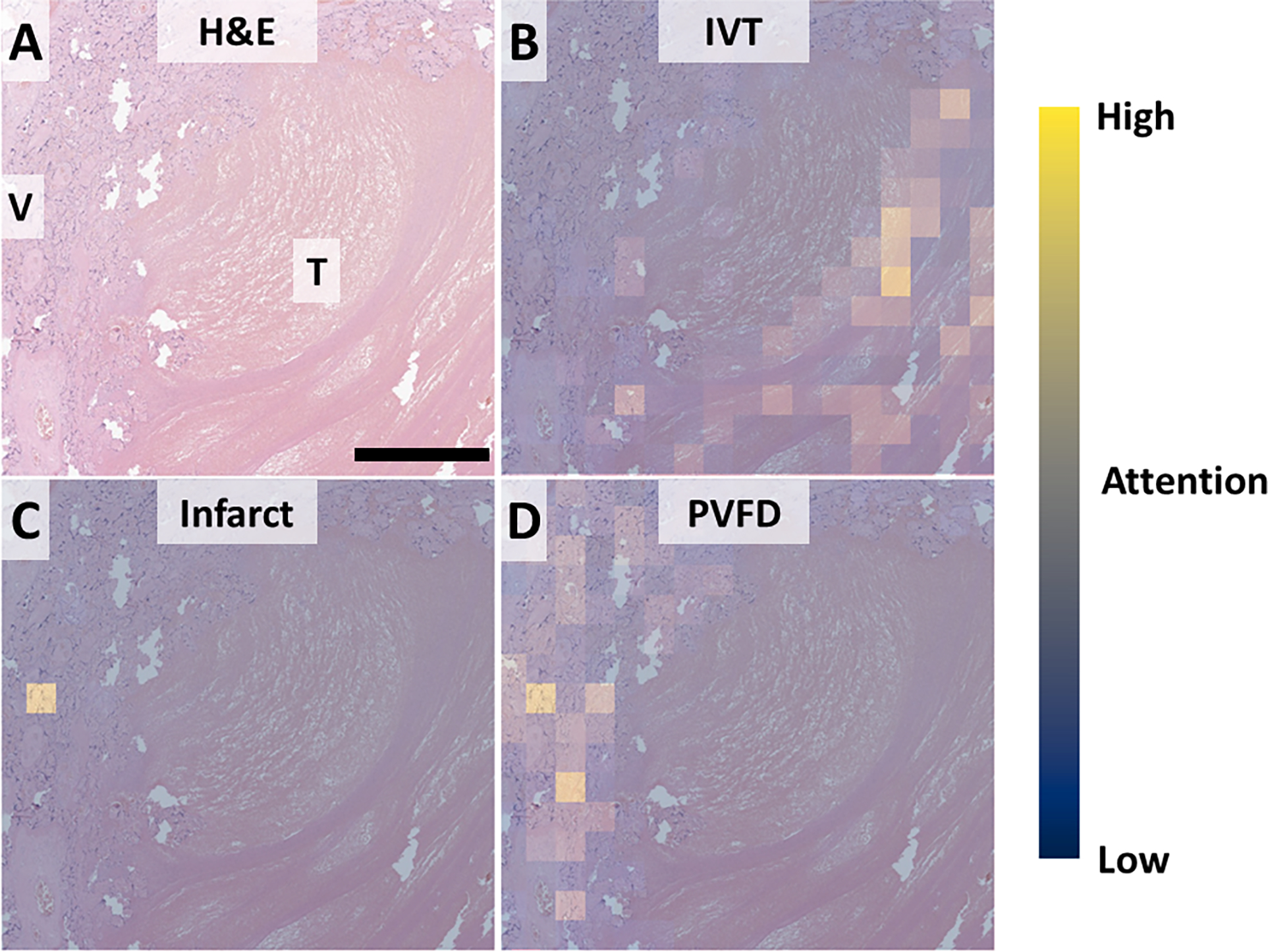
A: Low power view showing the interface between thrombus (IVT) and rim of compressed villi (V). B: Attention for IVT is appropriately highest within the thrombus and overlapping lines of Zahn. C: Attention for infarct highlighting expected a single very high-attention focus. D: Expected rim of PVFD between thrombus and normal tissue. Scale bar: 250 um. Attention: Blue – low, Yellow – high.
Limitations and future work
As a first attempt, we have chosen to focus on a subset of the most common lesions. Other parenchymal lesions include chorangiomas, retroplacental hematomas, subchorionic cysts, septal cysts, choriocarcinomas, macroscopic foci of avascular villi, and abscesses.[5,55–59] Additionally lesions may be seen together either coincidentally or in hybrid lesions, such as the infarction hematoma (infarct + retroplacental hematoma).[60,61] Future studies will expand the number and diversity of lesions studied.
Differences in staining over time and between institutions are a major barrier to generalizability of ML models.[62,63] Innovative approaches, including color normalization, color augmentation, and style transfer have been used to address this challenge.[63,64] In our study, images passed once through the feature extraction network, which did not undergo training. Thus, the opportunity for these approaches or other image augmentations (flip / rotation / noise etc.) was lost. Augmentation and tests of generalizability will be foci of future work.
Conclusion
In conclusion, we developed models to identify placental parenchymal lesions using un-annotated whole slide images.
Supplementary Material
A machine learning model can identify macroscopic placental lesions in whole slide images
Attention maps made by machine learning models highlight diagnostic areas of interest.
When models are wrong, attention maps can help interpret the model results.
ACKNOWLEDGEMENTS
JAG is supported by NIBIB K08EB030120 and the Walder Foundation Fund to Retain Clinician Scientists. LADC is supported by R01LM013523, and U01CA220401.
Footnotes
DISCLOSURES
The authors have no relevant disclosures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
BIBLIOGRAPHY
- [1].Maltepe E, Fisher SJ, Placenta: the forgotten organ, Annu. Rev. Cell Dev. Biol. 31 (2015) 523–552. 10.1146/annurev-cellbio-100814-125620. [DOI] [PubMed] [Google Scholar]
- [2].Dockery P, Bermingham J, Jenkins D, Structure-function relations in the human placenta, Biochem. Soc. Trans. 28 (2000) 202–208. 10.1042/bst0280202. [DOI] [PubMed] [Google Scholar]
- [3].Burton GJ, Jauniaux E, What is the placenta?, Am. J. Obstet. Gynecol. 213 (2015) S6.e1–S6.e4. 10.1016/j.ajog.2015.07.050. [DOI] [PubMed] [Google Scholar]
- [4].Baergen RN, Benirschke K, Manual of Benirschke and Kaufmann’s Pathology of the human placenta, Springer, New York, 2005. [Google Scholar]
- [5].Khong TY, Mooney EE, Ariel I, Balmus NCM, Boyd TK, Brundler M-A, Derricott H, Evans MJ, Faye-Petersen OM, Gillan JE, Heazell AEP, Heller DS, Jacques SM, Keating S, Kelehan P, Maes A, McKay EM, Morgan TK, Nikkels PGJ, Parks WT, Redline RW, Scheimberg I, Schoots MH, Sebire NJ, Timmer A, Turowski G, van der Voorn JP, van Lijnschoten I, Gordijn SJ, Sampling and Definitions of Placental Lesions: Amsterdam Placental Workshop Group Consensus Statement, Arch. Pathol. Lab. Med. 140 (2016) 698–713. 10.5858/arpa.2015-0225-CC. [DOI] [PubMed] [Google Scholar]
- [6].Proctor LK, Whittle WL, Keating S, Viero S, Kingdom JCP, Pathologic basis of echogenic cystic lesions in the human placenta: Role of ultrasound-guided wire localization, Placenta. 31 (2010) 1111–1115. 10.1016/j.placenta.2010.10.006. [DOI] [PubMed] [Google Scholar]
- [7].Blair E, de Groot J, Nelson KB, Placental infarction identified by macroscopic examination and risk of cerebral palsy in infants at 35 weeks of gestational age and over, Am. J. Obstet. Gynecol. 205 (2011) 124.e1–124.e7. 10.1016/j.ajog.2011.05.022. [DOI] [PubMed] [Google Scholar]
- [8].Vinnars M-T, Vollmer B, Nasiell J, Papadogiannakis N, Westgren M, Association between cerebral palsy and microscopically verified placental infarction in extremely preterm infants, Acta Obstet. Gynecol. Scand. 94 (2015) 976–982. 10.1111/aogs.12688. [DOI] [PubMed] [Google Scholar]
- [9].Vinnars M-T, Nasiell J, Ghazi S, Westgren M, Papadogiannakis N, The severity of clinical manifestations in preeclampsia correlates with the amount of placental infarction, Acta Obstet. Gynecol. Scand. 90 (2011) 19–25. 10.1111/j.1600-0412.2010.01012.x. [DOI] [PubMed] [Google Scholar]
- [10].Gibbins KJ, Silver RM, Pinar H, Reddy UM, Parker CB, Thorsten V, Willinger M, Dudley DJ, Bukowski R, Saade GR, Koch MA, Conway D, Hogue CJ, Stoll BJ, Goldenberg RL, Stillbirth, hypertensive disorders of pregnancy, and placental pathology, Placenta. 43 (2016) 61–68. 10.1016/j.placenta.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Roberts DJ, Post MD, The placenta in pre-eclampsia and intrauterine growth restriction, J. Clin. Pathol. 61 (2008) 1254–1260. 10.1136/jcp.2008.055236. [DOI] [PubMed] [Google Scholar]
- [12].Romero R, Kim YM, Pacora P, Kim CJ, Benshalom-Tirosh N, Jaiman S, Bhatti G, Kim J-S, Qureshi F, Jacques SM, Jung EJ, Yeo L, Panaitescu B, Maymon E, Hassan SS, Hsu C-D, Erez O, The frequency and type of placental histologic lesions in term pregnancies with normal outcome, J. Perinat. Med. 46 (2018) 613–630. 10.1515/jpm-2018-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Basnet KM, Bentley-Lewis R, Wexler DJ, Kilic F, Roberts DJ, Prevalence of Intervillous Thrombi Is Increased in Placentas from Pregnancies Complicated by Diabetes, Pediatr. Dev. Pathol. Off. J. Soc. Pediatr. Pathol. Paediatr. Pathol. Soc. 19 (2016) 502–505. 10.2350/15-11-1734-OA.1. [DOI] [PubMed] [Google Scholar]
- [14].Shanes ED, Miller ES, Otero S, Ebbott R, Aggarwal R, Willnow AS, Ozer EA, Mithal LB, Goldstein JA, Placental Pathology After SARS-CoV-2 Infection in the Pre-Variant of Concern, Alpha / Gamma, Delta, or Omicron Eras, Int. J. Surg. Pathol. (2022) 106689692211025. 10.1177/10668969221102534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Faye-Petersen OM, Ernst LM, Maternal Floor Infarction and Massive Perivillous Fibrin Deposition, Surg. Pathol. Clin. 6 (2013) 101–114. 10.1016/j.path.2012.10.002. [DOI] [PubMed] [Google Scholar]
- [16].Katzman PJ, Genest DR, Maternal Floor Infarction and Massive Perivillous Fibrin Deposition: Histological Definitions, Association with Intrauterine Fetal Growth Restriction, and Risk of Recurrence, Pediatr. Dev. Pathol. 5 (2002) 159–164. 10.1007/s10024001-0195-y. [DOI] [PubMed] [Google Scholar]
- [17].Romero R, Whitten A, Korzeniewski SJ, Than NG, Chaemsaithong P, Miranda J, Dong Z, Hassan SS, Chaiworapongsa T, Maternal Floor Infarction/Massive Perivillous Fibrin Deposition: A Manifestation of Maternal Antifetal Rejection?, Am. J. Reprod. Immunol. 70 (2013) 285–298. 10.1111/aji.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Becroft DMO, Thompson JMD, Mitchell EA, Placental Infarcts, Intervillous Fibrin Plaques, and Intervillous Thrombi: Incidences, Cooccurrences, and Epidemiological Associations, Pediatr. Dev. Pathol. 7 (2004) 26–34. 10.1007/s10024-003-4032-3. [DOI] [PubMed] [Google Scholar]
- [19].Redline RW, Extending the Spectrum of Massive Perivillous Fibrin Deposition (Maternal Floor Infarction), Pediatr. Dev. Pathol. 24 (2021) 10–11. 10.1177/1093526620964353. [DOI] [PubMed] [Google Scholar]
- [20].Cornish EF, McDonnell T, Williams DJ, Chronic Inflammatory Placental Disorders Associated With Recurrent Adverse Pregnancy Outcome, Front. Immunol. 13 (2022) 825075. 10.3389/fimmu.2022.825075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sun C-CJ, Revell VO, Belli AJ, Viscardi RM, Discrepancy in Pathologic Diagnosis of Placental Lesions, Arch. Pathol. Lab. Med. 126 (2002) 706–709. 10.5858/2002-126-0706-DIPDOP. [DOI] [PubMed] [Google Scholar]
- [22].Lipkova J, Chen TY, Lu MY, Chen RJ, Shady M, Williams M, Wang J, Noor Z, Mitchell RN, Turan M, Coskun G, Yilmaz F, Demir D, Nart D, Basak K, Turhan N, Ozkara S, Banz Y, Odening KE, Mahmood F, Deep learning-enabled assessment of cardiac allograft rejection from endomyocardial biopsies, Nat. Med. 28 (2022) 575–582. 10.1038/s41591-022-01709-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jayapandian CP, Chen Y, Janowczyk AR, Palmer MB, Cassol CA, Sekulic M, Hodgin JB, Zee J, Hewitt SM, O’Toole J, Toro P, Sedor JR, Barisoni L, Madabhushi A, Nephrotic Syndrome Study Network (NEPTUNE), Development and evaluation of deep learning-based segmentation of histologic structures in the kidney cortex with multiple histologic stains, Kidney Int. 99 (2021) 86–101. 10.1016/j.kint.2020.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mobadersany P, Yousefi S, Amgad M, Gutman DA, Barnholtz-Sloan JS, Velázquez Vega JE, Brat DJ, Cooper LAD, Predicting cancer outcomes from histology and genomics using convolutional networks, Proc. Natl. Acad. Sci. U. S. A. 115 (2018) E2970–E2979. 10.1073/pnas.1717139115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mobadersany P, Cooper LAD, Goldstein JA, GestAltNet: aggregation and attention to improve deep learning of gestational age from placental whole-slide images, Lab. Invest. 101 (2021) 942–951. 10.1038/s41374-021-00579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Pantanowitz L, Digital images and the future of digital pathology, J. Pathol. Inform. 1 (2010) 15. 10.4103/2153-3539.68332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Beebe LA, Cowan LD, Hyde SR, Altshuler G, Methods to improve the reliability of histopathological diagnoses in the placenta, Paediatr. Perinat. Epidemiol. 14 (2000) 172–178. 10.1046/j.1365-3016.2000.00253.x. [DOI] [PubMed] [Google Scholar]
- [28].Goldenberg RL, McClure EM, Bhutta ZA, Belizán JM, Reddy UM, Rubens CE, Mabeya H, Flenady V, Darmstadt GL, Stillbirths: the vision for 2020, The Lancet. 377 (2011) 1798–1805. 10.1016/S0140-6736(10)62235-0. [DOI] [PubMed] [Google Scholar]
- [29].Luchini C, Pantanowitz L, Adsay V, Asa SL, Antonini P, Girolami I, Veronese N, Nottegar A, Cingarlini S, Landoni L, Brosens LA, Verschuur AV, Mattiolo P, Pea A, Mafficini A, Milella M, Niazi MK, Gurcan MN, Eccher A, Cree IA, Scarpa A, Ki-67 assessment of pancreatic neuroendocrine neoplasms: Systematic review and meta-analysis of manual vs. digital pathology scoring, Mod. Pathol. 35 (2022) 712–720. 10.1038/s41379-022-01055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].López-Pérez M, Amgad M, Morales-Álvarez P, Ruiz P, Cooper LAD, Molina R, Katsaggelos AK, Learning from crowds in digital pathology using scalable variational Gaussian processes, Sci. Rep. 11 (2021) 11612. 10.1038/s41598-021-90821-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Madabhushi A, Lee G, Image analysis and machine learning in digital pathology: Challenges and opportunities, Med. Image Anal. 33 (2016) 170–175. 10.1016/j.media.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tizhoosh HR, Pantanowitz L, Artificial Intelligence and Digital Pathology: Challenges and Opportunities, J. Pathol. Inform. 9 (2018) 38. 10.4103/jpi.jpi_53_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Dietterich TG, Lathrop RH, Lozano-Pérez T, Solving the multiple instance problem with axis-parallel rectangles, Artif. Intell. 89 (1997) 31–71. 10.1016/S0004-3702(96)00034-3. [DOI] [Google Scholar]
- [34].Lu MY, Chen TY, Williamson DFK, Zhao M, Shady M, Lipkova J, Mahmood F, AI-based pathology predicts origins for cancers of unknown primary, Nature. 594 (2021) 106–110. 10.1038/s41586-021-03512-4. [DOI] [PubMed] [Google Scholar]
- [35].Lu MY, Williamson DFK, Chen TY, Chen RJ, Barbieri M, Mahmood F, Data-efficient and weakly supervised computational pathology on whole-slide images, Nat. Biomed. Eng. 5 (2021) 555–570. 10.1038/s41551-020-00682-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kraus OZ, Ba JL, Frey BJ, Classifying and segmenting microscopy images with deep multiple instance learning, Bioinformatics. 32 (2016) i52–i59. 10.1093/bioinformatics/btw252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gadermayr M, Tschuchnig M, Multiple Instance Learning for Digital Pathology: A Review on the State-of-the-Art, Limitations & Future Potential, (2022). http://arxiv.org/abs/2206.04425 (accessed September 28, 2022). [DOI] [PubMed] [Google Scholar]
- [38].Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, Brogi E, Reuter VE, Klimstra DS, Fuchs TJ, Clinical-grade computational pathology using weakly supervised deep learning on whole slide images, Nat. Med. 25 (2019) 1301–1309. 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tan M, Le QV, EfficientNetV2: Smaller Models and Faster Training, (n.d.) 11. [Google Scholar]
- [40].Deng J, Dong W, Socher R, Li L-J, Kai Li, Li Fei-Fei, ImageNet: A large-scale hierarchical image database, in: 2009 IEEE Conf. Comput. Vis. Pattern Recognit., IEEE, Miami, FL, 2009: pp. 248–255. 10.1109/CVPR.2009.5206848. [DOI] [Google Scholar]
- [41].Shanes ED, Mithal LB, Otero S, Azad HA, Miller ES, Goldstein JA, Placental Pathology in COVID-19, Am. J. Clin. Pathol. 154 (2020) 23–32. 10.1093/ajcp/aqaa089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chen A, Roberts DJ, Placental pathologic lesions with a significant recurrence risk - what not to miss!, APMIS. 126 (2018) 589–601. 10.1111/apm.12796. [DOI] [PubMed] [Google Scholar]
- [43].Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O’Neal L, McLeod L, Delacqua G, Delacqua F, Kirby J, Duda SN, The REDCap consortium: Building an international community of software platform partners, J. Biomed. Inform. 95 (2019) 103208. 10.1016/j.jbi.2019.103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gutman DA, Khalilia M, Lee S, Nalisnik M, Mullen Z, Beezley J, Chittajallu DR, Manthey D, Cooper LAD, The Digital Slide Archive: A Software Platform for Management, Integration, and Analysis of Histology for Cancer Research, Cancer Res. 77 (2017) e75–e78. 10.1158/0008-5472.CAN-17-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Krizhevsky A, Sutskever I, Hinton GE, ImageNet classification with deep convolutional neural networks, in: Proc. 25th Int. Conf. Neural Inf. Process. Syst. - Vol. 1, Curran Associates Inc., Red Hook, NY, USA, 2012: pp. 1097–1105. [Google Scholar]
- [46].Redline RW, Classification of placental lesions, Am. J. Obstet. Gynecol. 213 (2015) S21–S28. 10.1016/j.ajog.2015.05.056. [DOI] [PubMed] [Google Scholar]
- [47].Roescher AM, Timmer A, Erwich JJHM, Bos AF, Placental Pathology, Perinatal Death, Neonatal Outcome, and Neurological Development: A Systematic Review, PLoS ONE. 9 (2014) e89419. 10.1371/journal.pone.0089419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Heerema-McKenney A, Popek EJ, De Paepe ME, Ernst LM, eds., Placenta, 2. edition, Elsevier, Philadelphia, Pa, 2019. [Google Scholar]
- [49].Rudin C, Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead, Nat. Mach. Intell. 1 (2019) 206–215. 10.1038/s42256-019-0048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Amgad M, Atteya LA, Hussein H, Mohammed KH, Hafiz E, Elsebaie MAT, Alhusseiny AM, AlMoslemany MA, Elmatboly AM, Pappalardo PA, Sakr RA, Mobadersany P, Rachid A, Saad AM, Alkashash AM, Ruhban IA, Alrefai A, Elgazar NM, Abdulkarim A, Farag A-A, Etman A, Elsaeed AG, Alagha Y, Amer YA, Raslan AM, Nadim MK, Elsebaie MAT, Ayad A, Hanna LE, Gadallah A, Elkady M, Drumheller B, Jaye D, Manthey D, Gutman DA, Elfandy H, Cooper LAD, NuCLS: A scalable crowdsourcing, deep learning approach and dataset for nucleus classification, localization and segmentation, GigaScience. 11 (2022) giac037. 10.1093/gigascience/giac037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Evans T, Retzlaff CO, Geißler C, Kargl M, Plass M, Müller H, Kiehl T-R, Zerbe N, Holzinger A, The explainability paradox: Challenges for xAI in digital pathology, Future Gener. Comput. Syst. 133 (2022) 281–296. 10.1016/j.future.2022.03.009. [DOI] [Google Scholar]
- [52].Pocevičiūtė M, Eilertsen G, Lundström C, Survey of XAI in Digital Pathology, in: Holzinger A, Goebel R, Mengel M, Müller H (Eds.), Artif. Intell. Mach. Learn. Digit. Pathol, Springer International Publishing, Cham, 2020: pp. 56–88. 10.1007/978-3-030-50402-1_4. [DOI] [Google Scholar]
- [53].Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D, Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization, in: 2017 IEEE Int. Conf. Comput. Vis. ICCV, IEEE, Venice, 2017: pp. 618–626. 10.1109/ICCV.2017.74. [DOI] [Google Scholar]
- [54].Liu S, Kailkhura B, Loveland D, Han Y, Generative Counterfactual Introspection for Explainable Deep Learning, in: 2019 IEEE Glob. Conf. Signal Inf. Process. Glob., IEEE, Ottawa, ON, Canada, 2019: pp. 1–5. 10.1109/GlobalSIP45357.2019.8969491. [DOI] [Google Scholar]
- [55].Amer HZM, Heller DS, Chorangioma and Related Vascular Lesions of the Placenta — a Review, Fetal Pediatr. Pathol. 29 (2010) 199–206. 10.3109/15513815.2010.487009. [DOI] [PubMed] [Google Scholar]
- [56].Brewer JI, Mazur MT, Gestational choriocarcinoma. Its origin in the placenta during seemingly normal pregnancy, Am. J. Surg. Pathol. 5 (1981) 267–277. 10.1097/00000478-198104000-00007. [DOI] [PubMed] [Google Scholar]
- [57].Jiao L, Ghorani E, Sebire NJ, Seckl MJ, Intraplacental choriocarcinoma: Systematic review and management guidance, Gynecol. Oncol. 141 (2016) 624–631. 10.1016/j.ygyno.2016.03.026. [DOI] [PubMed] [Google Scholar]
- [58].Katzman PJ, Placental Disorders of Uncertain Etiology, in: Baergen RN, Burton GJ, Kaplan CG (Eds.), Benirschkes Pathol. Hum. Placenta, Springer International Publishing, Cham, 2022: pp. 729–751. 10.1007/978-3-030-84725-8_26. [DOI] [Google Scholar]
- [59].Khong TY, Extravillous Trophoblast Cyst, in: Khong TY, Mooney EE, Nikkels PGJ, Morgan TK, Gordijn SJ (Eds.), Pathol. Placenta, Springer International Publishing, Cham, 2019: pp. 97–99. 10.1007/978-3-319-97214-5_11. [DOI] [Google Scholar]
- [60].Bendon RW, Nosology: infarction hematoma, a placental infarction encasing a hematoma, Hum. Pathol. 43 (2012) 761–763. 10.1016/j.humpath.2011.07.023. [DOI] [PubMed] [Google Scholar]
- [61].Neville G, Russell N, O’Donoghue K, Fitzgerald B, Rounded intraplacental hematoma - A high risk placental lesion as illustrated by a prospective study of 26 consecutive cases, Placenta. 81 (2019) 18–24. 10.1016/j.placenta.2019.02.011. [DOI] [PubMed] [Google Scholar]
- [62].Schömig-Markiefka B, Pryalukhin A, Hulla W, Bychkov A, Fukuoka J, Madabhushi A, Achter V, Nieroda L, Büttner R, Quaas A, Tolkach Y, Quality control stress test for deep learning-based diagnostic model in digital pathology, Mod. Pathol. 34 (2021) 2098–2108. 10.1038/s41379-021-00859-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Shin SJ, You SC, Jeon H, Jung JW, An MH, Park RW, Roh J, Style transfer strategy for developing a generalizable deep learning application in digital pathology, Comput. Methods Programs Biomed. 198 (2021) 105815. 10.1016/j.cmpb.2020.105815. [DOI] [PubMed] [Google Scholar]
- [64].Tellez D, Litjens G, Bándi P, Bulten W, Bokhorst J-M, Ciompi F, van der Laak J, Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology, Med. Image Anal. 58 (2019) 101544. 10.1016/j.media.2019.101544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


