Abstract
High-grade serous carcinoma (HGSC) is the most common and lethal ovarian cancer subtype. PARP inhibitors (PARPi) have become the mainstay of HGSC-targeted therapy, given that these tumors are driven by a high degree of genomic instability (GI) and homologous recombination (HR) defects. Nonetheless, approximately 30% of patients initially respond to treatment, ultimately relapsing with resistant disease. Thus, despite recent advances in drug development and an increased understanding of genetic alterations driving HGSC progression, mortality has not declined, highlighting the need for novel therapies. Using a small-molecule activator of protein phosphatase 2A (PP2A; SMAP-061), we investigated the mechanism by which PP2A stabilization induces apoptosis in patient-derived HGSC cells and xenograft (PDX) models alone or in combination with PARPi. We uncovered that PP2A genes essential for cellular transformation (B56α, B56γ, and PR72) and basal phosphatase activity (PP2A-A and -C) are heterozygously lost in the majority of HGSC. Moreover, loss of these PP2A genes correlates with worse overall patient survival. We show that SMAP-061–induced stabilization of PP2A inhibits the HR output by targeting RAD51, leading to chronic accumulation of DNA damage and ultimately apoptosis. Furthermore, combination of SMAP-061 and PARPi leads to enhanced apoptosis in both HR-proficient and HR-deficient HGSC cells and PDX models. Our studies identify PP2A as a novel regulator of HR and indicate PP2A modulators as a therapeutic therapy for HGSC. In summary, our findings further emphasize the potential of PP2A modulators to overcome PARPi insensitivity, given that targeting RAD51 presents benefits in overcoming PARPi resistance driven by BRCA1/2 mutation reversions.
Watch the interview with Analisa DiFeo, PhD, recipient of the 2025 Molecular Cancer Therapeutics Award for Outstanding Journal Article: https://vimeo.com/1100470225
Introduction
High-grade serous carcinoma (HGSC) is the 5th leading cause of cancer-related deaths in women and the most common and lethal subtype of gynecological malignancies, accounting for approximately 60% of all ovarian carcinomas (1, 2). HGSC is most commonly diagnosed as late-staged metastatic cancer for which treatment options rely on aggressive surgical resection and either single or combinatorial approaches with platinum-based chemotherapies (3). Nevertheless, success rates remain highly unfavorable (4) and acquired chemoresistance is one of the factors contributing to HGSC relapse and metastasis (5). Therefore, there is an urgent need to develop more efficient therapies. PARP inhibitors (PARPi) have been recently approved to treat HGSC (6–8) given their success in treating HR-deficient patients with HGSC, most commonly germline and/or somatic mutations in either BRCA1 or BRCA2 (BRCA1/2). PARPi are a class of small molecules that target PARP1, blocking the repair of single-strand DNA (ssDNA) breaks, resulting in DNA double-strand breaks (DSB; ref. 9). In the absence of functional PARP1, HR-deficient cells accumulate DNA damage, leading to cell death in a process known as synthetic lethality. However, when HGSC tumors are BRCA1/2 wild-type, thus HR-proficient, PARPi-dependent DSB formation is resolved by the functional HR machinery, preventing cell death and promoting cancer cell survival. Therefore, finding targeted therapies to prevent and treat tumors that are not responsive to PARPi are of high priority.
Protein Phosphatase 2A is a large family of Serine/Threonine phosphatases consisting of three main subunits—PP2A-A, PP2A-B, and PP2A-C—that form an active trimeric holoenzyme that dephosphorylates substrate proteins (Fig. 1A; refs. 10, 11). The scaffold (PP2A-A) subunit is highly flexible and serves as the platforming surface for all components to bind and closely interact (12). The catalytic (PP2A-C) subunit provides the phosphatase with its enzymatic capability. Finally, the regulatory (PP2A-B) subunit grants the phosphatase substrate specificity, and is comprised of 16 different subunits with divergent substrate recognition properties (13). PP2A's function is impaired in the majority of human cancers, as its main tumor-suppressive role is to coordinately regulate essential cellular processes, by counteracting oncogenic signals that drive tumorigenesis (13–16). The MAPK (11, 17, 18) and Wnt (18, 19) pathways have been extensively studied as direct targets of PP2A. Recently, the impact of PP2A on DNA damage response (DDR) and homologous recombination (HR) has been established (20–24). Multiple PP2A heterotrimers are essential in directing successful HR-dependent DSBs repair (24) and the BRCA2–PP2A complex formation/stability was established as required for HR-mediated DNA repair efficiency in human cells (25). Therefore, developing selective therapies that stabilize and modulate the tumor-suppressive activities of PP2A represents an attractive therapeutic target strategy for the treatment of human cancers driven by a high degree of genomic instability (GI), including HGSC.
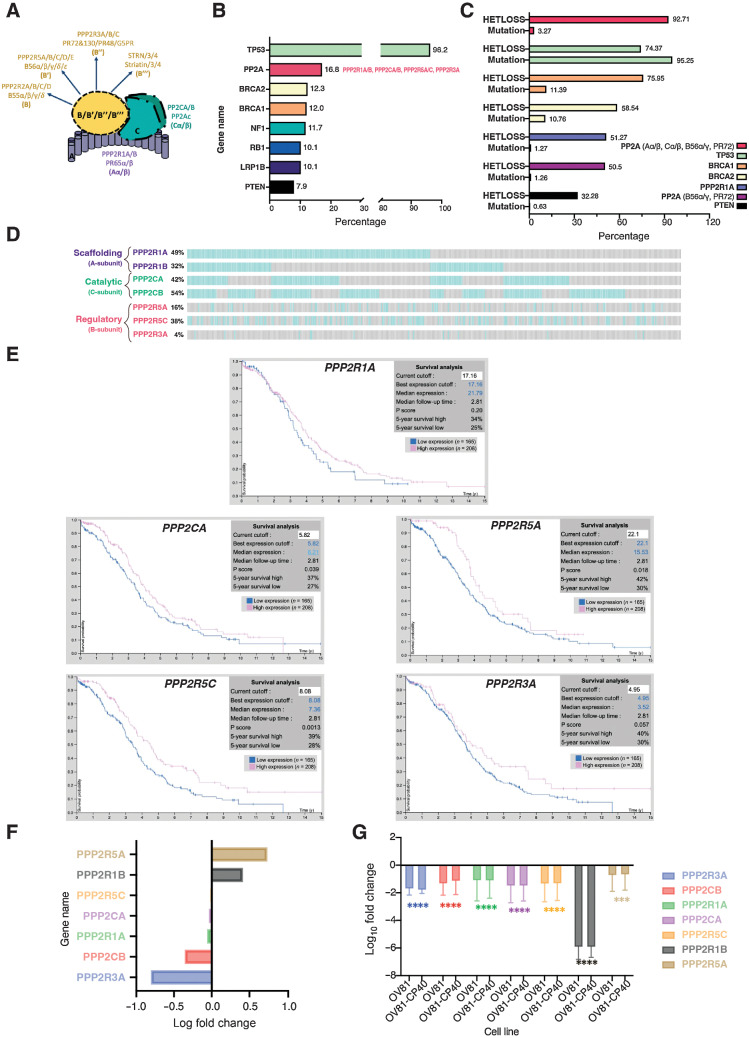
Figure 1.
Dysregulation of PP2A is a common event in HGSC. A, Schematic of the PP2A heterotrimeric structure, representing all possible combinations of the scaffolding (A), regulatory (B), and catalytic (C) subunit proteins (schematic created with BioRender.com). B, Top 8 most common altered genes in HGSC and respective genetic alterations frequency (mutation, amplification, and/or deletion). “PP2A” group includes PP2A-Aα/β, PP2A-Cα/β, PP2A-B56α/γ, and PP2A-PR72. Data adapted from cBioPortal. C, HGSC genes alteration type specificity: heterozygous/shallow deletion (Hetloss) or mutation; and respective frequencies (%). Data adapted from cBioPortal. D, Individual patient tumor data analysis showing frequency of heterozygous loss status for the PPP2R1A, PPP2R1B, PPP2CA, PPP2CB, PPP2R5A, PPP2R5C, and PPP2R3A genes. Co-occurrence profiles are also shown. Each bar represents an individual tumor, for which gray signifies negative and blue positive for Hetloss. E, Kaplan–Meier plots summarizing results from correlation analysis between mRNA expression level and patient survival for PP2A genes: PPP2R1A, PPP2CA, PPP2R5A, PPP2R5C, and PPP2R3A. Patients were divided according to expression levels into one of two groups “low” (under cutoff on table) or “high” (over cutoff on table). The x-axis shows time for survival (years), and the y-axis shows the probability of survival, where 1.0 corresponds to 100%. Data source: Protein Atlas. F, Log fold change of transcriptional expression of PP2A subunits between HGSC cell lines (OVSAHO, KURAMOCHI, and JHOS4) versus FTSEC cell lines (FT246, FT33, and FT194). Source: Elias et al. 2016 (45). G, Log10 fold change of fragments per kilobase of exon per million mapped fragments (FMPK) of the previously assessed PP2A genes (D) comparing expression of the FT246 nonmalignant with the OV81 and OV81-CP40 cell lines. Data presented as the mean ± SD (n = 3; unpaired Student t tests, comparing OV81 and OV81-CP40 relative with FT246; ***, P < 0.005; ****, P < 0.0001).
Targeting PP2A has been considered challenging given the multiple structurally distinct heterotrimers and the lack of detailed high-resolution structural data. A small-molecule activator of protein (SMAP-061; DT-061, as referenced by Leonard and colleagues; ref. 26) was shown to act as a molecular-glue that fits in a binding pocket created through the interaction between PP2A-A, PP2A-C, and PP2A-B subunits to effectively modulate PP2A holoenzymes (26), driving downstream dephosphorylation of oncogenic drivers. Cryo-EM results suggested that SMAP compounds preferentially recruit PP2A-B56α subunit to form a trimeric holoenzyme (26), despite alternative mechanisms postulating SMAP-061 stabilization properties of the B55α-PP2A heterotrimer (27). Nonetheless, multiple groups have evaluated the therapeutic potential of SMAP-mediated PP2A modulation and explored the molecular mechanisms behind their activity (26, 28–30). SMAP-061 shows potent anticancer properties in vitro and in vivo resulting in reduced tumor burden in various human cancer models without tolerability issues, even in long-term dosing studies in vivo (26, 28–32). Furthermore, preclinical studies using genetically engineered and orthotopic mouse models of cancer, demonstrate SMAP-061’s and other PP2A mediators’ potent activity in human cancers, including prostate (28), breast (31), lung (29, 30), and glioblastoma (32).
In this current study, we found that >90% of HGSC tumors display heterozygous loss of at least one of the PP2A genes essential for malignant transformation and/or holoenzyme formation. In addition, decreased expression of those genes correlate with worse overall HGSC patient survival rates. Here, we report this recurrent genetic perturbation in PP2A can be leveraged therapeutically using SMAP-061 in HGSC HR-proficient and -deficient tumors, by decreasing the expression and activity of multiple DDR and HR proteins. Furthermore, we confirmed that SMAP-061–induced RAD51 protein expression loss is necessary to inhibit RAD51-mediated filament formation, impairing the high-fidelity HR pathway and sensitizing HGSC cells to PARPi, ultimately inducing synthetic lethality in this malignant context. Finally, SMAP-061 works additively or synergistically with olaparib in patient-derived (PD) HGSC models with initial varying sensitivities to PARPi. Collectively, our data suggest that SMAP-061 represents a unique opportunity to expand the patient population that can benefit from PARPi, no longer having to rely only on tumors with HR deficiency (HRD).
Materials and Methods
Generation of patient cell lines and PDX models
Patient samples were collected under an approved University Hospitals of Cleveland Institutional Review Board (IRB). The IRB protocol included the prospective collection of discarded tissue and the generation of patient-derived xenograft (PDX) with written informed consent obtained from the study subjects (PI: A. DiFeo). As part of these IRB protocols, clinical and pathological data were gathered for some patients, and included age at diagnosis, race, tumor stage and grade, histological type, and overall survival. The PD cell lines (OV78, OV81, OV82, OV84, OV85, OV93, OV145, and OV262) were generated from primary human ovarian cancer tissue following protocols previously described (33). To generate PDX models, tumors or ascites were removed from patients and approximately 2 mm3 tumor implants were grafted, or cells were injected subcutaneously in NSG mice, respectively. Once tumors were detected, tumor volumes were measured weekly and were collected after reaching approximately 1,000 mm3. Tumors were then (i) re-implanted into another NSG mouse as a passaging tool, (ii) processed using IHC analysis to compare the pathology to the original patient tumor, (iii) processed for exome sequencing, and finally (iv) frozen in freezing media for future drug studies. All cell lines were tested for the presence of Mycoplasma using a PCR-based kit (Southern Biotech 13100–01) and all tested negative before proceeding with the experiment. These studies were approved by IACUC.
CRISPR Cas9 cell line generation
Guide RNAs to PPP2R5A (B56α) and PPP2R2A (B55α) sequences were obtained from GenScript that were deposited and licensed by the Feng Zheng laboratory at the Broad Institute. The guide RNA chosen for each gene had the following sequence: PPP2R5A (#sg1: TGATCAGAAATTCGTACAAC, sg#2: AACCCACGCTTGAGGCCTCT); PPP2R2A (#sg1: TAGAGTTGTCAT CTTTCAAC, #sg2: ACAGTAGAATTTAATCATTC). Guide RNAs were cloned into the vector pLentiCRISPRV2 using the CloneEZ method by GenScript. Plasmids were verified by sequencing through MCLAB. B56α, and B55α-LentiCRISPRV2 lentivirus was generated by the Vector Core at the University of Michigan. For infection, roughly 100,000 OV81 cells were incubated for 48 hours with 200 µL of 10X virus (2 mL total) for a 1X concentration. After infection, cells were selected with 1 µg/mL of Puromycin (Invivogen ant-pr-1) for 72-hours. Clones were collected and tested for the expression of B56α and B55α using Western blotting techniques. Three biological replicates were used to confirm knockout.
In vivo drug studies
Mice were randomized to one of 4 different treatment groups depending on their initial tumor size. Mice were enrolled when tumors reached approximately 100 mm3 and distributed in different treatment groups so that the initial tumor volume average was the same across each group. Mouse tumor volumes and body weight were measured every other day. SMAP-061 was diluted in 10% DMA (Sigma-Aldrich 271012–12), 10% Solutol (Sigma-Aldrich 42966), and 80% warm RNA-free water. DMA control and SMAP-061 were administered via oral gavage twice a day whereas PBS control and olaparib were injected via intraperitoneally, once a day. All the above stated studies were approved by IACUC and followed the appropriate guidelines.
Proliferation and colony forming assays
HGSC cells were seeded in a 12-well plate overnight to reach 70% confluency at the day of treatment. Cells were then treated with either DMSO (Thermo Fisher Scientific BP231–100), SMAP (5, 10, 20, 30 or 40 µmol/L), cisplatin (5, 10, 20, 40 or 80 µmol/L) or olaparib (25, 50, 100, 200, 600 µmol/L; Selleck Chemicals S1060) and incubated for 48 hours. Cell viability was measured by MTT assay using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide kit (Thermo Fisher Scientific M6494). For the isobologram assays, the combination of dose increments of SMAP and olaparib was used, and cell viability was measured after 48 hours of incubation with the drugs. To perform colony-forming assays, cells were plated at a low density (<100 cells/well) in biological triplicate in a 6-well plate. After 48 hours, cells were treated with DMSO, 6 or 10 µmol/L of SMAP, 100 nmol/L of olaparib or 6 µmol/L SMAP + 100 nmol/L olaparib and incubated for 10–12 days. Drug medium was replaced every three days. At day 12, cells were fixed and stained with 1% crystal violet solution. Image laboratory software was used to quantify colony-forming density, measured by pixel intensity.
Western blotting
Cells were harvested and pelleted for protein extraction using a cocktail consisting of 1X RIPA Lysis and Extraction Buffer (EMD Millipore 20–188), 5% glycerol, protease inhibitor (Thermo Fisher Scientific A32955) and phosphatase inhibitor (Thermo Fisher Scientific A32957). Quantification of the isolated proteins was performed using a Pierce BCA Protein Assay kit (Thermo Fisher Scientific 23227), run on a 12% or 4%–20% gradient SDS–PAGE (Bio-Rad 4568045 and 4568095, respectively). Proteins were transferred onto a nitrocellulose membrane (Bio-Rad 1704159) using the quick semi-wet transfer Trans-Blot Turbo transferring machine. Membranes were blocked using 5% non-fat milk (Thermo Fisher Scientific 50488785) made with 1% TBS Tween20 (TBST) buffer (AMRESCO 10791–792). Antibodies were purchased from (i) Cell Signaling Technology: p-Rb (8516), t-RB (9309 and 9313), Cyclin D1 (2922), PLK1 (4513), p-Cyclin B1 (4133), t-Cyclin B1 (4135), Cyclin E (4129), γH2Ax (9718), t-RPA (2208), BRCA1 (14823), BRCA2 (10741), p-ATM (13050), t-ATM (92356), p-CHK1 (2348), t-CHK1 (2360), p-CHK2 (2197), t-CHK2 (6334), RAD51 (8875), CDC25A (3652), Wee1 (13084), GAPDH (5174), Cleaved PARP (9541), Cleaved Caspase-3 (9661), Vinculin (13901); (ii) Bethyl Laboratory: p-RPA (A300–245A); (iii) Santa Cruz Biotechnology: Vinculin (sc-73614), GAPDH (sc-47724), B55α (sc-81606; iv) EMD Millipore BRCA2 (OP95), and (v) a gift from Dr. Egon Ogris’s Laboratory B56α-3A6D3.
Cell-cycle experiments
For cell-cycle analysis, cells were plated in 10-cm cell culture dishes at a density of 2 mol/L cells per plate in biological triplicate (approximately 70% confluency) and treated the next day. Cells were then treated with 2 mmol/L of Thymidine (Sigma-Aldrich T1895) and incubated for 24 hours to allow block of the cells in the S phase of the cell cycle. Cells were then released from the Thymidine block by replenishing with regular media for 6.5 hours to allow cells to reengage in cell-cycle progression. Following the 6.5 hours release, cells were then treated with either DMSO or 20 µmol/L SMAP and incubated for 6, 10, and 16 hours and subsequently harvested and fixed using 70% cold ethanol. PI (Thermo Fisher Scientific F10797) staining was performed according to FxCycle PI/RNAse Staining Solution protocol available online (www.thermofisher.com) and cell-cycle profiles were registered using the Bio-Rad Zed flow machine. Cell-cycle graphs were finalized using FlowJo 8.
Immunofluorescence
OV81 cells were grown on glass coverslips for 24 hours followed by 12 hours of exposure to 20 µmol/L SMAP-061 or 100 µmol/L of olaparib. Cells were then fixed using 4% paraformaldehyde, permeabilized with 0.1% TritonX-100, and blocked with 3% BSA diluted in 1X PBS. Cells were then incubated with γH2Ax (Cell Signaling Technology, 9718) or RAD51 primary antibody (Cell Signaling Technology, 8875) overnight at 4°C. The cells were washed with 1X PBS and incubated with fluorescent secondary antibody for 1-hour at room temperature. Coverslips were mounted on glass slides using mounting media with DAPI (Invitrogen, P36966). Images were acquired using a Leica DMI6000 inverted microscope and then analyzed using ImageJ.
Histology
Immunohistochemical staining was performed on the DAKO Autostainer (DAKO) using Rabbit-on-rodent HRP (BioCare Medical, Pachero, CA, RMR622) and diaminobenzadine (DAB) as the chromogen. Heat-induced epitope retrieval (10 mmol/L Tris/1 mmol/L EDTA pH9) was used before staining for both antibodies. De-paraffinized sections were then labeled with negative (no primary antibody) and positive controls (RAD51 Abcam, ab133534) and γH2Ax (Cell Signaling Technology, 9718), which were stained in parallel with each set of slides studied. γH2Ax foci were scored unbiasedly by a Pathologist and quantification was further verified using ImageJ.
Statistical analysis
Statistical methods used are described in each respective figure legend. Fold change controls and normalization methods are also specified for each one of the analyses.
The data generated in this study are available within the article and its Supplementary Data Files. TCGA data used in the current article are publicly available through cBioportal.
Results
Loss of PP2A is a frequent event in HGSC
Because of the heterogenous nature and high degree of GI of HGSC, validating and functionally identifying putative genes that drive its tumorigenesis remains challenging. Approximately 50% of HGSCs have genetic alterations in the HR pathway, with TP53, BRCA1/2, and RB1 at the top (Fig. 1B). The PP2A family of phosphatases is established as essential in directly dictating dephosphorylation within the DDR and HR pathways (22, 34). Furthermore, Karst and colleagues (35) has demonstrated that loss of PP2A is essential for the transformation of the Fallopian Tube Secretory Epithelial Cells (FTSEC or FT) model, suggesting an important role for PP2A in HGSC carcinogenesis. Interestingly, opposite to other gynecological malignancies, such as serous endometrial cancers (36–38), literature shows that PPP2R1A somatic mutations in ovarian cancers are rare across most histological subtypes (39), suggesting that the inactivation mechanism of PP2A in this tissue type is accomplished differently. Nevertheless, gene alterations to PP2A and their direct impact on HGSC development and progression remain unexplored. Thus, we first sought to define the differential expression of PP2A genes previously shown to drive tumorigenesis and correlate it with clinical outcomes in HGSC. We identified that multiple genes in the PP2A family significantly more altered in HGSC than the majority of the well-established HR genes (Fig. 1B and C). According to TCGA, PPP2R1A is heterozygously deleted/lost (HetLoss) in approximately 50% of HGSC, and at least 1 of the 3 B-subunits previously established as essential for malignant transformation (B56α, B56γ, and PR72; refs. 40–42) was also approximately 50% heterozygously lost (Fig. 1C). Looking at the probability of the minimal essential genes for PP2A enzymatic/phosphatase basal activity (i.e., PP2A-A and PP2A-C, essential core dimer proteins, although lacking specificity) and the B-subunit genes with transformative potential (B56α, B56γ, and PR72), these genetic alterations were present in 92.7% HGSC tumors (Fig. 1C, PPP2R1A/B, PPP2CA/B, PPP2R5A/C, and PPP2R3A). Interestingly, such alterations are not mutually exclusive, as certain patients harbor multiple shallow deletions in more than one of these PP2A genes (Fig. 1D). All B-subunits alterations were individually analyzed (Supplementary Table S1A). Interestingly, somatic mutations are rare (3.48%; Fig. 1C; Supplementary Table S1A), suggesting that optimizing PP2A heterotrimer formation and biasing the existing pools toward a more tumor-suppressive holoenzymatic structure can be a promising strategy to produce optimal anticancer effects in HGSC tumors. Further analysis shows that copy-number alterations proportionally correlate with their respective mRNA levels (Supplementary Fig. S1A–S1F), where low mRNA expression of PPP2R1A, PPP2CA, PPP2R5A, PPP2R5C, and PPP2R3A directly correlates with decreased patient outcome (Fig. 1E). These data suggest these specific PP2A genetic vulnerabilities may be contributing to patient survival. Individual patient tumor data also demonstrate that Hetloss is not mutually exclusive within the PP2A genes (Supplementary Fig. S1G and Supplementary Table S1B), and its co-occurrence is prevalent with similar combination patterns between the scaffolding, catalytic, and regulatory subunits (Supplementary Fig. S1H). In addition, PP2A-A is haploinsufficient in cellular and in vivo model systems (40, 43, 44), further supporting that these highly recurrent alterations in HGSC might play pathogenic roles in disease development and progression. Moreover, as these genes are rarely mutated, selective deletion of specific pools of PP2A’s activity is suggested to be critical for HGSC tumorigenesis. Thus, therapeutic reactivation and modulation of the remaining PP2A heterotrimers pool might be sufficient to trigger HGSC cell death.
SMAP-061 treatment induces apoptotic cell death in HGSC
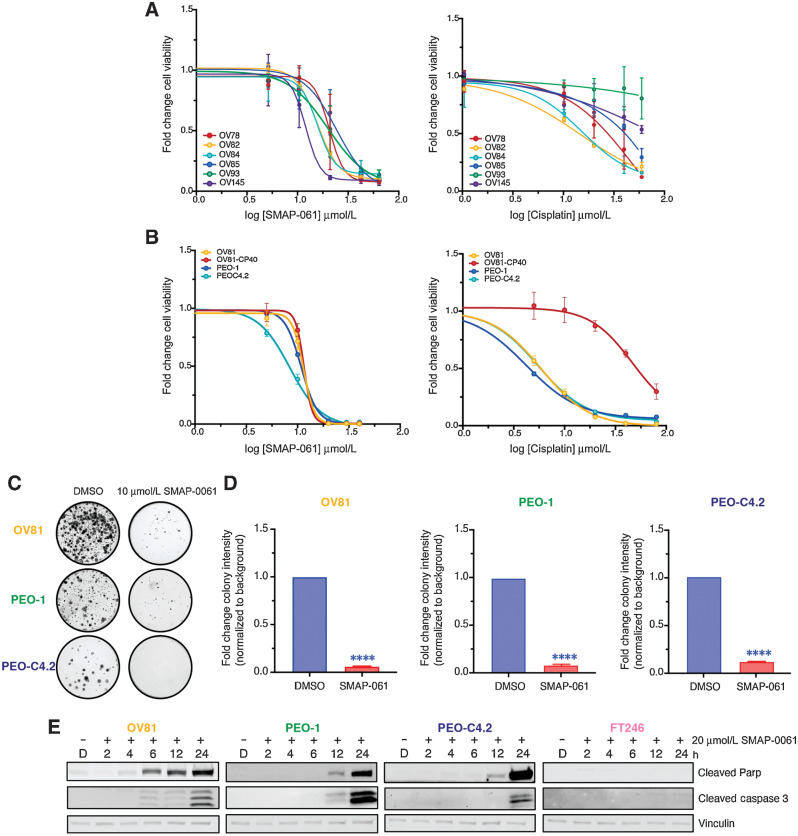
Given that a large majority of HGSC tumors displayed heterozygous loss of at least one of the PP2A genes and loss of PP2A is required for HGSC transformation (41, 42), we next investigated whether PP2A modulation using SMAP-061 affects HGSC PD (Fig. 2A, left and Supplementary Table S2A) and cisplatin-sensitive/resistant isogenic cell lines (Fig. 2B, left and Supplementary Table S2B). We first confirmed that the subunits commonly lost in HGSC tumors were significantly reduced in the HGSC models compared with FT non-malignant lines (Fig. 1F and G; ref. 45). All HGSC lines showed sensitivity to SMAP-061, with EC50 values ranging between 10 and 20 µmol/L, despite the wide range of responses with cisplatin treatment, which is the first-line chemotherapy for HGSC (Fig. 2A, right and Supplementary Table S3A). To further assess whether cisplatin sensitivity and/or HR defects dictated SMAP-061 response, we used isogenic cisplatin-sensitive and -resistant lines (Fig. 2B; Supplementary Table S2B). OV81 and its cisplatin-resistant (CR) pair OV81-CP40 showed significant distinct responses to cisplatin given the continuous growth in low cisplatin concentrations for 40 passages; however, their response to SMAP-061 remained the same (Fig. 2B, left). PEO-1 and its isogenic pair PEO-C4.2 showed similar responses to SMAP-061; however, higher sensitivity to cisplatin treatment due to being exposed to cisplatin for only 2 passages. However, the unique feature of this pair is that PEO-1 is HR-deficient as it harbors a BRCA2 mutation 5193C>G(Y1655X) whereas PEO-C4.2 acquired an BRCA2 reversion mutation after cisplatin treatment, thus making it HR-proficient (ref. 46; Supplementary Table S2B; Fig. 2B, right). Interestingly, SMAP-061 shows that similar potency profiles despite differences in platinum sensitivities and HR status (Fig. 2A and B). To further investigate the impact of HR proficiency on the molecular response of HGSC lines to SMAP-061, the PD line (OV81) and the isogenic HR-deficient and proficient pair (PEO-1 and PEO-C4.2) were selected. Long-term effects of SMAP-061 treatment on these cells were assessed using colony-forming assays and SMAP-061 exposure resulted in significantly fewer colonies (Fig. 2C and D). Moreover, SMAP-061 led to a significant induction of apoptosis in a time-dependent manner, as indicated by the increase of cleaved PARP and cleaved caspase-3 in all three HGSC cell lines (Fig. 2E). Given that HGSCs are derived from the fallopian tube epithelium, we wanted to further investigate whether these apoptotic effects would also be observed in the non-malignant precursor cells or if they were specific to their malignant counterparts. We found that the benign FTSEC 246 (FT246) was less sensitive to SMAP-061 when compared with HGSC cells (Supplementary Fig. S3D; EC50: 22.97 µmol/L vs. EC50: 11.38 µmol/L, respectively) and apoptotic cues were not triggered when equal doses of SMAP-061 were used as in the cancer cells (Fig. 2E). Together, these data suggest that stabilization of PP2A leads to robust apoptotic effects in clinically and genetically diverse PD primary and established HGSC cells.
Figure 2.
HGSC cells are sensitive to SMAP-061 and have similar responses independent of genetic background. A, A wide panel of HGSC PDX cell lines were exposed to increasing doses of either SMAP-061 (left) or cisplatin (right) treatment, and cell viability was measured at 48 hours by MTT analysis, to generate an EC50 curve. Data are presented as the mean ± SD (n = 3). B, OV81 and its cisplatin-resistant pair, OV81-CP40, and two isogenic lines PEO-1 and PEO-C4.2, were treated with increasing doses of either SMAP-061 (right) or cisplatin (right) and cell viability was measured at 48 hours by MTT analysis, to generate an EC50 curve. Data presented as the mean ° SD (n = 3). C, Clonogenic assay of OV81, PEO-1, and PEO-C4.2 cells treated with DMSO or 10 µmol/L of SMAP-061 for 2 weeks. D, Quantification of 2C. Data presented as the mean ± SD (n = 3; unpaired Student t tests, comparing SMAP-061 treatment relative with DMSO, ****, P < 0.0001). E, OV81, PEO-1, PEO-C4.2, and FT246 treated with DMSO or SMAP-061, and harvested at 2, 4, 6, 12, and 24 hours for Western blot analysis of cell death markers (cleaved Parp and cleaved Caspase-3). Vinculin-housekeeping gene used as loading control.
SMAP-061–induced apoptosis is due to inhibition of HR and DDR signaling output and unresolved DNA damage
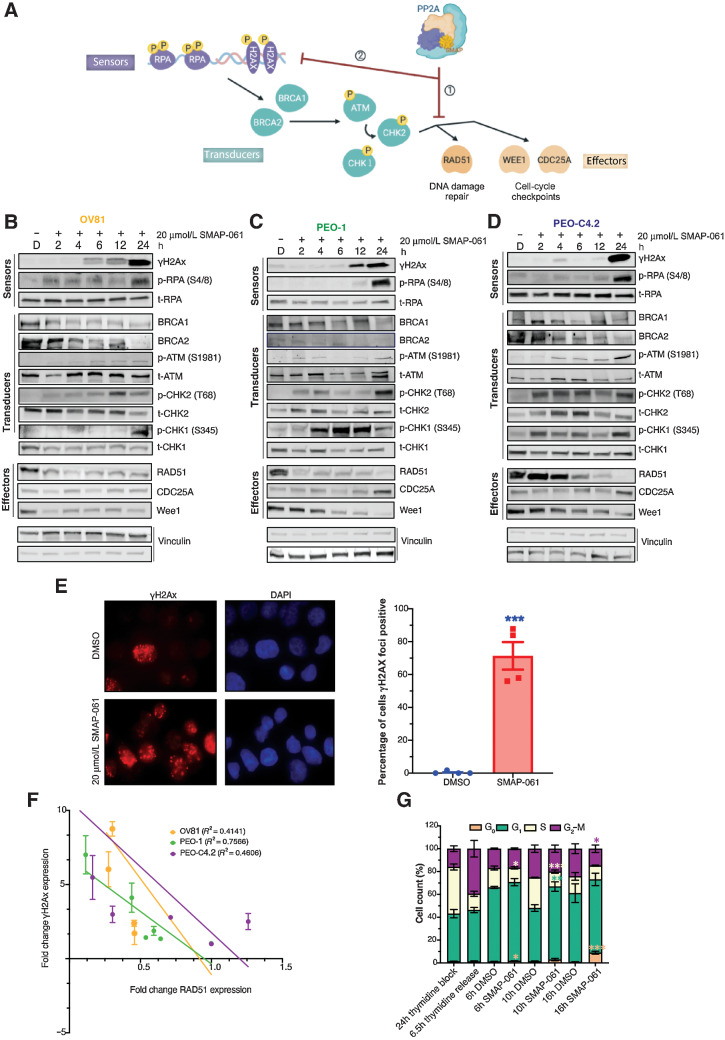
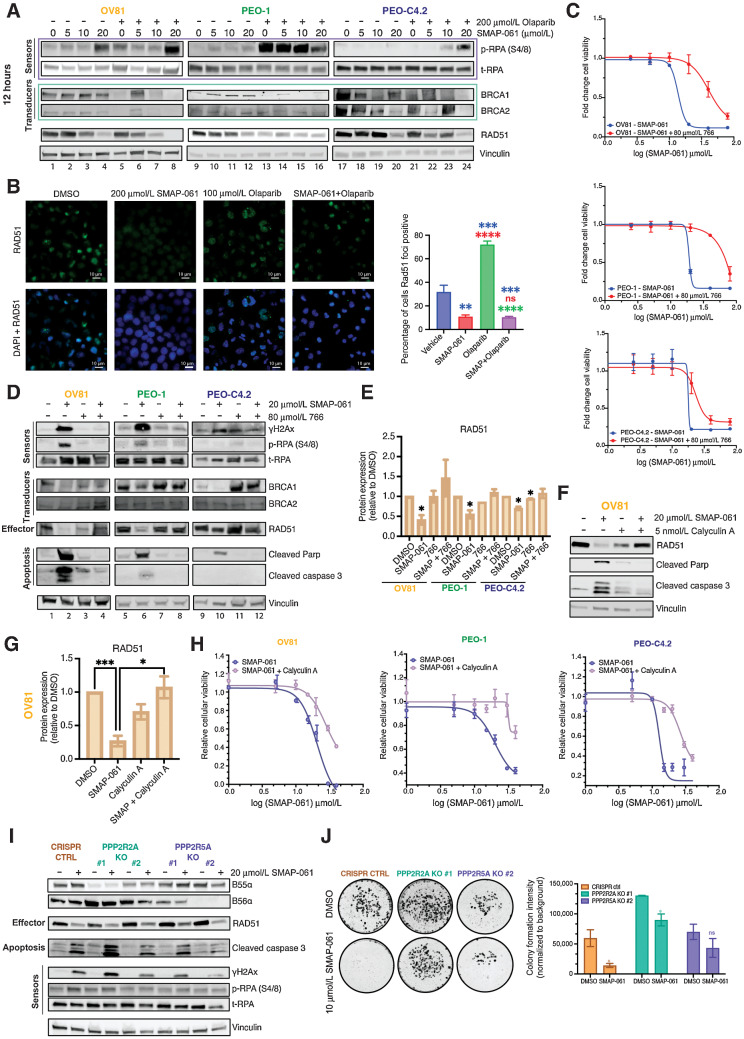
A hallmark of HGSC cells is their ability to withstand high levels of DNA damage, due to defects in HR that lead to high degrees of GI (47). These HR-deficient HGSC cells continue to rely on and benefit from the minimally functional HR pathway, to maintain sufficient levels of DNA repair that can sustain baseline cell survival. Given that SMAP-061 induces cell death in a wide-array of HR-proficient and -deficient HGSC cell lines and that PP2A regulates several components of the HR pathway, we next wanted to examine whether SMAP-061 was affecting HR signaling (Fig. 3A). Interestingly, unlike what is commonly seen with DDR signaling in that the sensor proteins are first induced followed by the activation of the transducers and effectors to repair the damage (Fig. 3A), we found that loss of RAD51 and WEE1 occurred significantly earlier (<2 hours) and before the accumulation of γH2Ax and p-RPA, only seen at approximately 6–12 hours post-treatment (Fig. 3B–D; Supplementary Fig. S2A–S2L). As expected, the downstream phosphorylation and activation of direct targets such as ATM and CHK1/2 proteins increased, attempting to fully activate HR and proceed to repairing the DNA insults. By remaining unresolved, such damage accumulation ultimately triggered apoptosis, as previously observed (Fig. 2E). SMAP-061 treatment increased γH2Ax foci formation by 70% compared with DMSO, after 12 hours of treatment (Fig. 3E), suggesting that SMAP-061 promotes unresolved DNA damage accumulation. Furthermore, a significant inverse correlation between RAD51 and γH2Ax expression for all three treated HGSC cell lines was observed (Fig. 3F), correlating the loss of RAD51 with the accumulation of γH2Ax. Interestingly, the observed phenotype is independent of HGSC BRCA genes’ status and HR genetic background, as all three cell lines show similar molecular signatures with SMAP-061, except for the RAD51 degradation and γH2Ax accumulation being delayed in PEO-C4.2 cells, which harbor a BRCA2 reversion mutation (Fig. 3B–D). These results suggest that SMAP-061 could be inducing synthetic lethality through the regulation of crucial HR pathway proteins, thus leading to the accumulation of unresolved DNA damage and concomitant cell death (Fig. 3A).
Figure 3.
SMAP-061–induced baseline DNA damage accumulation is a result of the inhibition of HR signaling output, including DNA damage repair and cell-cycle regulation mechanisms. A, Schematic of the HR pathway, representing the DNA sensors (RPA and H2Ax) that detect damage, the transducers (BRCA1/2, ATM, and CHK1/2) that can amplify and transmit those damage signals to the effectors (RAD51, WEE1, and CDC25A), which ultimately regulate DNA damage repair and cell-cycle checkpoint activity. Ultimately, when the HR output is successfully transmitted to the sensors by RAD51, HGSC baseline DNA damage is kept low and homeostasis is restored, allowing cells to survive long-term with inherent DNA errors due to genomic instability (GI). However, when SMAPs are added, PP2A gets activated, allowing for (i) RAD51 expression to be inhibited and (ii) leading to the HR effectors’ function to be chronically impaired. This results in the incapacity of the cells to restore cell-cycle progression and DNA damage repair, eventually dying to self-triggered apoptosis (schematic created with BioRender.com). Western blot analysis of OV81 (B), PEO-1 (C), and PEO-C4.2 (D) cells after 2, 4, 6, 12, and 24 hours of SMAP-061 treatment evaluates the expression of sensor, transducer, and effector proteins illustrated in the schematic of A. E, γH2Ax foci imaging comparing DMSO and 20 µmol/L SMAP-061–treated OV81 cells after 12 hours of incubation and their respective quantification. Data presented as the mean ± SEM (n = 3; unpaired Student t tests, comparing the SMAP-061 treatment group relative with DMSO; ***, P < 0.001). F, Correlation analysis graph and R2 values comparing the expression of RAD51 with γH2Ax in OV81, PEO-1, and PEO-C4.2 during 2, 4, 6, 12, and 24 hours of SMAP-061 treatment. Data presented as the mean ± SEM (n = 3). G, Statistical and quantification analysis of cell-cycle flow (from Supplementary Fig. S2O) for cells after incubation with 2 mmol/L thymidine for 24 hours (first bar), at 6.5 hours thymidine released (to allow cells to reengage cell-cycle progression; second bar), and consecutive treatments with either DMSO or SMAP-061 for 6 hours (third and fourth bars), 10 hours (fifth and sixth bars), and 16 hours (seventh and eighth bars), respectively, using FlowJo. Data presented as the mean ± SD (n = 3; unpaired Student t tests, comparing treatment groups with each other, for each stage of the cell-cycle, *, P < 0.05; **, P < 0.01; ***, P < 0.001).
SMAP-061 treatment results in G1 arrest and reduction of cyclin D1 protein expression
Given that previous studies in multiple myeloma cells showed that RAD51 inhibition leads to similar increases in unrepaired DDR due to a G1 cell-cycle arrest (48), we wanted to assess SMAP-061’s role in cell-cycle progression. After synchronizing HGSC cells using thymidine to undergo a full cycle, our data revealed that SMAP-061 induced G1 arrest, followed by an accumulation of apoptotic populations (G0; Fig. 3G; Supplementary Fig. S2M and S2N). Specifically, cells treated with DMSO 10 hours after thymidine release, were able to normally transition into S-phase (S-phase: <25%; Supplementary Fig. S2N); however, HGSC cells treated with SMAP-061 were unable to transition into S-phase after 10 and 16 hours of treatment (S-phase: <11% and 12%, respectively; Fig. 3G; Supplementary Fig. S2N, top). These results suggest that SMAP-061–treated cells were continuously halted at the G1 checkpoint and unable to reengage or progress through the different stages of the cell-cycle (Supplementary Fig. S2N, bottom). Furthermore, SMAP-061–treated cells inability to leave G1 led to their accumulation in sub G1, indicating that these cells were dying. Western blots showed that Cyclin D1 expression, an essential cyclin that functions as a G1 cell-cycle checkpoint gatekeeper, decreases significantly and as early as 2 hours after SMAP-061 (Supplementary Fig. S2O–S2Q), possibly explaining the observed G1 arrest phenotype. Expression levels of different cyclins and other cell-cycle proteins were moderately unaffected comparatively to D-type cyclins in SMAP-061–treated cells, indicating that SMAP-061 may be specifically targeting E2F responsive genes (Cyclin D1, RAD51, BRCA1, and BRCA2) that are known to decrease in G1-arrested cells. Moreover, mitotic-specific proteins could not reinstate protein turnover over time, given the downregulation of PLK1, Cyclin B1 and Securin (Supplementary Fig. S2O–S2Q).
These results indicate that SMAP-061 inhibits cell-cycle transition by impairing HGSC cells from progressing through G1. More importantly, by reducing Cyclin D1 expression, SMAP-061 prevents cells from transitioning into S-phase, where most HR proteins are commonly expressed, thus halting the HR pathway from repairing damaged DNA. This process slowly leads to a chronic accumulation of γH2Ax, ultimately inducing programmed cell death due to synthetic lethality.
SMAP-061 treatment potentiates PARPi effects in vitro and leverages synthetic lethality
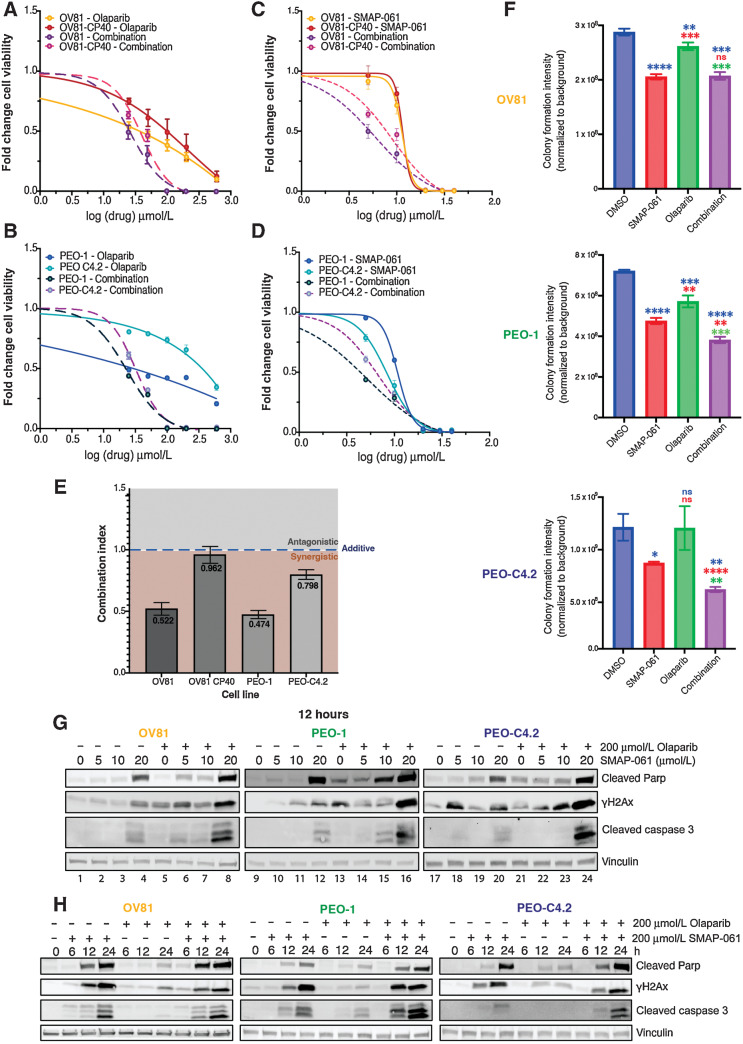
Given SMAP-061’s ability to regulate HR protein expression, we next assessed whether combining PP2A activators with PARPi could effectively produce synthetic lethal relationships in patients lacking HRD and/or potentiate PARPi’s effects in patients that are insensitive to PARPi. PARPis are a mainstay maintenance treatment option for patients with HGSC, after platinum-based regimens. However, the mechanism behind response and sensitivity of tumors to PARPi before and after acquired CR remains unexplored. Interestingly, the cisplatin-naïve OV81 and PEO-1 lines displayed greater sensitivity to the PARPi olaparib comparatively to their respective CR counterpart (Fig. 4A–D; Supplementary Table S3B), potentially indicating an overlap in resistance mechanisms between PARPi and cisplatin. To evaluate whether SMAP-061 could potentiate PARPi response in resistant models, we performed cell viability assays using increasing doses of SMAP-061 and olaparib, as single or combination agents, in both platinum-naïve and resistant cell lines. OV81 (Fig. 4A and C) and PEO1 (Fig. 4B and D) pairs showed increased cell death when the two drugs were combined comparatively to each agent alone, decreasing SMAP-061 EC50 value more than 2-fold (Supplementary Table S3B). This was further confirmed by conducting isobologram analysis and determining the Combination Index (CI) for SMAP-061 and olaparib (Fig. 4E) in our HGSC lines. We found that the CI for all cell lines was <1, indicating that the combination of the two drugs induces synergistic effects, although to a lesser extent in the cisplatin-resistant cell lines, as their cisplatin-naïve counterparts have a significantly lower CI. To determine whether the synergistic impact of SMAP-061 was exclusive to combinations with PARPi, we examined SMAP-061’s combination potential with platinum-based therapies, which remain the first-line treatment option for HGSC (Supplementary Fig. S3A–S3C and Supplementary Table S3C). Interestingly, despite SMAP-061 not sensitizing CR cells to cisplatin treatment, it still induced synergistic cell death in the cisplatin-naïve PEO-1 cells (CI = 0.6; Supplementary Fig. S3B and Supplementary Table S3C). In addition, SMAP-061–sensitized FT246 cells to PARPi; however, it required significantly higher doses to induce death comparatively to cancer cells (Fig. 2E; Supplementary Fig. S3E and S3F). Looking at colony formation, we observed that in PEO-1 and PEO-C4.2, SMAP-061+PARPi combination was significantly more effective than either drug alone, showing an additive potential. Surprisingly, no synergy or additivity was observed in OV81 at the selected doses (Fig. 4F; Supplementary Fig. S4A). Furthermore, protein analysis of cellular death (cleaved PARP and cleaved Caspase-3) and DNA damage markers (γH2Ax) by Western blot correlated with these significant effects in a dose (Fig. 4G; Supplementary Fig. S4B, S4D, and S4F) and a time (Fig. 4H; Supplementary Fig. S4C, S4E, and S4G)-dependent manner.
Figure 4.
SMAP-061 shows synergistic efficacy in targeting HGSC for cell death when used in combination with PARPi in vitro. OV81 and OV81-CP40 (A) and PEO-1 and PEO-C4.2 (B) cells were treated with increasing doses of olaparib alone or in combination with SMAP-061, and SMAP-061 alone or combination of both OV81 and OV81-CP40 (C), and PEO-1 and PEO-C4.2 (D), to measure cell viability at 48 hours using MTT. Data presented as the mean ± SD (n = 3). E, Combination index (CI) was calculated using the proliferation combinatorial assays conducted in (A) for all four cell lines. CI > 1: antagonistic; CI = 1: additive; CI < 1: synergistic. Formula: (EC50_combination/EC50_SMAP-061) + (EC50_combination/EC50_PARPi). Data presented as the mean ± SD (n = 3; one-way ANOVA compares all treatment groups; ***, P < 0.001). F, Clonogenic assay of OV81, PEO-1, and PEO-C4.2 was performed and quantified for cells treated with 6 µmol/L SMAP-061, 100 nmol/L olaparib and the combination of both, after 2 weeks. Data presented as the mean ± SD (n = 3; unpaired Student t tests, comparing each treatment group relative with each other, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Raw data represented in Supplementary Fig. S4A. Western blot analysis assessing cell death markers (cleaved Parp and cleaved Caspase-3) and DNA damage (γH2Ax) expression to further evaluate dose (G) and time-dependency profiles in OV81, PEO-1, and PEO-C4.2 cells treated with SMAP-061, olaparib, or combination (H).
SMAP-061 potentiates PARPi-induced cell death by decreasing RAD51 and preventing DNA repair
Previously, we showed that SMAP-061 modulation of PP2A leads to significant downregulation of DDR proteins, specifically, RAD51, ultimately triggering synthetic lethality (Fig. 3B–D). Furthermore, SMAP-061 potentiates PARPi effects in cells with varying HR-status and platinum sensitivity profiles (Fig. 4). However, the mechanisms driving these outcomes are not clear. SMAP-061 alone increased phosphorylation of RPA (Fig. 5A; Supplementary Fig. S5A–S5F) and H2Ax (Fig. 4G and H) in a dose-dependent manner, indicating DNA damage signaling activation. The combinatorial effects on p-RPA and γH2Ax were most significant in OV81 and PEO-C4.2, as these cell lines are resistant to PARPi alone due to HR-proficiency. Thus, SMAP-061 sensitized these lines to olaparib and led to more robust p-RPA and γH2Ax induction, than either treatment alone. However, PEO-1 harbors a BRCA2 mutation, rendering activation of DNA damage with PARPi alone significantly higher and thus combinatorial potential of SMAP-061+PARPi having insignificant effects (Fig. 5A). A decrease in protein expression of BRCA1/2, RAD51 (Supplementary Fig. S5G–S5O) and the cell-cycle regulators Rb, WEE1, and Securin (Supplementary Fig. S5P–S5U) was also observed. Interestingly, comparing SMAP-061 alone with SMAP+olaparib (Fig. 5A, lanes 2–4 vs. 6–8; 10–12 vs. 14–16; and 18–20 vs. 22–24), RAD51 protein levels were substantially decreased in the combo groups, even when olaparib was used with SMAP-0612s lowest dose, in all lines (Fig. 5A; Supplementary Fig. S5G–S5I). These data further suggest SMAP-061 can exert equally efficient tumor-suppressive and anti-proliferative effects at lower doses when in combination with olaparib (Supplementary Table S3B), compared with higher doses as a single agent. Other transducers and effector DDR targets presented similar expression patterns (Supplementary Fig. S5S–S5U and Supplementary Fig. S5J–S5O). These results further support the global additive effects of SMAP+olaparib combination on several proteins downstream the DDR/HR pathways (Supplementary Fig. S5A–S5U). Previous studies report olaparib's clinically achievable concentration to be approximately 100 µmol/L (49, 50), which in our models slightly increased DNA damage signals after 24 hours. Despite SMAP-0612s higher potency, SMAP+olaparib showed a more robust effect than each agent alone (Supplementary Fig. S5V). Therefore, these data suggest that DNA damage and apoptosis were also efficiently induced at longer time points when using clinically achievable concentrations, supporting previous observations. To further confirm HR repair efficiency, we evaluated the ability of OV81 cells treated with SMAP-061 to form RAD51 foci. Our data show OV81 expresses baseline levels of RAD51 foci (Fig. 5B) due to higher GI associated with HGS cancers. However, upon SMAP-061 treatment, RAD51 foci were reduced in 50% of cells and more significantly in combination with PARPi. Interestingly, olaparib alone significantly increased RAD51 foci, as treatment with PARPi leads to inefficient ssDNA DNA repair and recruitment of RAD51that is needed for HR-repair.
Figure 5.
Combination of SMAP-061 and PARPi synergistically engages the DDR pathway and prevents DNA repair by specifically targeting and downregulating RAD51. A, Western blot analysis evaluating dose-dependency effects of SMAP-061, olaparib, or combination on DDR protein expression and activity. Sensor, transducer, and effector proteins (from Fig. 3A schematic) were analyzed in OV81, PEO-1, and PEO-C42. B, RAD51 foci imaging comparing DMSO with 20 µmol/L SMAP-061, 100 µmol/L olaparib or combination of SMAP+olaparib in OV81 cells treated after 24 hours of incubation (left) and their respective quantification bar graph (right). Data presented as the mean ± SEM (n = 3; unpaired Student t tests, comparing each group with each other, **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). C, OV81, PEO-1, and PEO-C4.2 cell lines were exposed to increasing doses of either SMAP-061 alone (blue) or in combination with 80 µmol/L of 766 (a biologically inactive analogue of SMAP-061; red), and cell viability was measured at 48 hours by MTT analysis, to generate an EC50 curve. Data are presented as the mean ± SD (n = 3). D, Rescue experiments were performed on OV81, PEO-1, and PEO-C42 cells treated with DMSO, SMAP-061, 766, or SMAP + 766. Rescue profile analysis of DDR (BRCA1/2 and RAD51) protein expression as well as recovery in cell death markers (cleaved Parp and cleaved Caspase-3) and DNA damage signaling capacity (γH2Ax and p-RPA) after 12 hours of treatment are analyzed by Western blotting. Values underneath RAD51 blot represent the fold change of protein expression to its DMSO control sample, after Vinculin normalization. E, Quantification of RAD51 protein expression, with data presented as the mean ± SEM (n = 3; unpaired Student t tests, comparing each treatment group relative with its respective DMSO; *, P < 0.05). F, Calyculin-A (PP2A Catalytic subunit inhibitor) rescue experiments were performed using Western blotting (and respective quantification of RAD51 in G). H, MTT techniques to assess cellular viability and an EC50 value shift for OV81, PEO-1, and PEO-C4.2. 5 nmol/L of Calyculin-A was preincubated for 1 hour, followed by SMAP-061 treatment. Data presented as the mean ± SD (n = 3; unpaired Student t tests, comparing each treatment group relative with each other, *, P < 0.05; ***, P < 0.001). I, CRISPR Cas9 KO of 2 specific B-subunits (PPP2R2A and PPP2R5A) rescue experiments were performed using Western blotting looking at HR sensors (γH2Ax and p-RPA), RAD51 effector, and apoptotic markers (cleaved PARP and cleaved Caspase3). Representative quantification is located under each blot and its respective lane. Each target was normalized to Vinculin. p/t-RPA ratio was calculated before normalization to Vinculin. J, Clonogenic assay of OV81 CTRL CRISPR, PPP2R2A KO #1, and PPP2R5A KO #2. Cells were treated with DMSO or 10 µmol/L SMAP-061 for 2 weeks (n = 3; Left). Quantification the clonogenic assay comparing OV81 control (CTRL CRISPR), PPP2R2A KO #1, and PPP2R5A KO #2. Data presented as the mean ± SD (n = 3; unpaired Student t tests, comparing each treatment group with its own DMSO control, *, P < 0.05; Right).
Next, we wanted to confirm whether the SMAP-061 effects observed were mediated through the modulation of PP2A. Competition studies were conducted using a biologically inactive SMAP analog, 766 (51, 52), previously shown to have similar chemical structure and bind the same pocket in PP2A as our active analogs. Yet, this molecule does not stabilize or activate trimeric PP2A, resulting in no changes in cell viability. We treated cells with DMSO, SMAP-061, 766, or SMAP-061+766 and analyzed essential DDR and cell-cycle molecular targets. To ensure successful competition and reversibility of PP2A's activity and stability, we used 4X the concentration of 766 as the active SMAP-061, enabling us to significantly compete off phosphatase binding and reverse cell death profiles (Fig. 5C). Interestingly, the effects of SMAP-061 on DNA damage–sensing mechanisms (γH2Ax and p-RPA expression) were fully reversed in SMAP-061+766 combination, after 12 hours (Fig. 5D, lanes 2 vs. 4; 6 vs. 8; and 10 vs. 12, and Supplementary Fig. S6A–S6C). In addition, the protein expression levels of Cyclin D1, cleaved PARP, and cleaved Caspase-3, were also reversed in the co-treated cells when compared with SMAP-061 alone (Fig. 5D and E; Supplementary Fig. S6D). However, BRCA1/2 expression was not rescued in all cellular models (Fig. 5D; Supplementary Fig. S6E and S6F), indicating that these proteins are not the driving factors of SMAP-061–mediated apoptosis. Notably, the RAD51 protein loss completely reversed, suggesting this target is necessary for SMAP-061–triggered cell death. In support of the latter statement, comparing RAD51 expression intensity in SMAP-061 alone conditions (Fig. 5D, lanes 2, 6, and 10), we showed that OV81 and PEO-1 cells had a stronger decrease in RAD51 expression when compared with PEO-C4.2 (Fig. 5E). Therefore, the degree of RAD51 protein expression was significantly and inversely correlated with γH2Ax, p-RPA, cleaved PARP and cleaved Caspase-3 expression levels for all three cell lines, as confirmed in our correlation studies (Supplementary Fig. S6G).
To further confirm SMAP-0612s specificity to PP2A, we used Calyculin-A, a highly specific PP2A and PP1 Catalytic subunit inhibitor (53). Using similar rescue experiments as for 766, we demonstrated that SMAP-061–mediated effects were directly dependent on PP2A modulation (Fig. 5F and G). Calyculin-A shifted the EC50 value of SMAP-061 after 1-hour preincubation, priming PP2A to remain inactive despite of SMAP-061 presence. Calyculin-A rescues cell death by 40% at SMAP-061’s highest concentration (40 µmol/L), which alone leads to 100% cell death (Fig. 5H). We observed that for all three cell lines, SMAP-061 activity is effectively impaired by Calyculin-A preincubation, as observed by the cell death markers and RAD51 protein expression rescue (Fig. 5F).
Given Leonard and colleagues (26) previous findings, we next wanted to evaluate the dependency profiles of SMAP-061’s activity and the preferential stability of the PPP2R5A(B56α) subunit in HGSC models. Thus, we generated PPP2R5A-KO CRISPR lines and evaluated their ability in rescuing SMAP-061–induced apoptosis (Fig. 5I; Supplementary Fig. S5W) and survival (Fig. 5J). PPP2R2A(B55α)-KO was used as a control given its previous postulated stabilization properties by SMAP-061 (27). Our KO models show successful decrease in protein expression of either B55α or B56α, respectively (Fig. 5I; Supplementary Fig. S5W). When PPP2R5A is KO (only in sg#2), cell viability and cleaved Caspase-3, p-RPA, and γH2Ax markers decrease significantly with SMAP-061 compared with CRISPR control (CRISPR CTRL; Fig. 5I and J), suggesting that apoptotic signals are in part dependent on the PP2A-B56α holoenzyme. No significant rescue was observed in viability or cleaved Caspase-3, p-RPA, and γH2Ax in PPP2R2A-KO cells treated with SMAP-061 (Fig. 5I and J). Surprisingly, neither of the B-subunits affected the effect of SMAP-061 on RAD51 protein expression after 24 hours of exposure, possibly indicating that other PP2A regulatory subunits may be compensating for the loss of B56α interaction (Fig. 5I). To evaluate such hypothesis, functional CRISPR screens with varying combinations of PP2A-B-KO subunits must be performed for stronger correlation and dependency interpretations to be drawn, assessing compensatory mechanisms among different B-subunits of PP2A.
In sum, we provide evidence that SMAP-061 activates PP2A specifically and downregulates RAD51; however, the direct role of PPP2R5A (B56α) in regulating RAD51 needs to be further examined.
SMAP-061 improves in vivo survival and significantly decreases tumor progression in PDX mouse models, as a single agent and in combination with PARPi
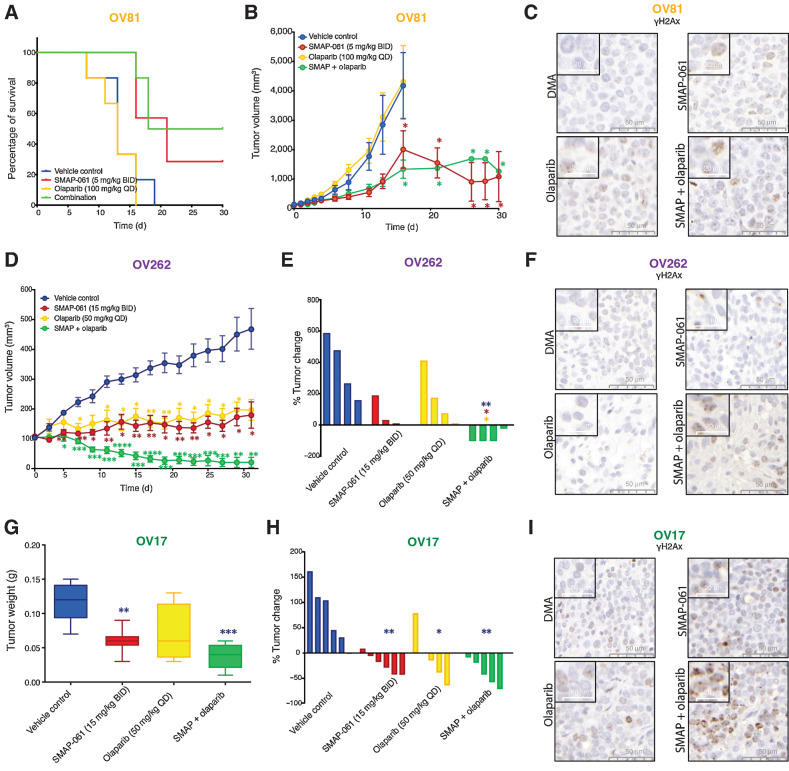
PP2A stabilization leads to apoptotic effects in vitro, through targeting essential DDR, HR, and cell-cycle proteins, resulting in an irreversible “BRCAness” phenotype. In combination with PARPi, SMAP-061’s anticancer activity is enhanced, triggering synthetic lethality in HGSC models, independently of HR status. Given our recent discoveries and the clinical promise these compounds hold, we evaluated their efficacy in vivo. OV81, OV262, and OV17 PDX models (Supplementary Table S2C), were used to conduct efficacy studies, evaluate tumor growth and survival. As expected, olaparib alone was ineffective in improving survival (Fig. 6A) or decreasing tumor burden (Fig. 6B) in OV81, as this PDX is HR-proficient. However, SMAP-061 alone and with olaparib showed significant improvement in median survival (Fig. 6A), consistent with significant tumor growth inhibition in both groups (Fig. 6B). Interestingly, the synergistic effects observed in our in vitro models were not replicated in vivo, indicating the need to adjust dosing and schedule regimens, although SMAP-061+olaparib induced the most γH2Ax staining (Fig. 6C; Supplementary Fig. S7D). OV262 PDX HGSC model harbors a germline BRCA2 mutation (p.N3124I), responded as efficiently to SMAP-061 alone as to olaparib single agent (Fig. 6D and E). However, the combination resulted in significant tumor regression, which was not achieved by either compound alone. These results highlight that the SMAP-061+olaparib combination strategy has synergistic effects in reducing tumor burden in HR-deficient HGSC tumor models. γH2Ax staining reveals that neither SMAP-061 or olaparib alone can significantly increase foci formation; however, SMAP+olaparib combination significantly increases its accumulation (Fig. 6F; Supplementary Fig. S7E). The lack of γH2Ax staining in these tumors treated with single agents could be due to inherent ability of these cells in resolving DNA damage before exacerbated accumulation. However, in the combination treatment groups, the tumor cells do not overcome the chronically accumulated DNA insults, resulting in tumor regression. Finally, OV17 harbors a BRCA1 germline mutation (p.R762fs), for which SMAP-061 alone had higher efficacy in affecting tumor weight (Fig. 6G) and the percentage of tumor change (Fig. 6H) when compared with the single-agent olaparib. Importantly, despite significant tumor growth inhibition in both single-agent groups, SMAP-061+olaparib combination induced tumor regression (Fig. 6H). Histology shows that γH2Ax foci are significantly induced in OV17 PDX tumors treated with SMAP-061 alone or SMAP-061+olaparib combination (Fig. 6I; Supplementary Fig. S7F). In all treatment studies no significant weight loss was observed in any of the treated groups (Supplementary Fig. S7A–S7C), representing no associated toxicity.
Figure 6.
Effects of SMAP-061 in vivo show improved survival and significant tumor regression, as a single agent and in combination with PARPi, in both BRCA1/2 wild-type and mutant HGSC PDX tumors. OV81 PDX studies were conducted with tumors implanted in the right flank of NSG mice and allowed to grow between approximately 100 and 200 mm3 before enrollment in one of 4 treatment groups: Vehicle control (n = 6), 5 mg/kg SMAP-061 (n = 6), 100 mg/kg olaparib (n = 6), or SMAP + olaparib combination (n = 6). Tumors were measured every other day. Data plotted as a function of mouse survival (A) and tumor volume (efficacy; B) over time. Survival threshold was selected to be 1,000 mm3 (tumors < 1,000 mm3: survival; tumors > 1,000 mm3: no survival). Data presented as mean ± SEM (Student t tests, comparing each treatment group relative with vehicle control; *, P < 0.05). C, Histology slides with tumor samples stained for γH2Ax (brown), representative of each individual treatment group in the OV81 PDX in vivo study. 50- and 20-µm scales are included for each respective picture. D, Efficacy study of OV262 PDX tumors, implanted in the right flank of NSG mice and allowed to grow between approximately 90–120 mm3 before enrollment in one of 4 treatment groups: vehicle control (n = 4), 15 mg/kg SMAP-061 (n = 3), 50 mg/kg olaparib (n = 4), or SMAP + olaparib combination (n = 4). Tumors were measured every other day. Tumor volume was calculated and plotted over time (31 days). Data presented as mean ± SEM (Student t tests, comparing each treatment group relative with vehicle control, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). E, Waterfall plot of OV262 tumor volume change (from days 0 to 31) comparing all treatment groups with each other. Data presented as mean ± SEM (unpaired Student t tests, comparing each individual treatment group relative with each other, *, P < 0.05; **, P < 0.01). F, Histology slides with tumor samples stained for γH2Ax (brown), representative of each individual treatment group in the OV262 PDX in vivo study. 50- and 20-µm scales are included for each respective picture. G and H, OV17 PDX studies were conducted with tumors implanted in the right flank of NSG mice and allowed to grow between approximately 80 and 250 mm3 before enrollment in one of 4 treatment groups: vehicle control (n = 6), 15 mg/kg SMAP-061 (n = 6), 50 mg/kg olaparib (n = 5), or SMAP + olaparib combination (n = 5). Tumors were measured every other day. Data plotted as a function of tumor weight post sacrifice (G) and tumor change percentage (waterfall plot; H) for each treatment group. Data presented as mean ± SEM (unpaired Student t tests, comparing each individual treatment group relative with each other, *, P < 0.05; **, P < 0.01; ***, P < 0.001). I, Histology slides with tumor samples stained for γH2Ax (brown), representative of each individual treatment group in the OV17 PDX in vivo study. 50- and 20-µm scales are included for each respective picture.
Together, these results further support that SMAP-061 effectively inhibits tumor growth in BRCA1/2 mutant and wild-type tumors in vitro and in vivo. Furthermore, when combined with PARPi, SMAP-061 significantly induces tumor regression in HRD HGSC tumors. Importantly, these data highlight the therapeutic potential of SMAP-061 for the treatment of HGSC and strongly sustains the possibility of PP2A modulation in inducing synthetic lethality, expanding the patient population that can benefit from PARPi therapies regardless of HR status.
Discussion
Although the discovery of the BRCA genes revolutionized ovarian cancer screening and prevention, allowing PARPi to thrive clinically (6–8), only 21% of patients with HGSC harbor germline (15%) or somatic (6%) BRCA1/2 mutations (54), resulting in a specific subset of patients with HGSC that benefit from these therapies. Therefore, it is essential to identify unique targetable genetic drivers of HGSC and introduce novel targeted therapeutic opportunities that take advantage of HGSC tumors HR dependency.
PP2A is a major tumor-suppressor negative regulator of multiple oncogenic pathways. Its phosphatase activity dephosphorylates a vast array of downstream substrates to regulate cellular homeostasis. Although targeted reduction of PP2A causes transformation of the non-malignant human FTSEC model (35), the genetic and molecular impact each PP2A family member has in HGSC tumorigenesis is currently unknown. Unlike most tumor suppressors that are functionally inactivated in cancer via genetic mutations, such as p53, PP2A components are not commonly mutated in human malignancies and its inactivation primarily involves non-genetic mechanisms (15). In this article, we uncovered that >90% of HGSC tumors harbor heterozygous loss and decreased mRNA expression of PP2A genes important to malignant transformation and/or holoenzymatic core formation, correlating with decreased overall patient survival. Thus, suggesting that PP2A tumor-suppressor activation represents an attractive therapeutic alternative to treat HGSC. Protein phosphatases have been understudied as a drug development target, mostly due to their perceived “undruggable” nature. Here, we uncover that SMAP-061, an established PP2A modulator, induces apoptosis in numerous PDX HGSC models and potentiates PARPi-mediated cell death independent of platinum sensitivity and HR-status. SMAP molecules directly bind to and promote the formation and stabilization of PP2A tumor-suppressive holoenzymes, ultimately modulating PP2A activity (28–51). Disruption of its binding pocket on PP2A-A induces cellular resistance to SMAP-061 and rescues tumor growth, suggesting PP2A specificity (29, 55, 56). Our research studies provide multiple experimental evidence that SMAP-061 specifically modulates the HR/DDR pathway and apoptotic induction through PP2A activation. First, we used a biologically inactive SMAP-061 analog, 766, for competition assays, where we rescued SMAP-061–induced DNA damage and apoptosis in the combination group. Second, the specificity of the SMAP-061 in modulating PP2A’s activity and the role of RAD51 in cell death were confirmed using Calyculin-A, a highly specific PP2A and PP1 (previously confirmed to be unaffected by SMAP-061) Catalytic subunit inhibitor (53). We found that SMAP-061–dependent decrease of RAD51 is prevented by Calyculin-A preincubation, abrogating apoptosis. Our third method assessed which regulatory subunit was reducing RAD51 levels and further confirmed the dependency of SMAP-061 on PP2A, using CRISPR/Cas9 to target B55α or B56α. Our results confirmed that SMAP-061–induced apoptosis was only reduced in B56α-KO cells, although RAD51 expression was unaffected, suggesting possible compensatory PP2A holoenzymes formation. Thus, further evaluation of the direct role of PP2A on RAD51 protein stability and regulation must be pursued. In sum, we provide evidence that SMAP-061 works through the activation of PP2A; however, further studies are required to better understand which regulatory subunit(s) of PP2A drive the anticancer effects, to leverage their therapeutic potential.
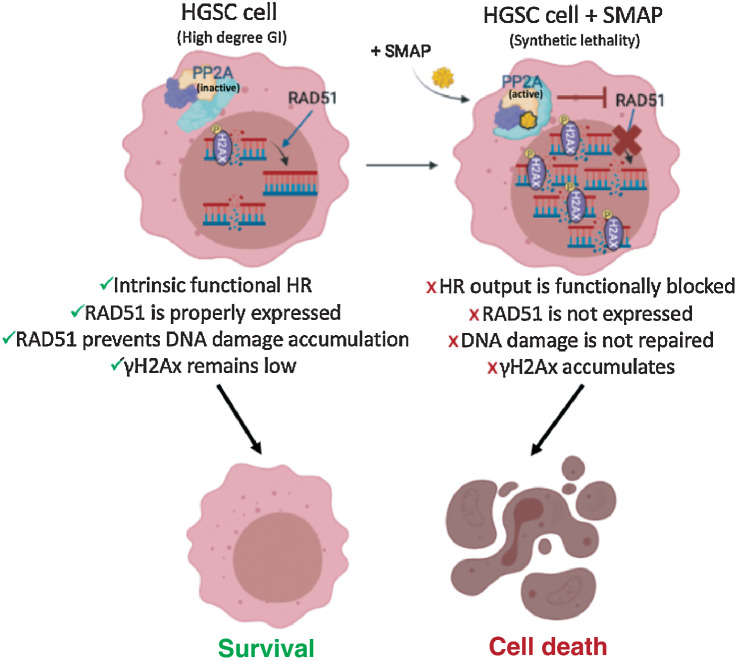
As >70% of HGSC tumors are characterized by HR defects, RAD51 is critical to their tumorigenesis processes, being an essential HR component that modulates DNA strand exchange in DSB-dependent repair. RAD51 inhibition leads to synthetic lethality through γH2Ax accumulation, thus spearheading the clinical development of RAD51 inhibitors (48, 57–59). Our studies introduce a unique mechanism of synthetic lethality, through a small-molecule mediator of PP2A that potently reduces RAD51 expression, leading to concomitant apoptosis (Fig. 7). These results suggest that SMAP-061 is effective against HR-proficient tumors and PARPi-resistant tumors, given that RAD51 upregulation is a common mechanism of resistance. Although the mechanism by which SMAP-061 leads to RAD51 expression reduction remains unclear, published evidence has demonstrated that PP2A plays an indirect role on RAD51’s activity and stability. The BRCA2–PP2A-B56 complex is essential for the efficient RAD51 filament formation at the sites of DNA damage (25). Furthermore, PLK1, which directly phosphorylates and activates RAD51’s recruitment (60), is dephosphorylated and inactivated by PP2A (61, 62). Thus, further studies are necessary to assess the mechanism by which PP2A and SMAP-061 regulate RAD51 (Fig. 7). Our current studies show that neither B-subunits previously postulated to be recruited upon SMAP-061 interaction affected RAD51 levels. Our colony-forming assays show that long-term incubation with lower doses of SMAP-061 rescued PPP2R5A-KO cells’ ability to form colonies. Such observations suggest that B56α may not be the exclusive B-subunit responsible for SMAP-061’s anticancer properties in HGSC models to regulate HR, as previously suggested by literature. Perhaps, inhibiting all B-subunits responsible for RAD51 regulation, thus rescuing RAD51 levels, is functionally necessary to fully drive more significant viability and biological relevant phenotypes. This is an important consideration as redundancy may occur due to overlapping roles and downstream substrates of different B-subunits: In the absence/downregulation of certain B-subunits, others can be upregulated as a compensatory mechanism, rendering dependency difficult to evaluate.
Figure 7.
RAD51 inhibition leads to the chronic accumulation of γH2Ax and DNA errors inherent from genomic instability (GI), an ovarian cancer trait. HGSC cells harbor a high degree of GI that results in small nicks and breaks in the DNA over time. Sensor proteins such as γH2Ax are recruited to the site of damage to prevent DNA degradation and remain in check until a repair mechanism is triggered. The minimally functional HR machinery is activated and prompts the recruitment of the main downstream effector protein, RAD51, which by keeping those damaging signals at bay, allows cancer cells to survive, even if with relatively higher baseline levels of γH2Ax when compared with normal cells. For this reason, ovarian cancer cells are known to functionally tolerate and survive high baseline levels of DNA damage while avoiding apoptotic cues (left). When SMAP-061 is added to the cell, PP2A’s antitumor activity is functionally restored, and synthetic lethality is induced (right). RAD51 expression is inhibited by PP2A, preventing the HR output to be efficiently propagated. This results in the incapacity of the cells to repair their inherent DNA damage, thereby leading to the accumulation of γH2Ax signals in the nucleus, and ultimately, triggering apoptosis (schematic created with BioRender.com).
Furthermore, the functional implications of the majority of HGSC tumors with loss of specific PP2A genes remain unclear. Thus, the effect of PP2A’s loss on DDR during HGSC development and response to PARPi needs to be further explored. RAD51 is essential to HR’s success in resolving DNA damage; and without it, homology search and strand invasion characteristic of HR cannot occur. Unlike RAD51 and other HR regulators, BRCA1/2 mutation carriers are significantly more prevalent, and pathogenic variants of these genes as well as their functional output have been best studied in HGSC. Mutations in BRCA1/2 genes are functionally relevant for tumor development and resistance, as they lead to HRD, due to the failure to accomplish homology search and strand invasion of the sister chromatid, being ultimately incapable of forming RAD51 filaments (59). Thus, targeting proteins downstream the BRCA genes in the HR pathway, specifically RAD51, may represent better strategies to tackle clinical challenges and overcome PARPi resistance. Targeting RAD51 has proven biologically efficacious for the inhibition of human cancer progression (51, 57–59, 63, 64). Data show that RAD51 antisense oligonucleotides, RNAi and specific small-molecule inhibitors reducing RAD51 expression, re-sensitized multiple cancer types to chemo- and radiation therapies, proving RAD51 to be a suitable therapeutic target. As viable potent, direct, and specific inhibitors of RAD51 are currently lacking, our PP2A modulators are effective alternatives that regulate RAD51 expression and/or stability, presenting a great promise to be used in the clinic as indirect RAD51 inhibitors. Beyond HGSC, these studies also lay the foundation for PP2A modulators as treatment options for other cancers harboring HRD or RAD51-dependency profiles, commonly seen in breast cancers (52, 65). Distinct from HGSC, patients with breast cancer with BRCA1/2 mutations have not shown as promising response to PARPi; therefore, PP2A modulation alone or in combination with PARPi may represent a more effective therapeutic approach (66).
In summary, a deeper molecular and mechanistic understanding of the antitumor effects of SMAP-061 allowed us to introduce novel pharmacodynamic markers of PP2A target engagement and therapeutic efficacy, such as RAD51. We believe that introducing SMAP-061 or other PP2A modulators as a novel targeted therapy for HGSC can optimize treatment selection, patient stratification, and, in the future, help leverage drug development and discovery for the treatment of HGSC to prevent intrinsic and acquired PARPi resistance and treat HR-proficient tumors.
Supplementary Material
Fig. S1
Fig. S2
Fig. S3
Fig. S4
Fig. S5
Fig. S6
Fig. S7
Table S1
Table S2
Table S3
Acknowledgments
This work was supported by grants from the National Cancer Institute, R01CA197780 (to A. DiFeo), R01CA240374 (to J. Zhang), Department of Defense, OC150553 and OC210149, The Young Scientist Foundation (to A. DiFeoD), T32 CA009676 (to J.A. Colina), and Dr. Eleanor Lewis award from Rackham Graduate School (to R.A. Avelar). The authors would like to thank several core facilities within the Case Comprehensive Cancer Center (NIH P30CA043703) including the Athymic Animal and Preclinical Therapeutics Core. The authors would like to thank the Tissue and Molecular Pathology at Rogel Cancer Center at the University of Michigan (NIH P30 CA04659229) as well as the Cancer Center's pilot funds in support of this project. In addition, we would like to thank all the generous donors and foundations who have supported the DiFeo laboratory and strive to improve the outcomes of patients with ovarian cancer, including C. Norma and Albert I. Geller, The Silver Family Foundation, and Jacqueline E. Bayley (JEB) Foundation. Further acknowledgments are extended to Dr. T. Taniguchi for providing us with the PEO-1 and PEO-C4.2 isogenic cell lines.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors’ Disclosures
C.M. O’Connor reports personal fees from RAPPTA Therapeutics outside the submitted work. G. Narla reports grants from NIH, Department of Defense, and RAPPTA Therapeutics during the conduct of the study; as well as grants and personal fees from RAPPTA Therapeutics outside the submitted work; and reports a patent for compositions and methods for treating cancer issued. A. DiFeo reports other support from RAPPTA Therapeutics during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
R.A. Avelar: Investigation, writing–original draft, project administration. A.J. Armstrong: Data curation, formal analysis. G. Carvette: Data curation. R. Gupta: Data curation. N. Puleo: Data curation. J.A. Colina: Data curation, methodology. P. Joseph: Data curation. A.M. Sobeck: Data curation, methodology. C.M. O’Connor: Formal analysis, writing–review and editing. B. Raines: Resources, data curation. A. Gandhi: Data curation. M.L. Dziubinski: Data curation. D.S. Ma: Resources, data curation. K. Resnick: Resources, data curation, writing–review and editing. S. Singh: Resources, data curation, writing–review and editing. K. Zanotti: Resources, writing–review and editing. C. Nagel: Resources, writing–review and editing. S. Waggoner: Resources, data curation, methodology, writing–review and editing. D.G. Thomas: Data curation, formal analysis, methodology, writing–review and editing. S.L. Skala: Resources, data curation, formal analysis, writing–review and editing. J. Zhang: Resources, data curation, formal analysis, writing–review and editing. G. Narla: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, methodology, project administration, writing–review and editing. A. DiFeo: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, methodology, project administration, writing–review and editing.
References
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 2. Vaughan S, Coward JI, Bast RC, Berchuck A, Berek JS, Brenton JD, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer 2011;11:719–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lisio M-A, Fu L, Goyeneche A, Gao Z, Telleria C. High-grade serous ovarian cancer: basic sciences, clinical, and therapeutic standpoints. Int J Mol Sci 2019;20:952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Heintz APM, Odicino F, Maisonneuve P, Quinn MA, Benedet JL, Creasman WT, et al. Carcinoma of the ovary. Int J Gynecol Obstet 2006;95:S161–92. [DOI] [PubMed] [Google Scholar]
- 5. Davis A, Tinker AV, Friedlander M. “Platinum resistant” ovarian cancer: what is it, who to treat and how to measure benefit? Gynecol Oncol 2014;133:624–31. [DOI] [PubMed] [Google Scholar]
- 6. Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet North Am Ed 2010;376:245–51. [DOI] [PubMed] [Google Scholar]
- 7. Swisher EM, Lin KK, Oza AM, Scott CL, Giordano H, Sun J, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol 2017;18:75–87. [DOI] [PubMed] [Google Scholar]
- 8. Gelmon KA, Tischkowitz M, Mackay H, Swenerton K, Robidoux A, Tonkin K, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol 2011;12:852–61. [DOI] [PubMed] [Google Scholar]
- 9. Loizzi V, Ranieri G, Laforgia M, Gadaleta C, Gargano G, Kardhashi A, et al. PARP inhibitors and epithelial ovarian cancer: molecular mechanisms, clinical development and future prospective. Oncol Lett 2020;20:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kiely M, Kiely PA. PP2A: the wolf in sheep's clothing? Cancers 2015;7:648–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ruvolo PP. The broken “Off” switch in cancer signaling: PP2A as a regulator of tumorigenesis, drug resistance, and immune surveillance. BBA Clin 2016;6:87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grinthal A, Adamovic I, Weiner B, Karplus M, Kleckner N. PR65, the HEAT-repeat scaffold of phosphatase PP2A, is an elastic connector that links force and catalysis. Proc Natl Acad Sci 2010;107:2467–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cho US, Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 2007;445:53–57. [DOI] [PubMed] [Google Scholar]
- 14. Grech G, Baldacchino S, Saliba C, Grixti MP, Gauci R, Petroni V, et al. Deregulation of the protein phosphatase 2A, PP2A in cancer: complexity and therapeutic options. Tumor Biol 2016;37:11691–700. [DOI] [PubMed] [Google Scholar]
- 15. Kauko O, Westermarck J. Non-genomic mechanisms of protein phosphatase 2A (PP2A) regulation in cancer. Int J Biochem Cell Biol 2018;96:157–64. [DOI] [PubMed] [Google Scholar]
- 16. Longin S, Zwaenepoel K, Louis JV, Dilworth S, Goris J, Janssens V. Selection of protein phosphatase 2A regulatory subunits is mediated by the C terminus of the catalytic subunit. J Biol Chem 2007;282:26971–80. [DOI] [PubMed] [Google Scholar]
- 17. Letourneux C, Rocher G, Porteu F. B56-containing PP2A dephosphorylate ERK and their activity is controlled by the early gene IEX-1 and ERK. EMBO J 2006;25:727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kumar A, Pandurangan AK, Lu F, Fyrst H, Zhang M, Byun H-S, et al. Chemopreventive sphingadienes downregulate Wnt signaling via a PP2A/Akt/GSK3β pathway in colon cancer. Carcinogenesis 2012;33:1726–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thompson JJ, Williams CS. Protein phosphatase 2A in the regulation of Wnt signaling, stem cells, and cancer. Genes 2018;9:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Campos A, Clemente-Blanco A. Cell cycle and DNA repair regulation in the damage response: protein phosphatases take over the reins. Int J Mol Sci 2020;21:446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramos F, Villoria MT, Alonso-Rodríguez E, Clemente-Blanco A. Role of protein phosphatases PP1, PP2A, PP4, and Cdc14 in the DNA damage response. Cell Stress 2019;3:70–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Freeman AK, Monteiro AN. Phosphatases in the cellular response to DNA damage. Cell Commun Signal 2010;8:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peng A, Maller JL. Serine/threonine phosphatases in the DNA damage response and cancer. Oncogene 2010;29:5977–88. [DOI] [PubMed] [Google Scholar]
- 24. Kalev P, Simicek M, Vazquez I, Munck S, Chen L, Soin T, et al. Loss of PPP2R2A inhibits homologous recombination DNA repair and predicts tumor sensitivity to PARP inhibition. Cancer Res 2012;72:6414–24. [DOI] [PubMed] [Google Scholar]
- 25. Ambjoern SM, Duxin JP, Hertz EPT, Nasa I, Duro J, Kruse T, et al. A complex of BRCA2 and PP2A-B56 is required for DNA repair by homologous recombination. Nat Commun 2021;12:5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leonard D, Huang W, Izadmehr S, O'Connor CM, Wiredja DD, Wang Z, et al. Selective PP2A enhancement through biased heterotrimer stabilization. Cell 2020;181:688–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morita K, He S, Nowak RP, Wang J, Zimmerman MW, Fu C, et al. Allosteric Activators of protein phosphatase 2A display broad antitumor activity mediated by dephosphorylation of MYBL2. Cell 2020;181:702–15. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28. McClinch K, Avelar RA, Callejas D, Izadmehr S, Wiredja D, Perl A, et al. Small-molecule activators of protein phosphatase 2A for the treatment of castration-resistant prostate cancer. Cancer Res 2018;78:2065–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sangodkar J, Perl A, Tohme R, Kiselar J, Kastrinsky DB, Zaware N, et al. Activation of tumor suppressor protein PP2A inhibits KRAS-driven tumor growth. J Clin Invest 2017;127:2081–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tohmé R, Sudeh I, Sai G, Giancarlo T, Sanjay V, Ava T, et al. Direct activation of PP2A for the treatment of tyrosine kinase inhibitor–resistant lung adenocarcinoma. JCI Insight 2019;4:e125693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Risom T, Wang X, Liang J, Zhang X, Pelz C, Campbell LG, et al. Deregulating MYC in a model of HER2+ breast cancer mimics human intertumoral heterogeneity. J Clin Invest 2020;130:231–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Merisaari J, Denisova OV, Doroszko M, Le Joncour V, Johansson P, Leenders WPJ, et al. Monotherapy efficacy of blood–brain barrier permeable small molecule reactivators of protein phosphatase 2A in glioblastoma. Brain Commun 2020;2:fcaa002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nagaraj AB, Joseph P, Kovalenko O, Singh S, Armstrong A, Redline R, et al. Critical role of Wnt/β-catenin signaling in driving epithelial ovarian cancer platinum resistance. Oncotarget 2015;6:23720–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wlodarchak N, Xing Y. PP2A as a master regulator of the cell cycle. Crit Rev Biochem Mol Biol 2016;51:162–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karst AM, Levanon K, Drapkin R. Modeling high-grade serous ovarian carcinogenesis from the fallopian tube. Proc Natl Acad Sci U S A 2011;108:7547–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shih I-M, Wang T-L. Mutation of PPP2R1A: a new clue in unveiling the pathogenesis of uterine serous carcinoma. J Pathol 2011;224:1–4. [DOI] [PubMed] [Google Scholar]
- 37. McConechy MK, Anglesio MS, Kalloger SE, Yang W, Senz J, Chow C, et al. Subtype-specific mutation of PPP2R1A in endometrial and ovarian carcinomas. J Pathol 2011;223:567–73. [DOI] [PubMed] [Google Scholar]
- 38. Nagendra DC, Burke J III, Maxwell GL, Risinger JI. PPP2R1A mutations are common in the serous type of endometrial cancer. Mol Carcinog 2012;51:826–31. [DOI] [PubMed] [Google Scholar]
- 39. Rahman M, Nakayama K, Rahman MT, Nakayama N, Katagiri H, Katagiri A, et al. PPP2R1A mutation is a rare event in ovarian carcinoma across histological subtypes. Anticancer Res 2013;33:113–8. [PubMed] [Google Scholar]
- 40. Sablina AA, Hahn WC. The role of PP2A a subunits in tumor suppression. Cell Adh Migr 2007;1:140–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen W, Possemato R, Campbell KT, Plattner CA, Pallas DC, Hahn WC. Identification of specific PP2A complexes involved in human cell transformation. Cancer Cell 2004;5:127–36. [DOI] [PubMed] [Google Scholar]
- 42. Arroyo JD, Hahn WC. Involvement of PP2A in viral and cellular transformation. Oncogene 2005;24:7746–55. [DOI] [PubMed] [Google Scholar]
- 43. Chen W, Arroyo JD, Timmons JC, Possemato R, Hahn WC. Cancer-associated PP2A Aα subunits induce functional haploinsufficiency and tumorigenicity. Cancer Res 2005;65:8183–92. [DOI] [PubMed] [Google Scholar]
- 44. Sents W, Meeusen B, Kalev P, Radaelli E, Sagaert X, Miermans E, et al. PP2A inactivation mediated by PPP2R4 haploinsufficiency promotes cancer development. Cancer Res 2017;77:6825–37. [DOI] [PubMed] [Google Scholar]
- 45. Elias KM, Emori MM, Westerling T, Long H, Budina-Kolomets A, Li F, et al. Epigenetic remodeling regulates transcriptional changes between ovarian cancer and benign precursors. JCI Insight 2016;1:e87988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 2008;451:1116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang ZC, Birkbak NJ, Culhane AC, Drapkin R, Fatima A, Tian R, et al. Profiles of genomic instability in high-grade serous ovarian cancer predict treatment outcome. Clin Cancer Res 2012;18:5806–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alagpulinsa DA, Ayyadevara S, Shmookler Reis RJ. A small-molecule inhibitor of RAD51 reduces homologous recombination and sensitizes multiple myeloma cells to doxorubicin. Front Oncol 2014;4:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liston DR, Davis M. Clinically relevant concentrations of anticancer drugs: a guide for nonclinical studies. Clin Cancer Res 2017;23:3489–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brill E, Yokoyama T, Nair J, Yu M, Ahn Y-R, Lee J-M. Prexasertib, a cell-cycle checkpoint kinases 1 and 2 inhibitor, increases in vitro toxicity of PARP inhibition by preventing Rad51 foci formation in BRCA wild type high-grade serous ovarian cancer. Oncotarget 2017;8:111026–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mueller AC, Sun D, Dutta A. The miR-99 family regulates the DNA damage response through its target SNF2H. Oncogene 2013;32:1164–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rebbeck TR, Friebel TM, Friedman E, Hamann U, Huo D, Kwong A, et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum Mutat 2018;39:593–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Favre B, Turowski P, Hemmings BA. Differential inhibition and posttranslational modification of protein phosphatase 1 and 2A in MCF7 cells treated with calyculin-A, okadaic acid, and tautomycin. J Biol Chem 1997;272:13856–63. [DOI] [PubMed] [Google Scholar]
- 54. Jiang X, Li X, Li W, Bai H, Zhang Z. PARP inhibitors in ovarian cancer: sensitivity prediction and resistance mechanisms. J Cell Mol Med 2019;23:2303–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Westermarck J, Neel BG. Piecing together a broken tumor suppressor phosphatase for cancer therapy. Cell 2020;181:514–7. [DOI] [PubMed] [Google Scholar]
- 56. Shenolikar S. A SMAP in the face for cancer. J Clin Invest 2017;127:2048–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen Q, Cai D, Li M, Wu X. The homologous recombination protein RAD51 is a promising therapeutic target for cervical carcinoma. Oncol Rep 2017;38:767–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Budke B, Lv W, Kozikowski AP, Connell PP. Recent developments using small molecules to target RAD51: how to best modulate RAD51 for anticancer therapy? ChemMedChem 2016;11:2468–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Grundy MK, Buckanovich RJ, Bernstein KA. Regulation and pharmacological targeting of RAD51 in cancer. NAR Cancer 2020;2:zcaa024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yata K, Lloyd J, Maslen S, Bleuyard J-Y, Skehel M, Smerdon SJ, et al. Plk1 and CK2 act in concert to regulate Rad51 during DNA double-strand break repair. Mol Cell 2012;45:371–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hyun S-Y, Hwan H-I, Jang Y-J. Polo-like kinase-1 in DNA damage response. BMB Rep 2014;47:249–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lee H-J, Hwang H-I, Jang Y-J. Mitotic DNA damage response: Polo-like kinase-1 is dephosphorylated through ATM–Chk1 pathway. Cell Cycle 2010;9:2389–98. [DOI] [PubMed] [Google Scholar]
- 63. Gavande NS, VanderVere-Carozza PS, Hinshaw HD, Jalal SI, Sears CR, Pawelczak KS, et al. DNA repair targeted therapy: the past or future of cancer treatment? Pharmacol Ther 2016;160:65–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lodovichi S, Cervelli T, Pellicioli A, Galli A. Inhibition of DNA repair in cancer therapy: toward a multi-target approach. IJMS 2020;21:6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Surowy HM, Sutter C, Mittnacht M, Klaes R, Schaefer D, Evers C, et al. Clinical and molecular characterization of the BRCA2 p.Asn3124Ile variant reveals substantial evidence for pathogenic significance. Breast Cancer Res Treat 2014;145:451–60. [DOI] [PubMed] [Google Scholar]
- 66. Swisher EM, Kwan TT, Oza AM, Tinker AV, Ray-Coquard I, Oaknin A, et al. Molecular and clinical determinants of response and resistance to rucaparib for recurrent ovarian cancer treatment in ARIEL2 (Parts 1 and 2). Nat Commun 2021;12:2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1
Fig. S2
Fig. S3
Fig. S4
Fig. S5
Fig. S6
Fig. S7
Table S1
Table S2
Table S3