Abstract
Onset of rheumatoid arthritis (RA) is widely believed to be preceded by exposure to some environmental trigger such as bacterial infectious agents. The influence of bacteria on RA disease onset or pathology has to date been controversial, due to inconsistencies between groups in the report of bacterial species isolated from RA disease tissue. Using a modified technique of reverse transcriptase-PCR amplification, we have detected bacterial rRNA in the synovial tissue of late-stage RA and non-RA arthritis controls. This may be suggestive of the presence of live bacteria. Sequencing of cloned complementary rDNA (crDNA) products revealed a number of bacterial sequences in joint tissue from each patient, and from these analyses a comprehensive profile of the organisms present was compiled. This revealed a number of different organisms in each patient, some of which are common to both RA and non-RA controls and are probably opportunistic colonizers of previously diseased tissue and others which are unique species. These latter organisms may be candidates for a specific role in disease pathology and require further investigation to exclude them as causative agents in the complex bacterial millieu. In addition, many of the detected bacterial species have not been identified previously from synovial tissue or fluid from arthritis patients. These may not be easily cultivable, since they were not revealed in previous studies using conventional in vitro bacterial culture methods. In situ hybridization analyses have revealed the joint-associated bacterial rRNA to be both intra- and extracellular. The role of viable bacteria or their nucleic acids as triggers in disease onset or pathology in either RA or non-RA arthritis controls is unclear and requires further investigation.
In addition to the influence of innate susceptibility factors, most notably certain HLA class II alleles, onset of rheumatoid arthritis (RA) is widely believed to be preceded by exposure to some environmental trigger. The precise nature of this initiating factor has not yet been elucidated despite much study. There has been considerable interest in a possible role for bacterial infectious agents in disease onset (20, 35, 38), since there are certain similarities between RA and other inflammatory arthritides, e.g., reactive arthritis (ReA) and Lyme arthritis (LyA). These latter conditions are known to be preceded by bacterial infection at a site distant from the involved joint and also show an association with HLA alleles, ReA with HLA-B27 (40) and LyA more weakly with HLA-DR4 (30). ReA can be triggered by gastrointestinal or genitourinary infection with a number of different bacterial species including Yersinia, Salmonella, Campylobacter, and Chlamydia (12), and LyA is triggered by infection with the tick-borne spirochaete Borrelia burgdorferi (12).
There is an accumulating body of evidence suggesting that these conditions, previously thought to be sterile arthropathies, may be perpetuated by small numbers of persistent organisms which have trafficked to the affected joint. Spirochetes can occasionally be recovered by culture of synovial fluid from individuals with LyA but are only detected by PCR or electron microscopy in synovial tissue (12, 15, 28, 48). Live organisms have not reproducibly been recovered by culture from ReA-affected joints; however, DNA from Yersinia and Chlamydia species has been detected by PCR in the synovial fluid of some patients with ReA (12, 62, 63). Bacterial rRNA has also been detected (18, 23), which is suggestive of the presence of live replicating organisms due to the relative lability of rRNA compared to DNA in nonviable organisms (59). No such evidence of specific infection as a trigger for arthritis onset has been uncovered in RA. Early culture studies of synovial fluid yielded a variety of different bacterial species (26, 58). The organisms identified varied substantially between different investigative groups, suggesting the absence of common culturable etiological agents involved in disease pathology. In addition, contamination during sample handling and culture procedures often could not be ruled out, and the organisms could not be associated directly with the disease process.
In more recent molecular studies, Melief et al. (43) found intestinal flora-derived peptidoglycan polysaccharides within macrophages and dendritic cells from synovial tissue of RA patients. It is not clear whether bacterial antigen was derived from material carried by mononuclear cells from other body sites or from live organisms within the joint. Evidence for the presence of live bacteria in the synovial tissue of RA patients has come from the work of Medrano and Galbete, who observed cell-associated unidentified bacilli in 33 of 34 synovial membrane explant cultures (41). The bacilli in this study were not characterized; therefore it is not known whether these organisms are similar to those identified in early culture studies. Medrano and Galbete made the observation that the bacteria appeared to exist in a partially cell wall-deficient (CWD) form and were difficult to culture. This may suggest the possible involvement of uncultivable or “difficult-to-grow” organisms in RA. CWD (or L-form) bacteria are notoriously difficult to culture owing to their osmotic sensitivity and have been implicated in other diseases of unknown etiology like Crohn's disease (60) and sarcoidosis (29). However, owing to the lack of reproducibility by other workers, these observations are still the subject of some controversy.
The detection of bacteria by conventional culture methods, staining, or species-specific PCR is not perhaps the most sensitive or comprehensive means of assessing the range of bacteria that could be present in disease tissue. With the advent of PCR-based detection techniques based on bacterial rDNA (49), a number of other conditions of unknown etiology have been found to be caused by previously unidentified and uncultivable bacteria, e.g. Tropheryma whippelii in Whipple's disease (50). It is conceivable that uncultivable or difficult-to-grow bacteria could be involved in RA. Wilbrink et al. have demonstrated the presence of bacterial DNA in synovial biopsy specimens from individuals with septic and inflammatory arthropathies by PCR of rDNA with universal primers (61). By rDNA sequencing, this group were able to partially characterize the bacteria found in joints of four individuals with undifferentiated arthritis (UA), some to the species level. Multiple bacterial species were observed in each, suggesting colonization with more than one organism. It is not known from this study whether these tissues contained any previously unidentified microorganisms.
Here we present data demonstrating the presence of multiple bacterial species in joint tissue of both late-stage RA patients and non-RA arthritis controls, using the similar technique of reverse transcriptase-PCR (RT-PCR) of bacterial rRNA. We have used this adaptation of established DNA-based techniques because bacteria have multiple copies of rRNA compared with their rRNA genes (16); thus, RT-PCR of rRNA may offer a severalfold-increased sensitivity over rDNA PCR. This technique has been used to detect bacterial rRNA in arthritis joint tissue, suggesting the presence of viable organisms, and to carry out detailed characterization of the bacteria present by sequencing of complementary rRNA (crDNA) products. These analyses have revealed the presence of both previously characterized and novel bacterial species. The in situ localization of these microorganisms has also been investigated by conventional bacteriological staining of tissue sections and hybridization with digoxigenin-labeled rDNA oligonucleotides. Microorganisms present in these apparently subclinically infected joints appear to be both cell associated and extracellular.
MATERIALS AND METHODS
Patients.
RA and osteoarthritis (OA) synovial tissue specimens were collected with patient consent at surgery for joint replacement, with the exception of the specimens from RA patient 2, which was obtained by needle biopsy, and OA patient 20, which was collected at the first metatarsal-phalangeal joint MTP surgery. Normal synovial tissues from patients 22 and 23 were collected by needle biopsy, and normal synovial tissue from patient 21 was collected at arthroscopy for unexplained knee pain (clinical details of all patients are given in Table 1). Normal controls were not age and sex matched to the RA patient group, but trauma specimens were unlikely to have features of joint disease pathology in common with arthritis patients of many years duration. These tissues were used as process controls and were run with each study sample. All RA patients were classified according to the American College of Rheumatology criteria (5) and had late-stage disease, i.e., disease of many years duration with joint destruction requiring arthroplasty. A classification of UA was made on the basis of mono- or oligoarthritis where all other diseases had been excluded.
TABLE 1.
Details of patient tissues used in this study
| Patient | Diagnosis | Sex/age (yr)/tissue detailsa |
|---|---|---|
| 1 | RA | DNG/DNG/TKR |
| 2 | RA | M/DNG/needle biopsy (knee) |
| 3 | RA | F/58/TKR |
| 4 | RA | M/84/THR |
| 5 | RA | F/63/TKR |
| 6 | RA | M/54/TKR |
| 7 | RA | M/72/THR |
| 8 | RA | F/60/TKR |
| 9 | RA | F/60/TKR |
| 10 | OA | F/76/THR |
| 11 | OA | M/64/TKR |
| 12 | OA | M/79/TKR |
| 13 | OA | M/57/TKR |
| 14 | OA | F/64/THR |
| 19 | OA | M/77/THR |
| 20 | OA | F/28/1st MTP surgery |
| 15 | UA | M/79/TKR |
| 16 | UA | F/71/TKR |
| 17 | UA | F/88/TKR |
| 18 | UA | M/66/TKR |
| 21 | Normal synovium | M/DNG/knee trauma, synovium obtained at menisectomy |
| 22 | Normal synovium | M/DNG/normal needle biopsy (knee) |
| 23 | Normal synovium | M/DNG/normal needle biopsy (knee) |
DNG, patient details not given; THP, total hip replacement; TKR, total knee replacement; 1st MTP surgery, first metatarsal-phalangeal joint surgery.
Materials.
All chemicals including Gram stain reagents were purchased from Sigma-Aldrich Co. Ltd., Poole, England. TB Carbolfuschein staining reagents were supplied by Difco Laboratories, West Molesey, England. Faramount aqueous mountant medium and nitroblue tetrazolium–5-bromo-4-chloro-3-indolyl phosphate–iodonitrotetrazolium violet (NBT-BCIP-INT) were purchased from Dako, Ely, England. The Hybaid Ribolyser kit was purchased from Hybaid, Teddington, England. Amplitaq Taq polymerase and buffers were supplied by Perkin-Elmer, Warrington, England. Oligonucleotide primers for RT-PCR were purchased from GibcoBRL Life Technologies, Paisley, Scotland. Dual 5′ and 3′ digoxygenin-end-labeled oligonucleotides were purchased from Sigma-Genosys Ltd., Pampisford, England. Deoxyribonucleotides and anti-digoxigenin-coupled alkaline phosphatase were purchased from Roche Diagnostics, Lewes, England. The Novagen pT7-blue PCR cloning kit was purchased from Cambridge Bioscience, Cambridge, England.
Tissue handling and RNA isolation.
Resected synovial tissue samples collected at surgery were immediately frozen in hexane on dry ice and then stored at −80°C prior to use. Synovial biopsy specimens were placed immediately into 500 μl of guanidinium isothiocyanate extraction buffer (GIEB). Total RNA was isolated from late-stage RA, OA and UA synovial tissue and normal control tissue using a modification of the Hybaid RiboLyser guanidinium isothiocyanate-acid phenol extraction method, in which buffer A was replaced with fresh GIEB (39). In short, approximately 0.1 g of resected synovial tissue was thawed in 500 μl of GIEB, chopped finely and extracted using shear lysis in the presence of 500 μl of phenol (pH 4.0) and 100 μl of chloroform–isoamyl alcohol in a Hybaid RiboLyser, as specified by the manufacturer. Total RNA was recovered by precipitation with propan-2-ol, dried under appropriate sterile conditions, and dissolved in 50 μl of diethylpyrocarbonate-treated water containing 0.1 mM EDTA.
RT-PCR amplification of bacterial crDNA from bacterial and synovial tissue RNAs.
To eliminate the risk of contamination with bacterial nucleic acids from external sources, all reagents were prepared using distilled water irradiated with UV at 254 nm for 2 min. Rigorous controls were instigated at each stage of the RT-PCR procedure to ensure no contamination of samples during protocol implementation. Samples of control bacterial rRNAs (25 ng) or total tissue RNA (100 ng) were reverse transcribed by the same method, in 4 μl of buffer containing 50 mM Tris-Cl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 40 μM deoxynucleoside triphosphates, and 20 μM primer R2 (all primer sequences are given in Table 2). This mixture was heated to 65°C for 1 min and then cooled to room temperature for 3 min. Superscript RT (200 U) was added, and the mixture was incubated at 37°C for 1 h. The reaction was stopped by incubation at 65°C for 10 min.
TABLE 2.
Oligonucleotide primers used in RT-PCR and in situ hybridization analyses
| Oligonucleotide primer | Sequence | Position in E. coli rRNA gene sequence |
|---|---|---|
| R1 | 5′ AGAGTTTGATCCTGGCTCAG 3′ | 8–27 |
| R2 | 5′ ACTGCTGCCTCCCGTAGGAG 3′ | 339–358 |
| ISH1 | 5′ ATTCCCCACTGCTGCCTCCCGTAGGAGT 3′ | 338–365 |
| ISH2 | 5′ GACTTGACGTCATCCCCACCTTCCTCC 3′ | 1175–1300 |
| ISH3 | 5′ CGGGCGGTGTGTACAAGGCCCGGGAACG 3′ | 1378–1405 |
| B40F | 5′ GTTTTCCCAGTCACGAC 3′ | NAa |
| B40R | 5′ AGCGGATAACAATTTCACACAGGA 3′ | NA |
NA, not applicable.
Bacterial rDNA fragments were amplified from total cDNA and bacterial genomic DNA by PCR amplification using universal bacterial rRNA-specific oligonucleotide primers R1 and R2. RT mix (1 μl) or genomic DNA (5 ng) was used as template in a PCR mixture containing 1× Amplitaq PCR buffer, 0.2 μM deoxynucleoside triphosphates, 0.2 μM PCR primers R1 and R2, 1.5 mM MgCl2, and 2.5 U of Amplitaq Taq polymerase. These were amplified at 94°C for 4 min and then for 30 cycles of 58°C for 1 min, 72°C for 3 min, and 94°C for 1 min. PCR products were visualized by electrophoresis on a 2% agarose gel.
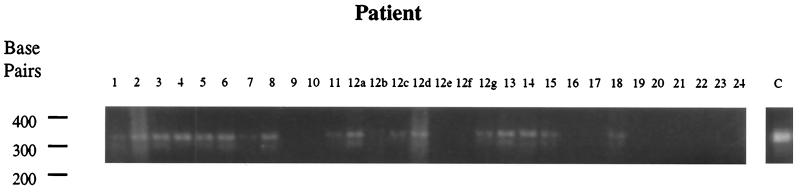
In samples from many patients, bacterial 16S crDNA bands could be seen (Fig. 1); some were diffuse, suggesting mixed bacterial crDNA products and the possible presence of more than one organism. PCR products were therefore cloned into the PCR product cloning vector pT7-blue (Novagen), as specified by the manufacturer, for isolation and sequencing. Individual clones were inoculated into 96-well plates and grown overnight, and then a small amount of bacterial suspension was transferred into 96-well PCR plates containing 25 μl of PCR mixture as above but containing 3 mM MgCl2 and PCR primers which amplify the cloned fragment using flanking plasmid primer binding sites (primers B40F and B40R). These were amplified at 94°C for 4 min and then for 25 cycles of 60°C for 1 min, 72°C for 3 min, and 94°C for 1 min. Individual PCR products were diluted, sequenced with primer B40F on an ABI automated sequencer, and analyzed using the search algorithm BLAST (3) on database sequences and compared horizontally using the Genetics Computer Group (GCG) algorithm PileUp (18a).
FIG. 1.
Results of RT-PCR of bacterial 16S rRNA from patient RNA samples, amplification products visualized by agarose gel electrophoresis, and ethidium bromide staining.
Detection of tissue-localized bacteria by conventional staining and in situ hybridization.
Frozen arthritis and control tissues were embedded in OCT (Agar Scientific), and 0.3-μm sections were produced on a Shandon Cryotome cryostat. Conventional bacterial staining was performed on selected sections using Gram stain and Carbolfuschein reagents as specified by the manufacturers. For in situ hybridization, sections were rinsed in phosphate-buffered saline (PBS), rehydrated by immersion in 0.2% Triton X-100–PBS for 15 min, and washed twice in PBS. Half the slides were treated with 200 μl of RNase solution (10 mg/ml in 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) for 30 min at 37°C as a control. RNase-treated slides were washed twice in 2× SSC and then digested at 37°C for 17 min with a solution containing 0.1 M Tris-Cl, 50 mM EDTA (pH 8.0), and 5 μg of proteinase K per ml. These were then immersed in 0.1 M glycine in PBS and fixed with 4% paraformaldehyde in PBS. After being rinsed with PBS and treated in 0.25% acetic anhydride and 0.1 M triethanolamine solution (pH 8.0), all slides were incubated in 20% acetic acid at 4°C, washed three times in PBS, dehydrated through alcohol, and dried.
For oligonucleotide annealing, sections were prehybridized at 37°C for 30 min in 50 μl of buffer containing 36% formamide, 5× SSC, 10% dextran Sulfate, 5% Denhardt's solution, 0.5% sodium dodecyl sulfate (SDS), 100 μg of sheared herring sperm DNA per ml. A 40-μl volume of fresh buffer was then added containing a mix of 5 ng of digoxigenin-labeled probes ISH1 to ISH3 per μl. The sections were incubated at 37°C overnight and then washed four times in 2× SSC–0.1% SDS at 45°C, twice in 0.1× SSC–0.1% SDS at 45°C, and twice in 2× SSC at room temperature. They were then treated with 10 μg of RNase per ml in 2× SSC at 37°C for 15 min. For visualization of the digoxigenin-labeled probe, the slides were rinsed with Tris-buffered saline (TBS) and then incubated for 30 min at room temperature in TBS buffer containing 10% bovine serum albumin and 0.5% Triton X-100 and for 5 min with TBS containing 2% normal sheep serum and 0.5% Triton X-100. The TBS was removed, 100 μl of buffer was added containing antidigoxigenin immunoglobulin G conjugated to alkaline phosphatase diluted 1:100 in TBS–2% normal sheep serum–0.5% Triton X-100, and the sections were incubated for 2 h. The slides were washed three times in TBS, and bound alkaline phosphatase was visualized by incubation for 16 h in NBT-BCIP-INT solution. The slides were rinsed in water, counterstained in Mayers hematoxylin solution (Pioneer Research Chemicals Ltd.), and mounted in Mountant. All the sections were visualized by light microscopy, and images were captured either using electronic imaging or on 35-mm Kodak Ektachrome 64T film.
RESULTS
RT-PCR amplification and sequencing of bacterial crDNA fragments from total synovial RNA.
crDNA amplification products from total synovial tissue RNAs were observed in 8 of 9 RA specimens, 6 of 11 non-RA arthritis controls (i.e., 4 of 7 OA specimens and 2 of 4 UA specimens), and 0 of 3 normal specimens (Fig. 1). Due to the high risk of contamination, great care was taken in sample handling, buffer generation, and RT-PCR analysis with the implementation of appropriate RT and PCR negative controls; these were consistently negative. Since the normal control samples were also consistently negative, it was concluded that the RT-PCR signal in positive tissues was derived from tissue-associated bacterial rRNA and not contamination from skin or other environmental sources, e.g., introduced during surgical removal of tissue.
DNase treatment was not conducted on all samples, to preserve the total signal obtained from both RNA and DNA. However, when tested on a larger cohort of patient samples than presented in this study, DNase treatment of total synovial RNA prior to RT-PCR did not abolish the signal, suggesting the presence of live bacteria in these tissues (K. Kempsell and C. Cox, unpublished data). The intensity of the PCR signals varied between samples; this cannot be related directly to total bacterial numbers since the number of rRNA transcripts can vary enormously in bacterial cells according to rates of growth (16).
The bacterial crDNA fragments from each positive sample were cloned into the vector pT7-Blue, and the clones were sequenced. At least 46 individual clones were sequenced, and where bacterial species determination proved difficult due to cloning of nonspecific or partial crDNA products, additional sequencing was conducted (Table 3). In general, samples that consistently gave a very strong PCR signal yielded good recovery of bacterial crDNA-containing clones. Tissues with weaker RT-PCR signals gave less efficient recovery of bacterial crDNA-containing clones; therefore more clones were sequenced to generate an accurate bacterial species profile. Tissues which consistently gave very weak signals, e.g., patient 12 samples c, e, and f, were not cloned, since the recovery of bacterial crDNA-containing clones was expected to be very poor. Clones were sequenced on one pass only, which was estimated to be more than 97% accurate. Data analysis using the BLAST algorithm on database sequences revealed a number of bacterial species, most of which could be identified to near species level. These are outlined in Table 4, along with the percent similarity to the best-fit database sequence. Further comparisons were made between the cloned sequences to determine the overall similarity of bacterial species between patient samples. Figures 2 and 3 and Table 4 give graphic color-coded depictions of the profile of bacterial species found in each patient.
TABLE 3.
Summary of RT-PCR analysis of patient RNAs, histologic tests, and in situ hybridization with digoxigenin-labeled bacterial 16S rRNA oligonucleotidesa
| Disease and patient | Total no. of tissue extractions | Total no. of PCR tests | Mean PCR intensity score | Total no. of clones sequenced | No. of bacterial sequences | In situ intensity score | Comparative histology and in situ profile |
|---|---|---|---|---|---|---|---|
| RA | |||||||
| 1 | 1 | 2 | ++ | 46 | 46 | ND | |
| 2 | 2 | 4 | +++ | 46 | 43 | ND | |
| 3 | 2 | 4 | ++ | 46 | 42 | + | Diffuse inflammatory infiltrate, diffuse staining through section |
| 4 | 3 | 8 | ++ | 46 | 46 | ND | |
| 5 | 1 | 2 | + | 250 | 10 | ± | Diffuse inflammatory infiltrate, little staining, tissue fragile and fibrous |
| 6 | 2 | 3 | ++ | 46 | 46 | +++ | Diffuse inflammatory infiltrate, diffuse intense staining throughout section |
| 7 | 1 | 2 | + | 150 | 46 | +++ | Discrete foci of inflammatory cells, strong staining associated with these |
| 8 | 1 | 2 | ++ | 150 | 35 | ± | Diffuse inflammatory infiltrate with some small foci of inflammatory cells around vessels, little general staining but some associated with isolated cells |
| 9 | 1 | 2 | − | NDb | ND | ||
| OA | |||||||
| 10 | 2 | 4 | − | ND | − | Diffuse cellular infiltrate, no staining. | |
| 11 | 2 | 4 | + | 150 | 15 | ± | Large inflammatory infiltrate mainly at margin of tissue, little staining |
| 12a | 4 | 8 | ++ | 46 | 46 | ++ | Some isolated pockets of inflammatory infiltrate, strong staining associated with these, remaining tissue fragile and fibrous |
| 12b | 2 | 4 | − | ND | ± | Large inflammatory infiltrate with a small number of inflammatory foci, little staining | |
| 12c | 1 | 3 | ± | ND | ± | Large inflammatory infiltrate with a very small number of inflammatory foci, little staining | |
| 12d | 2 | 4 | +++ | 150 | 32 | +++ | Inflammatory infiltrate concentrated at margins, intense staining associated with this, remaining tissue fragile and fibrous |
| 12e | 2 | 4 | ± | ND | − | Large inflammatory infiltrate, no staining | |
| 12f | 2 | 4 | ± | ND | ± | Large inflammatory infiltrate, some weak staining associated with inflammatory cells | |
| 12g | 1 | 2 | + | 150 | 23 | ± | Large inflammatory infiltrate mainly at margins, little staining |
| 13 | 2 | 4 | ++ | 46 | 40 | + | Unusual appearance compared with other OA tissues, highly vascularized with large vessels and large fat deposits, diffuse inflammatory infiltrate, some diffuse staining associated with this |
| 14 | 1 | 2 | ++ | 150 | 38 | ND | |
| 19 | 1 | 1 | − | ND | ND | ||
| 20 | 1 | 2 | − | ND | ND | ||
| UA | |||||||
| 15 | 1 | 3 | + | 150 | 26 | ± | Diffuse inflammatory infiltrate plus small foci of inflammatory cells, staining associated with foci |
| 16 | 1 | 3 | − | ND | − | Diffuse inflammatory infiltrate throughout plus large inflammatory infiltrate at margins, no staining | |
| 17 | 1 | 3 | − | ND | ± | Diffuse inflammatory infiltrate, very weak staining | |
| 18 | 1 | 3 | + | 250 | 1 | ND | |
| Nonec | |||||||
| 21 | 1 | 3 | − | ND | ND | ||
| 22 | 1 | 3 | − | ND | ND | ||
| 23 | 1 | 3 | − | ND | ND |
Each patient was analyzed for bacterial crDNA amplification products in a number of independent experiments, and the results were collated to gain an overall profile of tissue positivity.
ND, not done.
None, normal controls.
TABLE 4.
Bacterial species identified by sequencing of amplified crDNA ampliconsa
| Code | SpeciesAccession no.b | % Similarity | Accession no.c | Species | Accession no. | % Similarity | Accession no. | ||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
 |
Accession number of the bacterial species in the database.
Accession number of the sequence isolated in this study. The percent similarity between the sequence isolated and the bacterial species in the database is given in the column between the two accession numbers.
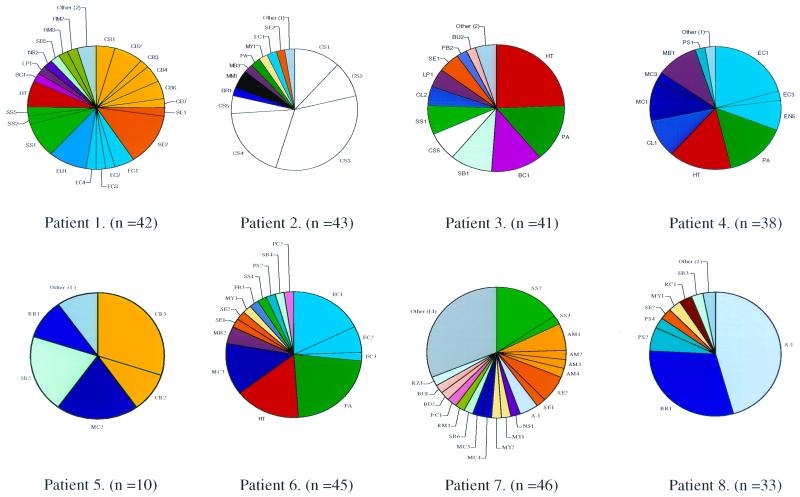
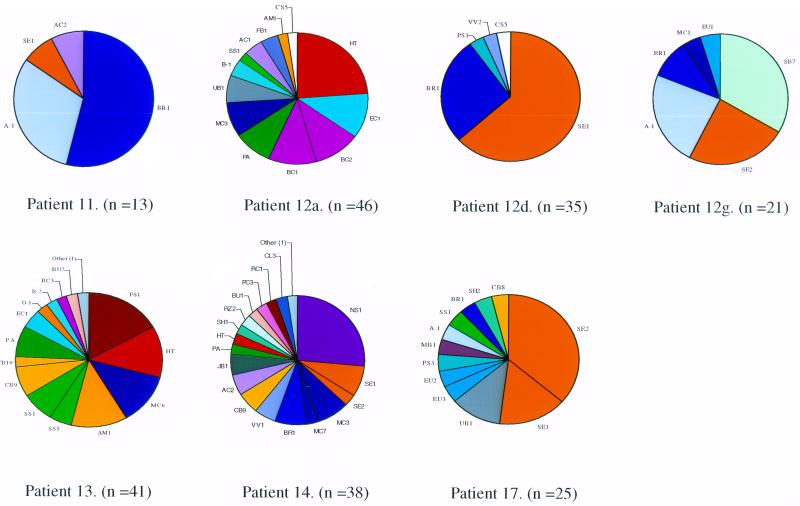
FIG. 2.
Diagrammatic representation of the bacterial species identified in RA patients by sequencing of cloned 16S crDNA amplicons. Each genus is represented by colour coding, and species are depicted by initials derived from abbreviated species names (Table 4). Section sizes are representative of the total number of sequences for that species in each patient.
FIG. 3.
Diagrammatic representation of the bacterial species identified in the OA and UA patients. The outline for genus and species representation is given in Fig. 2.
It can be seen from these analyses that there were a number of different bacteria in each patient tissue, with almost unique complements of bacterial species in each. Some species were unique to individuals, while others were shared with other patients in the study. The profiles were distinct and highly variable, indicating no bacterial contamination of the sample either at the clinical source or during processing. A number of well-characterized bacterial species were found, distributed across both disease groups. Most of these are of commensal origin, in particular from the skin and gastrointestinal tract. These include most notably Staphylococcus epidermidis, Propionibacterium acnes, and Escherichia coli, as well as other coliforms. Bacterial species that may also be derived from members of the endogenous microflora include streptococci, actinomycetes, and neisseriae. Some of these organisms have opportunistically infectious or pathogenic potential, and their presence is of note. Many of the bacterial sequences detected are, however, unique and have not been previously characterized by sequencing of rRNA since they show less than 97% similarity to known database sequences; these may represent new species. It is not clear what the source of these organisms is, but they could be either unidentified commensal organisms or environmental in origin. The only indication of the presence of a potentially pathogenic organism is the finding of Mycobacterium tuberculosis group (MTG) crDNA sequences in RA patient 6. The rRNA genes of the MTG, which also contains the vaccine strain M. bovis BCG, are more than 99.9% similar (31), so that the presence of these sequences may not indicate clinical infection with pathogenic M. tuberculosis.
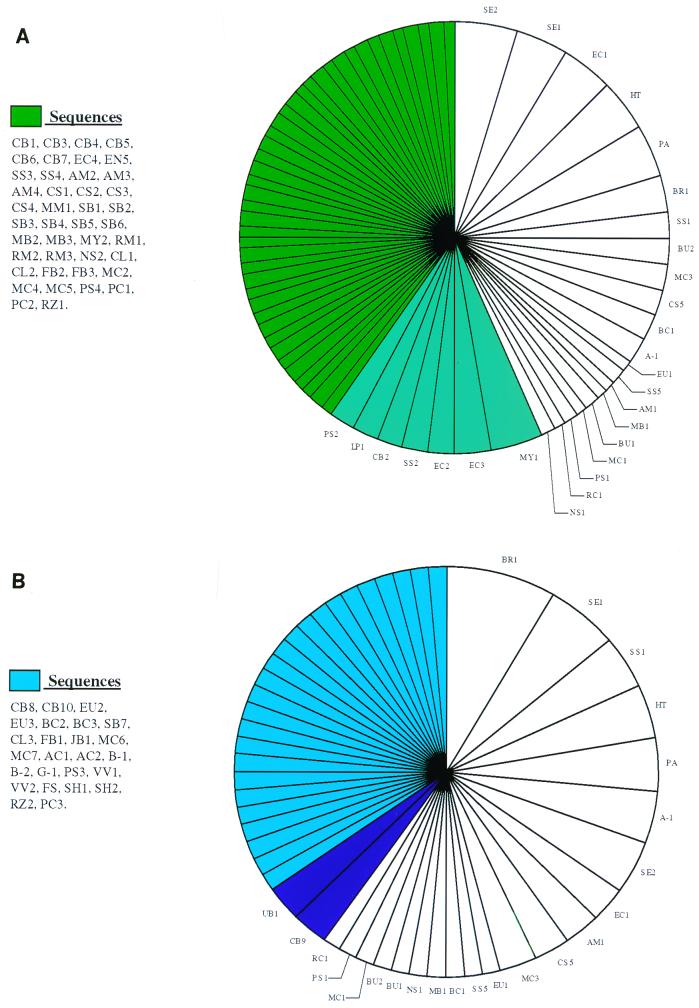
A total of 92 individual species were found in the RA group and 50 were found in the non-RA control group, implying that RA-affected joints, in addition to having a greater bacterial load as indicated by their RT-PCR signal, contain a greater number of species. Overall, the RA and non-RA control groups shared 21 species; therefore, these organisms are probably opportunistic colonizers of diseased synovium. Seven species in the RA group were unique (Fig. 4) and were found in more than one patient. These were Corynebacterium species 2, E. coli species 2 and 3, Streptococcus species 2, Pseudomonas species 2, Leptospira species 1, and Methylobacterium species 1. A total of 42 other identifiable bacterial species were unique to this group but in one individual only. In the non-RA group (Fig. 4), two species were unique and were found in more than one individual (Corynebacterium species 9 and unidentified bacterium species 1); a further 25 were unique and appeared in only one individual.
FIG. 4.
Diagrammatic representation of the total numbers of bacterial species unique to each patient group. (A) RA patients; (B) non-RA patients. Dark-shaded segments indicated species unique to that disease and found in more than one patient. Lighter-shaded segments indicate species unique to that disease and found in one patient only. Blank segments indicate species common to both patient groups.
Synovial tissue samples are not uniform with respect to bacterial species colonization.
In addition to species variation among patients, intrasample variation was found in tissue samples from the same patient. There appeared to be signal variation between individually analyzed samples from the same source. Samples 12 a to g, taken from the same knee of a patient with OA, were found to be substantially different in the intensity of their RT-PCR signals (Fig. 1), suggesting that not all parts of the same tissue have the same bacterial load. In addition to variation in RT-PCR signal intensity among samples, patient 12 had a different complement of bacterial species in the three strongly positive tissue samples sequenced, samples a, d, and g. This suggests that, since not all parts of the same tissue specimen have a similar bacterial load, they also do not contain the same species, suggesting microcolonization of different tissue areas.
In situ Localization of bacteria in synovial tissue sections.
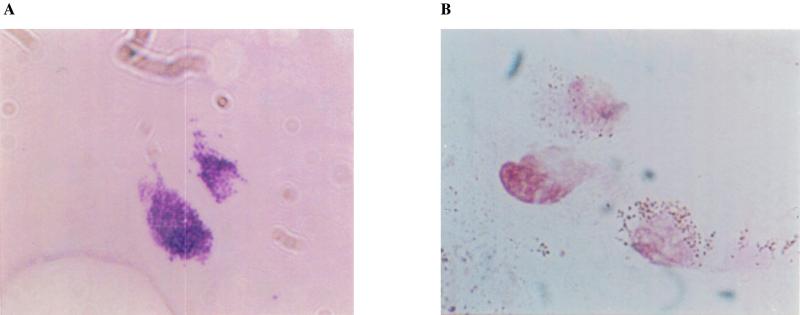
Since no organisms were found in normal synovial specimens, it is not thought that the presence of bacterial rRNA sequences in the test specimens is due to contamination from the surgical procedure or during subsequent processing and analysis. Staining of bacterial crDNA-positive tissue sections by conventional Gram stain did reveal some gram-positive organisms (Fig. 5), which appeared to be staphylococci, and other bacterium-like bodies, which did not give the expected positive (purple), or negative (pink) Gram stain result. This implies that bacteria present may not stain conventionally with these reagents, perhaps due to alterations in cell morphology. Conventional staining may not be appropriate in tissues where bacteria may exist in a CWD form.
FIG. 5.
Cryostat tissue sections from patient 8, stained with bacterial Gram stain. (A) Extracellular microcolony of staphylococci. Magnification, ×100. (B) cell-associated bacteria staining unconventionally brown by this staining technique. Magnification, ×40.
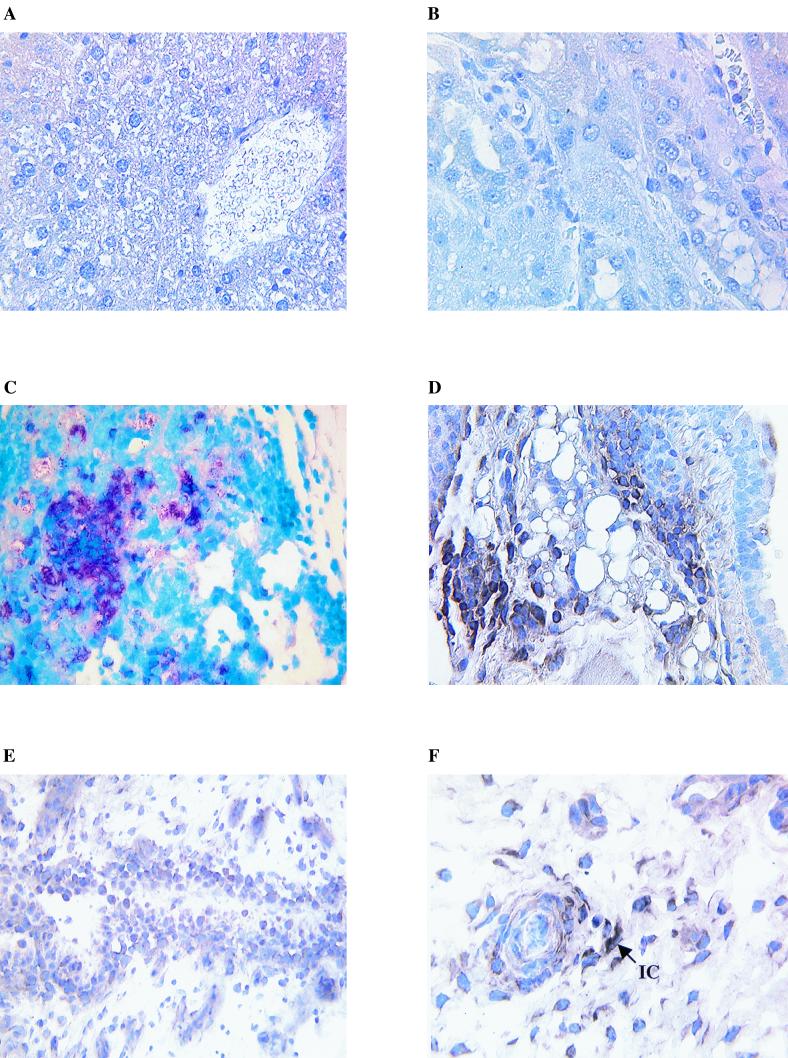
To validate our observations obtained by RT-PCR and to determine the in situ localization of the bacteria previously detected, we conducted in situ hybridization experiments with digoxigenin-labeled universal bacterial rRNA-specific oligonucleotides. These are complementary to bacterial rRNA and will bind directly to ribosome-associated rRNA. This technique proved to be extremely sensitive in the detection of bacteria in infected tissues (Fig. 6). M. tuberculosis-infected mouse lung tissue stained by conventional Ziel-Nielson Carbolfuschein methods and by in situ hybridization gave signals of comparable intensity. Control human and mouse tissues were negative. A UA sample that was previously negative by RT-PCR also proved negative by in situ hybridization. A weakly positive UA sample showed sparse localized intracellular staining around what appeared to be inflammatory foci.
FIG. 6.
Control and undifferentiated arthritis cryostat tissue sections stained with bacterium-specific stains or by in situ hybridization using digoxgenin-labeled bacterial 16S rRNA oligonucleotides ISH1 to ISH3. (A) Control mouse liver stained by in situ hybridization. (B) Control human kidney stained by in situ hybridization. (C) M. tuberculosis-infected mouse lung stained with mycobacterium-specific Ziehl-Nielson Carbolfuschein. (D) M. tuberculosis-infected mouse was stained by in situ hybridization. (E) Section from negative undifferentiated arthritis patient 16 stained by in situ hybridization. (F) Section from undifferentiated arthritis patient 15 stained by in situ hybridization. Note the intracellular staining (IC) within a focus of inflammatory cells. Magnifications, ×20 (A), ×40 (B and F), and ×10 (C to E).
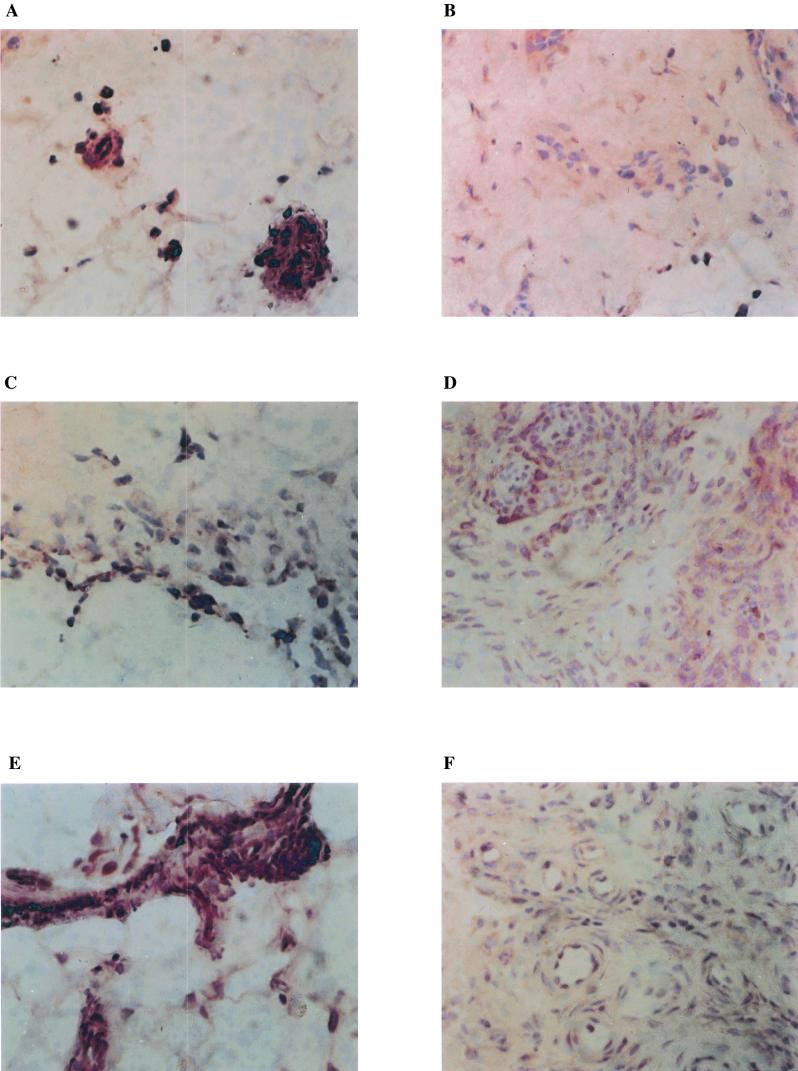
Human synovial tissues that had previously been found positive by RT-PCR gave strong hybridization signals with these probes (Fig. 7). Negative tissues did not, and the relative intensity of section staining appeared to correlate with the result obtained by RT-PCR (Table 3). This was particularly evident for samples from patient 12, where RT-PCR-negative tissue sections b and f gave weak in situ hybridization signals whereas sections a and d stained strongly. In most of the strongly staining sections, the staining appeared to be synovial cell associated, implying an intracellular location for much of the tissue-associated bacterial rRNA. Some small regions of extracellular staining may be associated with bacterial microcolonies, but these are less easy to distinguish by light microscopy. Single organisms were not readily detectable even at higher magnifications.
FIG. 7.
RA and OA arthritis cryostat tissue sections stained by in situ hybridization using digoxigenin-labeled bacterial 16S rRNA oligonucleotides ISH1 to ISH3. (A) RA patient 7. Note the heavy staining within a focus of what appear to be inflammatory cells. (B) RA patient 8. Note the sparse staining in isolated cells. (C) Patient 12 sample a. (D) Patient 12 sample b. (E) Patient 12 sample g. (F) Patient 12 sample e. Signals correlate with those obtained by RT-PCR. Magnifications, ×20.
DISCUSSION
The presence of a wide variety of bacterial species in both RA and other forms of chronic arthritis was an unexpected and novel discovery and indicates that arthritic joints are not sterile, as thought previously. Some of the species we identified are known organisms, but many are novel and may represent organisms previously uncharacterized by rRNA sequencing. The source of these organisms is not known, but they may be derived from environmental sources or from the indigenous microflora, since it is thought that in the gut at least, only a proportion of resident commensal microorganisms have been identified (9). Since many of the bacteria we identified in synovium have not been characterized previously in other studies of either environmental or clinical material, it may suggest that they are not readily cultivable. Certainly the majority of bacterial species identified in this study have not previously been found in the synovium.
Many species, e.g., P. acnes and S. epidermidis, were found in both the RA and non-RA patient groups, implying that their presence in synovium is not disease specific and that they are likely to be opportunistic colonizers of already diseased and compromised tissue. P. acnes is part of the normal skin microflora and has previously been isolated by culture from RA synovial fluid (6). Antigen from this organism has also been detected in synovial fluid leukocytes (7), implying an intracellular location. S. epidermidis has not previously been isolated from arthritic joints, other than in overt septic arthritis; however, since both organisms have pathogenic potential (reviewed in references 17, 25, and 27), particularly S. epidermidis, their presence may be significant. Since many of the other species found in both patient groups are also normal commensal residents of the skin or gastrointestinal tract, the role that any of these bacteria play in joint pathology must remain uncertain.
The presence of commensal organisms suggests trafficking from sites such as the gut; it has previously been suggested that gut permeability and mucosal competence is impaired in RA, although other studies have implicated nonsteroidal drugs in the causation of these abnormalities (24, 42), and all of the arthritic patients in this study, including those with OA, are likely to have been exposed to these agents. In a previous PCR study, Wilbrink et al. did not report evidence of bacteria in OA-affected synovium (61). However, the duration of disease in their OA patients was less than 12 months, compared to many years in our patients coming to joint replacement surgery. In this former study it also appears that the synovia of UA patients with a disease duration of more than 12 months are more often positive for bacteria by PCR (4 of 4 as opposed to 4 of 16). This would also suggest increasing colonization of arthritis tissue over time, irrespective of the cause, and may in part explain differences in bacterial positivity in OA patients between the two studies.
Many of the OA-affected synovia in our study showed histological features of late-stage disease and contained a substantial inflammatory infiltrate. For example, in synovial tissue from OA patient 12, in situ hybridization analyses showed large numbers of both intracellular and extracellular bacteria associated with inflammatory cells. Therefore, we conclude that any chronic synovitis may be colonized by commensal bacteria, which most probably reached the joint from the gut and skin within phagocytic cells, particularly macrophages, which are continuously recruited to the synovium. However, in general a large number of the RA-affected joint tissues were positive for bacteria and each contained a larger number of individual species, consistent with the greater degree of inflammation present in this disease.
Some species of bacteria were found only in RA, and while in many cases a particular organism was seen in only a single RA patient, some organisms were seen in more than one. These included corynebacteria and streptococci, which have been isolated from the synovial fluid of RA patients previously by culture (26, 58). Whether any of these organisms could play a role in RA specifically awaits further investigation, but it is noteworthy that previous studies of ReA have demonstrated the causative organism in only a proportion of affected synovia, even when the diagnosis has been firmly established. Thus, a significant etiologic agent would not necessarily be detected in all RA-affected synovia. This may be due in part to the limits in sensitivity of any rDNA amplification and sequencing technique. Studies such as this are problematical due to inherent sampling errors and the prohibitive sequencing effort that must be conducted to collate meaningful results. Low-copy-number sequences in these mixed crDNA pools are often overlooked; these can be seen by specific nested PCR but are not observed by large-scale sequencing (Kempsell and Cox, unpublished). Thus, large numbers of nonspecific organisms can obscure any etiological agent in low abundance.
Also conspicuously absent from the list of bacteria identified in RA in this study are bacterial species which have captured interest in recent years as possible etiological agents of RA, including mycoplasmas (52, 53) and Proteus mirabilis (64). M. tuberculosis (10, 51) was also not found, with the exception of MTG crDNA sequences in patient 6. In addition, we found no evidence of bacterial species that commonly cause ReA in RA-affected joint tissue, whereas these organisms can be identified in ReA-affected synovium (12, 18, 23, 62, 63).
If bacteria are involved in the pathology of RA, the condition might be expected to respond to antibacterial therapy. Antibiotic trials have been conducted in RA with different degrees of success (reviewed in reference 47), but little evidence of a general efficacy of antimicrobials has emerged. Caruso and coworkers have reported striking results using high-dose intra-articular injection of rifamycin SV (14); however these studies have not been corroborated by other workers. Some effect of tetracyclines has also been reported (1, 33, 57), but since these and other efficacious antibiotics have profound anti-inflammatory properties (19, 34, 56), the mechanism of any effect seen in RA remains unclear. However, given the evidence of subclinical bacterial colonization presented in this paper, part of their mode of action might well be antibacterial. If this were the case, the results of treatment might be expected to be variable since not all colonizing species would be sensitive to the particular antibiotic under trial. In addition, some of the organisms identified in this study are notoriously refractory to antibiotic treatment; in S. epidermidis, this is due to the production of protective biofilm matrices (17). Streptococci can also persist in tissues and evade killing by sequestration inside host cells (45). Since some of the bacteria appeared to be intracellular, this would be another reason to explain the lack of efficacy of some antimicrobials. ReA (55) and LyA (28, 43) are also relatively refractory to antimicrobial therapy, particularly in chronic disease, even though there is no doubt about the causative organism. Again, intractability to antimicrobials may be due to the persistence of slow-growing or latent bacteria. Thus, even if bacteria are involved in the primary pathogenesis of RA, the disease might not respond to conventional antimicrobial therapy.
Bacteria could cause or influence inflammatory joint disease in a number of ways including (i) persistent infection (35); (ii) induction of autoimmune pathology, perhaps through molecular mimicry (2, 35); (iii) production of bacterial superantigens (46, 54); and (iv) induction of immune dysfunction through other mechanisms (25). Which of these mechanisms is responsible for ReA or LyA remains obscure, and several may be involved. In LyA there is evidence of both persistent infection (27) and induction of immune dysfunction or autoimmune pathology, the latter arising from the autoreactive potential of T cells recognizing the B. burgdorferi outer membrane protein OspA (11, 21, 32, 37). Whether any of the bacterial species identified in RA could induce disease pathology by similar means remains unclear.
Other bacterially mediated autoimmune diseases in which persistent infection and induction of autoimmune pathology have been postulated to contribute to disease pathology include gastric inflammation (Helicobacter pylori) (4, 44) and arteriosclerosis (Chlamydia pneumoniae) (22, 36). In the former case, the organism is clearly implicated, since the disease responds to elimination of the organism, whereas in the latter case, the organism may either contribute to the pathogenesis of the atherosclerotic plaque or merely act as a colonizer of the diseased and compromised tissue. The present study raises the same questions with regard to the role of synovial bacteria in chronic arthritis. While the general colonization of the synovium which is a feature of all chronic synovitis may exacerbate inflammation irrespective of the cause of the arthritis, particular organisms may play an initiating role in diseases such as RA. It will be particularly informative to compare the spectrum of bacterial species isolated from long-established chronic synovitis and from acute disease, since in the latter case nonspecific colonization may not yet have occurred and any organisms isolated at this early stage would be more likely to be relevant to pathogenesis as initiators of disease.
ACKNOWLEDGMENTS
We thank Gabriel Panayi, Dorian Haskard, Peter Sewell, George Southgate, Gill Pountain, Andrew Hassell, and Sally Roberts for the kind provision of clinical material for use in this work. We also thank Alan Lewis for help with sequence analysis and other technical assistance.
We thank Glaxo Wellcome UK for sponsoring this project and for financial support of Charles Cox.
REFERENCES
- 1.Alarcón G S, Mikhail I S. Antimicrobials in the treatment of rheumatoid arthritis and other arthritides: a clinical perspective. Am J Med Sci. 1994;308:201–209. doi: 10.1097/00000441-199409000-00017. [DOI] [PubMed] [Google Scholar]
- 2.Albani S, Carson D A. A multistep molecular mimicry hypothesis for the pathogenesis of rheumatoid arthritis. Immunol Today. 1996;17:466–470. doi: 10.1016/0167-5699(96)20029-g. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appelmelk B J, Faller G, Claeys D, Kirchner T, Vandenbrouke-Grauls C M J E. Bugs on trial: the case of Helicobacter pylori and autoimmunity. Immunol Today. 1998;19:296–299. doi: 10.1016/s0167-5699(98)01281-x. [DOI] [PubMed] [Google Scholar]
- 5.Arnett F C, Edworth S M, Bloch D A, Shane D J, Fries J F, Cooper N S. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 6.Bartholemew L E, Nelson F R. Corynebacterium acnes in rheumatoid arthritis. I. Isolation and antibody studies. Ann Rheum Dis. 1972;31:22–27. doi: 10.1136/ard.31.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartholemew L E, Nelson F R. Corynebacterium acnes in rheumatoid arthritis. II. Identification of antigen in synovial fluid leukocytes. Ann Rheum Dis. 1972;31:28–33. doi: 10.1136/ard.31.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behar S M, Porcelli S A. Mechanisms of autoimmune disease induction: the role of the immune response to microbial pathogens. Arthritis Rheum. 1995;38:458–476. doi: 10.1002/art.1780380403. [DOI] [PubMed] [Google Scholar]
- 9.Berg R D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 10.Billingham M E J. Adjuvant arthritis: the first model. In: Henderson B, Edwards J C W, Pettipher E R, editors. Mechanisms and models in rheumatoid arthritis. London, United Kingdom: Academic Press Ltd.; 1995. pp. 25–46. [Google Scholar]
- 11.Brightbill H D, Libraty D H, Krutzik S R, Yang R-B, Belisle J T, Bleharski J B, Maitland M, Norgard M V, Plevy S E, Smale S T, Brennan P J, Bloom B R, Godowski P J, Modlin R L. Host defence mechanisms triggered by microbial lipoproteins through Toll-like receptors. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 12.Burmester G R, Daser A, Kamradt T, Krause A, Mitchison N A, Seiper J. Immunology of reactive arthritides. Annu Rev Immunol. 1995;13:229–250. doi: 10.1146/annurev.iy.13.040195.001305. [DOI] [PubMed] [Google Scholar]
- 13.Calleja C, Pascussi J M, Mani J C, Maurel P, Vilarem M J. The antibiotic rifampicin is a nonsteroidal ligand and activator of the human glucocorticoid receptor. Nat Med. 1998;4:92–96. doi: 10.1038/nm0198-092. [DOI] [PubMed] [Google Scholar]
- 14.Caruso I. Twenty years of experience with intra-articular rifamycin for chronic arthritides. J Int Med Res. 1997;25:307–317. doi: 10.1177/030006059702500601. [DOI] [PubMed] [Google Scholar]
- 15.Chary-Valckenaere I, Jaulhac B, Champigneulle J, Piedmont Y, Mainard D, Pourel J. Ultrastructural demonstration of intracellular localisation of Borrelia burgdorferi in Lyme arthritis. Br J Rheumatol. 1998;37:468–469. doi: 10.1093/rheumatology/37.4.468. [DOI] [PubMed] [Google Scholar]
- 16.Condon C, Squires C, Squires C L. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costerton J W, Stewart P S, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 18.Gaston J S H, Cox C J, Granfors K. Clinical and experimental evidence for persistent Yersinia infection in reactive arthritis. Arthritis Rheum. 1999;42:2239–2242. doi: 10.1002/1529-0131(199910)42:10<2239::AID-ANR29>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 18a.Genetics Computer Group. Wisconsin package version 10.0. Madison, Wis: Genetics Computer Group; 1999. [Google Scholar]
- 19.Golub L M, Suomalainen K, Sorsa T. Host modulation with tetracyclines and their chemically modified analogues. Curr Opin Dent. 1992;2:80–90. [PubMed] [Google Scholar]
- 20.Griffiths M M. Arthritis induced by bacteria and viruses. In: Henderson B, Edwards J C W, Pettipher E R, editors. Mechanisms and models in rheumatoid arthritis. London, United Kingdom: Academic Press Ltd.; 1995. pp. 411–430. [Google Scholar]
- 21.Gross D M, Forsthuber T, Tary-Lehmann M, Etling C, Ito K, Nagy Z A, Field J A, Steere A C, Huber B T. Identification of LFA-1 as a candidate autoantigen in treatment-resistent Lyme arthritis. Science. 1998;281:703–706. doi: 10.1126/science.281.5377.703. [DOI] [PubMed] [Google Scholar]
- 22.Guru T. Infections: a cause of artery-clogging plaques. Science. 1998;281:35–37. doi: 10.1126/science.281.5373.35. [DOI] [PubMed] [Google Scholar]
- 23.Hammer M, Nettelnbreker E, Hopf S, Schmitz E, Porshke K, Zeidler H. Chlamydial rRNA in the joints of patients with Chlamydia-induced arthritis and undifferentiated arthritis. Clin Exp Rheumatol. 1992;10:63–66. [PubMed] [Google Scholar]
- 24.Hernandez-Cruz B, Cardiel M H, Villa A R, Alcocer-Varela J. Development, recurrence, and severity of infections in Mexican patients with rheumatoid arthritis. A nested case-control study. J Rheumatol. 1998;25:1900–1907. [PubMed] [Google Scholar]
- 25.Holland K T, Aldana O, Bojar R A, Cunliffe W J, Eady E A, Holland D B, Ingham E, McGeown C, Till A, Walters C. Propionibacterium acnes and acne. Dermatology. 1998;196:67–68. doi: 10.1159/000017870. [DOI] [PubMed] [Google Scholar]
- 26.Hollander J L. History of rheumatoid arthritis: an american perspective. In: Utsinger P D, Zvaifler N J, Erlich G E J B, editors. Rheumatoid arthritis: etiology: diagnosis: management'. J. B. Philadelphia, Pa: Lippincott Co.; 1985. pp. 11–17. [Google Scholar]
- 27.Jakab E, Zbinden R, Gubler J, Ruef C, von Graevenitz A, Krause M. Severe infections caused by Propionibacterium acnes: an underestimated pathogen in late postoperative infections. Yale J Biol Med. 1996;69:477–482. [PMC free article] [PubMed] [Google Scholar]
- 28.Jaulhac B, Sibilia J, Pourel J, Kuntz J-L. Borrelia burgdorferi in Lyme and undifferentiated arthritis. Rev Rhum. 1999;66(Suppl.):20S–22S. [PubMed] [Google Scholar]
- 29.Jones R E, Chatham W W. Update on sarcoidosis. Curr Opin Rheumatol. 1999;11:83–87. doi: 10.1097/00002281-199901000-00015. [DOI] [PubMed] [Google Scholar]
- 30.Kalish R A, Leong J M, Steere A C. Association of treatment-resistant chronic Lyme arthritis with HLA-DR4 and antibody reactivity to OspA and OspB of Borrelia burgdorferi. Infect Immun. 1993;61:2774–2779. doi: 10.1128/iai.61.7.2774-2779.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kempsell K E, Ji Y-E, Estrada I C E, Colston M J, Cox R A. The nucleotide sequence of the promoter, 16S rRNA and spacer region of the ribosomal RNA operon of Mycobacterium tuberculosis and comparison with Mycobacterium leprae precursor rRNA. J Gen Microbiol. 1992;138:1717–1727. doi: 10.1099/00221287-138-8-1717. [DOI] [PubMed] [Google Scholar]
- 32.Kerksiek K M, Pamer E G. T cell responses to bacterial infection. Curr Opin Immunol. 1999;11:400–405. doi: 10.1016/S0952-7915(99)80067-3. [DOI] [PubMed] [Google Scholar]
- 33.Kloppenberg M, Dijkmans B A C, Breedveld F C. Antimicrobial therapy for rheumatoid arthritis. Baillières Clin Rheumatol. 1995;9:759–769. doi: 10.1016/s0950-3579(05)80312-7. [DOI] [PubMed] [Google Scholar]
- 34.Kloppenburg M, Verweij C L, Miltenburg A M, Verhoeven A J, Daha M R, Dijkmans B A, Breedveld F C. The influence of tetracyclines on T cell activation. Clin Exp Immunol. 1995;102:635–641. doi: 10.1111/j.1365-2249.1995.tb03864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krause A, Kamradt T, Burmester G. Potential infectious agents in the Induction of Arthritides. Curr Opin Rheumatol. 1996;8:203–209. doi: 10.1097/00002281-199605000-00007. [DOI] [PubMed] [Google Scholar]
- 36.Kuo C-C, Campbell L A. Is infection with Chlamydia pneumoniae a causative agent in atheriosclerosis. Mol Med Today. 1998;Octr:426–430. doi: 10.1016/s1357-4310(98)01351-3. [DOI] [PubMed] [Google Scholar]
- 37.Lengl-Janssen B, Strauss A F, Steere A C, Kamradt T. The T helper response in Lyme arthritis: differential recognition of Borrelia burgdorferi outer surface protein A in patients with treatment resistent or treatment-responsive Lyme arthritis. J Exp Med. 1994;180:2069–2078. doi: 10.1084/jem.180.6.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maini R N, Chu C Q, Feldmann M. Aetiopathogenesis of rheumatoid arthritis. In: Henderson B, Edwards J C W, Pettipher E R, editors. Mechanisms and models in rheumatoid arthritis. London, United Kingdom: Academic Press Ltd.; 1995. pp. 25–46. [Google Scholar]
- 39.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 40.Marker-Hermann E, Hohler T. Pathogenesis of human leukocyte antigen B27-positive arthritis. Information from clinical materials. Rheum Dis Clin North Am. 1998;24:865–881. doi: 10.1016/s0889-857x(05)70046-6. [DOI] [PubMed] [Google Scholar]
- 41.Medrano J M M, Galbete J C V. Evidencia morfológica dela presencia de un bacilo en cultivos celulares de membrana sinovial de enfermos de artritis reumatoide. Rev Clin Esp. 1990;187:329–333. [PubMed] [Google Scholar]
- 42.Mielants H, De Vos M, Goemaere S, Schelstraete K, Cuvelier C, Goethals K, Maertens M, Ackerman C, Veys E M. Intestinal mucosal permeability in inflammatory rheumatic diseases. II. Role of disease. J Rheumatol. 1991;18:394–400. [PubMed] [Google Scholar]
- 43.Melief M J, Hoijer M A, Van Paassen H C, Hazenberg M P. Presence of bacterial flora-derived antigen in synovial tissue macrophages and dendritic cells. Br J Rheumatol. 1995;34:1112–1116. doi: 10.1093/rheumatology/34.12.1112. [DOI] [PubMed] [Google Scholar]
- 44.Morshed M G, Karita M, Konishi H, Okita K, Nakazawa T. Growth medium containing cyclodextrin and low concentration of horse serum for cultivation of Helicobacter pylori. Microbiol Immunol. 1994;38:897–900. doi: 10.1111/j.1348-0421.1994.tb02143.x. [DOI] [PubMed] [Google Scholar]
- 45.Neeman R, Keller N, Barzilai A, Korenman Z, Sela S. Prevalence of internalisation-associated gene, prtF1, among persisting group-A streptococcus strains isolated from asymptomatic carriers. Lancet. 1998;352:1974–1977. doi: 10.1016/S0140-6736(97)12452-7. [DOI] [PubMed] [Google Scholar]
- 46.Paliard X, West S G, Lafferty J A, Clements J R, Kappler J W, Marrack P, Kotzin B L. Evidence for the effects of a superantigen in rheumatoid arthritis. Science. 1991;253:325–329. doi: 10.1126/science.1857971. [DOI] [PubMed] [Google Scholar]
- 47.Panush R S, Longley S. Therapies of potential but unproven benefit. In: Utsinger P D, Zvaifler N J, Erlich G E J B, editors. Rheumatoid arthritis: etiology: diagnosis: management. J. B. Philadelphia, Pa: Lippincott Co.; 1985. pp. 695–709. [Google Scholar]
- 48.Priem S, Burmester G R, Kamradt T, Wolbart K, Rittig M G, Krause A. Detection of Borrellia burgdorferi by polymerase chain reaction in synovial membrane, but not in synovial fluid from patients with persisting Lyme arthritis after antibiotic therapy. Ann Rheum Dis. 1998;57:118–121. doi: 10.1136/ard.57.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Relman D A. The search for unrecognised pathogens. Science. 1999;284:1308–1310. doi: 10.1126/science.284.5418.1308. [DOI] [PubMed] [Google Scholar]
- 50.Relman D A, Schmit T M, McDermott R P, Falkow S. Identification of the uncultured bacillus of Whipples disease. N Engl J Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 51.Rook G, Lydyard P, Stanford J. Mycobacteria and rheumatoid arthritis. Arthritis Rheum. 1990;33:431–435. doi: 10.1002/art.1780330319. [DOI] [PubMed] [Google Scholar]
- 52.Schaeverbeke T, Gilroy C B, Bébéar C, Dehais J, Taylor-Robinson D. Mycoplasma fermentans, but not M. penetrans detected by PCR assays in synovium from patients with rheumatoid arthritis and other rheumatic disorders. J Clin Pathol. 1996;49:824–828. doi: 10.1136/jcp.49.10.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaeverbeke T, Clerc M, Lequen L, Charron A, Bébéar C, de Barbeyrac B, Bannwarth B, Dehais J, Bébéar C. Genotypic characterization of seven strains of Mycoplasma fermentans isolated from synovial fluids of patients with arthritis. J Clin Microbiol. 1998;36:1226–1231. doi: 10.1128/jcm.36.5.1226-1231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schiffenbauer J, Soos J, Johnson H. The possible role of bacterial superantigens in the pathogenesis of autoimmune disorders. Immunol Today. 1998;19:117–120. doi: 10.1016/s0167-5699(97)01199-7. [DOI] [PubMed] [Google Scholar]
- 55.Sieper J, Braun J. Treatment of reactive arthritis with antibiotics. Br J Rheumatol. 1998;37:717–720. doi: 10.1093/rheumatology/37.7.717. [DOI] [PubMed] [Google Scholar]
- 56.Spisani S, Dovigo L, Corazzo G, Carletti R, Traniello S. The effect of Rifamycin SV on neutraphil functions in patients with rheumatoid arthritis. Scand J Rheumatol. 1982;11:65–69. doi: 10.3109/03009748209098164. [DOI] [PubMed] [Google Scholar]
- 57.Trentham D E, Dynesius-Trentham R A. Antibiotic therapy for rheumatoid arthritis: scientific and anecdotal appraisals. Rheum Dis Clin North Am. 1995;21:817–834. [PubMed] [Google Scholar]
- 58.Utsinger P D, Zvaifler N J, Weiner S B. Etiology. In: Utsinger P D, Zvaifler N J, Erlich G E J B, editors. Rheumatoid arthritis: etiology: diagnosis: management. J. B. Philadelphia, Pa: Lippincott Co.; 1985. pp. 21–48. [Google Scholar]
- 59.van der Vliet G M E, Schlepers P, Schukkink R A, van Gemen B, Klatser P R. Assessment of mycobacterial viability by RNA amplification. Antimicrob Agents Chemother. 1994;38:1959–1965. doi: 10.1128/aac.38.9.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wall S, Kunze Z M, Saboor S, Soufleri I, Seechurn P, Chiodini R, McFadden J J. Identification of spheroplast-like agents isolated from tissues of patients with Crohn's disease and control tissues by polymerase chain reaction. J Clin Microbiol. 1993;31:1241–1245. doi: 10.1128/jcm.31.5.1241-1245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilbrink B, van der Heijden I M, Schouls L M, van Embden J D A, Hazes J M W, Breedveld F C, Tak P P. Detection of bacterial DNA in joint samples from patients with undifferentiated arthritis and reactive arthritis using polymerase chain reaction with universal 16S ribosomal RNA primers. Arthritis Rheum. 1998;41:535–543. doi: 10.1002/1529-0131(199803)41:3<535::AID-ART20>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 62.Wilkinson N Z, Kingsley G H, Jones H W, Sieper J, Braun J, Ward M E. The detection of DNA from a range of bacterial species in the joints of patients with a variety of arthritides using a nested, broad-range polymerase chain reaction. Rheumatology. 1999;38:260–266. doi: 10.1093/rheumatology/38.3.260. [DOI] [PubMed] [Google Scholar]
- 63.Wilkinson N Z, Kingsley G H, Sieper J, Braun J, Ward M E. Lack of correlation between the detection of Chlamydia trachomatis DNA in synovial fluid from patients with a range of rheumatic disease and the presence of an antichlamydial immune response. Arthritis Rheum. 1998;41:845–854. doi: 10.1002/1529-0131(199805)41:5<845::AID-ART11>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 64.Wilson C, Thakore A, Isenberg D, Ebringer A. Correlation between anti-Proteus antibodies and isolation rates of Proteus mirabilis in rheumatoid arthritis. Rheumatol Int. 1997;16:187–189. doi: 10.1007/BF01330294. [DOI] [PubMed] [Google Scholar]