Abstract
Streptococcus equi and Streptococcus zooepidemicus are major etiological agents of upper and lower airway disease in horses. Despite the considerable animal suffering and economic burden associated with these diseases, the factors that contribute to the virulence of these equine pathogens have not been extensively investigated. Here we demonstrate the presence of a homologue of the Streptococcus pneumoniae PsaA protein in both of these equine pathogens. Inhibition of signal peptide processing by the antibiotic globomycin confirmed the lipoprotein nature of the mature proteins, and surface exposure was confirmed by their release from intact cells by mild trypsinolysis.
Streptococcus equi subsp. equi, the etiological agent of strangles, has been estimated to be responsible for nearly 30% of all reported equine infections worldwide (6). Strangles is characterized by pharyngeal constriction in the horse's upper respiratory tract as a consequence of lymph node swelling and is often accompanied by abscessation. The very closely related organism Streptococcus zooepidemicus (S. equi subsp. zooepidemicus) is a significant cause of equine lower airway disease, foal pneumonia, endometritis, and abortion (6). Despite the considerable animal suffering and economic burden associated with these diseases, there is little information regarding the molecular basis of virulence of these two streptococci, and there are presently no effective vaccines against either organism (6). Most studies have focused on the M-like proteins of these streptococci (36, 37), and that of S. equi has been shown to be a fibrinogen-binding protein (31). Recently, other studies have focused on a streptolysin S-like toxin (17), a fibronectin-binding protein (29), and a hyaluronate-associated protein conferring partial protection in murine models of S. equi and S. zooepidemicus infection (9). The pyogenic streptococci are highly host adapted, so that pathogenicity is likely to depend on many biochemical, immunological, and cellular interactions. Interference with a critical combination of these may be important in the development of protective immunity (6). The characterization of bacterial cell surface proteins vital for host-pathogen interactions is an essential step toward identifying components which are likely to elicit protective immune responses.
Studies have identified a class of at least eight highly homologous (ca. 70% or greater amino acid identity) 35- to 37-kDa proteins in streptococci, including the PsaA protein of Streptococcus pneumoniae, the FimA protein of Streptococcus parasanguis, and the MtsA protein of Streptococcus pyogenes (4, 15, 22, 33). The genes encoding these proteins are located within operons encoding components of putative ATP-binding cassette (ABC) transport systems (13, 16, 22, 24–26, 32, 34). Moreover, these proteins appear to be a subfamily of a larger family of substrate-binding proteins (cluster 9) involved in the transport of metal ions such as iron, manganese, and zinc (2, 3, 10, 13, 14, 18, 22, 25, 28). A characteristic component of the importer ABC systems of gram-positive bacteria is a solute-binding lipoprotein (35), and consistent with this, the streptococcal 35- to 37-kDa proteins are all putative lipoproteins.
A stable nomenclature has yet to be adopted for these streptococcal proteins, so we refer to them herein as metal binding lipoproteins (MBLs). These lipoproteins may be of considerable importance in the physiology and pathogenicity of streptococci, since MBL-deficient mutants of Streptococcus mutans, S. parasanguis, and S. pneumoniae were significantly less virulent than their wild-type parent strains in animal models of disease (4, 5, 24). Consequently, we have investigated the presence of homologous proteins in S. equi and S. zooepidemicus, because they may also have significance as virulence determinants.
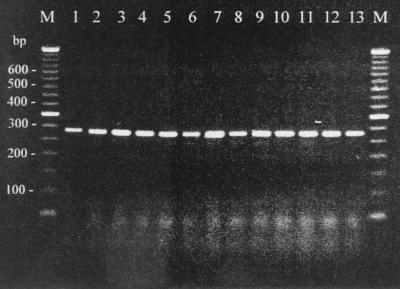
Initially, degenerate PCR primers were designed based upon the highly conserved regions EDPHAW and WEINTE within the published streptococcal MBL sequences corresponding to amino acids 136 to 141 and 223 to 228, respectively, in the pneumococcal PsaA protein (4, 33). PCR with these primers amplified DNA fragments from S. equi NCTC 9682 and S. zooepidemicus NCTC 7023 that comigrated with a psaA fragment amplified with the same primers from S. pneumoniae DNA (Fig. 1). Furthermore, amplimers of the same size were also obtained from five disparate clinical isolates of S. equi and five disparate isolates of S. zooepidemicus (Fig. 1). The S. zooepidemicus isolates were selected on the basis of differences in the polymorphisms of their 16S to 23S RNA gene intergenic spacers (Table 1). Since S. equi has just one intergenic spacer type (7), strains were selected on the basis of temporal and geographical differences in isolation (Table 1).
FIG. 1.
PCR amplification of fragments of putative MBL genes from S. equi and S. zooepidemicus and of a psaA gene fragment from S. pneumoniae. Lanes: M, 50-bp standard ladder (Gibco, Paisley, United Kingdom); 1, S. pneumoniae NCTC 11910; 2, S. equi NCTC 9682; 3, S. equi 1742; 4, S. equi 2112; 5, S. equi CF32; 6, S. equi 4047; 7, S. equi 1026; 8, S. zooepidemicus NCTC 7023; 9, S. zooepidemicus 2809; 10, S. zooepidemicus 3682; 11, S. zooepidemicus 3685; 12, S. zooepidemicus K3; 13, S. zooepidemicus 461.
TABLE 1.
Bacterial strains used in this study
| Straina | 16S–23S RNA gene intergenic spacer type | Host and site of isolation | Geographical location, date of isolation | Reference(s) |
|---|---|---|---|---|
| S. equi | ||||
| NCTC 9682T | NTb | |||
| CF32 | D1 | Equine abscess | New York, 1981 | 37 |
| 4047 | D1 | Equine abscess | Northamptonshire, United Kingdom, 1990 | 7, 8 |
| 1026 | D1 | Equine nasopharynx | Suffolk, United Kingdom, 1995 | 7 |
| 1742 | D1 | Equine nasopharynx | Buckinghamshire, United Kingdom, 1995 | 7 |
| 2112 | D1 | Equine nasopharynx | Kent, United Kingdom, 1997 | This study |
| S. zooepidemicus | ||||
| NCTC 7023T | NT | |||
| 2809 | A1 | Equine trachea | Berkshire, United Kingdom, 1993 | 7 |
| 3682 | A2 | Equine trachea | Sussex, United Kingdom, 1992 | 7 |
| 3685 | B1 | Equine trachea | Sussex, United Kingdom, 1992 | 7 |
| 461 | C1 | Equine nasopharynx | Suffolk, United Kingdom, 1994 | 7 |
| K3 | D2 | Equine lung | Suffolk, United Kingdom, 1993 | 7 |
| S. pneumoniae NCTC 11910 (serotype 23F) | Not applicable |
NCTC, National Collection of Type Cultures, London, United Kingdom.
NT, not tested.
Sequencing of the amplified fragments from S. equi NCTC 9682 and from S. zooepidemicus NCTC 7023 afforded 243 nucleotides of DNA sequence for each organism. The sequences were 97% identical at the nucleotide level, with 100% homology at the translated amino acid level. Homologues of the 81 amino acids derived from these nucleotide sequences were identified by a BLAST search (1) using the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/BLAST). The translated sequence showed significant homology to internal sequences of all proteins in the MBL family, with greatest homology to MtsA from S. pyogenes (22). The DNA sequence of the S. equi PCR product was also 100% identical to a contig sequence within the unfinished S. equi strain 4047 genome (http://www.sanger.ac.uk/Projects/S_equi/). The contig within which this sequence was located contained a putative open reading frame (ORF) encoding a protein of 310 amino acids with 89% identity to MtsA of S. pyogenes. To verify the presence of the mbl gene in S. equi, contig sequences corresponding to the MBL signal peptide region and to the 5′ end of the adjacent, downstream ORF were used to design PCR primers which allowed the amplification and sequencing of a DNA fragment encoding the entire mature MBL. The gene contains a “lipobox” motif (MLVAC↓S) conforming to that directing lipoprotein cysteine modification in gram-positive bacteria (35). The 290 amino acids deduced for the mature protein sequence (starting from the N-terminal lipobox cysteine) were used in a BLAST search of the available databases. Highly significant homologies (72 to 92% identity) were found with the streptococcal MBLs, most notably with MtsA of S. pyogenes (22), and lower homologies (28 to 58% identity) were found with the more distant relatives within the cluster 9 binding proteins (Table 2). Secondary structure analysis using PSIPRED (23; http://insulin.brunel.ac.uk/psipred/) predicted that the major helix or strand features of PsaA (28) are also conserved in the S. equi MBL.
TABLE 2.
Homology of the mature S. equi MBL sequence with other streptococcal MBLs and members of the cluster 9 solute binding protein familya
| Organism and protein | % Identity | % Similarity | Length (amino acids)b | BLAST E value | Reference or accession no.c |
|---|---|---|---|---|---|
| Streptococcus pyogenes MtsA | 92 | 95 | 290 | 1 × 10−153 | 22 |
| Streptococcus sanguis SsaB | 78 | 88 | 290 | 1 × 10−133 | 19 |
| Streptococcus oralis PsaA | 77 | 88 | 290 | 1 × 10−132 | AF248237 |
| Streptococcus anginosus PsaA | 77 | 88 | 290 | 1 × 10−132 | AF248235 |
| Streptococcus pneumoniae PsaA | 76 | 87 | 290 | 1 × 10−131 | 4 |
| Streptococcus sp. “PsaA”d | 76 | 88 | 290 | 1 × 10−131 | 34 |
| Streptococcus crista ScbA | 77 | 88 | 289 | 1 × 10−130 | 11 |
| Streptococcus mitis PsaA | 76 | 88 | 290 | 1 × 10−130 | AF248236 |
| Streptococcus parasanguis FimA | 75 | 88 | 290 | 1 × 10−130 | 15 |
| Streptococcus gordonii ScaA | 76 | 88 | 289 | 1 × 10−130 | 26 |
| Streptococcus mutans LraI | 72 | 84 | 285 | 1 × 10−119 | 24 |
| Enterococcus faecalis EfaA | 58 | 75 | 285 | 1 × 10−96 | 30 |
| Staphylococcus epidermidis SitC | 51 | 69 | 288 | 2 × 10−79 | 10 |
| Yersinia pestis YfeA | 35 | 56 | 285 | 1 × 10−48 | 3 |
| Synechocystis sp. MntC | 32 | 57 | 277 | 2 × 10−45 | 2 |
| Bacillus subtilis YcdH | 30 | 48 | 303 | 4 × 10−30 | 18, 27 |
| Streptococcus pneumoniae AdcA | 28 | 48 | 294 | 1 × 10−27 | 14 |
BLAST searches of the S. equi genome database with the sequences of the S. pyogenes MtsB and MtsC proteins revealed homologues of each (80% identity over 197 amino acids for MtsB; 89% identity over 275 amino acids for MtsC), with the MtsB sequence located downstream of mbl on the same contig. The nucleotide sequence in the region between the mbl gene and the mtsB homologue in S. equi was also highly homologous to that found between mtsA and mtsB of S. pyogenes and which was previously identified as a putative stem-loop transcription terminator for the mtsA gene (22). This therefore suggests that mbl, like mtsA, is transcribed both individually and as part of a polycistronic message.
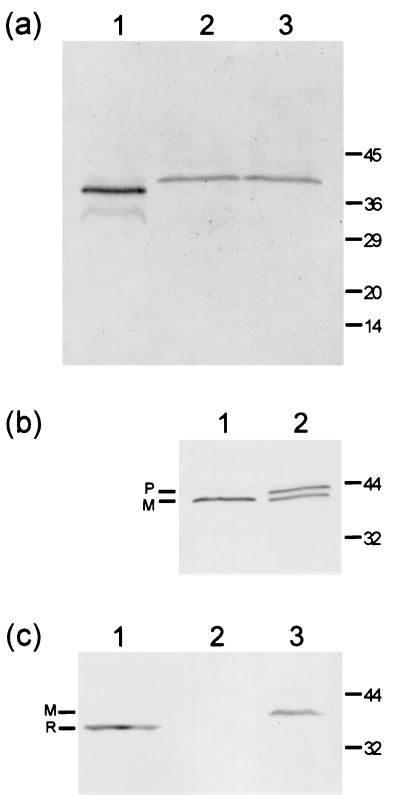
Expression of the MBL homologues in S. equi and S. zooepidemicus was analyzed by Western blotting as previously described (20). Rabbit polyclonal anti-PsaA antiserum (12) cross-reacted with a polypeptide of ca. 38 kDa in extracts of both S. equi and S. zooepidemicus (Fig. 2a, lanes 2 and 3), whereas a strong reaction with PsaA was detected at ca. 36 kDa in the S. pneumoniae control lane (Fig. 2a, lane 1). Approximately twice as much S. equi cell extract was needed to produce band intensities comparable to that of S. zooepidemicus. Because the MBL sequences from S. equi and S. zooepidemicus are nearly identical, the weaker reaction in the S. equi extract may be due to lower expression of this protein under the growth conditions employed. Growth of S. equi in different culture media did not result in increased recovery of the MBL (data not shown). The anti-PsaA antiserum cross-reacted with polypeptides of ca. 38 kDa in extracts of all of the strains of S. equi and S. zooepidemicus listed in Table 1 (data not shown). Antiserum raised against ScaA, the Streptococcus gordonii MBL (26), also cross-reacted with S. equi and S. zooepidemicus polypeptides with the same molecular masses as those detected with the anti-PsaA antiserum (data not shown). Cumulatively, these results strongly indicate the expression of MBL homologues in S. equi and S. zooepidemicus.
FIG. 2.
Western blot analyses demonstrating S. equi and S. zooepidemicus MBL expression (a), lipoprotein modification (b), and localization (c) using an anti-PsaA antibody. Cell extracts or supernatants were separated by SDS-PAGE, blotted onto nitrocellulose, and probed with polyclonal anti-PsaA antibody. The positions of molecular mass markers are shown (in kilodaltons) at the right-hand side of each panel. (a) Lanes: 1, S. pneumoniae-positive control; 2, S. equi; 3, S. zooepidemicus. (b) Lanes: 1, S. zooepidemicus control growth; 2, S. zooepidemicus grown in the presence of globomycin. P, prolipoprotein; M, mature lipoprotein. (c) Lanes: 1, supernatant from cells of S. zooepidemicus treated with trypsin; 2, supernatant from S. zooepidemicus cells incubated without trypsin; 3, SDS extract of untreated S. zooepidemicus cells. R, released protein; M, mature lipoprotein.
To confirm the predicted lipoprotein modification of their MBLs, S. equi and S. zooepidemicus were grown in the presence of the antibiotic globomycin, which specifically inhibits cleavage of lipoprotein signal peptides by signal peptidase II (21). As expected, globomycin treatment of S. zooepidemicus resulted in the appearance of an additional, slightly larger polypeptide (Fig. 2b) that cross-reacted with the polyclonal anti-PsaA antiserum and is attributed to the accumulation of the prolipoprotein form of the MBL. Comparable results were obtained for S. equi (data not shown). To confirm the surface accessibility of the MBL, whole bacterial cells were subjected to mild digestion with trypsin as previously described (20). As shown in Fig. 2c, truncated forms of the anti-PsaA cross-reactive protein were released from cells of S. zooepidemicus in the presence, but not the absence, of trypsin. The size of the released product is consistent with cleavage of the protein at one of the several potential trypsin cleavage sites close to the N terminus of the mature MBL. Similar experiments have shown the release by papain of a truncated form of MtsA from S. pyogenes (22).
The results presented here thus provide genetic and biochemical evidence that a selection of disparate isolates of S. equi and S. zooepidemicus all produce proteins homologous to the PsaA protein of S. pneumoniae and other related MBLs. The expression and surface accessibility of this lipoprotein were confirmed serologically in both organisms. Because these proteins are thought to be substrate-binding lipoproteins participating in metal transport systems (13, 22) it seems likely that the new members of this family described herein are also encoded by genes located within operons for ABC transport systems, and this is further supported by the identification of homologues of MtsB and MtsC of S. pyogenes within the unfinished S. equi genome. These transport systems could play vital roles in the acquisition of nutrients in the equine host. Because these putative virulence factors may represent novel therapeutic targets in S. equi and S. zooepidemicus, further studies to characterize their function are now in progress.
Nucleotide sequence accession number.
The EMBL accession number for the nucleotide sequence described in this work is AJ249889.
Acknowledgments
We are grateful to Jacquelyn Sampson, National Center for Infectious Diseases, Atlanta, Ga., for supplying the rabbit polyclonal anti-PsaA antiserum; John Timoney, Gluck Equine Research Centre, University of Kentucky, Lexington, for supplying S. equi strain CF32; Howard Jenkinson, Department of Oral Biology, University of Bristol, Bristol, United Kingdom, and Paul Kolenbrander, Oral Infection and Immunity Branch, National Institute of Dental Research, Bethesda, Md., for providing anti-ScaA antiserum; and Shunichi Miyakoshi (Sankyo Chemical Co., Tokyo, Japan) who kindly supplied the globomycin. DNA sequencing was carried out by the Molecular Biology Unit at the University of Newcastle upon Tyne, Newcastle upon Tyne, United Kingdom.
This work was supported by project grant 056042 from The Wellcome Trust.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programmes. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartsevich V V, Pakrasi H B. Molecular identification of an ABC transporter complex for manganese: analysis of a cyanobacterial mutant strain impaired in the photosynthetic oxygen evolution process. EMBO J. 1995;14:1845–1853. doi: 10.1002/j.1460-2075.1995.tb07176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bearden S W, Staggs T M, Perry R D. An ABC transporter system of Yersinia pestis allows utilization of chelated iron by Escherichia coli SAB11. J Bacteriol. 1998;180:1135–1147. doi: 10.1128/jb.180.5.1135-1147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry A M, Paton J C. Sequence heterogeneity of PsaA, a 37-kilodalton putative adhesin essential for virulence of Streptococcus pneumoniae. Infect Immun. 1996;64:5255–5262. doi: 10.1128/iai.64.12.5255-5262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnette-Curley D, Wells V, Viscount H, Munro C L, Fenno J C, Fives-Taylor P, Macrina F L. FimA, a major virulence factor associated with Streptococcus sanguis endocarditis. Infect Immun. 1995;63:4669–4674. doi: 10.1128/iai.63.12.4669-4674.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chanter N. Streptococci and enterococci as animal pathogens. J Appl Microbiol Symp Suppl. 1997;83:100S–109S. doi: 10.1046/j.1365-2672.83.s1.11.x. [DOI] [PubMed] [Google Scholar]
- 7.Chanter N, Collin N, Holmes N, Binns M, Mumford J. Characterization of the Lancefield group C streptococcus 16S-23S RNA gene intergenic spacer and its potential for identification and sub-specific typing. Epidemiol Infect. 1997;118:125–135. doi: 10.1017/s0950268896007285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chanter N, Smith K C, Mumford J A. Equine strangles modelled in mice. Vet Microbiol. 1995;43:209–218. doi: 10.1016/0378-1135(94)00090-j. [DOI] [PubMed] [Google Scholar]
- 9.Chanter N, Ward C L, Collin N C, Flanagan J A, Binns M, Houghton S B, Smith K C, Mumford J A. Recombinant hyaluronate associated protein as a protective immunogen against Streptococcus equi and Streptococcus zooepidemicus challenge in mice. Microb Pathog. 1999;27:133–143. doi: 10.1006/mpat.1999.0290. [DOI] [PubMed] [Google Scholar]
- 10.Cockayne A, Hill P J, Powell N B L, Bishop K, Sims C, Williams P. Molecular cloning of a 32-kilodalton lipoprotein component of a novel iron-regulated Staphylococcus epidermidis ABC transporter. Infect Immun. 1998;66:3767–3774. doi: 10.1128/iai.66.8.3767-3774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Correia F F, DiRienzo J M, McKay T L, Rosan B. scbA from Streptococcus crista CC5A: an atypical member of the IraI gene family. Infect Immun. 1996;64:2114–2121. doi: 10.1128/iai.64.6.2114-2121.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crook J, Tharpe J A, Johnson S E, Williams D B, Stinson A R, Facklam R R, Ades E W, Carlone G M, Sampson J S. Immunoreactivity of five monoclonal antibodies against the 37-kilodalton common cell wall protein (PsaA) of Streptococcus pneumoniae. Clin Diagn Lab Immunol. 1998;5:205–210. doi: 10.1128/cdli.5.2.205-210.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dintilhac A, Alloing G, Granadel C, Claverys J-P. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol. 1997;25:727–739. doi: 10.1046/j.1365-2958.1997.5111879.x. [DOI] [PubMed] [Google Scholar]
- 14.Dintilhac A, Claverys J-P. The adc locus, which affects competence for genetic transformation in Streptococcus pneumoniae encodes an ABC transporter with a putative lipoprotein homologous to a family of streptococcal adhesins. Res Microbiol. 1997;148:119–131. doi: 10.1016/S0923-2508(97)87643-7. [DOI] [PubMed] [Google Scholar]
- 15.Fenno J C, LeBlanc D J, Fives-Taylor P. Nucleotide sequence analysis of a type 1 fimbrial gene of Streptococcus sanguis FW213. Infect Immun. 1989;57:3527–3533. doi: 10.1128/iai.57.11.3527-3533.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenno J C, Shaikh A, Spatafora G, Fives-Taylor P. The fimA locus of Streptococcus parasanguis encodes an ATP-binding membrane transport system. Mol Microbiol. 1995;15:849–863. doi: 10.1111/j.1365-2958.1995.tb02355.x. [DOI] [PubMed] [Google Scholar]
- 17.Flanagan J, Collin N, Timoney J, Mitchell T, Mumford J A, Chanter N. Characterization of the haemolytic activity of Streptococcus equi. Microb Pathog. 1998;24:211–221. doi: 10.1006/mpat.1997.0190. [DOI] [PubMed] [Google Scholar]
- 18.Gaballa A, Helmann J D. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol. 1998;180:5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganeshkumar N, Hannam P M, Kolenbrander P E, McBride B C. Nucleotide sequence of a gene coding for a saliva-binding protein (SsaB) from Streptococcus sanguis 12 and possible role of the protein in coaggregation with Actinomyces. Infect Immun. 1991;59:1093–1099. doi: 10.1128/iai.59.3.1093-1099.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamilton, A., D. Harrington, and I. C. Sutcliffe. Characterisation of acid phosphatase activities in the equine pathogen Streptococcus equi. Syst. Appl. Microbiol., in press. [DOI] [PubMed]
- 21.Inukai M, Takeuchi M, Shimizu K, Arai M. Mechanism of action of globomycin. J Antibiot. 1978;31:1203–1205. doi: 10.7164/antibiotics.31.1203. [DOI] [PubMed] [Google Scholar]
- 22.Janulczyk R, Pallon J, Björck L. Identification and characterization of a Streptococcus pyogenes ABC transporter with multiple specificity for metal cations. Mol Microbiol. 1999;34:596–606. doi: 10.1046/j.1365-2958.1999.01626.x. [DOI] [PubMed] [Google Scholar]
- 23.Jones D T. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 24.Kitten T, Munro C L, Michalek S M, Macrina F L. Genetic characterization of a Streptococcus mutans LraI family operon and role in virulence. Infect Immun. 2000;68:4441–4451. doi: 10.1128/iai.68.8.4441-4451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolenbrander P E, Andersen R N, Baker R A, Jenkinson H F. The adhesion-associated sca operon in Streptococcus gordonii encodes an inducible high-affinity ABC transporter for Mn2+ uptake. J Bacteriol. 1998;180:290–295. doi: 10.1128/jb.180.2.290-295.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolenbrander P E, Andersen R N, Ganeshkumar N. Nucleotide sequence of the Streptococcus gordonii PK488 coaggregation adhesin gene, scaA, and ATP-binding cassette. Infect Immun. 1994;62:4469–4480. doi: 10.1128/iai.62.10.4469-4480.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumano M, Tamakoshi A, Yamane K. A 32 kb nucleotide sequence from the region of the lincomycin-resistance gene (22°–25°) of the Bacillus subtilis chromosome and identification of the site of the lin-2 mutation. Microbiology. 1997;143:2775–2782. doi: 10.1099/00221287-143-8-2775. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence M C, Pilling P A, Epa V C, Berry A M, Ogunniyi A D, Paton J C. The crystal structure of pneumococcal surface antigen PsaA reveals a metal-binding site and a novel structure for a putative ABC-type binding protein. Structure. 1998;6:1553–1561. doi: 10.1016/s0969-2126(98)00153-1. [DOI] [PubMed] [Google Scholar]
- 29.Lindmark H, Guss B. SFS, a novel fibronectin-binding protein from Streptococcus equi, inhibits the binding between fibronectin and collagen. Infect Immun. 1999;67:2383–2388. doi: 10.1128/iai.67.5.2383-2388.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowe A M, Lambert P A, Smith A W. Cloning of an Enterococcus faecalis endocarditis antigen: homology with adhesins from some oral streptococci. Infect Immun. 1995;63:703–706. doi: 10.1128/iai.63.2.703-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meehan M, Nowlam P, Owen P. Affinity purification and characterization of a fibrinogen-binding protein complex which protects mice against lethal challenge with Streptococcus equi subsp. equi. Microbiology. 1998;144:993–1003. doi: 10.1099/00221287-144-4-993. [DOI] [PubMed] [Google Scholar]
- 32.Novak R, Braun J S, Charpentier E, Tuomanen E. Penicillin tolerance genes of Streptococcus pneumoniae: the ABC-type manganese permease complex Psa. Mol Microbiol. 1998;29:1285–1296. doi: 10.1046/j.1365-2958.1998.01016.x. [DOI] [PubMed] [Google Scholar]
- 33.Sampson J S, Furlow Z, Whitney A M, Williams D, Facklam R, Carlone G M. Limited diversity of Streptococcus pneumoniae psaA among pneumococcal vaccine serotypes. Infect Immun. 1997;65:1967–1971. doi: 10.1128/iai.65.5.1967-1971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sampson J S, O'Connor S P, Stinson A R, Tharpe J A, Russell H. Cloning and nucleotide sequence analysis of psaA, the Streptococcus pneumoniae gene encoding a 37-kilodalton protein homologous to previously reported Streptococcus sp. adhesins. Infect Immun. 1994;62:319–324. doi: 10.1128/iai.62.1.319-324.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutcliffe I C, Russell R R B. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Timoney J F, Artiushin S C, Boschwitz J S. Comparison of the sequences and functions of Streptococcus equi M-like proteins SeM and SzPSe. Infect Immun. 1997;65:3600–3605. doi: 10.1128/iai.65.9.3600-3605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timoney J F, Trachman J. Immunologically reactive proteins of Streptococcus equi. Infect Immun. 1985;48:29–34. doi: 10.1128/iai.48.1.29-34.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]