Abstract
Objective
Advanced cervical cancer is still difficult to treat and in the case of recurrent cancer, it is desirable to utilize personalized treatment rather than uniform treatment because the type of recurrence is different for each individual. Therefore, this study aimed to establish a patient-derived organoid (PDO) platform to determine the effects of chemotherapy, radiation therapy, and targeted therapy in cervical cancer.
Methods
We established organoids from 4 patients with various types of cervical cancer. The histopathological and gene profiles of these organoid models were compared to determine their characteristics and the maintenance of the patient phenotype. Each type of organoid was also subjected to anticancer drug screening and radiation therapy to evaluate its sensitivity.
Results
We established PDOs to recapitulate the main elements of the original patient tumors, including the DNA copy number and mutational profile. We selected 7 drugs that showed growth inhibition in cervical cancer organoids out of 171 using an Food and Drug Administration -approved drug library. Moreover, adenocarcinoma and large-cell neuroendocrine carcinoma showed resistance to radiation therapy. whereas squamous cell carcinoma and villoglandular carcinoma showed a significant response to radiotherapy.
Conclusion
Our results showed that patient-derived cervical cancer organoids can be used as a platform for drug and radiation sensitivity testing. These findings suggest that patient-derived cervical cancer organoids could be used as a personalized medicine platform and may provide the best treatment options for patients with various subtypes of cervical cancer.
Keywords: Organoids, Cervical Cancer, Radiation, Tumor-Specific Drug Screening Tests
Synopsis
Patient-derived cervical cancer organoids recapitulated the main elements of the original patient tumor. Drug screening and radiotherapy of organoids facilitate access to patient tumor radiation sensitivity and chemotherapy prediction. Patient-derived cervical cancer organoids could be used as a personalized medicine confirmation tool.
Graphical Abstract
INTRODUCTION
Advanced cervical cancer remains difficult to treat. There is also no standard treatment method for recurrent cervical cancer. Cervical cancer is the fourth most common cancer in women worldwide. The cause of the majority of cervical cancers is an infection of the human papillomavirus (HPV) [1,2]. Two HPV types (16 and 18) are responsible for nearly 70% of high-grade cervical cancers [3,4,5]. The 2 major histological subtypes of cervical cancer are squamous cell carcinoma (SqCa) and adenocarcinoma (AdCa), accounting for 75%–90% and 10%–25% of all cervical cancer cases, respectively [6]. AdCa is more challenging to detect than SqCa and has a poorer prognosis [7].
Villoglandular carcinoma (VGA) is an invasive tumor that occurs in young women and presents extrinsic growth with minimal vascular space involvement, and generally has a better prognosis than other types of cervical AdCa [8,9]. Neuroendocrine cervical cancer (NECC) is a malignant tumor derived from neuroendocrine cells that often accompanies metastasis and has a high recurrence rate even after surgery and radiation therapy [10,11]. Although studies are being conducted on these rare types of cancer, the low incidence makes it difficult to plan prospective cohort studies and determine the optimal treatment.
Currently, chemotherapy, targeted therapy, and immunotherapeutic agents are used for cervical cancer treatment, but biomarkers that can determine effectiveness in patients are lacking [12,13]. Therefore, a new preclinical model is needed to predict the response to treatment according to the characteristics of tumor types.
Patient-derived organoids (PDOs) are being considered as models of the new preclinical models. Organoids can recapitulate patient cancer cells and the tissue environment of patients through self-organization. Tumor organoids can be compared to normal cell organoids, and individual differences in the mechanisms by which tumors develop at various stages of cancer can be studied [14,15]. As a result, organoids are being evaluated as basic and clinically applied cancer research tools in which histological complexity and genetic diversity can be realized [16,17].
Although there have been few reports of radiation sensitivity testing for cancer organoids including rectal, colon, and nasopharyngeal cancer [18,19,20,21], to the author’s knowledge, this is the first study to establish the preclinical in vitro model to predict radiotherapy response using cervical cancer organoids. Optimizing radiation sensitivity through these PDOs is an essential strategy to increase the long-term survival of cervical cancer.
Therefore, this study aimed to establish if patient-derived cervical cancer organoids can recapitulate patient tumors and determine their efficacy as an in vitro platform for drug and radiation sensitivity testing in cervical cancer.
MATERIALS AND METHODS
1. Human data
The study was conducted with the approval of the Institutional Review Board (IRB) at Yonsei University Medical Center (IRB No. 4-2017-0516), and signed consent was obtained from each patient. Cervical cancer tissue was obtained according to the guidelines from Ethical Committee. Clinical data for the individuals are presented in Table 1.
Table 1. Clinical characteristics of patients with cervical cancer (n=6).
| Study information | Sample information | |||||
|---|---|---|---|---|---|---|
| Organoid lab name | Line name in publication | Patient age | Sample event | Radiation (Yes/No) | Sample pathology at diagnosis | HPV |
| CC011 | CC001 | 49 | Punch | Y | Invasive SqCa | HPV 16 |
| CC005 | CC002 | 42 | Punch | Y | AdCa | Negative |
| CC013 | CC003 | 33 | Punch | Y | Large cell neuroendocrine carcinoma | HPV 18 |
| CC027 | CC004 | 25 | Punch | Y | Villoglandular AdCa | Negative |
| CC048 | CC005 | 38 | Punch | Y | AdCa | HPV 16 |
| CC049 | CC006 | 39 | Radical hysterectomy | Y | Small cell neuroendocrine carcinoma | HPV 18 |
| CC089 | CC007 | 50 | Punch | Y | Invasive SqCa | HPV 16 |
AdCa, adenocarcinoma; CC, cervical cancer; HPV, human papillomavirus; SqCa, squamous cell carcinoma.
2. Tumor-derived organoid culture
Cervical tumor tissues were obtained from patients via punch biopsy. The tissues were digested in collagenase solution (2 mg/mL, C9407; Sigma, St. Louis, MO, USA) for 60–90 minutes at 37°C in a shaker. After digestion, cell suspensions were washed with AdDF+++ (Advanced DMEM/F12 containing 1× Glutamax, antibiotics, and 10 mM HEPES). The cells were filtered through a 100 μm nylon cell strainer (542070; Greiner bio-one, Frickenhausen, Germany) and collected via centrifugation for 5 minutes at 1,000 rpm. Next, the erythrocytes were lysed in the red blood cell lysis buffer (11814389001; Roche, Mannheim, Germany). Then, the cells were mixed with Matrigel (356231, Corning, NY, USA). For seeding, a volume of 20 μL was dispensed to pre-warmed 24-well plates (142475; Thermo Fisher Scientific, Waltham, MA, USA), and allowed to solidify at 37°C for 30 minutes. After 30 minutes, the media was added. The complete growth medium for cervical organoids consisted of AdDF+++, supplemented with Noggin (250-38; PeproTech, Rocky Hill, NJ, USA), RSPO1 conditioned medium (10%, made in-house), B27 supplement (1X, 12587010; Gibco, Grand Island, NY, USA), N2 supplement (1X, 17502048; Gibco), human EGF (50 ng/mL, E9644; Sigma), human HGF (50 ng/mL, 100-39; PeproTech), nicotinamide (2.5 mM, N0636; Sigma), N-acetylcysteine (1.25 mM, A9165; Sigma), ROCK inhibitor (10 μM, ALX-270-333; Enzo Life, Farmingdale, NY, USA), A83-01 (500 nM, 2939; Tocris, Minneapolis, MN, USA), and FGF10 (100 ng/mL, 100-26; PeproTech).
3. Histology
The tissues were fixed overnight in 4% paraformaldehyde (PFA, pc2031-050-00; Biosesang, Seongnam, Korea) at 4°C, followed by dehydration and paraffin embedding. Organoids including Matrigel were collected. Matrigel was then removed using cell harvest solution (3700-100-01; R&D Systems, Minneapolis, MN, USA). Then, organoids were fixed with 4% PFA and washed with phosphate-buffered saline (PBS). Finally, the processed organoids were embedded in paraffin blocks. Sections were cut and hydrated before staining. These sections were subjected to hematoxylin and eosin (H&E) or immunohistochemical staining. The p16INK4a (1:500, ab108349; Abcam, Cambridge, UK) antibody was used for immunohistochemical staining. Images were acquired using the DM4000 microscope (Leica Microsystems, Wetzlar, Germany) and processed using Leica LAS X software.
4. Preparation of DNA
DNA was extracted from formalin-fixed paraffin-embedded (FFPE), cells, and tissues using the QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA, USA), DNeasy Blood & Tissue Kit (Qiagen), and the Gentra Puregene Tissue Kit (Qiagen), according to manufacturer instructions. Quantification of DNA was performed using a Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA).
5. Preparation of libraries and sequencing
DNA was sheared into fragments of a mean peak size of 180–200 bp using Adaptive Focused Acoustics (Covaris, Woburn, MA, USA). Libraries were prepared using the SureSelect XT protocol (Agilent Technologies, Santa Clara, CA, USA) with Axen Cancer Master (559 genes; Table S1) developed by Macrogen (Macrogen, Seoul, Korea). Their quality was checked using the 2100 Bioanalyzer (Agilent Technologies); the desired size of the DNA product was 200–400 bp. The libraries were then quantified using the Qubit dsDNA HS Assay Kit and the Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA). Finally, the libraries were sequenced paired-end (2 x ×150 bp) on a NextSeq500 instrument (Illumina, San Diego, CA, USA) with high output using sequencing by synthesis chemistry to a depth of approximately 2,000 × coverage.
6. Analysis of DNA sequences
The adapter sequences were removed using Cutadapt [22]. Trimmed reads were aligned to the reference genome (GRCh37/hg19) using BWA-MEM [23]. Poorly mapped reads with mapping quality (MAPQ) below 20 were removed using Samtools version 1.3.1. [24]. Duplicated reads were discarded using Picard MarkDuplicates (version 2.2.4). The base quality of deduplicated reads was recalibrated using a GATK BaseRecalibrator. Somatic mutations, including single nucleotide variants (SNVs), small insertions, and deletions (INDELs), were identified using the MuTect2 algorithm [25]. False-positive variant calls originating from oxoG artifacts were excluded. Additionally, mutations below 2% variant allele frequency (VAF) and 100X total depth were excluded. Germline variants were excluded when minor allele frequency (MAF) ≥5% in ExAC_EAS or MAF ≥5% in the Macrogen Korean Population Database. All the remaining variants were annotated using SnpEff & SnpSift v4.3i [26,27] with dbNSFP v2.9.3 [28]. The microsatellite instability (MSI) is the calculated ratio of unstable sites over QC-passed regions of the predefined ones of MSI-related sites using mSINGS [29]. The tumor mutational burden is reported as the number of mutations per megabase (mut/Mb) of the passed missense mutations [30].
7. In vitro drug screen
Cells containing Matrigel were seeded at 100 μL/well in a 96-well plate (3474, Corning). Three days after plating the cells, the drugs were added using the Xplorer electronic pipettor (NB.01; Eppendorf, Hauppauge, NY, USA). Paclitaxel (T7402; Sigma), cisplatin (232120; Sigma), LY290042 (S1105; Selleckchem, Houston, TX, USA), and rapamycin (S1039; Sigma) were dissolved in dimethyl sulfoxide (DMSO). Carboplatin (C2538; Sigma) was dissolved in PBS. All wells were normalized to each vehicle solvent used. Drug treatment was performed in triplicate for each concentration.
8. Drug library screen
Cells from the organoid plate were plated (3×103 cells) and cultured for 3 days, and the drug compounds were added to the culture medium at a final concentration of 1 µM. After 4 days of treatment with the compounds, cell viability was examined using the CellTiter-Glo® assay. The suppressive effect was considered significant when the ratio of the average cell viability in the presence of the compounds compared to the control (0.1% DMSO, n=1) was <0.1. The drug library was approved by the Food and Drug Administration.
9. Cell proliferation assay
Four days (96 hours) after adding the drugs, adenosine triphosphate (ATP) levels were measured using the CellTiter-Glo™ 3D cell viability assay (G9683; Promega, Madison, WI, USA). CellTiter-Glo was subjected to temperature equilibration of the plate to room temperature for 30 minutes. Celltiter-Glo reagent was added to each well in a 1:1 ratio with the medium amount and treated at room temperature for 10 minutes. And luminescence was measured using a Centro XS3 LB 960 microplate reader (Berthold Technologies, Oak Ridge, TN, USA). The results represent relative cell viability expressed as % control (drug untreated cell viability=100%). Data analyses were performed using GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA); the values of IC50 and the area under the receiver operating characteristic curve were calculated using nonlinear regression (curve fit) for log [inhibitor] vs. normalized response (variable slope).
10. In vitro organoid radiation studies
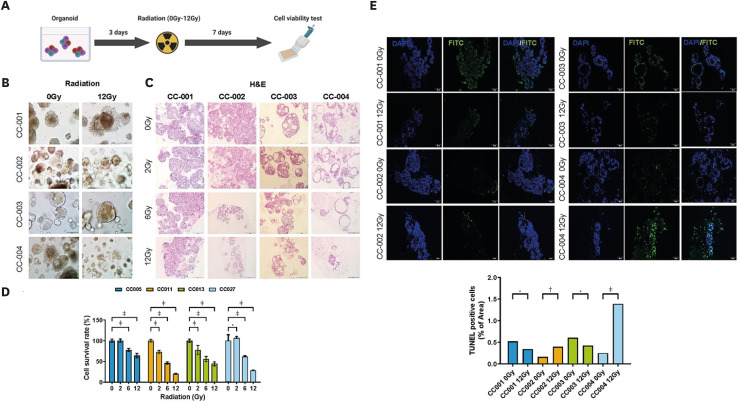
The organoids were plated in 24-well culture plates and left to grow for 3 days. Subsequently, the organoids were exposed to single-fraction radiation (range, 0–12 gray [Gy]). Brightfield imaging of the organoids after radiation was performed using an Olympus IX73 inverted microscope at a 4X objective before irradiation treatment. After imaging, the organoids were exposed to single-fraction radiation (range, 2–12 Gy). The ATP activity of the surviving organoids was compared with that of the unirradiated sample using the CellTiter-Glo™ 3D cell viability assay. All experiments were performed in triplicates.
11. TUNEL assay
Cell death was evaluated by TUNEL assay after 7 days of incubation following radiation treatment at each dose (0, 2, 6, 12 Gy). Paraffin-embedded sections were treated with TUNEL kit reagents (G3250; Promega) according to the manufacturer’s instructions. After incubation with the staining reagents, the samples were washed 3 times with PBS. The images were acquired using an Olympus, BX53 confocal microscope. Cell viability was quantitatively analyzed by using ImageJ (National Institutes of Health, Bethesda, MD, USA) to calculate the percentage of TUNEL-positive cells.
12. Data analysis
Values are expressed as the mean ± standard deviation. Statistical analyses were performed using GraphPad Prism 7 (GraphPad Software). All experiments were repeated in triplicate in 3 independent experiments. One-way analysis of variance and independent t-tests were performed to compare the means of different values. The p-values < 0.05 were considered significant.
RESULTS
1. Establishment of organoids from a cervical biopsy sample
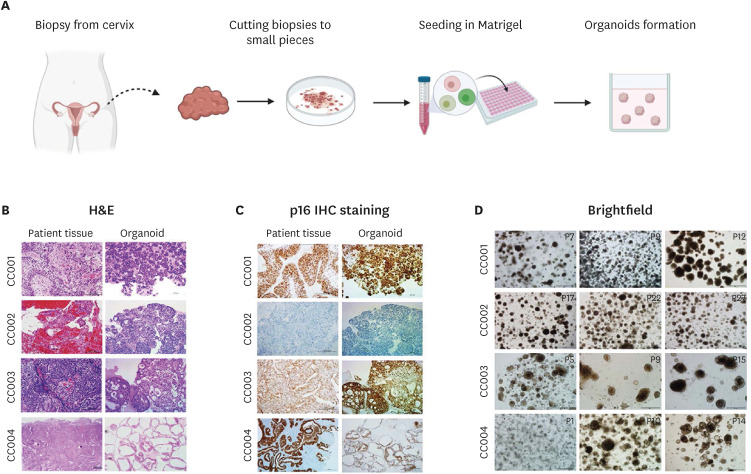
To establish PDOs, we obtained tissues from consented patients with cervical cancer via punch biopsy (Fig. 1A). We dissociated and isolated the cancer tissue using collagenase. After digestion, the cells were embedded into the Matrigel matrix and submerged in the organoid culture medium for growing cervical cancer cells. Seventeen PDOs could be established in the cervical organoid culture medium (34% success rate) from 50 tumor tissues. However, it was difficult to establish organoids from the SqCa type (22.2% success rate) (Table S2). To elucidate the pathological characteristics of the organoids derived from these cervical tissues, we selected 4 types of established organoids SqCa (CC001), AdCa (CC002), NECC (CC003), and VGA (CC004). Clinical data of the patients are presented in Table 1. We first confirmed by H&E analysis that the organoids derived from the patients’ tissues exhibited the same characteristics as the original tumors (Fig. 1B). As shown in the H&E staining results, the morphological features of the organoids are similar to those of the original tumor. The pathology was confirmed by staining for a diagnostic marker of cervical neoplasia p16 was expressed at a high concordance rate in the organoids and patient tumors (CC001, CC003, and CC004) (Fig. 1C). However, the p16 expression of the organoids derived from the AdCa (CC002) was different from that of the original tumors. We also confirmed that the PDOs could be propagated for at least 6 months and that they tolerated a cycle of freeze and thaw (Fig. 1D). These results demonstrate the reliable expression of the pathological pattern of the original patient tumor samples by the established organoids and the feasibility of maintaining the pathological patterns in long-term cultures.
Fig. 1. The establishment of cervical cancer organoids from patient tumor tissues. Patient-derived organoids can be established from a variety of cervical tumor types and can characterize the tissues of the original tumor. (A) Schematic of the cervical organoid culture procedure. (B) H&E staining image of various tumor tissues and the corresponding organoids. (C) IHC for p16 in tumor tissues and the corresponding organoid. (D) Organoid morphology was not changed after the freeze-thawing cycle. Scale bars, 200 μm (magnified images).
H&E, hematoxylin and eosin; IHC, immunohistochemistry.
2. Recapitulation of tumor-associated gene expression patterns by the PDOs
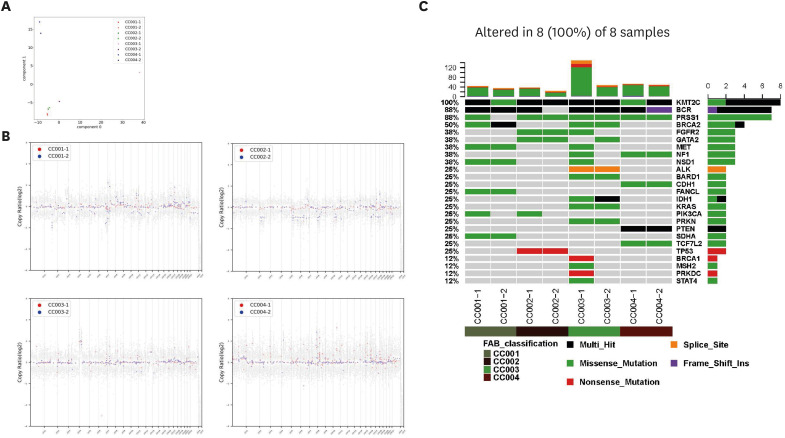
In many types of cancers, tumor-derived organoids have been shown to retain most of the somatic mutations, copy number variations (CNVs), and intratumoral heterogeneity observed in the original tumors. Therefore, we performed whole-genome sequencing on the biopsy-derived organoids and original tumor samples. A principal component analysis map also demonstrated that primary tumor tissue samples were segregated from cancer organoids (Fig. 2A). As shown in Fig. 2B, the copy number profiles showed a degree of concordance for PDOs derived from the same patient. Moreover, organoids derived from NECC (CC003) and VGA (CC004) tumors displayed high CNVs, whereas organoids derived from SqCa (CC001) and AdCa (CC002) showed relatively few CNVs. Next, we characterized oncogenic events in the genes (point mutations, insertions and deletions, amplifications) associated with cervical cancer (Fig. 2C). A missense mutation in the KRAS gene, frequently found in cervical cancer, was identified in CC003. In addition, organoids derived from the CC002 tumor exhibited a nonsense mutation in the TP53 gene. Importantly, KMT2C multi-hit and PRSS1 missense mutations were observed in all tumor types. Taken together, these results indicated that the organoid samples maintained the gene expression pattern after their isolation from patient sample tissues.
Fig. 2. Genomic profiles of cervical patient-derived organoids recapitulating genetic alterations commonly found in primary tumor tissues. (A) The principal component analysis map for gene expression profiles of organoids and cancer tissues from patients. (B) Scatterplots showing the common losses and gains of chromosome number between the patient tissue sample (red) and the organoid (blue). The gains and losses were normalized against the mean genome ploidy level. (C) Somatic mutations and amplifications/deletions in the relevant genes of cervical cancer. For each sample, the tumor (No. -1) and organoid (No. -2) are paired and indicated by color coding (deep green, CC001; brown, CC002; green, CC003; deep red, CC004).
3. Differential drug responses of the cervical tumor organoids according to cancer subtypes
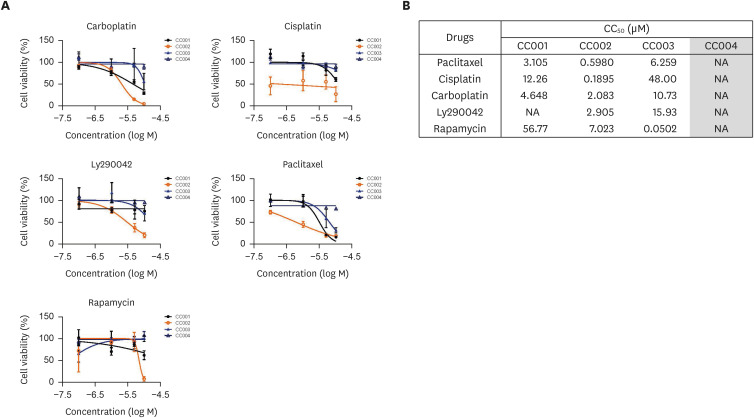
In addition to surgical intervention, radiotherapy and its combination with chemotherapy (i.e., chemoradiation) are commonly used for the treatment of cervical cancer. However, the exact benefit of combining these treatment options has remained controversial. Moreover, many patients suffer long-term adverse effects from chemoradiation. Therefore, we aimed to confirm the drug sensitivity of organoids derived from various cervical cancers. We tested tumor organoid sensitivity to several commonly used chemotherapy regimens, including paclitaxel, carboplatin, cisplatin, Ly290042, and rapamycin. These assays revealed differential drug responses of individual tumor organoid lines (Fig. 3A). In our analysis, paclitaxel had potent anticancer effects on organoids derived from all types of cervical cancer except VGA (CC004). Compared to other organoid types, CC002 organoids also showed high sensitivity to paclitaxel and cisplatin. Similar to other organoids, SqCa (CC001) was more resistant to Ly290042, a PI3K inhibitor, than to other drugs. NECC (CC003) organoids were sensitive to rapamycin, a mammalian target of rapamycin inhibitor. VGA (CC004) showed resistance to all drugs compared to other cancer types. The half-maximal cytotoxic concentration values for these drugs in all organoid lines are illustrated in Fig. 3B. These results show that PDOs can be used for drug screening assays to investigate the resistance and sensitivity of various cervical cancers to chemotherapy.
Fig. 3. The cervical organoids show differential drug responses. (A) The representative dose-response curves for carboplatin, Ly290042, cisplatin, paclitaxel, and rapamycin in PDOs. (B) The CC50 values for anticancer drugs in CC organoids. Data are expressed as the mean ± standard deviation.
CC, cervical cancer; CC50, half-maximal cytotoxic concentration; NA, not applicable; PDO, patient-derived organoid.
4. High-throughput drug screening in PDOs reveals sensitivities to therapeutic agents
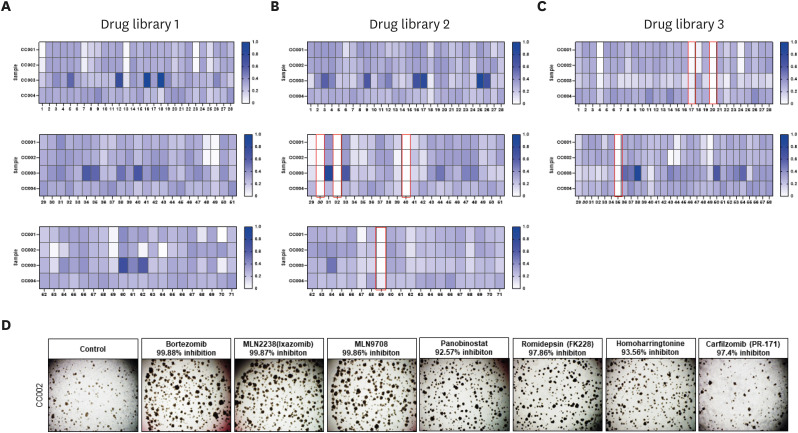
Based on the previous results, we next investigated the feasibility of using these established organoids in personalized therapeutic strategies. To this end, we screened a compound library of clinically used drugs for their ability to suppress the viability of organoids derived from cervical cancer tissue (Table S3). As shown in Fig. 4A, we successfully screened 7 compounds that can substantially suppress the viability of cervical cancer organoids. As expected, 4 compounds were proteasome inhibitors (bortezomib, MLN2238, MLN9708, carfilzomib) are proteasome inhibitors, 2 were histone deacetylase inhibitors (panobinostat, romidepsin) and one was a translation inhibitor (homoharringtonine [HHT]). Mutation of serine protease 1 (PRSS1) was commonly found in the 4 PDOs in this study, and bortezomib was confirmed to target PRSS1 [31]. Moreover, bortezomib has been reported as a potential drug for radiation sensitivity [32]. In the case of the organoids used in this paper, HPV (16,18) infection was confirmed in the patients. The virus is associated with the expression of 2 oncogenes, E6 and E7, which are also ubiquitin–proteasome system-related factors. E7 is known to bind to histone deacetylase (HDAC1/2). Moreover, it has been reported that combining a proteasome inhibitor and an HDAC inhibitor delays tumor growth [33,34]. Furthermore, translation inhibitors, such as HHT, are used for the treatment of patients with chronic myelogenous leukemia. It is an effective anti-melanoma agent that works by inhibiting myeloid leukemia-1 cells to inhibit protein synthesis, leading to DNA damage, apoptosis, and cell cycle arrest [35,36]. HHT has also been recognized as a potential drug with radiation sensitivity in cervical cancer. To confirm this result, we tested the individual drugs in the organoids and observed >90% inhibition in the cervical cancer organoids for all 7 drugs (Fig. 4B). These results suggest that PDOs can be useful for evaluating drug response and resistance to target drugs, and thus, predict patient outcomes.
Fig. 4. Drug library screening of PDOs showing sensitivity to various therapeutic agents. A total of 171 compounds were tested in 4 types of PDOs. The heatmap of cell survival change profile by the anticancer library shows that survival after drug treatment is normalized to a maximum of 1. Selected drugs are marked with a red box. (A) Drug library #1 test with various CC organoids. (B) Drug library #2 test with various CC organoids. (C) Drug library #3 test with various CC organoids. (D) Brightfield image of CC002 treated with selected compounds. Scale bars, 200 μm (magnified images).
CC, cervical cancer; PDO, patient-derived organoid.
5. PDOs response to radiotherapy
To validate the response of PDOs to irradiation in vitro, we performed a radiation dose-dependent (0–12 Gy) survival analysis of the PDOs (Fig. 5A). Fig. 5B shows the degradation of the irradiated organoids (12 Gy) compared to the control (0 Gy), with the radiation-induced degradation being dose-dependent for CC001 and CC004 organoids (Fig. 5C). Next, we validated the survival fraction using the 3D CellTiter-Glo assay. SqCa (CC001) and VGA (CC004) showed radiosensitive characteristics, whereas AdCa (CC002) and NECC (CC003) showed radioresistant characteristics (Fig. 5D). Apoptotic cells were visualized via TUNEL staining of the organoids following radiation exposure (12 Gy). Radiation (12 Gy) induced an increase in TUNEL-positive apoptotic cells only in CC004 (Fig. 5E).
Fig. 5. Cell survival of human cervical cancer organoids after irradiation. (A) Schematic diagram of the irradiation procedure of cervical organoids. (B) Brightfield image of PDOs treated with 2 points (0, 12 Gy) of radiation. (C) PDOs are treated with indicated doses (0/2/6/12 Gy) of radiation, followed by H&E staining. Scale bars, 200 μm (magnified images). (D) Cell viability of PDOs according to radiation dose (0/2/6/12 Gy). Data are expressed as the mean ± standard deviation.
Gy, gray; H&E, hematoxylin and eosin; PDO, patient-derived organoid.
*p=0.010–0.050; †p=0.001–0.010; ‡p<0.001.
DISCUSSION
Cervical cancer is a heterogeneous disease that needs an appropriate tumor model for the development of novel therapeutics. PDOs are developed in various cancer types including colon, stomach, lung cancer et al. PDOs have appeared promising as a platform to better understand the mechanisms of tumor initiation, progression, and metastasis in a variety of cancer types.
Several studies have reported the establishment of patient-derived cervical cancer organoids (Table S4). Lõhmussaar et al. [37] modeled the normal human cervix organoids and used the organoid-based platform to study the HPV, which is one of the main causes of cervical cancer. Gurumurthy et al. [38] reported the formation of endocervical and ectocervical organoids derived from mouse and human adult stem cells. Maru et al. [39,40] also succeeded in establishing organoids in the squamous columnar junction of the normal cervix, and first established in culturing organoids from cervix clear cell carcinoma (cCCC) and discovered that the MET gene is a therapeutic target in cCCC recurrence. Other studies have established organoids in the rare disease, small-cell neuroendocrine carcinoma and identified a specific set of genes that contribute to drug susceptibility [20]. A recent study reported the use of patient-derived ectocervical organoids to model cancer-associated sexually transmitted bacterial pathogen infection and its effect on co-infection with HPV E6E7 and chlamydia [38,41]. Collectively, these suggest that the organoid system can recapitulate cervix cancer tumor properties and be used as models of disease pathogenesis. However, further research is needed to determine whether a screened companion diagnostic option for clinical trials can be developed.
We first established 4 types of organoids from different subtypes of cervical cancer and confirmed that these organoids retained the properties of the patient primary tissues. No specific cause was found for organoids that could not be established. We speculated that the condition and size of the specimens and the differences in the cervix from where the specimens were collected may have influenced organoid growth. Next, we analyzed the characteristics of the established organoids according to the type of cervical cancer. We identified the specific genes through gene profiling analysis of the organoids and patient sample tissues. The KMT2C gene is the third-most frequent gene mutation in cervical cancer and is also associated with a high tumor mutational burden [42]. Our results suggest that the mutated KMT2C gene in all carcinoma types of cervical cancer has the potential to be a predictor of targeted treatment response and prognosis.
To assess the potential of this system to identify or validate effective therapies for cervical cancer, the generated PDOs were exposed to an anticancer library and targeted agents. As a result of comparing the effects of anticancer drugs on various cervical cancer organoids, most PDOs were sensitive to proteasome inhibitors and histone deacetylase inhibitors, but other anticancer drugs showed different reactivity by type. This also indicates the importance of heterogeneity within cancer in cancer treatment resistance and the need for personalized medicine. Although we have shown that organoid response can predict the clinical outcome of anticancer drugs, we performed a radiation sensitivity test for organoids because radiation is the main treatment option for cervical cancer. Recently, after radiotherapy in rectal cancer organoids, sensitivity, specificity, and accuracy were evaluated through matching analysis with the patient’s histological tumor regression grade to evaluate organoids as clinical predictors [20]. And, in the glioblastoma study, established the approach to identify novel therapeutic targets to enhance radiation therapy using CRISPR-based technologies [43]. However, no studies have been conducted on cervical cancer. The radiation sensitivity test results showed that SqCa and VGA were sensitive to radiation therapy and AdCa and large-cell neuroendocrine carcinoma showed resistance to radiation therapy consistent with clinical data [7,44]. In addition, we are optimizing the radiation treatment method for more patients (Fig. S1). Altogether, our results are the first paper to apply radiation therapy in cervical cancer organoids and indicate the potential of cervical cancer organoids as a tool for radiation therapy.
In conclusion, we established patient-derived cervical cancer organoids that recapitulated the patients’ tumors in various histologic types. Cervical cancer organoids showed differential responses to drugs and radiation test and have a strong potential to be a pre-clinical model to identify personalized treatment options and develop new drugs in the era when animal testing is increasingly banned.
ACKNOWLEDGEMENTS
Cartoons in Figs. 1A, 5A and graphical abstract were created with BioRender.com.
Footnotes
Funding: This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT & Future Planning (2020R1A2B5B0100237113) and faculty research grant of Yonsei University College of Medicine (6-2019-0166, 6-2018-0053).
Conflict of Interest: No potential conflict of interest relevant to this article was reported.
- Conceptualization: S.H.S., N.E.J.
- Data curation: S.H.S., O.J.H., C.E., K.S.
- Formal analysis: S.H.S., O.J.H., C.E., K.S.
- Methodology: S.H.S., O.J.H., C.E., K.S., K.H.
- Project administration: N.E.J.
- Supervision: K.H., N.E.J.
- Writing - original draft: S.H.S., O.J.H.
- Writing - review & editing: S.H.S., O.J.H., N.E.J.
SUPPLEMENTARY MATERIALS
Genes analyzed by Axen Cancer Master
Success rates of the organoid culture model classified by cervical cancer types
Compounds list of drug library kits
Summary of reports for the establishment of human cervix organoids and cervical cancer organoids
The response of PDOs to radiation. (A) Brightfield image of PDOs treated with 4 points (0, 2, 4, 8 Gy) of radiation. Scale bars, 200 μm (magnified images). (B) Cell viability of PDOs according to radiation dose (0, 2, 4, 8 Gy). Data are expressed as the mean ± standard deviation.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16:1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaliff M, Sorbe B, Mordhorst LB, Helenius G, Karlsson MG, Lillsunde-Larsson G. Findings of multiple HPV genotypes in cervical carcinoma are associated with poor cancer-specific survival in a Swedish cohort of cervical cancer primarily treated with radiotherapy. Oncotarget. 2018;9:18786–18796. doi: 10.18632/oncotarget.24666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burger RA, Monk BJ, Kurosaki T, Anton-Culver H, Vasilev SA, Berman ML, et al. Human papillomavirus type 18: association with poor prognosis in early stage cervical cancer. J Natl Cancer Inst. 1996;88:1361–1368. doi: 10.1093/jnci/88.19.1361. [DOI] [PubMed] [Google Scholar]
- 5.Arbyn M, Weiderpass E, Bruni L, de Sanjosé S, Saraiya M, Ferlay J, et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob Health. 2020;8:e191–e203. doi: 10.1016/S2214-109X(19)30482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watson M, Saraiya M, Benard V, Coughlin SS, Flowers L, Cokkinides V, et al. Burden of cervical cancer in the United States, 1998-2003. Cancer. 2008;113:2855–2864. doi: 10.1002/cncr.23756. [DOI] [PubMed] [Google Scholar]
- 7.Hu K, Wang W, Liu X, Meng Q, Zhang F. Comparison of treatment outcomes between squamous cell carcinoma and adenocarcinoma of cervix after definitive radiotherapy or concurrent chemoradiotherapy. Radiat Oncol. 2018;13:249. doi: 10.1186/s13014-018-1197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo P, Liu P, Yang J, Ren T, Xiang Y. Villoglandular adenocarcinoma of cervix: pathologic features, clinical management, and outcome. Cancer Manag Res. 2018;10:3955–3961. doi: 10.2147/CMAR.S165817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salek G, Lalya I, Rahali DM, Dehayni M. Villoglandular papillary adenocarcinoma: case report. Pan Afr Med J. 2016;25:232. doi: 10.11604/pamj.2016.25.232.10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tempfer CB, Tischoff I, Dogan A, Hilal Z, Schultheis B, Kern P, et al. Neuroendocrine carcinoma of the cervix: a systematic review of the literature. BMC Cancer. 2018;18:530. doi: 10.1186/s12885-018-4447-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SW, Nam JH, Kim DY, Kim JH, Kim KR, Kim YM, et al. Unfavorable prognosis of small cell neuroendocrine carcinoma of the uterine cervix: a retrospective matched case-control study. Int J Gynecol Cancer. 2010;20:411–416. doi: 10.1111/IGC.0b013e3181ce427b. [DOI] [PubMed] [Google Scholar]
- 12.Baalbergen A, Veenstra Y, Stalpers LL, Ansink AC. Primary surgery versus primary radiation therapy with or without chemotherapy for early adenocarcinoma of the uterine cervix. Cochrane Database Syst Rev. 2010:CD006248. doi: 10.1002/14651858.CD006248.pub2. [DOI] [PubMed] [Google Scholar]
- 13.Bansal N, Herzog TJ, Shaw RE, Burke WM, Deutsch I, Wright JD. Primary therapy for early-stage cervical cancer: radical hysterectomy vs radiation. Am J Obstet Gynecol. 2009;201:485.e1–485.e9. doi: 10.1016/j.ajog.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 14.Hofer M, Lutolf MP. Engineering organoids. Nat Rev Mater. 2021;6:402–420. doi: 10.1038/s41578-021-00279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 16.Artegiani B, Clevers H. Use and application of 3D-organoid technology. Hum Mol Genet. 2018;27:R99–107. doi: 10.1093/hmg/ddy187. [DOI] [PubMed] [Google Scholar]
- 17.Bose S, Clevers H, Shen X. Promises and challenges of organoid-guided precision medicine. Med (N Y) 2021;2:1011–1026. doi: 10.1016/j.medj.2021.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jee J, Park JH, Im JH, Kim MS, Park E, Lim T, et al. Functional recovery by colon organoid transplantation in a mouse model of radiation proctitis. Biomaterials. 2021;275:120925. doi: 10.1016/j.biomaterials.2021.120925. [DOI] [PubMed] [Google Scholar]
- 19.Tirado FR, Bhanja P, Castro-Nallar E, Olea XD, Salamanca C, Saha S. Radiation-induced toxicity in rectal epithelial stem cell contributes to acute radiation injury in rectum. Stem Cell Res Ther. 2021;12:63. doi: 10.1186/s13287-020-02111-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao Y, Xu X, Yang L, Zhu J, Wan J, Shen L, et al. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell. 2020;26:17–26.e6. doi: 10.1016/j.stem.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 21.Lucky SS, Law M, Lui MH, Mong J, Shi J, Yu S, et al. Patient-derived nasopharyngeal cancer organoids for disease modeling and radiation dose optimization. Front Oncol. 2021;11:622244. doi: 10.3389/fonc.2021.622244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 23.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 May 26; doi: 10.48550/arXiv.1303.3997. [DOI] [Google Scholar]
- 24.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cingolani P, Platts A, Wang L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, et al. Using drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet. 2012;3:35. doi: 10.3389/fgene.2012.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC. Microsatellite instability detection by next generation sequencing. Clin Chem. 2014;60:1192–1199. doi: 10.1373/clinchem.2014.223677. [DOI] [PubMed] [Google Scholar]
- 30.Chang H, Sasson A, Srinivasan S, Golhar R, Greenawalt DM, Geese WJ, et al. Bioinformatic methods and bridging of assay results for reliable tumor mutational burden assessment in non-small-cell lung cancer. Mol Diagn Ther. 2019;23:507–520. doi: 10.1007/s40291-019-00408-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deadman JJ, Elgendy S, Goodwin CA, Green D, Baban JA, Patel G, et al. Characterization of a class of peptide boronates with neutral P1 side chains as highly selective inhibitors of thrombin. J Med Chem. 1995;38:1511–1522. doi: 10.1021/jm00009a012. [DOI] [PubMed] [Google Scholar]
- 32.Cui H, Qin Q, Yang M, Zhang H, Liu Z, Yang Y, et al. Bortezomib enhances the radiosensitivity of hypoxic cervical cancer cells by inhibiting HIF-1α expression. Int J Clin Exp Pathol. 2015;8:9032–9041. [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Z, Bazzaro M, Wang MC, Chan KC, Peng S, Roden RB. Combination of proteasome and HDAC inhibitors for uterine cervical cancer treatment. Clin Cancer Res. 2009;15:570–577. doi: 10.1158/1078-0432.CCR-08-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Z, Peng S, Knoff J, Lee SY, Yang B, Wu TC, et al. Combination of proteasome and HDAC inhibitor enhances HPV16 E7-specific CD8+ T cell immune response and antitumor effects in a preclinical cervical cancer model. J Biomed Sci. 2015;22:7. doi: 10.1186/s12929-014-0111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lü S, Wang J. Homoharringtonine and omacetaxine for myeloid hematological malignancies. J Hematol Oncol. 2014;7:2. doi: 10.1186/1756-8722-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang JF, Li GL, Zhang T, Du YM, Huang SY, Ran JH, et al. Homoharringtonine inhibits melanoma cells proliferation in vitro and vivo by inducing DNA damage, apoptosis, and G2/M cell cycle arrest. Arch Biochem Biophys. 2021;700:108774. doi: 10.1016/j.abb.2021.108774. [DOI] [PubMed] [Google Scholar]
- 37.Lõhmussaar K, Oka R, Espejo Valle-Inclan J, Smits MH, Wardak H, Korving J, et al. Patient-derived organoids model cervical tissue dynamics and viral oncogenesis in cervical cancer. Cell Stem Cell. 2021;28:1380–1396.e6. doi: 10.1016/j.stem.2021.03.012. [DOI] [PubMed] [Google Scholar]
- 38.Gurumurthy RK, Koster S, Kumar N, Meyer TF, Chumduri C. Patient-derived and mouse endo-ectocervical organoid generation, genetic manipulation and applications to model infection. Nat Protoc. 2022;17:1658–1690. doi: 10.1038/s41596-022-00695-6. [DOI] [PubMed] [Google Scholar]
- 39.Maru Y, Kawata A, Taguchi A, Ishii Y, Baba S, Mori M, et al. Establishment and molecular phenotyping of organoids from the squamocolumnar junction region of the uterine cervix. Cancers (Basel) 2020;12:694. doi: 10.3390/cancers12030694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maru Y, Tanaka N, Ebisawa K, Odaka A, Sugiyama T, Itami M, et al. Establishment and characterization of patient-derived organoids from a young patient with cervical clear cell carcinoma. Cancer Sci. 2019;110:2992–3005. doi: 10.1111/cas.14119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koster S, Gurumurthy RK, Kumar N, Prakash PG, Dhanraj J, Bayer S, et al. Modelling Chlamydia and HPV co-infection in patient-derived ectocervix organoids reveals distinct cellular reprogramming. Nat Commun. 2022;13:1030. doi: 10.1038/s41467-022-28569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, Li Z, Lu T, Pan J, Li L, Song Y, et al. Genomic landscape, immune characteristics and prognostic mutation signature of cervical cancer in China. BMC Med Genomics. 2022;15:231. doi: 10.1186/s12920-022-01376-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu SJ, Malatesta M, Lien BV, Saha P, Thombare SS, Hong SJ, et al. CRISPRi-based radiation modifier screen identifies long non-coding RNA therapeutic targets in glioma. Genome Biol. 2020;21:83. doi: 10.1186/s13059-020-01995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao G, Qiu J, Zhu F, Wang X. Survival of patients with cervical cancer treated with definitive radiotherapy or concurrent chemoradiotherapy according to histological subtype: a systematic review and meta-analysis. Front Med (Lausanne) 2022;9:843262. doi: 10.3389/fmed.2022.843262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes analyzed by Axen Cancer Master
Success rates of the organoid culture model classified by cervical cancer types
Compounds list of drug library kits
Summary of reports for the establishment of human cervix organoids and cervical cancer organoids
The response of PDOs to radiation. (A) Brightfield image of PDOs treated with 4 points (0, 2, 4, 8 Gy) of radiation. Scale bars, 200 μm (magnified images). (B) Cell viability of PDOs according to radiation dose (0, 2, 4, 8 Gy). Data are expressed as the mean ± standard deviation.