Abstract
Although substantial progress has been made in cancer biology and treatment, the prognosis of oral squamous cell carcinoma (OSCC) is still not satisfactory because of local tumor invasion and frequent lymph node metastasis. The tumor microenvironment (TME) is a potential target in which cancer-associated fibroblasts (CAFs) are of great significance due to their interactions with cancer cells. However, the exact mechanism is still unclear. Therefore, we focus on the crosstalk between cancer cells and CAFs and discover that CAFs are the main source of TGF-β1. Transwell assays and western blot analysis further prove that CAFs activate the TGF-β1/Smad pathway to promote OSCC invasion. Through survival analysis, we confirm that CAF overexpression is correlated with poor overall survival in OSCC. To further elucidate the origin and role of CAFs in OSCC, we analyze single-cell RNA sequencing (scRNA-seq) data from 14 OSCC tumor samples and identify four distinct cell types, including CAFs, in the TME, indicating high intratumoral heterogeneity. Then, two subtypes of CAFs, namely, myofibroblasts (mCAFs) and inflammatory CAFs (iCAFs), are further distinguished. Based on the differentially upregulated genes of mCAFs and iCAFs, GO enrichment analysis reveals their different roles in OSCC progression. Furthermore, the gene expression pattern is dynamically altered across pseudotime, potentially taking part in the transformation from epithelial to mCAFs or iCAFs through the epithelial to mesenchymal transition.
Keywords: oral squamous cell carcinoma, cancer-associated fibroblast, myofibroblast, TGF-β, single cell
Introduction
Oral squamous cell carcinoma (OSCC) is the most frequent entity among head and neck cancers [1]. Unfortunately, the prognosis of OSCC is poor, with a five-year survival rate of less than 50% due to local tumor invasion and frequent lymph node metastasis [2]. The current treatment strategies are generally limited to three aspects: surgery, chemotherapy and radiotherapy. Therefore, it is urgent to develop novel targeted therapies to improve the lagging status of OSCC.
The tumor stroma is believed to play a key role in tumor progression [3]. Cancer-associated fibroblasts (CAFs) are a major population of tumor stromal cells. Two distinct CAF subtypes with either a matrix-producing contractile phenotype or an immunomodulating secretome that have been found in disparate locations relative to cancer cells are often termed myofibroblasts (mCAFs) and inflammatory CAFs (iCAFs), respectively. iCAFs are detected in the desmoplastic areas of tumors far from cancer cells, while mCAFs are predominantly found around cancer cells [4]. Increasing studies have shed light on the critical roles of mCAFs and iCAFs in the tumor microenvironment (TME) of various cancers, such as lung cancer [5], prostate cancer [6], colorectal cancer [7], breast cancer [8] and head and neck cancer [9], indicating that CAFs have important effects on tumorigenesis, invasion and metastasis. In addition, some studies have also implied that CAFs can promote OSCC progression [ 10‒ 12] , but the molecular mechanism is still unclear.
Transforming growth factor β (TGF-β) signaling plays a dual role in tumor biology, suppressing tumor initiation while promoting tumor progression [13]. In the TME, CAFs are the major source of TGF-β1, which is the most commonly upregulated TGF-β ligand in tumor cells [14]. TGF-β1 initiates signaling by assembling receptor complexes formed by TGF-β type I and type II receptors (TβRI and TβRII, respectively) to activate Smad transcription factors [15]. The function of TβRII is to activate TβRI, and then activated TβRI phosphorylates the downstream mediators Smad2 and Smad3. Phosphorylated Smad2 and Smad3 (p-Smad2 and p-Smad3, respectively) bind with Smad4 to enter the nucleus to modulate gene transcription [ 14, 16] . Furthermore, TGF-β1 is essential for CAF formation and maintenance [ 17‒ 19] . Therefore, TGF-β1 acts as a mediator to regulate CAF functions and influence tumor biology by phosphorylating Smads to propagate signaling.
Single-cell RNA sequencing (scRNA-seq) is a rising technology that can be used to understand the TME of OSCC at a high resolution [ 20, 21] .
In the present study, we focused on the role of the CAF-induced TGF-β1/Smad2/3 pathway in OSCC progression. Our study revealed that CAFs secreted a specific amount of TGF-β1, which promoted OSCC invasion in vitro. In addition, the role and origin of different CAF subtypes in OSCC were also explored. These results allow us to better understand the heterogeneity and progression of OSCC and provide new ideas for individualized treatment for OSCC.
Materials and Methods
Data acquisition and preprocessing
After obtaining OSCC scRNA-seq data (GSE172577 and GSE164690) from the Gene Expression Omnibus (GEO) [22], the R packages Seurat and Harmony were used for quality control, removal of the batch effect and integration of data among 14 different samples (GSM5258385, GSM5258386, GSM5258387, GSM5258388, GSM5258389, GSM5258390, GSM5017023, GSM5017032, GSM5017035, GSM5017041, GSM5017044, GSM5017047, GSM5017050, and GSM5017062). These methods were performed as previously described [23]. Finally, 57,926 cells remained and were used in downstream analysis. Using the FindVariableGenes function in the Seurat package, the top 2000 variable genes were chosen, and then principal component analysis (PCA) was performed, followed by dimensionality reduction and visualization of uniform manifold approximation and projection (UMAP) or t-distributed stochastic neighbor embedding (t-SNE). We detected various cell groups using previously described marker genes. The Cancer Genome Atlas (TCGA) database was used to obtain RNA transcriptome data in the Fragments Per Kilobase Per Million (FPKM) format, as well as clinical and survival data for OSCC patients.
Differential gene analysis and enrichment analysis
The FindMarkers function of the Seurat package was used to calculate differential genes between distinct clusters using scRNA-seq data. A log2-fold change >0.5 and an adjusted P value <0.05 were set as the significance thresholds. The internet website Metascape [24] was used to conduct Gene Ontology (GO) analysis. The list of upregulated genes was imported, after which the most important pathways were enriched, and the relationship between them was depicted in the network graph.
Pseudotime analysis
The Monocle2 program was used to determine the differentiation trajectory of tumor cells to create a single-cell trajectory for each sample. We utilized the plot genes in the pseudotime function in the Monocle2 package to show the changes in classic tumor marker genes in the differentiation trajectory after determining the pseudotime value arrangement and differentiation trajectory. The plot pseudotime heatmap function was then used to display a clustered heatmap of the hub genes′ expression patterns.
Cell-cell interaction analysis
The CellChat package was used to investigate interactions between distinct cell populations. We chose many critical ligand–receptor interactions that govern epithelial-CAF crosstalk after calculating and analyzing cell-cell communication in OSCC, and the results are depicted in a bubble graph.
Patient samples
The current study was approved by the Ethics Committee of West China Hospital of Stomatology, Sichuan University. The patients were informed that the residual material from surgical procedures would be used for research purposes with full consent. Surgical samples were obtained from four individual oral cancer patients.
Isolation of primary fibroblasts
CAFs were isolated from surgically excised tongue squamous cell carcinoma tissue materials, whereas normal fibroblasts (NFs) were derived from normal tongue tissues. Fibrous areas of normal tongue tissue and tongue tumors were cut into small pieces of approximately 1 mm 3 and separated solely at the bottom of inversed 25 cm 2 culture flasks. After 2 h of culture, the culture flasks were turned back right, and 2 mL full DMEM (HyClone, Logan, USA) supplemented with 10% FBS (GIBCO, Carlsbad, USA) and 1% penicillin-streptomycin (Life Technologies, Boston, USA) was added. Another 3 mL of full DMEM was added slowly after 12 h of incubation. The culture medium was changed every 2 days as usual. CAFs and NFs were digested with 0.05% EDTA-Trypsin (Life Technologies) and passed to new sterile flasks.
Cell culture
The human tongue cancer cell line Cal27 (obtained from State Key Laboratory of Oral Disease, Sichuan University, Chengdu, China) was available for cell culture experiments. CAFs and NFs were collected from the tongue tumor and normal tissues of patients. All cells were cultured at 37°C, with 5% CO 2 and 95% humidity. Fibroblasts were purified after the third passage for use, but no more than tenth passage.
Immunofluorescence (IF) staining
CAFs and NFs were seeded into 6-well plates separately and fixed with 4% paraformaldehyde (PFA) after three washes with 1×PBS. Then, the cells were incubated with primary antibodies, including anti-ACTA2 (mouse monoclonal; 1:100; ZSGB-BIO, Beijing, China) and anti-vimentin (mouse monoclonal; 1:100; ZSGB-BIO), overnight at 4°C. The secondary antibody anti-mouse IgG-TR (red florescence) (goat; 1:200; BOSTER, Wuhan, China) was used. The nuclei were stained with DAPI. Photograph images were captured using an inverted microscope (OLYMPUS, Tokyo, Japan).
Conditioned medium (CM)
Purified CAFs and NFs were cultured in 25-cm 2 culture flasks with 5 mL of full DMEM containing 10% FBS. After the cell density reached 80%, the culture medium was changed to FBS-free DMEM, and the CM was harvested after 24 h. Both CAF-CM and NF-CM were stored at ‒80 °C until use.
Enzyme-linked immunosorbent assay (ELISA)
TGF-β1 ELISA kit (Uscn Life, Wuhan, China) was used to detect the secreted TGF-β1 amount in CAF-CM and NF-CM. Both CAF-CM and NF-CM were evaluated simultaneously under the same conditions.
Transwell assay
BioCoat TM Growth Factor Reduced (GFR) Invasion Chambers with 8 μm pore inserts in 24-well plates (BD, Franklin Lakes, USA) were used for the Transwell assay to detect the invasive ability of the cells. The chambers were coated with Matrigel (Corning, New York, USA) in advance. Inserts were removed, and CAF-CM and NF-CM (500 μL per well) were evenly added in duplicate to the bottom chambers. Inserts were relocated, and 200 μL of serum-free cell suspension containing 1×10 5 Cal27 cells was added to the inserts. Chambers were incubated for 48 h at 37°C. Noninvaded cells were removed from the top surface of inserts by scrubbing with swabs, while invaded cells were fixed on the membrane. The invaded cells were stained with crystal violet and counted on slides after being fixed on the membrane with 70% ethanol for 30 min at 4°C and washed twice with PBS. The average number of invaded cells per chamber was determined by counting from three fields at 20× view under a light microscope.
Western blot analysis
Cells were rinsed with precooled PBS and harvested in lysis buffer (KeyGEN BioTECH, Nanjing, China) containing 0.5% phosphatase inhibitor, 0.1% protease inhibitor, 0.5% 100 mM PMSF, 50 mM NaF, 1 mM sodium vanadate, 2.5 mM sodium pyrophosphate and 25 mM sodium glycerophosphate. The protein concentration of each sample was detected by using a BCA protein assay kit (Cell Signaling Technology, Beverly, USA). The cell lysates were prepared in denaturing SDS sample loading buffer at a proportion of 1:4, subject to SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, USA). After being blocked with 5% BSA for 1 h, the membranes were incubated with primary antibodies overnight at 4°C and then with secondary antibodies for 1 h at room temperature. The following antibodies were used: Smad2/3 (D7G7; rabbit monoclonal IgG; 1:1000; Cell Signaling Technology) and p-Smad2 (Ser465/467)/Smad3 (Ser423/425) (D27F4; rabbit monoclonal IgG; 1:1000; Cell Signaling Technology). Secondary antibody was used with goat anti-rabbit IgG, 1:2500 (ZSGB-BIO). The TGF-βRI kinase inhibitor SB431542 (Sigma, St Louis, USA) was used to inhibit TGF-βRI.
Statistical analysis
Unless otherwise indicated, statistical analysis was performed using a two-tailed, unpaired Student’s t test, assuming equal variances using GraphPad Prism (version 5.01) and R software (version 4.0.2). P<0.05 was considered to be statistically significant.
Results
CAFs activate the TGF-β1/Smad2/3 signaling pathway to promote OSCC invasion
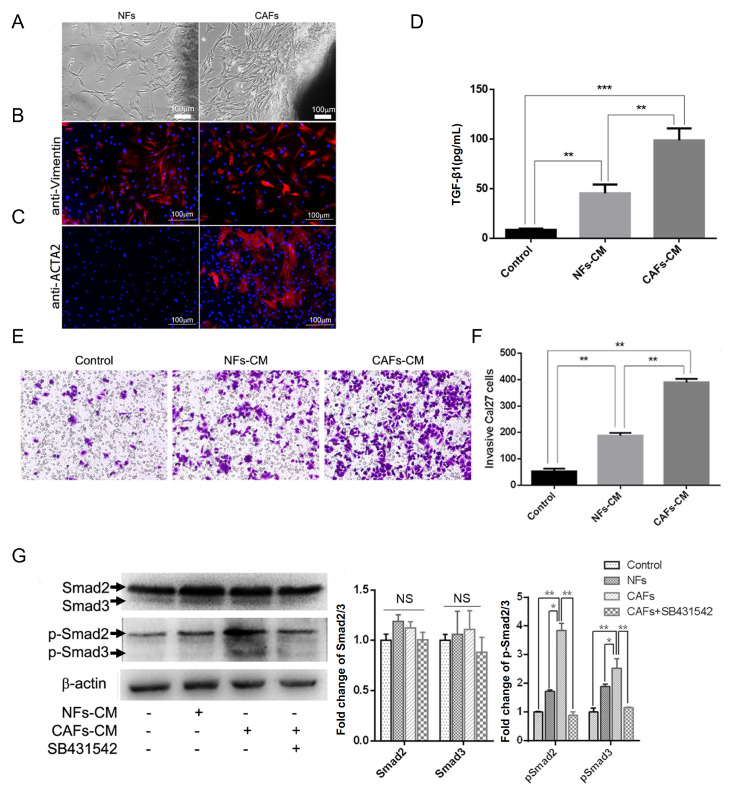
CAFs and their paired NFs were successfully isolated from four primary OSCCs by short-term primary culture in full DMEM supplemented with 10% FBS and 1% penicillin-streptomycin ( Figure 1A). The fibroblast marker vimentin and CAF marker ACTA2 were used to confirm that CAFs derived from tumor tissue were pure without epithelial cell and tumor cell contamination. Both CAFs and NFs were positive for vimentin (red) ( Figure 1B). CAFs were positive for ACTA2 (red), whereas NFs were not ( Figure 1C). Secreted TGF-β1 in NF-CM and CAF-CM was detected separately. Normal DMEM culture medium was used as a control. In the control group, the amount of TGF-β1 secreted in the culture medium was approximately 10 pg/mL on average. TGF-β1 from NFs was 42.8 pg/mL, whereas the secreted TGF-β1 from CAFs was 86.4 pg/mL, which was over 2 folds than that from NFs on average ( Figure 1D). CAF-CM and NF-CM was evenly added to the bottom chamber, and Cal27 cells were cultured in the upper chamber coated with Matrigel. Invaded cells were counted after 48 h from areas that were selected at random under a microscope ( Figure 1E). Our results showed that the number of invaded cancer cells in the presence of CAF-CM was the highest, while Cal27 cell invasion ability was limited in normal culture medium, which was consistent with the concentration changes of TGF-β1 secreted from NFs and CAFs ( Figure 1F). Furthermore, total protein was collected separately, and protein from untreated Cal27 cells was used as a control. The Smad2/3 and p-Smad2/3 expression levels were detected by western blot analysis, which demonstrated that the expression of total Smad2 and Smad3 was not significantly different. However, the expression levels of p-Smad2 and p-Smad3 were upregulated after treatment with CAF-CM and were inhibited by the TGF-βRI inhibitor SB431542 ( Figure 1G). Taken together, these results prove that CAFs promote OSCC invasion by activating the TGF-β1/Smad2/3 signaling pathway.
Figure 1 .
CAFs promote OSCC invasion through the TGF-β1/Smad2/3 pathway
(A) NFs were derived from normal tongue tissue. CAFs were derived from tongue carcinoma. (B) NFs and CAFs were both positive for vimentin (red). (C) CAFs were positive for ACTA2 (red), whereas NFs were not. (D) The amount of secreted TGF-β1 from control cells, NFs and CAFs after 24 h was detected. (E) Crystal violet staining of invaded Cal27 cells in control, NF-CM and CAF-CM. (F) Histogram showing that CAFs prominently promoted Cal27 cell invasion. (G) Western blot analysis of Smad2/3 and p-Smad2/3 proteins in Cal27 cells with different stimulations by NF-CM, CAF-CM or CAF-CM with SB431542. * P<0.01, ** P<0.001 and *** P<0.0001. Scale bar: 100 μm. Data are presented the mean±standard deviation.
Identification of four different cell types in OSCC
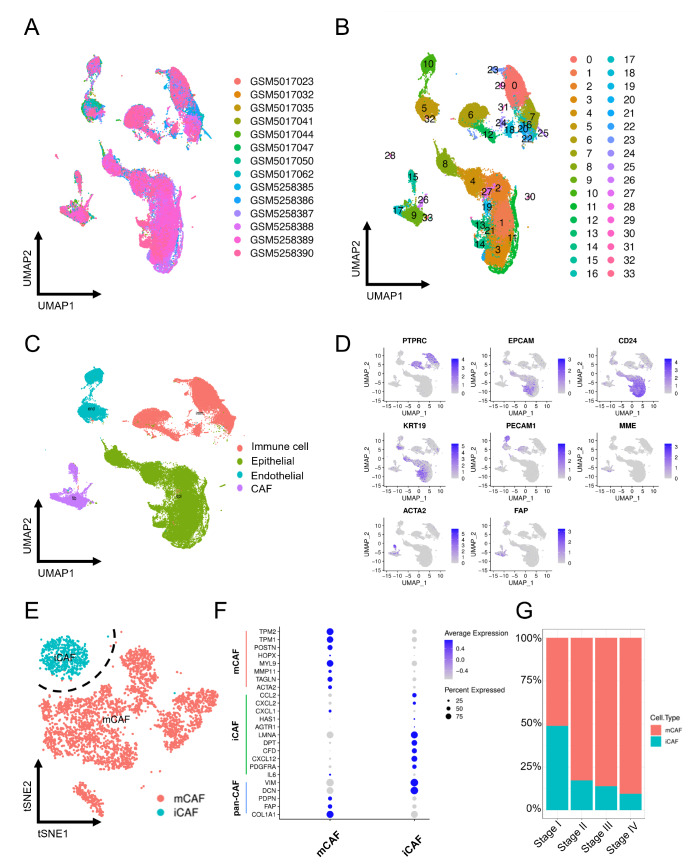
After quality control ( Supplementary Figure S1A,B) and removal of the batch effect between different samples ( Figure 2A), we divided 57,926 cells into 33 clusters ( Figure 2B) to comprehensively classify the cells present in OSCC using the downloaded scRNA-seq data GSE172577 and GSE164690 from the GEO database. Then, four major cell types ( Figure 2C) were identified with cluster-specific markers reported in prior research: PTPRC (immune cell), EPCAM, CD24, KRT19 (epithelial cell), PECAM1 (endothelial cell), MME, ACTA2, and FAP (CAF) ( Figure 2D and Supplementary Figure S1C,D). In accordance with previous studies, CAFs can be further classified into two distinct subtypes ( Figure 2E). One that highly expressed chemokines such as PDGFRA, CXCL12, CFD, DPT, LMNA, CXCL2 and CCL2 was identified as iCAFs, while the other was identified as mCAFs due to the high expression of ACTA2, TAGLN, MMP11, MYL9, POSTN, TPM1 and TPM2 ( Figure 2F and Supplementary Figure S1E). The bar graph revealed that the proportion of CAF subtypes varies among different stages. The mCAF/iCAF ratio improved significantly from stage I to stage IV, indicating that the alteration of the mCAF/iCAF ratio is potentially related to OSCC progression ( Figure 2G).
Figure 2 .
Single-cell analysis demonstrates two CAF subpopulations in OSCC
(A) UMAP plot of the single-cell profile colored by patient. (B) UMAP plot of the single-cell profile colored by cluster. (C) UMAP plot of the single-cell profile colored by major cell types. (D) The expression levels of marker genes were projected onto the UMAP atlas. (E) t-SNE plot of CAFs colored by cell type. (F) The bubble plot shows selected cell type-specific markers of iCAFs and mCAFs. The size of the dots represents the fraction of cells expressing a particular marker, and the intensity of the color indicates the level of mean expression. (G) Proportions of mCAFs and iCAFs in stage I–IV samples.
Functions of mCAFs and iCAFs
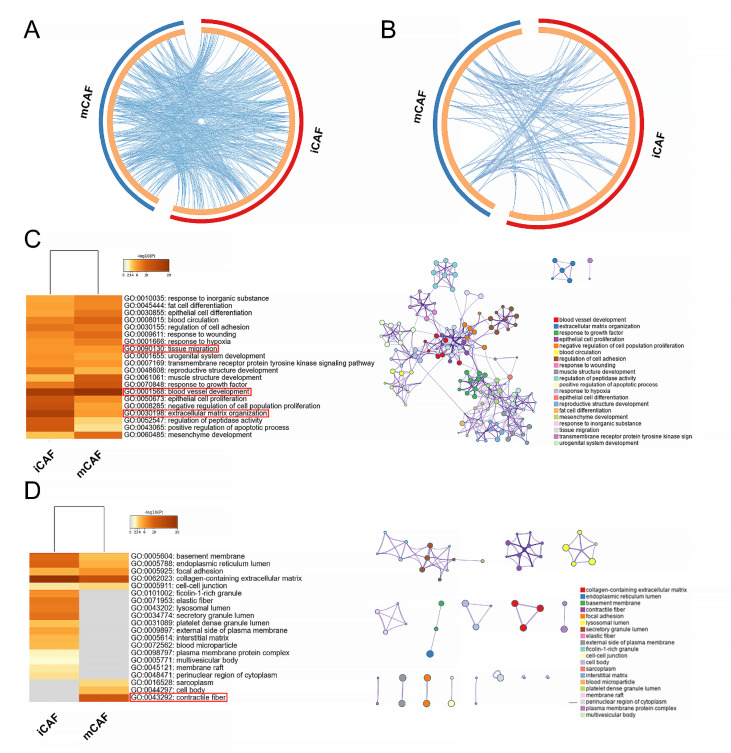
Based on the differentially upregulated genes of iCAFs and mCAFs, we used GO enrichment analysis to investigate the biological process ( Figure 3A,C) and cellular component ( Figure 3B,D) of each group. These genes were both enriched in two closely related biological processes: blood vessel development (GO: 0001568) and tissue migration (GO: 0090130) ( Figure 3C), suggesting the potential role of CAFs in promoting tumor migration. In addition, extracellular matrix (ECM) organization (GO: 0030198) was also significantly enriched in both subtypes ( Figure 3C), which was regulated by some enzymes. These enzymes can degrade ECM components to make space for cell growth and release various ECM-bound growth factors and cytokines, such as TGF-β, to further facilitate tumor growth and stimulate CAFs to synthesize ECM molecules. Such dynamic ECM alterations have a direct and ongoing impact on cancer metastasis [25]. However, contractile fiber (GO: 0043292), which can increase tissue stiffness, was specifically enriched in mCAFs ( Figure 3D). ECM stiffness can stretch the preexisting gaps within the basement membrane, reducing its integrity and thus providing a permissive basement membrane for cancer invasion [ 26‒ 28] . It also activates TGF-β signaling, which mediates epithelial to mesenchymal transition (EMT) in cancer cells [29]. Either stiffening or degradation is tumorigenic, and they are inextricably linked.
Figure 3 .
GO enrichment analysis of iCAFs and mCAFs
The Circos plot shows how differentially upregulated genes between two CAF subtypes overlap in GO biological process (A) and cellular component (B). On the outside, each arc represents the identity of each gene list. On the inside, each arc represents a gene list, where each gene has a spot on the arc. Blue lines link the different genes where they fall into the same ontology term, indicating the amount of functional overlap among the input gene lists. (C) GO biological process enrichment analysis of iCAFs and mCAFs with a network of representative terms shown on the right. (D) GO cellular component enrichment analysis of iCAFs and mCAFs with a network of representative terms shown on the right.
Conversion between epithelial cells and CAFs
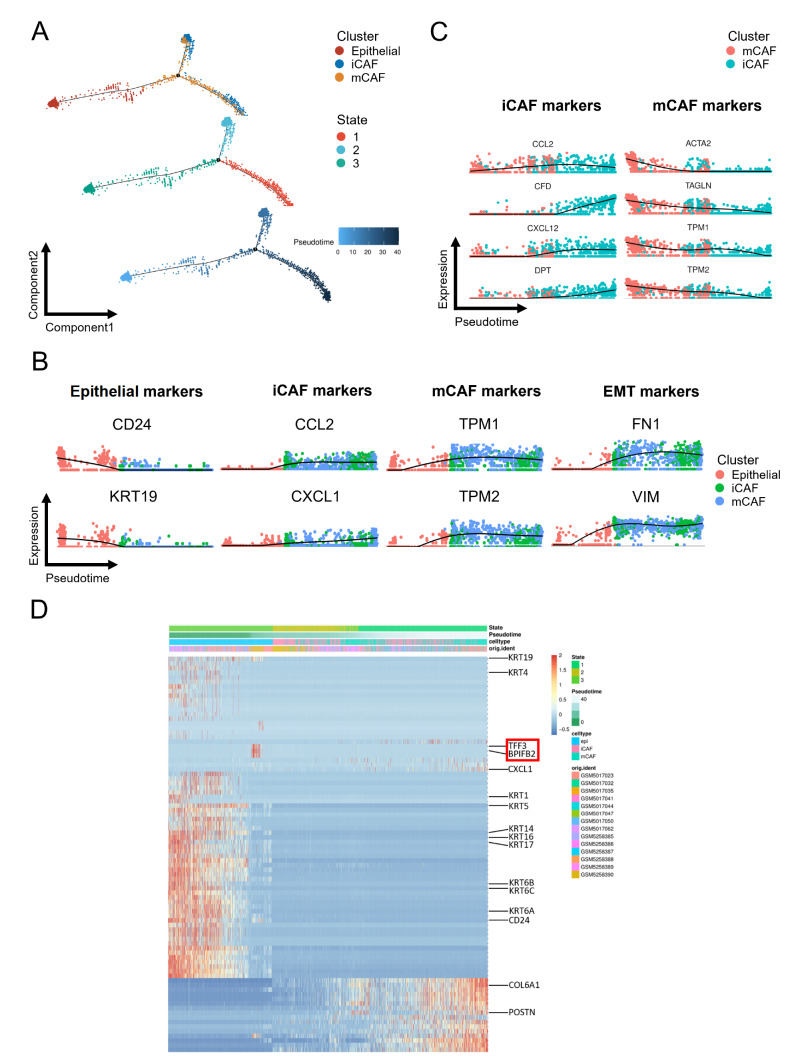
Based on the gene expression dynamics of epithelial cells and CAFs, we constructed a pseudotime developmental tree and determined three independent branches with epithelial cells, mCAFs and iCAFs scattered on it. State 3, which mainly represented epithelial cells, had the lowest pseudotime value and was located at the initial position of the developmental tree, indicating the potential conversion from epithelial cells to CAFs through EMT [30] ( Figure 4A). The gene expression pattern across pseudotime showed that the decrease in epithelial markers (CD24 and KRT19) was accompanied by an increase in CAF-related genes ( CCL2, CXCL1, TPM1 and TPM2). The expression of EMT-related genes ( FN1 and VIM) was also altered correspondingly ( Figure 4B). Furthermore, CAF-epithelial crosstalk analysis revealed that EMT was potentially induced by the PTN pathway ( Figure 5H), which was consistent with a previous study [31]. In CAFs, the expression curves of the representative genes showed that the decrease in mCAF-related genes ( ACTA2, TAGLN, TPM1, and TPM2) was accompanied by an increase in iCAF-related genes ( CCL2, CFD, CXCL12, and DPT), supporting the idea that CAF subtypes might interconvert [32] ( Figure 4C). The heatmap of genes with the most significant changes across pseudotime further supported the potential conversion from epithelial cells to CAFs through EMT, and their gene expression dynamics were consistent with the above analysis. Intriguingly, TFF3 and BPIFB2, which have been proven to mediate EMT in papillary thyroid cancer and gastric cancer, were specifically overexpressed when the cell type was transformed from epithelial to CAF [ 33, 34] ( Figure 4D).
Figure 4 .
Pseudotime analysis of epithelial, iCAF and mCAF populations
(A) Trajectory order of epithelial, iCAF and mCAF populations colored by cell type, state and pseudotime. (B) Relative expression alteration of marker genes for epithelial, iCAF, mCAF and EMT across pseudotime. (C) Relative expression alteration of marker genes for iCAF and mCAF populations across pseudotime. (D) Heatmap of differentially expressed genes ordered by state, pseudotime, cell type and patient.
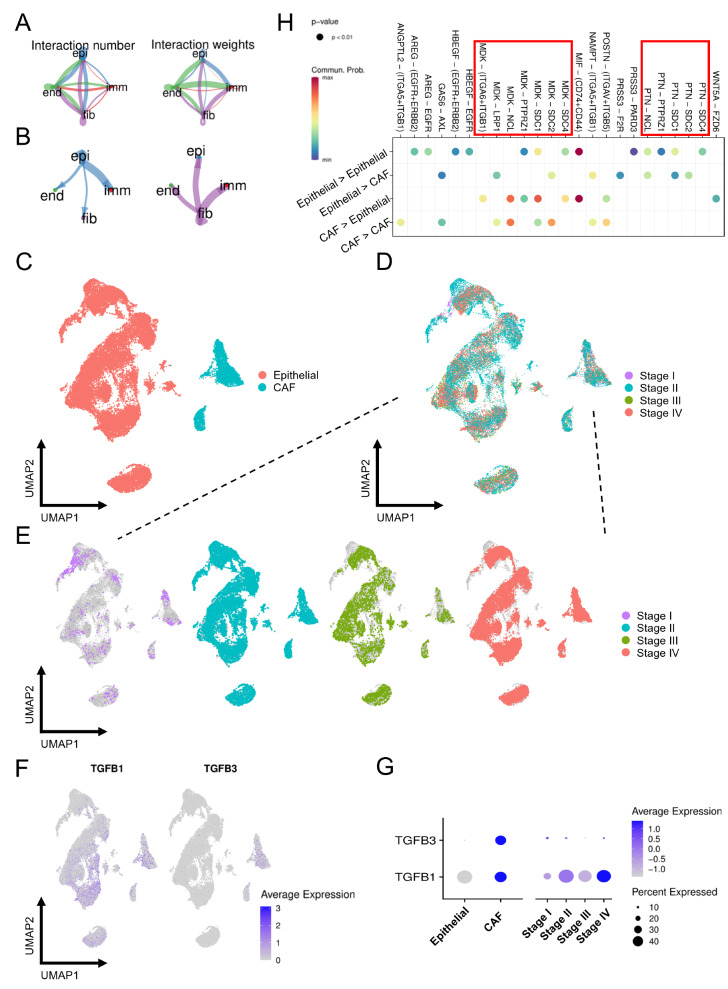
Figure 5 .
Identification of cell-cell interactions in OSCC
(A) Network diagram of the cell-cell interactions of different cells in OSCC. The color scale indicates the number of interactions and interaction weights. (B) The size of the circle represents the number of interactions with all other types of cells, and the thickness of the line represents the interaction number of cells between the lines. (C) UMAP plot of epithelial cells and CAFs colored by cell type. (D) UMAP plot of epithelial cells and CAFs colored by stage. (E) Epithelial cells and CAFs colored by four stages were projected onto the UMAP atlas separately. (F) The expression levels of TGF-β1 and TGF-β3 were projected onto the UMAP atlas. (G) Bubble plot shows the average and percent expression of TGF-β1 and TGF-β3. The size of the dots represents the fraction of cells expressing a particular marker, and the intensity of the color indicates the level of mean expression. (H) Bubble plot of the CAF-epithelial crosstalk pathway. epi: epithelial cell, imm: immune cell, fib: CAF, end: endothelial cell.
Identification of cell-cell interactions in OSCC
We discovered a close ligand/receptor-based relationship among different cell populations, and several biological functions were correlated with CAFs ( Figure 5A,B). To further elucidate the regulatory effect, we focused on CAF-epithelial crosstalk. We found that TGF-β1 and TGF-β3 were primarily expressed in CAFs ( Figure 5C,F,G) and were most abundant in stage IV samples ( Figure 5D‒G), implying that TGF-β is an important factor influencing tumor malignancy and that such function may be dominated by CAFs. In addition to TGF-β, various cytokines involved in CAF-epithelial crosstalk were plotted, and MDK was found to be one of the most important factors, taking part in several interactions ( Figure 5H). MDK, a growth factor that is overexpressed in a variety of human cancers [35], is involved in the acquisition of multiple cancer characteristics that promote tumor proliferation, transformation and EMT [ 36‒ 38] . In addition, it has been linked to chemoresistance [39].
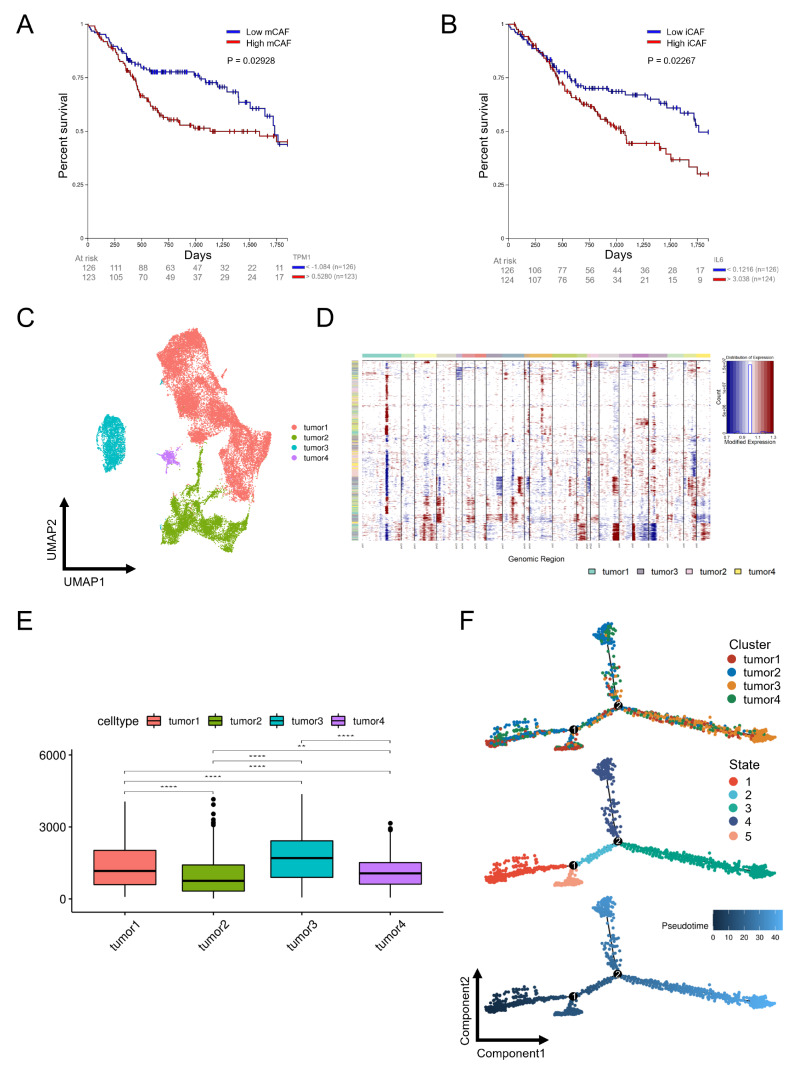
The relationship between OSCC malignancy and patient survival
TPM1 and IL-6 are specific markers of mCAFs ( Figure 6A) and iCAFs ( Figure 6B), respectively, and higher expression levels were significantly correlated with poor overall survival (OS) in OSCC. Next, we performed copy number variation (CNV) analysis to detect epithelial cells that underwent CNV in OSCC by comparing them to immune and endothelial cells. Then, large-scale CNV was identified in epithelial cells ( Supplementary Figure S2A), whose CNV scores were significantly higher than those of CAFs ( Supplementary Figure S2B). To further explore the relationship between tumor malignancy and OS, we divided epithelial cells into four distinct subclusters, namely, tumors 1‒4 ( Figure 6C), and frequent CNV events with different extents were confirmed ( Figure 6D), among which tumor 3 showed the highest CNV scores ( Figure 6E), indicating that tumor 3 was the most malignant component in the epithelial population. Furthermore, tumor 3 was located at the end position of the developmental tree, revealing the potential tumor progression from the low CNV group to the high CNV group in OSCC ( Figure 6F and Supplementary Figure S2C).
Figure 6 .
The relationship between OSCC malignancy and patient survival
Survival analysis of mCAFs (A) and iCAFs (B) in OSCC. (C) UMAP distribution of four tumor subclusters. (D) Large-scale CNVs of four tumor subclusters were identified. (E) CNV scores of four tumor subclusters. (F) Trajectory order of four tumor subclusters colored by cell type, state and pseudotime.
Discussion
Cancer initiation and progression are believed to be associated with the TME, which contains various cell types, including fibroblasts, immune cells, neoplastic epithelial cells, endothelial cells and pericytes [ 17, 40] . The crosstalk between cancer cells and matrix cells plays a critical role in tumor promotion and suppression. Multiple cytokines and factors, such as TGF-β, vascular endothelial growth factor A (VEGFA), hepatocyte growth factor (HGF), epidermal growth factor (EGF), tumor necrosis factor (TNF), interferon-γ (IFN-γ), C-X-C motif chemokine ligand 5 (CXCL5) and C-C motif chemokine ligand 5 (CCL5), act as mediators in crosstalk [17]. Fibroblasts are the most abundant component in the TME and can be activated to become CAFs. In many different coculture experiments, CAFs were proven to give rise to tumorigenesis compared with NFs [ 41, 42] . TGF-β signaling is one of the major pathways controlling cell and tissue proliferation, development, homeostasis and regeneration [ 43‒ 45] . A large number of studies have proven TGF-β to be associated with tumorigenesis, tumor growth, invasion and metastasis in various cancer types [ 41, 46‒ 48] and to be a promising target for cancer therapy [49]. Increasing evidence indicates that TGF-β plays a dual role not only in stimulating NFs to become CAFs but also in enhancing tumorigenesis and progression. Furthermore, activated CAFs tend to secrete more TGF-β to act back on tumor cells.
In our study, we revealed that TGF-β1 can be secreted by CAFs to promote OSCC invasion in vitro and investigated the effects of distinct CAF subtypes on OSCC. We believed that mCAFs are related to tumor adhesion, whereas iCAFs are linked to immunosuppression, and both contribute to OSCC progression. After we compared the mCAF/iCAF ratio from stage I to IV, we found that the ratio was increased with OSCC progression, implicating that mCAF is closely related to the malignant level of OSCC, which is consistent with the close correlation between ACTA2 (mCAF marker) overexpression and poor OS of OSCC reported in a previous study [50]. Moreover, mCAFs are associated with pathological features related to tumor aggressiveness, including the depth of invasion, lymphatic invasion and extranodal metastatic spread. Following GO analysis, we discovered that mCAFs could increase the matrix stiffness of the TME, which stimulated the transformation from normal epithelial cells to malignant cancer cells [51]. Furthermore, hard stiffness has the potential to breach the basement membrane barrier and loosen cancer cell adhesion, thereby promoting tumor migration [27]. In addition, the stiffness of the ECM and its degradation are inextricably linked, which occurs when the ECM is highly stiffened [25]. iCAFs have the potential to degrade the matrix and release growth factors, potentially promoting tumor progression [52]. Therefore, we speculate that the hardness of the matrix may influence CAF subcluster transition and is involved in tumor progression at different stages, and both stiffness and matrix degradation can promote tumor migration and invasion.
To investigate the relationship among different cells, we conducted pseudotime and cell-cell interaction analyses. Pseudotime analysis demonstrated that epithelial cells could transform into CAFs via EMT, which could be regulated by TFFS and BPIFB2. In addition, the potential interconversion between mCAFs and iCAFs was also explored. Further investigation in cytokines revealed that TGF-β is most abundant in CAFs and is closely related to tumor malignancy. Cell-cell interaction analysis revealed that CAFs can regulate epithelial cells via the MDK-mediated pathway, apart from the TGF-β pathway. In addition, our research also demonstrated the link between OSCC malignancy and patient survival. High mCAFs and iCAFs both predicted poor OS in OSCC. Epithelial cells can be further classified into four distinct subclusters with varying CNVs, and the high heterogeneity leads to potential tumor progression from the low CNV group to the high CNV group in OSCC.
In summary, our study demonstrated that CAFs can promote OSCC invasion through the activation of the TGF-β1/Smad2/3 signaling pathway, and different types of CAFs undergo complex conversion and play different regulatory roles in the TME, which further promotes OSCC progression.
Supplementary Data
Supplementary data is available at Acta Biochimica et Biophysica Sinica online.
Supporting information
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the Research and Develop Program, West China Hospital of Stomatology Sichuan University (No. LCYJ2019-1 to J.P.), the Interdisciplinary and Intercollege Research Project of the State Key Laboratory of Sichuan University (No. 2021KXK0403 to J.P.), the Sichuan University-Panzhihua City 2021 Campus Cooperation Special Fund Project (No. 2021CDPZH-7 to W.Y.), and the College Students’ Innovative Entrepreneurial Training Plan Program (No. 202210610194 to S.Z.).
References
- 1.Berndt A, Büttner R, Gühne S, Gleinig A, Richter P, Chen Y, Franz M, et al. Effects of activated fibroblasts on phenotype modulation, EGFR signalling and cell cycle regulation in OSCC cells. Exp Cell Res. . 2014;322:402–414. doi: 10.1016/j.yexcr.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 2.Fracalossi AC, Miranda SR, Oshima CT, Franco M, Ribeiro DA. The role of matrix metalloproteinases 2 and 9 during rat tongue carcinogenesis induced by 4-nitroquinoline 1-oxide. J Mol Hist. . 2010;41:19–25. doi: 10.1007/s10735-010-9258-6. [DOI] [PubMed] [Google Scholar]
- 3.Liu Z, Chen M, Zhao R, Huang Y, Liu F, Li B, Qin Y. CAF-induced placental growth factor facilitates neoangiogenesis in hepatocellular carcinoma. Acta Biochim Biophys Sin. . 2020;52:18–25. doi: 10.1093/abbs/gmz134. [DOI] [PubMed] [Google Scholar]
- 4.Öhlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, Corbo V, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. . 2017;214:579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.An J, Enomoto A, Weng L, Kato T, Iwakoshi A, Ushida K, Maeda K, et al. Significance of cancer-associated fibroblasts in the regulation of gene expression in the leading cells of invasive lung cancer. J Cancer Res Clin Oncol. . 2013;139:379–388. doi: 10.1007/s00432-012-1328-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ammirante M, Shalapour S, Kang Y, Jamieson CA, Karin M. Tissue injury and hypoxia promote malignant progression of prostate cancer by inducing CXCL13 expression in tumor myofibroblasts. Proc Natl Acad Sci USA. . 2014;111:14776–14781. doi: 10.1073/pnas.1416498111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akishima-Fukasawa Y, Ino Y, Nakanishi Y, Miura A, Moriya Y, Kondo T, Kanai Y, et al. Significance of PGP9.5 expression in cancer-associated fibroblasts for prognosis of colorectal carcinoma. Am J Clin Pathol. . 2010;134:71–79. doi: 10.1309/AJCPRJP39MIDSGBH. [DOI] [PubMed] [Google Scholar]
- 8.Al-Ansari MM, Aboussekhra A. Caffeine mediates sustained inactivation of breast cancer-associated myofibroblasts via up-regulation of tumor suppressor genes. PLoS ONE. . 2014;9:e90907. doi: 10.1371/journal.pone.0090907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curry JM, Sprandio J, Cognetti D, Luginbuhl A, Bar-ad V, Pribitkin E, Tuluc M. Tumor microenvironment in head and neck squamous cell carcinoma. Semin Oncol. . 2014;41:217–234. doi: 10.1053/j.seminoncol.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Kayamori K, Katsube KI, Sakamoto K, Ohyama Y, Hirai H, Yukimori A, Ohata Y, et al. NOTCH3 is induced in cancer-associated fibroblasts and promotes angiogenesis in oral squamous cell carcinoma. PLoS ONE. . 2016;11:e0154112. doi: 10.1371/journal.pone.0154112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klobukowska HJ, Munday JS. High numbers of stromal cancer-associated fibroblasts are associated with a shorter survival time in cats with oral squamous cell carcinoma. Vet Pathol. . 2016;53:1124–1130. doi: 10.1177/0300985816629713. [DOI] [PubMed] [Google Scholar]
- 12.Wu MH, Hong HC, Hong TM, Chiang WF, Jin YT, Chen YL. Targeting galectin-1 in carcinoma-associated fibroblasts inhibits oral squamous cell carcinoma metastasis by downregulating MCP-1/CCL2 expression. Clin Cancer Res. . 2011;17:1306–1316. doi: 10.1158/1078-0432.CCR-10-1824. [DOI] [PubMed] [Google Scholar]
- 13.Gu S, Feng XH. TGF-beta signaling in cancer. Acta Biochim Biophys Sin. . 2018;50:941–949. doi: 10.1093/abbs/gmy092. [DOI] [PubMed] [Google Scholar]
- 14.Yang L, Pang Y, Moses HL. TGF-β and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. . 2010;31:220–227. doi: 10.1016/j.it.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massagué J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. . 2000;103:295–309. doi: 10.1016/S0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 16.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature. . 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 17.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. . 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 18.Calon A, Tauriello DVF, Batlle E. TGF-beta in CAF-mediated tumor growth and metastasis. Semin Cancer Biol. . 2014;25:15–22. doi: 10.1016/j.semcancer.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Yang J, Lu Y, Lin YY, Zheng ZY, Fang JH, He S, Zhuang SM. Vascular mimicry formation is promoted by paracrine TGF-β and SDF1 of cancer-associated fibroblasts and inhibited by miR-101 in hepatocellular carcinoma. Cancer Lett. . 2016;383:18–27. doi: 10.1016/j.canlet.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 20.Cillo AR, Kürten CHL, Tabib T, Qi Z, Onkar S, Wang T, Liu A, et al. Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity. . 2020;52:183–199.e9. doi: 10.1016/j.immuni.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi Z, Barrett T, Parikh AS, Tirosh I, Puram SV. Single-cell sequencing and its applications in head and neck cancer. Oral Oncol. . 2019;99:104441. doi: 10.1016/j.oraloncology.2019.104441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng Y, Xiao L, Rong H, Ou Z, Cai T, Liu N, Li B, et al. Single-cell profiling of tumor-infiltrating TCF1/TCF7+ T cells reveals a T lymphocyte subset associated with tertiary lymphoid structures/organs and a superior prognosis in oral cancer. Oral Oncol. . 2021;119:105348. doi: 10.1016/j.oraloncology.2021.105348. [DOI] [PubMed] [Google Scholar]
- 23.Chen Z, Zhou L, Liu L, Hou Y, Xiong M, Yang Y, Hu J, et al. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat Commun. . 2020;11:5077. doi: 10.1038/s41467-020-18916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. . 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. . 2014;15:1243–1253. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, Harrington K, et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol. . 2013;15:637–646. doi: 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torok NJ. P300, a new player in mechanosensitivity and activation of cancer-associated fibroblasts. Gastroenterology. . 2018;154:2025–2026. doi: 10.1053/j.gastro.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itoh G, Chida S, Yanagihara K, Yashiro M, Aiba N, Tanaka M. Cancer-associated fibroblasts induce cancer cell apoptosis that regulates invasion mode of tumours. Oncogene. . 2017;36:4434–4444. doi: 10.1038/onc.2017.49. [DOI] [PubMed] [Google Scholar]
- 29.Bourboulia D, Stetler-Stevenson WG. Matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs): positive and negative regulators in tumor cell adhesion. Semin Cancer Biol. . 2010;20:161–168. doi: 10.1016/j.semcancer.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shelton M, Anene CA, Nsengimana J, Roberts W, Newton-Bishop J, Boyne JR. The role of CAF derived exosomal microRNAs in the tumour microenvironment of melanoma. Biochim Biophys Acta (BBA) - Rev Cancer. . 2021;1875:188456. doi: 10.1016/j.bbcan.2020.188456. [DOI] [PubMed] [Google Scholar]
- 31.Perez-Pinera P, Alcantara S, Dimitrov T, Vega JA, Deuel TF. Pleiotrophin disrupts calcium-dependent homophilic cell–cell adhesion and initiates an epithelial–mesenchymal transition. Proc Natl Acad Sci USA. . 2006;103:17795–17800. doi: 10.1073/pnas.0607299103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avery D, Govindaraju P, Jacob M, Todd L, Monslow J, Puré E. Extracellular matrix directs phenotypic heterogeneity of activated fibroblasts. Matrix Biol. . 2018;67:90–106. doi: 10.1016/j.matbio.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin X, Zhang H, Dai J, Zhang W, Zhang J, Xue G, Wu J. TFF3 contributes to epithelial-mesenchymal transition (EMT) in papillary thyroid carcinoma cells via the mapk/erk signaling pathway. J Cancer. . 2018;9:4430–4439. doi: 10.7150/jca.24361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren Q, Zhu P, Zhang H, Ye T, Liu D, Gong Z, Xia X. Identification and validation of stromal-tumor microenvironment-based subtypes tightly associated with PD-1/PD-L1 immunotherapy and outcomes in patients with gastric cancer. Cancer Cell Int. . 2020;20:92. doi: 10.1186/s12935-020-01173-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filippou PS, Karagiannis GS, Constantinidou A. Midkine (MDK) growth factor: a key player in cancer progression and a promising therapeutic target. Oncogene. . 2020;39:2040–2054. doi: 10.1038/s41388-019-1124-8. [DOI] [PubMed] [Google Scholar]
- 36.Muramatsu T. Structure and function of midkine as the basis of its pharmacological effects. Br J Pharmacol. . 2014;171:814–826. doi: 10.1111/bph.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadomatsu K, Kishida S, Tsubota S. The heparin-binding growth factor midkine: the biological activities and candidate receptors. J Biochem. . 2013;153:511–521. doi: 10.1093/jb/mvt035. [DOI] [PubMed] [Google Scholar]
- 38.Erguven M, Bilir A, Yazihan N, Ermis E, Sabanci A, Aktas E, Aras Y, et al. Decreased therapeutic effects of noscapine combined with imatinib mesylate on human glioblastoma in vitro and the effect of midkine. Cancer Cell Int. . 2011;11:18. doi: 10.1186/1475-2867-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Y, Yan B, Guo H, Qiu L, Sun X, Wang X, Shi Q, et al. Effect of midkine on gemcitabine resistance in biliary tract cancer. Int J Mol Med. . 2018 doi: 10.3892/ijmm.2018.3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen Z, Qin X, Yan M, Li R, Chen G, Zhang J, Chen W. Cancer-associated fibroblasts promote cancer cell growth through a miR-7-RASSF2-PAR-4 axis in the tumor microenvironment. Oncotarget. . 2017;8:1290–1303. doi: 10.18632/oncotarget.13609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. . 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 42.Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Cunha GR, Hein P, Tlsty TD. Carcinoma-associated fibroblasts stimulate tumor progression of initiated human epithelium. Breast Cancer Res. . 2000;2:S.19. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sulaiman W, Nguyen DH. Transforming growth factor beta 1, a cytokine with regenerative functions. Neural Regen Res. . 2016;11:1549–1552. doi: 10.4103/1673-5374.193223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moses HL, Coffey RJ, Leof EB, Lyons RM, Keski-Oja J. Transforming growth factor beta regulation of cell proliferation. J Cell Physiol. . 1987;133:1–7. doi: 10.1002/jcp.1041330403. [DOI] [PubMed] [Google Scholar]
- 45.Yang G, Zhou J, Teng Y, Xie J, Lin J, Guo X, Gao Y, et al. Mesenchymal TGF-β signaling orchestrates dental epithelial stem cell homeostasis through Wnt signaling. Stem Cells. . 2014;32:2939–2948. doi: 10.1002/stem.1772. [DOI] [PubMed] [Google Scholar]
- 46.Wei S, Li Q, Li Z, Wang L, Zhang L, Xu Z. miR-424-5p promotes proliferation of gastric cancer by targeting Smad3 through TGF-β signaling pathway. Oncotarget. . 2016;7:75185–75196. doi: 10.18632/oncotarget.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou FF, Drabsch Y, Dekker TJA, de Vinuesa AG, Li Y, Hawinkels LJA, Sheppard KA, et al. Nuclear receptor NR4A1 promotes breast cancer invasion and metastasis by activating TGF-β signalling. Nat Commun. . 2014;5:3388. doi: 10.1038/ncomms4388. [DOI] [PubMed] [Google Scholar]
- 48.Xue J, Lin X, Chiu WT, Chen YH, Yu G, Liu M, Feng XH, et al. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-β–dependent cancer metastasis. J Clin Invest. . 2014;124:564–579. doi: 10.1172/JCI71104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang S, Jin K, Li T, Zhou M, Yang W. Comprehensive analysis of INHBA: a biomarker for anti-TGFβ treatment in head and neck cancer. Exp Biol Med (Maywood) . 2022;247:1317–1329. doi: 10.1177/15353702221085203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bienkowska KJ, Hanley CJ, Thomas GJ. Cancer-associated fibroblasts in oral cancer: a current perspective on function and potential for therapeutic targeting. Front Oral Health. . 2021;2:686337. doi: 10.3389/froh.2021.686337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katara GK, Kulshrestha A, Mao L, Wang X, Sahoo M, Ibrahim S, Pamarthy S, et al. Mammary epithelium-specific inactivation of V-ATPase reduces stiffness of extracellular matrix and enhances metastasis of breast cancer. Mol Oncol. . 2018;12:208–223. doi: 10.1002/1878-0261.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol. . 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.