Abstract
Escherichia coli Sonnei has an O antigen identical to that of Plesiomonas shigelloides O17, and its O-antigen gene cluster is located on a plasmid. By sequencing the chromosomal O-antigen gene cluster of P. shigelloides O17 and comparing it with that of Sonnei, we showed that Sonnei gained its O-antigen genes recently.
In humans, Shigella spp. cause disease ranging from diarrhea to bacillary dysentery. Four species are officially recognized—Shigella dysenteriae, Shigella flexneri, Shigella boydii, and Shigella sonnei—but all are sufficiently similar to Escherichia coli to be placed in the same species (6, 10, 14, 35, 40). We will refer to Shigella sonnei as Sonnei, a clone of E. coli.
The O antigen, which contains many repeats of an oligosaccharide unit (O unit), is part of the lipopolysaccharide (LPS) present in the outer membrane of gram-negative bacteria. It contributes major antigenic variability to the cell surface and confers O-serotype specificity. There are 166 known O antigens recognized in the E. coli typing scheme (29), and there are 34 among the Shigella strains (11, 28). There is considerable overlap in O antigens of traditional E. coli and Shigella strains of E. coli, and the total number of unique O antigens in E. coli (including Shigella strains) is 190. The surface O antigen is subject to intense selection by the host immune system, which may account for the maintenance of many different O-antigen forms within species such as E. coli.
The genes for O-antigen synthesis are normally grouped together on the chromosome in a gene cluster which maps close to gnd in both E. coli and Salmonella enterica. We, among others, have undertaken an extensive study of the genetic basis of O-antigen variation by sequencing and identifying the O-antigen genes, mostly in S. enterica and E. coli (see reference 42 for a review). It has been found that, in general, O-antigen genes have a low GC content (usually less than 40%). We suggested that this deviation in GC content from that of typical S. enterica or E. coli genes (51%) indicates that the O-antigen DNA originated in a species other than S. enterica or E. coli and was captured by lateral transfer (21). We and others have previously studied DNA recombination events between O-antigen gene clusters within S. enterica (8, 59) and between E. coli and Klebsiella (49). In this study, we demonstrated interspecies transfer of an entire O-antigen gene cluster.
Sonnei has an O antigen (4, 23) not otherwise found in E. coli which is identical to that of serotype 17 of Plesiomonas shigelloides (41, 50). The O-antigen genes of Sonnei are unusual in that they occur on a plasmid, the invasion plasmid (Pinv), which is more than 180 kb in size and is essential for penetration of host epithelial cells (44, 56, 57). It has been shown that this O-antigen gene cluster can hybridize to the chromosomal O-antigen genes of P. shigelloides serotype O17 (52). Most of the plasmid-borne O-antigen gene cluster has been sequenced, and it has been shown that, for at least a few hundred base pairs, the Sonnei O-antigen gene cluster is identical in sequence to that of P. shigelloides O17 (19, 20).
Sonnei strains which have lost the Pinv plasmid lack O antigen (24), suggesting that O-antigen genes at the chromosomal site are nonfunctional. We have recently cloned and sequenced the chromosomal O-antigen genes from Sonnei and showed that this bacterium has a remnant O-antigen gene cluster on the chromosome; most of the original cluster apparently has been deleted by homologous recombination between manB genes in the adjacent O-antigen and colanic acid gene clusters (26). Sonnei, and indeed all Shigella and many other human-pathogenic clones of E. coli, is thought to have relatively recently adapted to the diarrhea-causing mode of pathogenesis in humans (12, 32). The finding that a presumably typical chromosomal O-antigen gene cluster was lost by deletion indicated that to adapt to a new niche, Sonnei gained new O-antigen genes on a plasmid and that this was followed by inactivation of the original chromosomal O-antigen gene cluster.
In this study, we sequenced the entire O-antigen gene cluster of P. shigelloides O17, as well as part of the Sonnei plasmid-borne O-gene cluster to obtain the full sequence. Comparison of the two sequences showed that they are almost identical, with the exception of the wzz and wbgZ genes. The high level of similarity indicates that the O-antigen gene cluster has recently transferred from one species to the other. We assume that because the Sonnei O-antigen genes are on a plasmid, the transfer has been from P. shigelloides to E. coli; the high level of similarity for most genes suggests that only recently did Sonnei gained its current O antigen. The level of difference between the two wzz and wbgZ genes is higher than that between other genes of the two gene clusters, and there is evidence that the wzz gene on the plasmid has been under selection for change in the new host.
Sequences of O-antigen gene clusters.
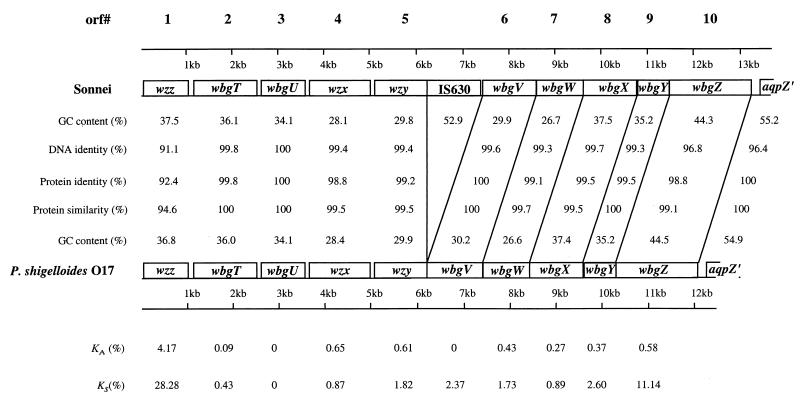
Figure 1 shows the O-antigen gene clusters of P. shigelloides O17 strain C27 (ATCC 14030) and the Pinv plasmid of E. coli Sonnei strain 53G1. The segment from positions 733 to 11750 of plasmid Pinv has already been sequenced (19), and the corresponding region in strain C27 (positions 733 to 10598 [Fig. 1]) was PCR amplified and sequenced by using oligonucleotide primers synthesized based on the published sequence of Pinv. Because these two sequences are highly similar (see below), we also resequenced this region from Pinv to make sure that variations observed were not due to sequencing errors. We found some discrepancies between our sequence of Pinv and the sequence obtained by Houng and Venkatesan (19). We have sequenced each of the relevant bases at least three times and are confident that our sequence is correct.
FIG. 1.
Comparison of the Sonnei plasmid-borne O-antigen gene cluster with that of P. shigelloides O17.
We have shown that a 39-bp element, termed the JUMPstart sequence, is present upstream of many polysaccharide gene clusters (18). To sequence the 5′ region of the O-antigen gene cluster (from positions 1 to 980 [Fig. 1]), oligonucleotides which bind to the middle of the JUMPstart sequence (no. 412; 5′-ATTGGTAGCTGTAAGCCAAGGGCGGTAGCGT) and positions 1024 to 1007 (no. 1628) within the O-antigen gene cluster were used to PCR amplify DNA fragments of about 1 kb from both strain C27 and the Pinv plasmid. Both PCR fragments were first sequenced from both ends, and then PCR walking was carried out to complete the sequence of each fragment.
The sequence of the 3′ region (positions 10599 to 12540 [Fig. 1]) of strain C27 was obtained by walking downstream from the sequenced region, using suppression PCR. This was done essentially as described by Siebert et al. (46). Adapters and adapter primers were those used previously by Lan and Reeves (27). Aliquots of chromosomal DNA were digested separately with one of several restriction enzymes (BglI, BglII, ClaI, DraI, HpaI, and KpnI), and adapters were ligated to the fragments. For the first round of amplification, oligonucleotide primer 2261 (5′-TTAGTGCACCGATGTGGT) was used in combination with suppression PCR primer 604 (5′-GGATGGTAATGAACCTCACTAATGCG). Amplified fragments were isolated from agarose gels and reamplified by using nested primer pair 2260 (5′-GCTGCAGCTATTCTTACC)-605 (5′-M13R-CTAATGCGGTCGAGCGGC). A fragment of about 1 kb was obtained from the KpnI digest, cloned into pGEM-T, and sequenced; this generated the sequence from positions 10430 to 11381. Two more rounds of amplification, using primers (3293 [5′-ACAGATGGTGATGAAGTC] and 3294 [5′-GTTGAGCATAATGTGGTG]) based on the newly obtained sequence in combination with the two suppression PCR primers, generated a fragment of about 1.1 kb from the BglI digest. This fragment was also cloned into pGEM-T and sequenced (positions 11350 to 12540).
Sequencing was performed with an Applied Biosystems model 377 automated DNA sequencer. Sequence data were assembled and analyzed by the Australian National Genomic Information Service, (A. H. Reisner, C. A. Bucholtz, J. Smelt, and S. McNeil, Proc. 26th Annu. Hawaii Int. Conf. Syst. Sci., p. 595–602, 1993). The region from positions 11751 to 12661 of PinV (Fig. 1) was sequenced by using primers based on the sequence of strain C27.
The O-antigen gene cluster is located between a JUMPstart sequence and an aquaporin gene.
The JUMPstart sequence is located upstream of O-antigen gene clusters. We used a primer which binds to it to amplify and sequence the 5′ ends of the gene clusters, and for both gene clusters the first base downstream of the JUMPstart sequence is treated as position 1.
The 3′ regions in both gene clusters (position 12259 to the end of strain C27 and position 13411 to the end of plasmid Pinv) showed a high degree of similarity (76% identity at the protein level) to the 5′ end of the E. coli aquaporin Z gene (GenBank accession no. AAC43518). The GC content for the regions downstream of positions 12258 and 13410 in strains C27 and plasmid Pinv, respectively, is relatively higher than that of the rest of the sequence (Fig. 1). We suggest that the O-antigen gene clusters end at positions 12258 and 13410 in strain C27 and plasmid Pinv, respectively.
O-antigen genes.
There are 10 genes in the gene cluster of strain C27, and there are 10 genes plus an insertion sequence (IS) in the gene cluster of plasmid Pinv; all genes have the same transcriptional direction from JUMPstart to the aquaporin gene (Fig. 1). The nucleotide and amino acid sequences were used to search available databases for indications of possible function. MULTICOMP software (43) was used to do pairwise comparisons of DNA and derived amino acid sequences and to estimate the number of synonymous substitutions per synonymous site (KS) and the number of nonsynonymous substitutions per nonsynonymous site (KA).
The O unit contains two sugars, 2-amino-2-deoxy-l-altruronic acid (2Ac-AltUA) and 2-acetamino-4-amino-2,4,6-trideoxy-d-galactose (4n-FucNAc) (4, 23). The gene cluster is predicted to contain two transferase genes, genes for the synthesis of the two deoxynucleoside triphosphate precursor sugars, an O-antigen flippase gene (wzx), an O-antigen polymerase gene (wzy), and a chain length determinant gene (wzz).
Comparison of corresponding genes in the two gene clusters showed levels of identity ranging from 91.1 to 100% at the DNA level and from 92.4 to 100% at the protein level. Thus, these corresponding genes must have the same function, and in the following discussion the percent similarity data are related to the genes of strain C27.
(i) orf1.
The deduced amino acid sequence of the protein encoded by orf1 exhibits about 46% similarity and 26% identity to that of the wzz gene of E. coli O111 (3). Wzz proteins are characterized by two transmembrane domains, located in the amino-terminal and carboxy-terminal regions, and a large hydrophilic central domain located in the periplasm (34). Orf1 has a similar hydrophobic profile, and we have named orf1 wzz.
(ii) orf2 and orf3.
The deduced amino acid sequences of the proteins encoded by orf2 and orf3 have significant homology to a variety of UDP-glucose/GDP-mannose dehydrogenase and nucleotide sugar epimerase genes, respectively. WcdA and WcdB of the S. enterica serovar Typhi Vi antigen gene cluster (16, 53) have the highest levels of identity to Orf2 (63%) and Orf3 (64%), respectively. orf2 and orf3, which are thought to be sugar pathway genes, have been named wbgT and wbgU, respectively.
(iii) orf4.
The orf4 gene is predicted to encode an integral inner-membrane protein with 12 transmembrane segments, suggesting that it is the wzx gene. The predicted protein also has the conserved motif found near the amino-terminal end of Wzx (48). orf4 has been named wzx.
(iv) orf5.
The orf5 gene encodes a protein containing eight predicted transmembrane segments with a large cytoplasmic loop (83 amino acids). This inner-membrane topology is a characteristic feature of all known O-antigen polymerases (33), and we believe that orf5 is the O-antigen polymerase gene, wzy.
(v) orf6.
The deduced amino acid sequence of the protein encoded by orf6 shows similarity to many NADH dehydrogenases. It is thought to be a sugar pathway gene and has been named wbgV.
(vi) orf7.
The deduced amino acid sequence of the orf7-encoded protein exhibits 54.2% similarity to WaaV, a glucosyl transferase involved in the synthesis of the LPS core in E. coli (17). It is highly likely that this gene, which has been named wbgW, encodes a transferase.
(vii) orf8.
The deduced amino acid sequence of the protein encoded by orf8 exhibits 75.5% similarity and 56.5% identity to that of the wlbF gene of the Bordetella pertussis O-antigen gene cluster, which shows similarity to a group of nucleotide sugar aminotransferase genes (2). orf8 is also similar to this group of genes, which includes the per gene (36.2% identity and 59.3% similarity at the amino acid level) of the E. coli O157 O-antigen gene cluster. This gene, which is highly likely to be involved in the synthesis of 4n-FucNAc (see below), has been named wbgX.
(viii) orf9.
The deduced amino acid sequence of the orf9-encoded protein shows 63% similarity and 39% identity to the C-terminal half of that of wbaP of the S. enterica O4 gene cluster (21). WbaP is the galactosyltransferase catalyzing the first step of S. enterica O4-antigen synthesis (21), and it has been shown that it is the C-terminal half that encodes the transferase function (54). We have shown that all or C-terminal half of WbaP exhibits similarity to a protein family whose members all catalyze the first step in polysaccharide synthesis (54). orf9, which has been named wbgY, is thought to encode the transferase catalyzing the first step of O-antigen synthesis.
(ix) orf10.
The deduced protein product of orf10 is homologous to the members of the WbcP family of proteins, which are required for the biosynthesis of surface polysaccharides in numerous bacterial species and may function as nucleotide sugar epimerases or dehydratases (9). Among the proteins of this family, WbcP of the Yersinia enterocolitica LPS outer-core gene cluster (47) is most similar to Orf10, exhibiting 82.4% similarity and 68.5% identity. orf10, a putative sugar pathway gene, has been named wbgZ.
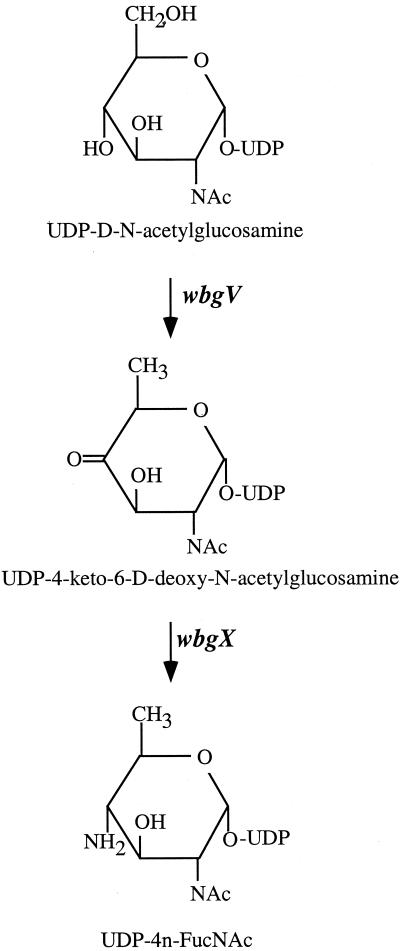
The two potential transferase genes are as expected based on the O-antigen structure with one (wbgY) having the characteristics expected for the first transferase. wbgV and wbgX may be involved in the synthesis of UDP-4-n-FucNAc from UDP-N-acetylglucosamine (UDP-GlcNAc); the putative biosynthetic pathway is shown in Fig. 2. WbgV is homologous to a variety of dehydrogenases, and we propose that it is a UDP-GlcNAc dehydrogenase. The reaction converting UDP-4-keto-6-deoxy-GlcNAc to UDP-4n-FucNAc is very similar to the last step of the biosynthesis of GDP-perosamine by the Per protein (55). WbgX is homologous to Per, and we believe that WbgX is required for this step. The biosynthetic pathway for 2Ac-AltUA is not clear, and we propose that wbgU and wbgZ (encoding two epimerase homologues) as well as wbgT (encoding a dehydrogenase homologue) are involved.
FIG. 2.
Proposed pathway for conversion of UDP-GlcNAc to UDP-4n- FucNAc.
The plasmid-borne Sonnei O-antigen gene cluster has an IS630 element.
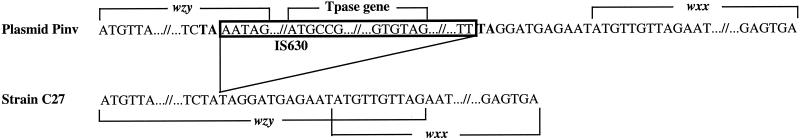
Houng and Venkatesan (19) found that between the wzy and wbgV genes in the O-antigen gene cluster of plasmid Pinv there is a 1,153-bp IS (positions 6215 to 7367) which is absent in the O-antigen gene cluster of strain C27 (Fig. 1). Comparison of the two gene clusters shows that the IS is within the sixth codon from the end of the wzy gene, making the wzy gene in plasmid Pinv four codons shorter than that in the C27 cluster and generating a duplication of the two bases (TA) upstream of the insertion site (Fig. 3). The IS, which is an IS630 element, is 1,153 bp in length and is almost identical (99.4% identity) to the IS630 element of Sonnei identified by Matsutani et al. (31). There is a transposase gene in the IS and a 29-bp inverted repeat at each end as described by Matsutani et al. (31).
FIG. 3.
The IS630 integration site within the Sonnei plasmid-borne O-antigen gene cluster. Tpase, transposase.
Relationship between the O-antigen gene clusters of P. shigelloides O17 and E. coli Sonnei.
The functional O-antigen genes of Sonnei are on a plasmid, and we have previously shown that this bacterium has a remnant O-antigen gene cluster on its chromosome (26). In the present study, we showed that the plasmid-borne Sonnei O-antigen gene cluster is almost identical to that of P. shigelloides O17. The gene order is conserved between the two gene clusters, except for the presence of an IS630 element between wzy and wbgV in Sonnei. The level of DNA identity is generally high between corresponding genes of the two gene clusters, ranging from 99.1 to 100%, except for wzz and wbgZ. Thus, the transfer of this gene cluster from one species to the other must have been a recent event. Because the gene cluster is located on a plasmid and has an IS630 element inserted in the case of Sonnei, it is highly likely that the direction of the transfer was from P. shigelloides to E. coli.
wzz and wbgZ, which show the highest level of difference (Fig. 1), are at the ends of the gene cluster. It has been observed that genes at the ends of polymorphic O-antigen and capsular gene clusters are often genes present in gene clusters for other forms of the O antigen or capsule in the species and are sites of recombination in intraspecies transfer of the gene cluster. A recent study of O-antigen rml genes in S. enterica (Li and P. R. Reeves, unpublished data) showed that this recombination can result in levels of variation for the genes at the end of a cluster similar to the intraspecies sequence variation seen with housekeeping genes, which is much higher than that for mutations in genes located in the center of the gene clusters, which are strongly conserved in different isolates with the same O antigen. The higher level of divergence in the Sonnei and O17 wzz and wbgZ sequences could be due to the genes in the P. shigelloides source of the gene cluster that is now in E. coli Sonnei being different from the corresponding genes in the gene cluster now selected from P. shigelloides for sequencing. This possibility is supported by the observation that there is very little variation for the first 675 bases of wbgZ (0.044%) compared to the rest of the gene (4.5%), indicative of a recombination event within the P. shigelloides wbgZ gene at about position 675 (position 10899 in the whole sequence). The divergence of 3.6% for the short segment of aqpZ sequenced is supportive of this possibility since it would reflect divergence in housekeeping genes in the two P. shigelloides strains involved in the recombination event.
The situation for wzz is different; there is a much higher level of divergence (8.9%) (Fig. 1) and a much higher KA than for wbgZ. The two wzz genes had a KS of 0.28, a KA of 0.042, and a KS/KA ratio of 7. Sharp (45) compared 67 housekeeping genes present in both E. coli K-12 and S. enterica serovar Typhimurium and found average values for KS and KA of 0.94 and 0.039, respectively, and an average KS/KA ratio of 24. The two wzz genes had a KS of 0.28, a KA of 0.042, and a KS/KA ratio of 7. The atypical value for KA and much lower KS/KA ratio of 7 relative to the corresponding values for housekeeping genes of E. coli and S. enterica, in which substitutions are assumed to be mainly due to genetic drift, indicate that there is selection pressure on wzz to change amino acids. Note that the KS/KA ratio for the 3′ end of wbgZ (positions 676 to 1917 in the gene) is 22, within the normal range for neutral mutations.
The chromosomal wzz gene in Sonnei.
The E. coli wzz gene is usually located outside of the O-antigen gene cluster in a region between the gnd gene and the his operon (3). We showed previously that most of the Sonnei chromosomal O-antigen gene cluster had been deleted, but we did not look at the wzz gene.
Fragments covering the wzz region were PCR amplified from Sonnei chromosomal DNA, using primer pairs 2126 (5′-GCKAACYTRATYCAGGCNCAGCGYGACTA)-465 (5′-CCCGGATCCTTATTTCGCGTTGTA) and 2127 (5′-ACGGTAATTGAGAACCTG)-466 (5′-CCCGAATTCATGAGAGTAGAAAAT). These two fragments were then sequenced using primers 466 and 465, respectively, and PCR walking was carried out until the full wzz gene was sequenced.
This wzz gene shows high level of similarity to many E. coli wzz genes, particularly to that of E. coli K-12 (5) and O157:H7 strain EDL933 (13) (99.8 and 96.4% DNA sequence identity, respectively). The basic O-antigen chain length of E. coli varies, generally being in the range of 10 to 18 O units (7, 25, 39). For convenience, the LPSs of E. coli have been divided into three groups, i.e., those having short (7 to 16 O units), intermediate (10 to 18 O units), or long (16 to 25 O units) chains (13). A comparison of Wzz proteins of 22 E. coli strains identified motifs specific for each group, and some of the residues involved have been shown by mutagenesis experiments to direct different chain lengths (13). Alignment of Wzz of the Sonnei chromosome with these 22 proteins shows that it belongs to the group conferring the intermediate chain length (data not shown).
Chain length of the Sonnei O antigen.
There is evidence of an association between O-antigen chain length and pathogenicity in E. coli, and it has been suggested that the specific chain length range of the O antigen may be an important virulence factor (13). Sonnei produces an LPS with a basic chain length of 18 to 25 O units (51). The Sonnei chromosomal wzz gene encodes a typical intermediate-length Wzz (for a chain length of 10 to 18 O units), and we believe that the O-antigen chain length is determined by the protein encoded by the plasmid wzz gene. Comparison of the Sonnei plasmid-borne wzz with that of P. shigelloides O17 showed a relatively high level of nonsynonymous substitutions. Sonnei has a shorter O-antigen chain length than P. shigelloides O17 (51), and we suggest that a shorter chain length better suits the niche occupied by Sonnei and that there has been selection pressure on the Sonnei plasmid-borne wzz gene to change.
Time of transfer of the O-antigen gene cluster to Sonnei.
The sequence variation at synonymous sites (Fig. 1) was used to estimate when the last common ancestor existed. wzz, wbgZ, and aqpZ were excluded for reasons given above. Two synonymous clock rates have been proposed for E. coli. Based on the proposal that E. coli and S. enterica last shared a common ancestor about 140 million years ago and that 95% of synonymous sites have been exchanged since then, Whittam calculated the rate of synonymous-polymorphism accumulation to be 6 × 10−9 per year (58). A higher rate of 3 × 10−8 was calculated by Guttman and Dykhuizen on the basis that the mutation rate is about 10−10 and that E. coli undergoes approximately 300 divisions per year under natural conditions (15). The mean KS value for these eight genes is 0.01339, and the date since divergence of the two O-antigen gene clusters was estimated to be 45,000 to 220,000 years, using the two clock rates. This estimate assumes similar clock rates for P. shigelloides and E. coli. Furthermore, the average generation time for Shigella may be different from that of other E. coli strains since Shigella forms have a rapid buildup of numbers during infection. Thus, the figures obtained here can only be thought of as estimates.
Diseases of humans caused by enteric bacterial infections are thought to have emerged after agricultural settlement, which occurred about 8000 B.C., because the nature of their infection and transmission makes them unlikely to have been successful in the previous hunter-gatherer society (12, 32). Thus, Sonnei is thought to have emerged as a human-pathogenic clone of E. coli in the last 10,000 years. One might expect that capture of the O-antigen gene cluster from P. shigelloides and loss of function of the original O-antigen gene cluster would have occurred in that period as part of the adaptation to a new niche. The synonymous-substitution data suggest an earlier data, but it is possible that there is some sequence variation within P. shigelloides O27, even in the O-antigen genes, which would account for part of the variation between the two sequences being compared. However, the data give an estimate for the earliest possible time of transfer of the Sonnei O-antigen genes into E. coli.
O-antigen variation is extensive in bacteria. It has been observed that some O-antigen forms are disproportionately represented in pathogenic clones of E. coli, and it has been concluded that O-antigen specificity is an important determinant of pathogenicity (36–38). P. shigelloides is pathogenic to humans, mainly causing diarrhea (22), and strains of serotype O17 are the most frequently isolated (1). Thus, the transfer of the P. shigelloides O17-antigen gene cluster into E. coli could have occurred as part of the evolution of the pathogenic Sonnei clone of E. coli.
It has been shown that Sonnei strains have more than six copies of IS630, but other strains of Enterobacteriaceae tested seldom have this IS (30). Thus, it is most likely that the IS630 element in the Sonnei O-antigen gene cluster was transposed to its current site from the Sonnei chromosome after entry of the Pinv plasmid into Sonnei. This is supported by the high level of DNA sequence identity between the plasmid-borne and chromosomal IS630 elements (GenBank accession no. SSIS630). The other possibility is that IS630 was brought into Sonnei from P. shigelloides via the Pinv plasmid and then moved into chromosomal sites. The position of IS630 in the middle of the gene cluster does not suggest any specific role in the mobilization of the plasmid.
Nucleotide sequence accession number.
The DNA sequences have been deposited in GenBank under accession no. AF285970 (the O-antigen gene cluster of P. shigelloides O17), AF285971 (the plasmid-borne O-antigen gene cluster of Sonnei), and AF295304 (the chromosomal wzz gene of Sonnei).
Acknowledgments
We thank Jean-Francois Viret for kindly supplying strains and Deborah Rothemund for excellent technical assistance.
This study was supported by the Australian Research Council.
REFERENCES
- 1.Aldova E. Serovars of Plesiomonas shigelloides. Zentbl Bakteriol. 1994;281:38–44. doi: 10.1016/s0934-8840(11)80635-3. [DOI] [PubMed] [Google Scholar]
- 2.Allen A G, Maskell D J. The identification, cloning and mutagenesis of a genetic locus required for lipopolysaccharide biosynthesis in Bordetella pertussis. Mol Microbiol. 1996;19:37–52. doi: 10.1046/j.1365-2958.1996.354877.x. [DOI] [PubMed] [Google Scholar]
- 3.Bastin D A, Brown P K, Haase A, Stevenson G, Reeves P R. Repeat unit polysaccharides of bacteria: a model for polymerisation resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol Microbiol. 1993;7:725–734. doi: 10.1111/j.1365-2958.1993.tb01163.x. [DOI] [PubMed] [Google Scholar]
- 4.Batta G, Liptak A, Schneerson R, Pozsgay V. Conformational stabilization of the altruronic acid residue in the O-specific polysaccharide of Shigella sonnei/Plesiomonas shigelloides. Carbohydr Res. 1998;305:93–99. doi: 10.1016/s0008-6215(97)00197-3. [DOI] [PubMed] [Google Scholar]
- 5.Blattner F R, Plunkett G I, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Brenner D J, Fanning G R, Miklos G V, Steigerwalt A G. Polynucleotide sequence relatedness among Shigella species. Int J Syst Bacteriol. 1973;23:1–7. [Google Scholar]
- 7.Brussow H, Sidoti J. Reactivity of human serum antibody with lipopolysaccharide O78 antigen from enterotoxigenic Escherichia coli. Epidemiol Infect. 1992;108:315–322. doi: 10.1017/s0950268800049785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curd H, Liu D, Reeves P R. Relationships among the O-antigen gene clusters of Salmonella enterica groups B, D1, D2, and D3. J Bacteriol. 1998;180:1002–1007. doi: 10.1128/jb.180.4.1002-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeShazer D, Brett P J, Woods D E. The type II O-antigen polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol Microbiol. 1998;30:1081–1100. doi: 10.1046/j.1365-2958.1998.01139.x. [DOI] [PubMed] [Google Scholar]
- 10.Ewing W H. Serological relationships between Shigella and coliform cultures. J Bacteriol. 1953;66:333–340. doi: 10.1128/jb.66.3.333-340.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewing W H. Edwards and Ewing's identification of the Enterobacteriaceae. Amsterdam, The Netherlands: Elsevier Science Publishers; 1986. [Google Scholar]
- 12.Fenner F. The effects of changing social organisation on the infectious diseases of man. In: Boyden S W, editor. The impact of civilization on the biology of man. Toronto, Ontario, Canada: University of Toronto Press; 1970. pp. 48–68. [Google Scholar]
- 13.Franco V A, Liu D, Reeves P R. The Wzz (Cld) protein in Escherichia coli: amino acid sequence variation determines O antigen chain length specificity. J Bacteriol. 1998;180:2670–2675. doi: 10.1128/jb.180.10.2670-2675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goullet P. Esterase electrophoretic pattern between Shigella species and Escherichia coli. J Gen Microbiol. 1980;117:493–500. doi: 10.1099/00221287-117-2-493. [DOI] [PubMed] [Google Scholar]
- 15.Guttman D S, Dykhuizen D E. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science. 1994;266:1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto Y, Li N, Yokoyama H, Ezaki T. Complete nucleotide sequence and molecular characterization of ViaB region encoding Vi antigen in Salmonella typhi. J Bacteriol. 1993;175:4456–4465. doi: 10.1128/jb.175.14.4456-4465.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinrichs D E, Monteiro M A M, Perry M B, Whitfield C. The assembly system for the lipopolysaccharide R2 core-type of Escherichia coli is a hydrid of those found in Escherichia coli K-12 and Salmonella enterica. J Biol Chem. 1998;273:8849–8859. doi: 10.1074/jbc.273.15.8849. [DOI] [PubMed] [Google Scholar]
- 18.Hobbs M, Reeves P R. The JUMPstart sequence: a 39 bp element common to several polysaccharide gene clusters. Mol Microbiol. 1994;12:855–856. doi: 10.1111/j.1365-2958.1994.tb01071.x. [DOI] [PubMed] [Google Scholar]
- 19.Houng H H, Venkatesan M M. Genetic analysis of Shigella sonnei form I antigen: identification of a novel IS630 as an essential element for the form I antigen expression. Microb Pathog. 1998;25:165–173. doi: 10.1006/mpat.1998.0222. [DOI] [PubMed] [Google Scholar]
- 20.Houng H H, Zapor M J, Hartman A B, Hale T L, Venkatesan M M. The roles of IS630 sequence in the expression of the form I antigen of Shigella sonnei: molecular and evolutionary aspects. In: van der Zeijst B A M, Hoekstra W P M, van Embden J D A, van Alphen A J W, editors. Ecology of pathogenic bacteria: molecular and evolutionary aspects. Amsterdam, The Netherlands: Elsevier; 1997. pp. 282–283. [Google Scholar]
- 21.Jiang X M, Neal B, Santiago F, Lee S J, Romana L K, Reeves P R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2) Mol Microbiol. 1991;5:695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 22.Kain K C, Kelly M T. Clinical features, epidemiology, and treatment of Plesiomonas shigelloides diarrhea. J Clin Microbiol. 1989;27:998–1001. doi: 10.1128/jcm.27.5.998-1001.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenne L, Lindberg B, Petersson K, Katzenellenbogen E, Romanowska E. Structural studies of the O-specific side-chains of Shigella sonnei phase I lipopolysaccharide. Carbohydr Res. 1980;78:119–126. [Google Scholar]
- 24.Kopecko D J, Washington O, Formal S B. Genetic and physical evidence for plasmid control of Shigella sonnei form I cell surface antigen. Infect Immun. 1980;29:207–214. doi: 10.1128/iai.29.1.207-214.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusecek B, Wloch H, Mercer A, Vaisanen V, Pluschke G, Korhonen T, Achtman M. Lipopolysaccharide, capsule, and fimbriae as virulence factors among O1, O7, O16, O18, or O75 and K1, K5, or K100 Escherichia coli. Infect Immun. 1984;43:368–379. doi: 10.1128/iai.43.1.368-379.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai V, Wang L, Reeves P R. Escherichia coli clone Sonnei (Shigella sonnei) had a chromosomal O-antigen gene cluster prior to gaining its current plasmid-borne O-antigen genes. J Bacteriol. 1998;180:2983–2986. doi: 10.1128/jb.180.11.2983-2986.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan R, Reeves P R. Recombination between rRNA operons created most of the ribotype variation observed in the seventh pandemic clone of Vibrio cholerae. Microbiology. 1998;144:1213–1221. doi: 10.1099/00221287-144-5-1213. [DOI] [PubMed] [Google Scholar]
- 28.Le Minor L, Richard C. Shigella. Paris, France: Institut Pasteur; 1993. pp. 72–78. . (In French.) [Google Scholar]
- 29.Lior H. Classification of Escherichia coli. In: Gyles C L, editor. Escherichia coli in domestic animals and humans. Wallingford, United Kingdom: CAB International; 1994. pp. 31–72. [Google Scholar]
- 30.Matsutani S, Ohtsubo E. Distribution of the Shigella sonnei insertion elements in Enterobacteriaceae. Gene. 1993;127:111–115. doi: 10.1016/0378-1119(93)90624-c. [DOI] [PubMed] [Google Scholar]
- 31.Matsutani S, Ohtsubo H, Maeda Y, Ohtsubo E. Isolation and characterization of IS elements repeated in the bacterial chromosome. J Mol Biol. 1987;196:445–455. doi: 10.1016/0022-2836(87)90023-4. [DOI] [PubMed] [Google Scholar]
- 32.McKeown T. The origins of human disease. Oxford, United Kingdom: Basil Blackwell; 1988. [Google Scholar]
- 33.Morona R, Mavris M, Fallarino A, Manning P A. Characterization of the rfc region of Shigella flexneri. J Bacteriol. 1994;176:733–747. doi: 10.1128/jb.176.3.733-747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morona R, Van Den Bosch L, Manning P. Molecular, genetic, and topological characterization of O antigen chain regulation in Shigella flexneri. J Bacteriol. 1995;177:1059–1068. doi: 10.1128/jb.177.4.1059-1068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochman H, Whittam T S, Caugant D A, Selander R K. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983;129:2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- 36.Ørskov F, Ørskov I. Special Escherichia coli serotypes among enterotoxigenic strains from diarrhoea in adults and children. Med Microbiol Immunol. 1976;162:73–80. doi: 10.1007/BF02121318. [DOI] [PubMed] [Google Scholar]
- 37.Ørskov F, Ørskov I. Special Escherichia coli serotypes from enteropathies in domestic animals and man. Fortschr Vetmed. 1979;529:7–14. [Google Scholar]
- 38.Ørskov I, Ørskov F. Special O:K:H serotypes among enterotoxigenic Escherichia coli strains from diarrhoea in adults and children. Med Microbiol Immunol. 1977;103:99–110. doi: 10.1007/BF02121318. [DOI] [PubMed] [Google Scholar]
- 39.Porat R, Johns M A, McCabe W R. Selective pressures and lipopolysaccharide subunits as determinants of resistance of clinical isolates of gram-negative bacilli to human serum. Infect Immun. 1987;55:320–328. doi: 10.1128/iai.55.2.320-328.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pupo G M, Karaolis D K R, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rauss K, Kontrohr T, Vertenyi A, Szendrei L. Serological and chemical studies of Shigella sonnei and Pseudomonas shigelloides strains C27. Acta Microbiol Acad Sci Hung. 1970;17:157–166. [PubMed] [Google Scholar]
- 42.Reeves P R. Evolution of Salmonella O antigen variation by interspecies gene transfer on a large scale. Trends Genet. 1993;9:17–22. doi: 10.1016/0168-9525(93)90067-R. [DOI] [PubMed] [Google Scholar]
- 43.Reeves P R, Farnell L, Lan R. MULTICOMP: a program for preparing sequence data for phylogenetic analysis. CABIOS. 1994;10:281–284. doi: 10.1093/bioinformatics/10.3.281. [DOI] [PubMed] [Google Scholar]
- 44.Sansonetti P J, Kopecko D J, Formal S B. Shigella sonnei plasmids: evidence that a large plasmid is necessary for virulence. Infect Immun. 1981;34:75–83. doi: 10.1128/iai.34.1.75-83.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharp P M. Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: codon usage, map position, and concerted evolution. J Mol Evol. 1991;33:23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- 46.Siebert P D, Chenchik A, Kellogg D E, Lukyanov A, Lukyanov S A. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skurnik M, Venho R, Toivanen P, Alhendy A. A novel locus of Yersinia enterocolitica serotype O-3 involved in lipopolysaccharide outer core biosynthesis. Mol Microbiol. 1995;17:575–594. doi: 10.1111/j.1365-2958.1995.mmi_17030575.x. [DOI] [PubMed] [Google Scholar]
- 48.Stevenson G, Andrianopoulos K, Hobbs H, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugiyama T, Kido N, Kato Y, Koide N, Yoshida T, Yokochi T. Generation of Escherichia coli O9a serotype, a subtype of E. coli O9, by transfer of the wb∗ gene cluster of Klebsiella O3 into E. coli via recombination. J Bacteriol. 1998;180:2775–2778. doi: 10.1128/jb.180.10.2775-2778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor D N, Trofa A C, Sadoff J, Chu C, Bryla D, Shiloach J, Cohen D, Ashkenazi S, Lerman Y, Egan W, Schneerson R, Robbins J B. Synthesis, characterization, and clinical evaluation of conjugate vaccines composed of the O-specific polysaccharides of Shigella dysenteriae type 1, Shigella flexneri type 2a, and Shigella sonnei (Plesiomonas shigelloides) bound to bacterial toxoids. Infect Immun. 1993;61:3678–3687. doi: 10.1128/iai.61.9.3678-3687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Viret J-F, Bruderer U, Lang A B. Characterization of the Shigella serotype D (S. sonnei) O polysaccharide and the enterobacterial R1 lipopolysaccharide core by use of mouse monoclonal antibodies. Infect Immun. 1992;60:2741–2747. doi: 10.1128/iai.60.7.2741-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Viret J-F, Cryz S J, Jr, Lang A B, Favre D. Molecular cloning and characterisation of the genetic determinants that express the complete Shigella serotype D (Shigella sonnei) lipopolysaccharide in heterologous live attenuated vaccine strains. Mol Microbiol. 1993;7:239–252. doi: 10.1111/j.1365-2958.1993.tb01115.x. [DOI] [PubMed] [Google Scholar]
- 53.Virlogeus I, Waxin H, Ecobichon C, Popoff M Y. Role of the viaB locus in synthesis, transport and expression of Salmonella typhi Vi antigen. Microbiology. 1995;141:3039–3047. doi: 10.1099/13500872-141-12-3039. [DOI] [PubMed] [Google Scholar]
- 54.Wang L, Liu D, Reeves P R. C-terminal half of Salmonella enterica WbaP (RfbP) is the galactosyl-1-phosphate transferase domain catalyzing the first step of O antigen synthesis. J Bacteriol. 1996;178:2598–2604. doi: 10.1128/jb.178.9.2598-2604.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Reeves P R. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect Immun. 1998;66:3545–3551. doi: 10.1128/iai.66.8.3545-3551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe H, Nakamura A. Large plasmids associated with virulence in Shigella species have a common function necessary for epithelial cell penetration. Infect Immun. 1985;48:260–262. doi: 10.1128/iai.48.1.260-262.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watanabe H, Nakamura A. Identification of Shigella sonnei form I plasmid genes necessary for cell invasion and their conservation among Shigella species and enteroinvasive Escherichia coli. Infect Immun. 1986;53:352–358. doi: 10.1128/iai.53.2.352-358.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whittam T S. Genetic variation and evolutionary processes in natural populations of Escherichia coli. In: Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 2708–2720. [Google Scholar]
- 59.Xiang S H, Hobbs M, Reeves P R. Molecular analysis of the rfb gene cluster of a group D2 Salmonella enterica strain: evidence for its origin from an insertion sequence-mediated recombination event between group E and D1 strains. J Bacteriol. 1994;176:4357–4365. doi: 10.1128/jb.176.14.4357-4365.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]