Abstract
Chickens were infected with a pathogenic strain of Mycoplasma gallisepticum, and the expression of pMGA, the major surface protein, was inferred by examination of colonies from ex vivo cells. Within 2 days postinfection, 40% of cells had ceased the expression of the original pMGA surface protein (pMGA1.1), and by day 6, the majority of recovered cells were in this category. The switch in pMGA phenotype which had occurred in vivo was reversible, since most colonies produced from ex vivo progenitors exhibited frequent pMGA1.1+ sectors. After prolonged in vivo habitation, increasing proportions of recovered cells gave rise to variant pMGA colonies which had switched from the expression of pMGA1.1 to another gene, pMGA1.2, concomitant with the acquisition of a (GAA)12 motif 5′ to its promoter. Collectively, the results suggest that changes in M. gallisepticum pMGA gene expression in vivo are normal, common, and possibly obligate events for successful colonization of the host. Surprisingly, the initial cessation of pMGA1.1 expression occurred in the absence of detectable pMGA antibodies and seemed to precede the adaptive immune response.
Mycoplasma gallisepticum cells express an abundant surface lipoprotein known as pMGA (12). Each M. gallisepticum strain usually expresses only one homogeneous, unique pMGA molecule (8), which in strain S6, the subject of this study, has been designated pMGA1.1 (13, 14). All strains thus far analyzed contain large pMGA multigene families ranging in size from 32 to 70 genes (2). In strain S6, all such genes contain a putative promoter motif (8) and many possess an uninterrupted open reading frame (14). Despite the presence of many pMGA genes, all but one either are transcriptionally silent or are transcribed at very low levels within individual field isolates of the organism (8). Previous work from this laboratory (15) has shown that the expression of pMGA1.1 by strain S6 cells ceased when cells were grown with a particular pMGA1.1-specific antibody (MAb66). Concomitant with the cessation of pMGA1.1 expression in these cells, the expression of a related lipoprotein, pMGA1.9, was switched on. Removal of antibody from culture medium then resulted in the reexpression of pMGA1.1 (15). The transcriptional switching between pMGA genes was shown to be unequivocally associated with changes in the length of a unique trinucleotide GAA repeat (9), a motif found to be common to all pMGA genes (2). Specifically, a (GAA)12 motif 5′ to a pMGA1.1 promoter was shown to be an obligate requirement for the expression of that gene (9). It was further shown that changes in pMGA gene expression occurred as a result of the inherent instability of GAA repeats in M. gallisepticum (9).
In vitro and, more importantly, in vivo, epitope switching has been observed for many M. gallisepticum surface molecules (3, 7, 11, 24) and in mollicutes generally (5, 21, 22, 25). This switching of surface epitopes may provide the organism with a means of avoiding the host immune response and/or of increasing tissue tropism. The surface pMGA1.1 lipoprotein is one of the main immune targets during M. gallisepticum strain S6 infection of chickens and is one of only approximately 10 proteins recognized in Western blot analysis using serum from chickens 2 weeks after infection (12). Given the proven importance of pMGA as an immune target and the potential of pMGA genes to be transcriptionally turned on or off by high-frequency alterations to their respective (GAA)n motifs, it was of interest to study pMGA gene expression during the course of a natural infection.
An experimental infection and sample collection procedure was therefore designed to determine whether this switching phenomenon occurred in the chicken and, if so, whether it was consequential to the production of host pMGA antibodies. The work herein confirms that switches in pMGA expression occur frequently during the course of a natural infection but that the elicitation of pMGA-specific antibodies is not required to mediate them.
MATERIALS AND METHODS
Mycoplasma, mycoplasma media, and immunological reagents.
A virulent clone of M. gallisepticum strain S6 which had been passaged through specific-pathogen-free turkeys (18) and had then undergone 14 in vitro passages (kindly supplied by Janet M. Bradbury) is referred to here as S6J. M. gallisepticum strain F (passage 16) was from S. H. Kleven (23). Mycoplasma broth (MB) (17) was a modified Frey medium (6) supplemented with 10% swine serum. Mycoplasma agar growth medium (MA) was the same as MB except that the medium was solidified with 1.0% (wt/vol) special Noble agar (Difco); glucose and phenol red were omitted. The monoclonal antibodies (MAbs), MAb66 and MAb86 to separate pMGA epitopes, and rabbit anti-pMGA1.1 have been previously described (12, 15).
Experimental infection of chickens and sample collection.
White Leghorn chickens were purchased from the Commonwealth Scientific and Industrial Research Organisation specific-pathogen-free flock, which were known to be free of M. gallisepticum. The chickens were housed in plastic bubble isolators under positive pressure and were removed from the isolators only for inoculation and sample collection purposes.
The chickens were experimentally infected by aerosol spray with a cloned isolate of S6J cells in two separate experiments using a 10-min exposure time in the aerosol chamber as previously described (20). The viable count of the inoculum (color-changing units [CCU] per milliliter) immediately before the inoculation of chickens was determined using the most-probable-number method (16). For one experiment, the chickens at the time of infection were 2 weeks old, the viable count of the starting culture was 1 × 108 CCU/ml, and the sample collection time points were 14, 28, and 42 days postinfection (p.i.). For a second experiment, the chickens at the time of infection were 6 weeks old, the viable count of the starting culture was 2.2 × 108 CCU/ml, and the sample collection time points were 1, 2, 4, 6, 8, 10, and 13 days p.i. Two chickens were selected at random at each time point p.i. and bled before being subjected to barbiturate euthanasia. Tracheal washes were obtained by excising the trachea with sterile scissors, attaching one end to a 5-ml syringe and placing the other end in 5 ml of MB, which was then aspirated through the trachea 10 times. Air sac samples were obtained by swabbing the left and right air sacs of each chicken using a cotton swab presoaked in 5 ml of MB. The swab was then replaced in the remaining MB and vortexed for 10 s before being removed. Viable-organism counts, determined as CFU per milliliter, from collected samples were obtained by appropriate duplicate dilutions of each sample spread onto MA, and colonies were counted after incubation at 37°C for 6 days.
pMGA1.1 ELISA.
The level of chicken pMGA antibodies in serum and tracheal wash samples was determined using a pMGA enzyme-linked immunosorbent assay (ELISA) as follows. Coating antigen (pMGA1.1) for the ELISA was obtained from a clone of M. gallisepticum S6 cells. In brief, the cells were lysed in Triton X-114 (4) and the lipophilic fraction containing pMGA1.1 was subjected to immunoaffinity chromatography using MAb86 coupled to Sepharose 4B as previously described (12). Individual wells of Nunc-Immuno MaxiSorp flat-bottom plates (Inter Med) were coated with 100 μl of purified pMGA1.1 (0.1 mg/ml) in carbonate buffer (0.032 M Na2CO3, 0.068 M NaHCO3), and the plates were incubated at 4°C overnight. The wells were blocked with 200 μl of phosphate-buffered saline (PBS)–1% bovine serum albumin (BSA) for 1 h at room temperature (RT) and then washed three times in PBS–0.05% (vol/vol) Tween 20 (PBS-T). Serum and tracheal or air sac washes were centrifuged at 16,000 × g for 30 min at 4°C and assayed as serial 10-fold dilutions using ELISA diluent (0.5 M NaCl, 1 mM disodium EDTA, 0.1 M Tris, pH 7.4 [HCl], containing 2% [wt/vol] BSA, 3% [vol/vol] Triton X-100, and 3% [vol/vol] Tween 20). Duplicate 100-μl samples were added to the wells and incubated for 3 h at RT. The plate was washed three times using PBS-T, 100 μl of rabbit anti-chicken immunoglobulin G (IgG) heavy plus light chains conjugated to horseradish peroxidase (Nordic Immunological Laboratories) diluted 1:2,000 in ELISA diluent was added to each well, and the plate was incubated for 1 h at RT. The plate was washed twice in PBS-T and once in PBS before 100 μl of prepared substrate (one tablet of 1 mg of 3,3′,5,5′-tetramethylbenzidine dihydrochloride [Sigma] reconstituted as recommended by the manufacturer) was added to each well. The reactions were stopped after 5 min by adding 25 μl of 2 M H2SO4. Absorbance was measured at 450 nm with a Titertek Multiskan MC automatic reader. Specific pMGA antibody titers were determined relative to a high-titer reference serum (day 14 p.i. with S6J cells) arbitrarily set to 1 U/ml.
Immunostaining of colony lifts.
Colonies either obtained from the starting inoculum or reisolated from birds, as described above, were lifted onto sterile nitrocellulose filters (Hybond C; Amersham) from plates (diameter, 82 mm) containing no more than 300 colonies, left for 3 min before being peeled off the plate, and allowed to air dry.
Colony lifts were blocked for 1 h at RT with Tris-buffered saline (TBS) (20 mM Tris, pH 7.4 [HCl], 0.9% [wt/vol] NaCl) containing 5% BSA, rinsed in TBS–0.05% (vol/vol) Tween 20 (TBS-T), and incubated for 1 h at RT with a 1:200 dilution of MAb66 in TBS-T containing 1% BSA. The filters were rinsed, washed twice in 0.1% BSA–TBS-T for 20 min, and incubated for 1 h at RT with a 1:2,000 dilution (in TBS-T–1% BSA) of sheep anti-mouse Ig conjugated to horseradish peroxidase (Silenus). The filters were washed as above and then rinsed in TBS before being developed in 3,3′-diaminobenzidine tetrahydrochloride (DAB tablets [D-5905; Sigma]) to give a brown color. Second-layer staining was performed essentially as for the first layer, using a 1:1,000 dilution of rabbit anti-pMGA (1 h) and a 1:2,000 dilution of swine anti-rabbit Ig conjugated to alkaline phosphatase (Dako Corp.). The final color development was carried out with Fast Red TR/naphthol AS-MX (FR tablets [F-4523; Sigma]) as specified by the manufacturer.
Derivation of colony clones C20.2A and C20.2D with homogeneous pMGA phenotypes.
The colony pMGA phenotype was determined by lifting the colony onto nitrocellulose filters and doubly immunostaining it as described above. A stained colony (well separated from neighboring colonies) with the desired phenotype was then located, removed from the original plate as a colony-agar plug, transferred to 5 ml of MB, and grown to late log phase at 37°C. These cells were grown on MA, and the above procedure was repeated.
Statistical analysis.
Statistical analysis of the data presented in Fig. 3 was done using a two-tailed test and the computer program SuperANOVA (Abacus Concepts, Inc., Berkeley, Calif.).
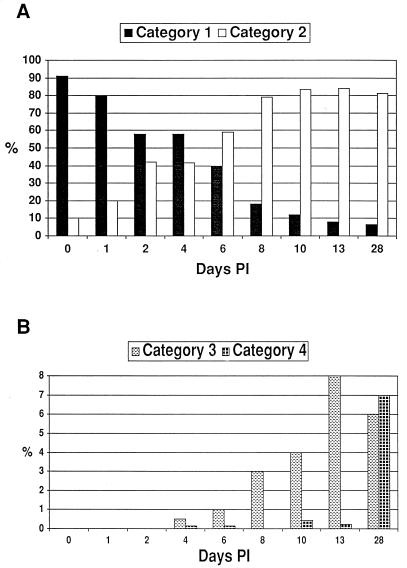
FIG. 3.
Changes in S6J colony pMGA1.1 and variant pMGA phenotypes with time p.i. (A) Bar graph comparing the average proportion of colonies in categories 1 and 2 (see Fig. 2) in the starting inoculum and from tracheal wash samples at various time points p.i. The total number of colonies counted for each time point ranged from 119 to 663. The mean proportions of category 1 and 2 colonies on days 2, 4, 6, 8, 13, and 28 were found to be significantly different from those on day 0 (P < 0.05). (B) Bar graph comparing the average proportion of colonies in categories 3 and 4 (see Fig. 2) in the starting inoculum and from tracheal wash samples at various time points p.i. The total number of colonies counted for each time point ranged from 119 to 663. The mean proportions of category 3 colonies on days 10, 13, and 28 were significantly different from the proportion seen on day 0 (P < 0.05). Only the mean proportion of category 4 colonies on day 28 was significantly different from that on day 0 (P < 0.05).
Amino-terminal sequencing.
Cellular proteins were partitioned into the TX-114 detergent phase, subjected to reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to polyvinylidene difluoride membrane, and the protein identified by the rabbit antisera was excised and sequenced as previously described (8).
Northern blot analysis.
Total RNA was prepared and subjected to Northern blot analysis as previously described (8). PCR probes specific for the pMGA1.1 or pMGA1.2 genes (both encompassed a region encoding 10 amino acids of the leader sequence followed by 45 [pMGA1.1] or 44 [pMGA1.2] amino acids of the mature polypeptide) were labeled and used under the same conditions as previously described (8).
Determination of the pMGA1.2 gene (GAA)n repeat lengths.
The region of the pMGA1.2 gene containing the pMGA1.2 (GAA)n repeat motif was amplified by two-round PCR as previously described, cloned into pGEM-T, and sequenced (9).
RESULTS
Confirmation of the pMGA phenotype of S6J.
The virulent S6J isolate expressed a pMGA molecule with an identical size to pMGA1.1 and possessed the same MAb epitopes as pMGA1.1 (MAb66 and MAb86 [data not shown]). The amino-terminal sequence of the expressed pMGA molecule from S6J cells was −TTPTPSPAHNPN, which is identical to pMGA1.1 (14) except for a single proline to histidine change at cycle 10. Using a rabbit antiserum to pMGA1.9 (9), S6J was also found to be expressing pMGA1.9 (data not shown).
Reisolation counts.
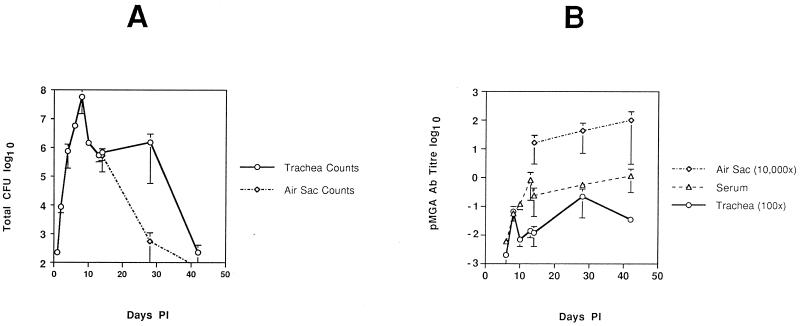
The average total number of S6J cells reisolated from two chickens at each time point p.i. is shown in Fig. 1A. There was little difference between the reisolation numbers from the two birds tested at most time points p.i. (indicated by the error bars in Fig. 1A). The average reisolation counts at the neighboring time points of days 13 and 14 p.i. from two separate experiments (Fig. 1A) agreed well with each other and indicate that the aerosol method of inoculation produced reproducible results between experiments. Figure 1A shows that after inoculation with S6J cells there was a rapid growth burst of organisms in the tracheas of the birds, peaking on day 8 p.i. (average, 5.8 × 107 CFU), which plateaued between days 10 and 28 p.i. (average, 106 CFU) and then fell to day 1 levels by day 42 p.i. (average, 225 CFU). The chickens appeared to clear S6J organisms from the air sacs earlier than from the trachea. Figure 1A shows that on day 14 p.i. the counts from the tracheal and air sac sites were comparable but by day 28 p.i. the numbers had rapidly dropped in the air sac while the tracheal numbers remained relatively constant.
FIG. 1.
Kinetics of in vivo S6J growth and pMGA antibody production in chickens p.i. (A) Semilogarithmic plot of the average number of S6J organisms (CFU) recovered from the tracheal washes from two birds at each time point p.i. (days 1, 2, 4, 6, 8, 10, 13, 14, 28, and 42 p.i.). (B) Semilogarithmic plot of the average pMGA antibody levels in serum and tracheal wash samples at all time points p.i. and from air sac samples on days 14, 28, and 42 p.i. Levels of pMGA-specific antibodies were calculated relative to a selected high-titer serum. Tracheal wash levels have been multiplied by 100, and air sac levels have been multiplied by 10,000. Error bars represent the range between the two samples tested for each point rather than the standard deviation.
pMGA antibody titer.
Antibody titers were measured in serum and tracheal wash samples (and at later time points in air sac samples) at each time point p.i. by using the pMGA1.1 ELISA. Most importantly, Fig. 1B shows that pMGA antibodies were not detectable on days 1, 2, and 4 p.i. for both tracheal wash and serum samples. Levels which were barely detectable were achieved by day 6.
pMGA phenotype proportions.
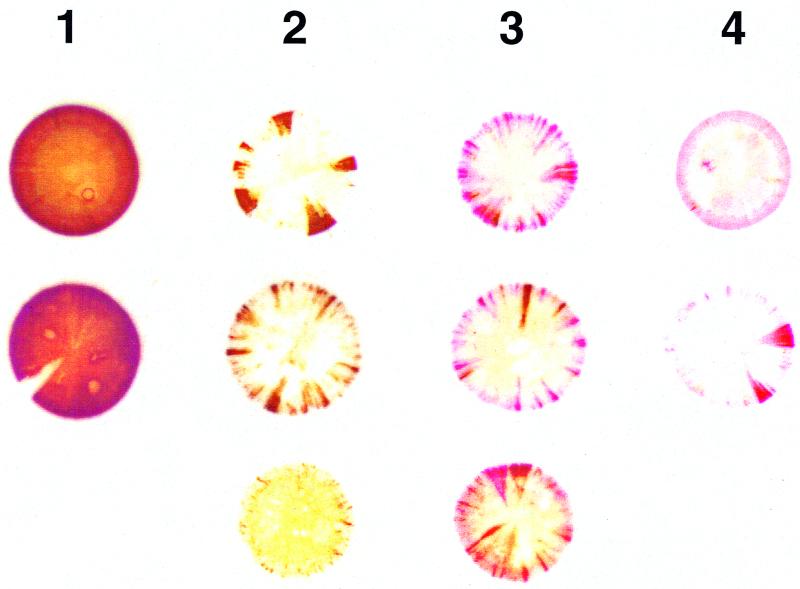
A two-stage immunostaining protocol was developed to identify colonies expressing (parental-type) pMGA1.1 expression (detected by MAb66) and those which expressed a variant form of pMGA which cross-reacted with rabbit hyperimmune serum to pMGA1.1 but did not possess the MAb66 epitope. Staining the colonies with MAb66 identified the expression of pMGA1.1 as brown (first-layer stain), and subsequent staining with rabbit anti-pMGA1.1 identified the expression of variant pMGA as red (second-layer stain). F strain (MAb66−, rabbit anti-pMGA1.1+) colony filter squares (subjected to first-layer staining) were used as a positive control during second-layer staining. Figure 2 shows the typical S6J pMGA colony phenotypes obtained from cells before passage through birds and from cells reisolated from infected birds. For ease of counting and graphical representation, all colonies were grouped into one of four categories. Category 1 colonies were predominantly or exclusively brown (MAb66+) and were assumed to be derived from pMGA1.1+ progenitor cells. Category 2 colonies had nonstained centers with brown-only sectors and were assumed to be derived from pMGA1.1− cells. Category 3 colonies also had white centers but contained both brown (MAb66+) and red (MAb66−; rabbit anti-pMGA1.1+) sectors. Category 4 colonies stained red only, either uniformly or sectorially, and were derived either from pMGA1.1− cells or from pMGA variant cells. The proportions of category 1 and category 2 colonies at various time points p.i. are shown in Fig. 3A. The population of S6J cells used for the infection was predominantly pMGA1.1+ (>90%), and there was no significant change in category proportions on day 1 p.i. compared to the starting inoculum. However, between 2 and 8 days p.i., the proportion of pMGA1.1+ cells rapidly decreased as they were replaced or overgrown by predominantly pMGA1.1− cells. Specifically, a significant change in categories 1 and 2 was observed on day 2 p.i., and cells recovered from one of the two birds sampled at this time point produced colonies, of which 51% were derived from pMGA1.1− cells. The percentages of pMGA1.1+ cells determined on days 4, 6, 8, 10 and 13 p.i. (category 1 colonies) were considered to be overestimates, since a substantial proportion of these colonies (between 25 and 90%, depending on the bird and time point) possessed an “affected” phenotype (i.e., predominantly but not entirely positive and containing a small nonstaining sector).
FIG. 2.
S6J ex vivo colony pMGA phenotype categories. Typical S6J colony pMGA phenotypes obtained before infection and after reisolation from infected chickens were identified by sequential staining using the double-staining protocol. Colony phenotypes after immunostaining were grouped into four categories. Category 1 colonies were predominantly brown (MAb66+) and were assumed to be derived from pMGA1.1+ cells. Category 2 colonies were predominantly white with only brown sectors and were assumed to be derived from pMGA1.1− cells. Category 3 colonies, also called mixed colonies, contained both brown (MAb66+) and red (rabbit anti-pMGA1.1+) sectors and were assumed to be derived from pMGA1.1− cells. Category 4 colonies stained red only and were derived either from pMGA1.1− cells or from pMGA variant cells.
Figure 3B shows the proportions of colonies belonging to the third and fourth categories at the same time points p.i. as in Fig. 3A. Colonies belonging to these two categories were not observed until day 4 p.i. and then were found at only a very low level (<0.5%). Until day 28 p.i., the major variant colony phenotype was mixed (category 3), ranging from about 0.5% on day 4 p.i. to 8% on day 13 p.i., and was significantly different from the starting inoculum only after day 10 p.i. (Fig. 3B). The proportion of red-only colonies (category 4) remained very low (between 0 and 0.5%) between days 4 and 13 p.i. Only on day 28 p.i. did the proportion of category 4 colonies significantly increase (7%) relative to the starting inoculum (Fig. 3B). Combining the third and fourth categories, the highest proportion of variant colonies was 13% on day 28.
Analysis of an ex vivo variant pMGA clone.
The loss within red-stained colonies of the MAb66 epitope (which is typical of the pMGA1.1 protein) could have resulted from either of two mechanisms: (i) a switch in pMGA gene expression (one pMGA gene turned off and another turned on) or (ii) a point mutation in the pMGA1.1 gene specifically affecting the MAb66 epitope. To distinguish between them, a variant S6J colony from a day 10 p.i. tracheal sample belonging to category 3 (mixed) and designated C20 was cloned. This colony contained an unstained center and many peripheral brown and red sectors. A uniformly red-staining colony was then cloned from C20 cells and designated C20.2A. The amino-terminal sequence of the pMGA molecule expressed by C20.2A cells was −TTPTPNPTPNPNPP; this was identical to the corresponding pMGA1.2 sequence and differed from the pMGA molecule of S6J cells (pMGA1.1).
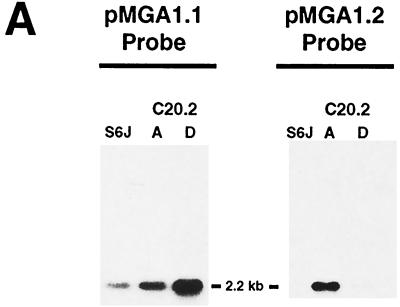
From the original C20 colony, another cloned variant, C20.2D, was obtained which bound MAb66 and which was therefore considered to express pMGA1.1. To confirm the pMGA gene expression patterns in clones C20.2A and C20.2D, Northern blot experiments were conducted. Specifically, RNA samples from parental S6J cells and from clones C20.2A and C20.2D were electrophoresed and probed for their expression of pMGA1.1 and pMGA1.2 (Fig. 4A). Parental S6J cells contained a 2.2-kb RNA species which bound the pMGA1.1 probe but not the pMGA1.2 probe. A corresponding mRNA was present in the C20.2D clone, which bound much more efficiently to the pMGA1.1 probe than to the pMGA1.2 probe. In contrast, a similarly sized RNA molecule from the C20.2A clone bound at least as efficiently to the pMGA1.2 probe as to the pMGA1.1 probe. The pMGA1.1 and pMGA1.2 probes are known to cross-hybridize to a small extent even when high-stringency hybridization conditions are used (8). The Northern blots of Fig. 4A are therefore highly compatible with the expression of the pMGA1.1 gene but not the pMGA1.2 gene in both S6J cells and the C20.2D clone. They are also compatible with the converse expression pattern in the C20.2A clone (pMGA1.1− pMGA1.2+), although the existence of mRNA molecules for both pMGA1.1 and pMGA1.2 in this clone could not be ruled out.
FIG. 4.
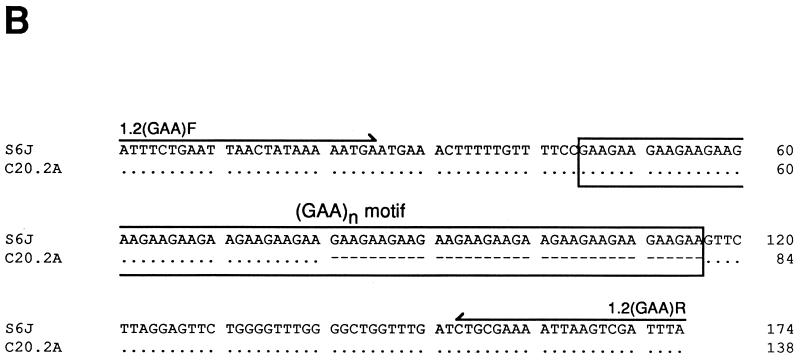
Detection of pMGA1.2 expression. (A) Northern analysis of 3 μg of total RNA from M. gallisepticum S6 cells and clones, described in the text, using a probe specific for the pMGA1.1 or pMGA1.2 gene as indicated. (B) Alignment of the DNA sequences of the 5′ region of the pMGA1.2 genes amplified from S6J and C20.2A cells. Dots represent identities to the S6J sequence, and dashes represent deletions. Arrows above the S6J sequence indicate the orientations and locations of the oligonucleotides [1.2(GAA)F and 1.2(GAA)R] used to amplify the PCR products.
Previous studies had demonstrated a link between the expression of a pMGA gene within a given clone and the presence of a (GAA)12 sequence 5′ to the promoter. It seemed important to investigate whether this sequence had been acquired 5′ to the pMGA1.2 gene in the C20.2A clone. Accordingly, DNA samples from S6J cells and from the C20.2A clone were subjected to PCR amplification of the region which contained the relevant part of the pMGA1.2 gene. The PCR product from S6J cells was approximately 170 bp, and that from C20.2A cells was approximately 130 bp. Sequence analysis (Fig. 4B) showed that S6J cells, which expressed little or no pMGA1.2, contained a (GAA)24 motif 5′ to this gene whereas the corresponding region from C20.2A cells contained a (GAA)12 motif. This reduction in the trinucleotide repeat number was the only sequence difference between the two PCR products.
DISCUSSION
It was previously established that when M. gallisepticum S6 cells were cultured in vitro in the presence of certain antibodies directed to pMGA1.1, they gave rise to a population which ceased to express that protein (15). This “antibody effect” on pMGA expression was selectively directed at the transcription of the pMGA1.1 gene (15). It was of interest to know whether this antibody-mediated pMGA switching phenomenon in M. gallisepticum had any relevance to pathogenesis. To achieve this aim, a virulent clone of strain S6, designated S6J, was used to infect chickens. Reisolation of S6J cells after inoculation of birds allowed the progress of the infection to be observed and showed a maximum mycoplasmal load in the chicken trachea on day 8 p.i. The growth of S6J cells during the early stages of infection was also found to be fairly uniform since there were only small differences in the number of cells reisolated from the two birds at each time point, at least until day 28 p.i. (Fig. 1A).
In addition to reisolation of M. gallisepticum cells, the pMGA antibody levels were measured in chicken sera and in tracheal washes at each time point p.i. to determine if there was a link between changes in S6 pMGA phenotype and the production of host pMGA antibodies. The secondary reagent used in the pMGA ELISA to assay antibody titers was directed to both heavy and light chains of chicken Ig and would therefore detect most or all chicken Ig species. The average titers in serum rapidly increased between days 6 and 14 p.i. to a plateau level. Titers within the air sac and tracheal wash samples increased more or less in proportion to those in serum between days 14 and 42 p.i. but at a 100-fold-lower level due to the dilution effect of sample collection. Most important was that pMGA antibodies in both tracheal wash and serum samples were not detectable until day 6 p.i. It was nevertheless evident (Fig. 3A) that pMGA1.1 expression within the in vivo expanding M. gallisepticum population had altered significantly by day 2 p.i., as judged by the phenotypes of the colonies from reisolated cells. The population of S6J cells used for the infection produced predominantly pMGA1.1+ colonies (>90%). After inoculation, the proportion of uniformly pMGA1.1+ colonies decreased, with a corresponding increase in sectored or uniformly pMGA1.1− colonies over the 8 days p.i. (Fig. 3A). By 2 days p.i., 40% of recovered cells produced colonies whose pMGA1.1 phenotype established their origins from pMGA1.1− progenitors (Fig. 3A). These numbers may underestimate the extent of the effect of in vivo passage on pMGA expression, since many of the colonies isolated from birds 4 days p.i. which belonged to category 1 exhibited unstained centers, and even if scored pMGA1.1+ according to the arbitrary classification criteria used to differentiate colony types (Fig. 2), such colonies were probably derived from pMGA− progenitors. Unexpectedly, this switching effect from the pMGA1.1+ to the pMGA1.1− state occurred in a significant proportion of cells by day 2 p.i., which was at least 2 days before pMGA antibodies were detectable. This would indicate that unlike the in vitro case, where growth with pMGA1.1 antibodies such as rabbit anti-pMGA1.1 and MAb66 produced the pMGA1.1− expression state, in vivo this phenomenon was initiated in the absence of host pMGA1.1 antibodies.
It was noted in the course of these studies that late in infection unusual variant cells arose which produced colonies of types 3 and 4. Such colonies, in part or in toto, lacked reactivity with MAb66 but retained reactivity with polyclonal rabbit anti-pMGA1.1 serum (Fig. 3B). The occurrence of such cells became significant from day 10 p.i. and accounted for 13% of the total colonies at the last time point of the analysis (day 28 p.i.). Few if any such cells were present within the starting S6J inoculum. The majority of the pMGA ex vivo variant colonies derived from these cells contained centers which were unstained, but they exhibited stained peripheral sectors. This implies that the cells from which they arose expressed neither pMGA1.1 nor any antigenically related variant in vivo. Cells which produced uniformly stained category 4 colonies (red only) were a small minority even on day 28 p.i., comprising between 1 and 2% of category 4 colonies. A typical category 3 colony (C20) was subcloned to derive two clonal isolates, C20.2A and C20.2D. The former clone expressed the pMGA1.2 polypeptide, as determined by its amino-terminal sequence and by Northern transfer, whereas the latter expressed pMGA1.1. The results of the Northern blot experiments which confirmed the pMGA phenotypes of S6J, C20.2A, and C20.2D are depicted in Fig. 4A. Sequence analysis of the region containing the (GAA)n motif 5′ to the pMGA1.2 gene (Fig. 4B) revealed that clone C20.2A had acquired the (GAA)12 repeat previously shown to be critical for the expression of a pMGA gene (9). It seems likely that during prolonged in vivo habitation, the antecedent of the C20 colony first lost pMGA1.1 expression by loss of its (GAA)12 motif and then, in vitro, within the C20 colony some cells reacquired (GAA)12 5′ to pMGA1.1 and others acquired this motif 5′ to pMGA1.2 by the deletion of 12 of the GAA repeats found 5′ to pMGA1.2 in the parental S6J cells (Fig. 4B). Notably, within the C20 colony and indeed within the majority of other category 3 and 4 colonies, multiple pink sectors were apparent. In contrast, within category 2 colonies, no red sectors at all were apparent. Thus, the acquisition of expression of pMGA1.2 (or other antigenically cross-reactive pMGA molecules) within colonies is nonrandom; instead, individual founder cells seem predisposed to produce descendants which express particular pMGA genes that are not expressed within the founder cells themselves. The predisposition phenomenon was previously established in populations of pMGA1.9+ pMGA1.1− cells, which invariably produced colonies with multiple pMGA1.1+ sectors (9). The predisposition of the descendants of founder cells to express specific pMGA genes within their colonies may relate to the molecular mechanism by which switching occurs, slipped-strand mispairing (9). This phenomenon is thought to account for the frequencies of expansion and contraction events which occur during the DNA replication of motifs such as tandemly repeated trinucleotides. In humans, DNA segments which contain such repeat elements sometimes exhibit length heterogeneity due to one or at most a few repeat differences between somatic cells (19), suggesting that changes in repeat numbers by slipped-strand mispairing may occur by addition or deletion of one repeat unit at a time. If this is right, then M. gallisepticum cells which have recently switched off one pMGA gene would be expected to retain (GAA)n motifs in which n is close to 12. Our previous studies confirm this prediction (9). If the gain or loss of only one or two GAA repeats is required to restore expression to a particular pMGA gene, it seems likely that variants expressing this gene would arise within a population much more frequently than for other pMGA genes which required a larger adjustment of repeat numbers. It may be significant in this regard that variants with a predisposition to express the pMGA1.2 gene product (or antigenically related polypeptides) did not arise in significant numbers until 8 to 10 days p.i. The S6J cells used to establish the infection on day 0 contained a pMGA1.2 gene with 24 GAA repeats: assuming that expansions and contractions are equally likely and that DNA synthesis is an obligate requirement for repeat length changes, many cell divisions would be required to reduce this relatively long GAA repeat motif to the vicinity of (GAA)12. In contrast, the cells which produced category 2 colonies (progeny predisposed to reexpress pMGA1.1) were predominant in frequency at earlier times p.i. because their GAA repeat lengths differed from 12 by only 1 or 2 units.
Collectively, these data support the notion that the pMGA gene family is actively involved in pMGA phenotypic variation in M. gallisepticum. The in vivo results showed that cells which were predominantly pMGA1.1+ rapidly convert to pMGA1.1− cells after growth in the host. Unlike the in vitro case, where this effect was caused by growth with MAb66 and rabbit anti-pMGA1.1 antibodies, in vivo this pMGA switching phenomenon was not brought about by the presence of detectable host pMGA1.1 antibodies. Instead, there seems to be some selective growth advantage for pMGA1.1− cells in the host and the expression of other pMGA molecules (which cannot be detected by the pMGA immunological reagents used in this work) endows these cells with an improved colonization ability. It is possible that this could improve or better stabilize microcolony formation at the tips of the microvilli, structures observed during the early stages (2 days p.i.) of tracheal colonization by M. gallisepticum (1, 10). This form of colonization might require adherence to “self” mediated by pMGA-pMGA binding interactions between cells. We have already noted the possibility that such homotypic pMGA-pMGA interactions might occur on the basis of other evidence (15). Alternatively, some host epithelial ligand may exhibit optimal affinity for cells expressing a pMGA molecule other than pMGA1.1. Whatever the notional nature of such a host ligand, it is unlikely to be pMGA-specific antibody, whose elicitation occurs after, rather than before, the change in population phenotype documented herein.
ACKNOWLEDGMENTS
We thank Jeff Gill, Craig Cunningham, and Jason Twohig for their constructive critiques of the manuscript.
This work was supported by an Australian Research Council grant (to I.D.W.). M.D.G. was supported in part by a scholarship from the Rural Industries Research and Development Corporation.
REFERENCES
- 1.Abu-Zahr M N, Butler M. Growth, cytopathogenicity and morphology of Mycoplasma gallisepticum and M. gallinarum in tracheal explants. J Comp Pathol. 1976;86:455–463. doi: 10.1016/0021-9975(76)90014-1. [DOI] [PubMed] [Google Scholar]
- 2.Baseggio N, Glew M D, Markham P F, Whithear K G, Browning G F. Size and genomic location of the pMGA multigene family of Mycoplasma gallisepticum. Microbiology. 1996;142:1429–1435. doi: 10.1099/13500872-142-6-1429. [DOI] [PubMed] [Google Scholar]
- 3.Bencina D, Kleven S H, Elfaki M G, Snoj A, Dove P, Dorrer D, Russ I. Variable expression of epitopes on the surface of Mycoplasma gallisepticum demonstrated with monoclonal antibodies. Avian Pathol. 1994;23:19–36. doi: 10.1080/03079459408418972. [DOI] [PubMed] [Google Scholar]
- 4.Bordier C. Phase separation of integral membrane proteins in Triton-X114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 5.Droesse M, Tangen G, Gummelt I, Kirchhoff H, Washburn L R, Rosengarten R. Major membrane proteins and lipoproteins as highly variable immunogenic surface components and strain-specific antigenic markers of Mycoplasma arthritidis. Microbiology. 1995;141:3207–3219. doi: 10.1099/13500872-141-12-3207. [DOI] [PubMed] [Google Scholar]
- 6.Frey M L, Hanson R P, Anderson D P. A medium for the isolation of avian mycoplasmas. Am J Vet Res. 1968;29:2163–2171. [PubMed] [Google Scholar]
- 7.García M, Elfaki M G, Kleven S H. Analysis of the variability in expression of Mycoplasma gallisepticum surface antigens. Vet Microbiol. 1994;42:147–158. doi: 10.1016/0378-1135(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 8.Glew M D, Markham P F, Browning G F, Walker I D. Expression studies on four members of the pMGA multigene family in Mycoplasma gallisepticum. Microbiology. 1995;141:3005–3014. doi: 10.1099/13500872-141-11-3005. [DOI] [PubMed] [Google Scholar]
- 9.Glew M D, Markham P F, Browning G F, Walker I D. Variation in the GAA trinucleotide repeat lengths within the 5′ non-coding regions of Mycoplasma gallisepticum pMGA genes controls their expression. Infect Immun. 1998;66:5833–5841. doi: 10.1128/iai.66.12.5833-5841.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levisohn S, Yegana Y, Hod I, Herz A. A correlative study of the surface morphology and colonisation of the chicken trachea infected by Mycoplasma gallisepticum. Avian Pathol. 1983;12:247–261. doi: 10.1080/03079458308436167. [DOI] [PubMed] [Google Scholar]
- 11.Levisohn S, Rosengarten R, Yogev D. In vivo variation of Mycoplasma gallisepticum antigen expression in experimentally infected chickens. Vet Microbiol. 1995;45:219–231. doi: 10.1016/0378-1135(95)00039-d. [DOI] [PubMed] [Google Scholar]
- 12.Markham P F, Glew M D, Brandon M R, Walker I D, Whithear K G. Characterization of a major hemagglutinin protein from Mycoplasma gallisepticum. Infect Immun. 1992;60:3885–3891. doi: 10.1128/iai.60.9.3885-3891.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markham P F, Glew M D, Whithear K G, Walker I D. Molecular cloning of a member of the gene family that encodes pMGA, a hemagglutinin of Mycoplasma gallisepticum. Infect Immun. 1993;61:903–909. doi: 10.1128/iai.61.3.903-909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markham P F, Glew M D, Sykes J E, Bowden T R, Pollocks T D, Browning G F, Whithear K G, Walker I D. The organisation of the multigene family which encodes the major cell surface protein, pMGA, of Mycoplasma gallisepticum. FEBS Lett. 1994;352:347–352. doi: 10.1016/0014-5793(94)00991-0. [DOI] [PubMed] [Google Scholar]
- 15.Markham P F, Glew M D, Browning G F, Whithear K G, Walker I D. Expression of two members of the pMGA gene family of Mycoplasma gallisepticum oscillates and is influenced by pMGA-specific antibodies. Infect Immun. 1998;66:2845–2853. doi: 10.1128/iai.66.6.2845-2853.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meynell G G, Meynell E. Theory and practice in experimental bacteriology. Cambridge, United Kingdom: Cambridge University Press; 1970. [Google Scholar]
- 17.Patten B E, Higgins P A, Whithear K G. A urease-ELISA for the detection of mycoplasma infections in poultry. Aust Vet J. 1984;61:151–154. doi: 10.1111/j.1751-0813.1984.tb07219.x. [DOI] [PubMed] [Google Scholar]
- 18.Power J, Jordan F T W. A comparison of the virulence of three strains of Mycoplasma gallisepticum and one strain of Mycoplasma gallinarum in chicks, turkey poults, tracheal organ cultures and embryonated fowl eggs. Res Vet Sci. 1976;21:41–46. [PubMed] [Google Scholar]
- 19.Ross C A, McInnis M G, Margolis R L, Li H-S. Genes with triplet repeats: candidate mediators of neuropsychiatric disorders. Trends Neurobiol. 1993;16:254–260. doi: 10.1016/0166-2236(93)90175-l. [DOI] [PubMed] [Google Scholar]
- 20.Whithear K G, Harrigan K E, Kleven S H. Standardized method of aerosol challenge for testing the efficacy of Mycoplasma gallisepticum vaccines. Avian Dis. 1996;40:654–660. [PubMed] [Google Scholar]
- 21.Wise K S. Adaptive surface variation in mycoplasmas. Trends Microbiol. 1993;1:59–63. doi: 10.1016/0966-842X(93)90034-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wise K S, Yogev D, Rosengarten R. Antigenic variation. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C.: American Society for Microbiology; 1992. pp. 473–489. [Google Scholar]
- 23.Yamamoto R, Adler H E. Characterization of pleuropneumonia-like organisms of avian origin. I. Antigenic analysis of seven strains and their comparative pathogenicity for birds. J Infect Dis. 1958;102:143–152. doi: 10.1093/infdis/102.2.143. [DOI] [PubMed] [Google Scholar]
- 24.Yogev D, Menaker D, Strutzberg K, Levisohn S, Kirchhoff H, Hinz K-H, Rosengarten R. A surface epitope undergoing high-frequency phase variation is shared by Mycoplasma gallisepticum and Mycoplasma bovis. Infect Immun. 1994;62:4962–4968. doi: 10.1128/iai.62.11.4962-4968.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng X, Teng L-J, Watson H L, Glass J I, Blanchard A, Cassell G H. Small repeating units within the Ureaplasma urealyticum MB antigen gene encode serovar specificity and are associated with antigen size variation. Infect Immun. 1995;63:891–898. doi: 10.1128/iai.63.3.891-898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]