Abstract
Transient intracellular expression of ExoT in CHO cells stimulated cell rounding and actin reorganization. Biochemical studies showed that ExoT was a GTPase-activating protein for RhoA, Rac1, and Cdc42. Together, these data show that ExoT interferes with Rho signal transduction pathways, which regulate actin organization, exocytosis, cell cycle progression, and phagocytosis.
Pseudomonas aeruginosa is a gram-negative opportunistic pathogen affecting patients with cystic fibrosis, AIDS, severe burns, neutropenia, and ocular infections. Recent studies have shown that P. aeruginosa possesses a type-III system to deliver cytotoxins directly into eukaryotic cells (3). To date, four type-III cytotoxins have been identified: ExoS, ExoT, ExoY, and ExoU.
The enzymatic activity of exoenzyme S was purified from the culture supernatant of P. aeruginosa 388 as an aggregate composed of two proteins with apparent molecular masses of 49 kDa (ExoS) and 53 kDa (ExoT). Subsequent studies showed ExoS (8) and ExoT (14) were encoded by separate genes and had 76% primary amino acid homology. ExoS is a bifunctional cytotoxin that encodes two independent catalytic activities. The carboxyl terminus encodes a cytotoxic FAS (factor activating exoenzyme S)-dependent ADP-ribosyltransferase activity (6) while the amino terminus stimulates the intrinsic GTPase activity of Rho GTPases (4). ExoT also catalyzes a FAS-dependent ADP ribosylation of eukaryotic proteins, but at a velocity that is only 0.2% of that of ExoS (9). Recently, Vallis et al. showed that delivery of ExoT by P. aeruginosa stimulates CHO cells to round (13).
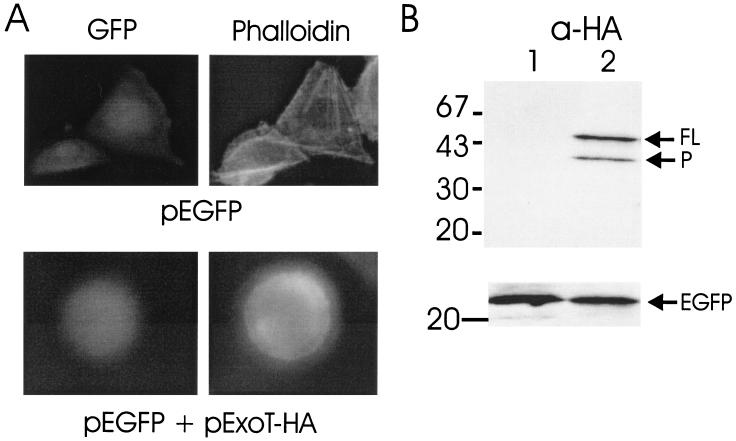
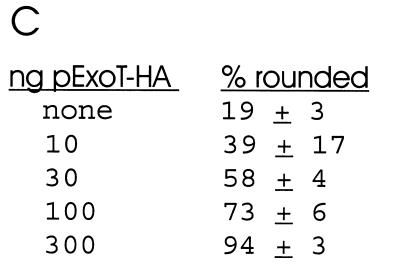
A transient transfection system was used to directly measure the effect of ExoT on eukaryotic cell physiology (10). Hemagglutinin (HA) epitope-tagged ExoT (ExoT-HA) was engineered to encode the entire open reading frame of ExoT fused at its carboxyl terminus to the amino acid sequence of the hemagglutinin epitope (11). ExoT-HA was subcloned into pEGFP-N1 and expressed under the control of a cytomegalovirus promoter. Chinese Hamster Ovary (CHO) K1 cells were seeded at 5 × 105 cells/dish in 100-mm dishes and 5 × 104 cells/dish in 12-well dishes the day before transfection. Cotransfections were performed with Lipofectamine-PLUS (Gibco-BRL) using 1 μg (100-mm dishes) or 50 ng (12-well dishes) of reporter plasmid (pEGFP-N1) and the indicated amount of effector DNA (pExoT-HA). Total DNA transfected was normalized with pCMV-10luc. Twenty-four hours posttransfection, pExoT-HA-transfected cells showed a rounded morphology that was dose responsive (Fig. 1). To associate the rounding phenotype with ExoT expression, Western blot analyses of cell lysates from pExoT-HA-transfected CHO cells were performed by enhanced chemiluminescence (ECL), using the rabbit anti-HA immunoglobulin (IgG) or mouse anti-green fluorescent protein (anti-GFP) IgG (1/1000 dilution; BABCO) as the primary antibody and the appropriate horseradish peroxidase-conjugated IgG (1:40,000; Pierce) (11). Lysates from pExoT-HA-transfected CHO cells possessed two bands indicating reactivity to HA with apparent molecular masses of 53 and 39 kDa (Fig. 1). The 53-kDa band migrated at the predicted molecular weight as full-length ExoT-HA, while the presence of the 39-kDa band suggested that ExoT-HA had been processed at its amino terminus, since the HA epitope was fused to the carboxyl terminus of ExoT. Coexpression of ExoT did not inhibit expression of the reporter protein, EGFP, indicating that ExoT was not cytotoxic to CHO cells.
FIG. 1.
Transfection of CHO cells with pExoT-HA. (A) CHO cells were cotransfected with 50 ng of pEGFP alone or with 300 ng of pExoT-HA. Twenty-four hours posttransfection, cells were fixed, stained with phalloidin-rhodamine, and visualized by fluorescence microscopy for GFP or rhodamine (phalloidin). (B) Lysates from CHO cells transfected with pEGFP (lane 1) or pEGFP and pExoT-HA (lane 2) were subjected to ECL Western blot analysis, using rabbit anti-HA IgG (upper section) or mouse anti-GFP IgG (lower section) as the primary antibody and HRP-conjugated goat anti-rabbit or mouse IgG as the secondary antibody. (C) CHO cells were transfected with the indicated amount of pExoT-HA and 50 ng of reporter plasmid (pEGFP) and, 24 h posttransfection, were scored for morphology. The percentages of rounded cells shown are averages from analysis of three fields of transfected cells (approximately 50 transfected cells per field).
The ability of ExoT-HA to stimulate the reorganization of actin in CHO cells was also determined (11). CHO cells were transfected with pEGFP (control) or pEGFP and pExoT-HA. Twenty-four hours posttransfection CHO cells were fixed with 1% paraformaldehyde in phosphate-buffered saline (PBS), permeabilized with 4% paraformaldehyde containing 0.1% Triton X-100 in H2O, and incubated with 0.2 μg of phalloidin-rhodamine conjugate (Sigma) per ml in PBS to stain the actin cytoskeleton. Fluorescence microscopy showed that control-transfected cells had typical cellular morphology with a well-defined actin cytoskeleton, while pExoT-HA transfected cells were rounded and did not have a defined actin cytoskeleton or stress fibers (Fig. 1). Thus, ExoT stimulates the reorganization of actin.
Other studies in our laboratory localized the GTPase-activating protein (GAP) domain of ExoS within residues 78 to 234 and showed that a deletion peptide comprising this region could be expressed in Escherichia coli as a soluble enzymatically active His(6) fusion protein (K. J. Pederson and J. T. Barbieri, unpublished data). Thus, a recombinant deletion peptide, which comprised residues 78 to 237 of ExoT, was engineered, subcloned into pET15b (pExoT78-237), and sequenced to confirm the absence of secondary mutations. ExoT(78-237) was expressed in E. coli BL21(DE3) as a His(6) fusion protein and purified by Ni-affinity chromatography as previously described (6). Purified ExoT(78-237) was dialyzed in 25 mM Tris, pH 7.6, plus 40% glycerol and stored at −20°C. ExoT(78-237) was determined to have an apparent molecular mass of 22 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis and purity of about 85%. ExoT(78-237) possessed limited immunoreactivity to a polyclonal antibody that was prepared against ExoS (7).
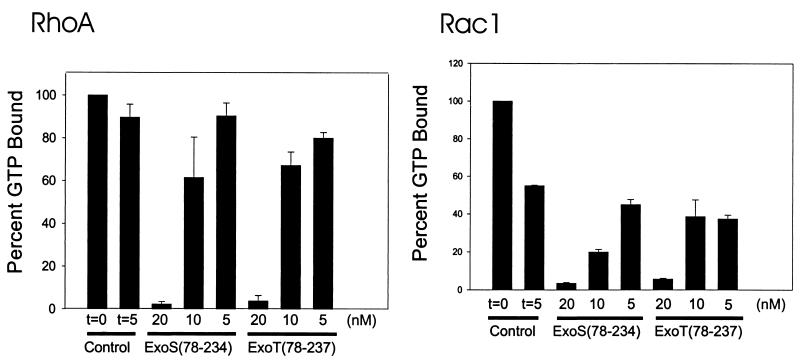
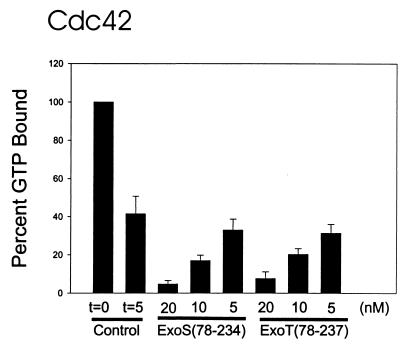
The GAP activity of ExoT(78-237) was determined with a filter-binding assay (4). Rho GTPases were loaded with 10 μM [γ-32P]GTP. Intrinsic GTPase activity was initiated with the addition 3 mM MgCl2 and 1 mM GTP. After 5 min, the reaction mixtures were spotted onto a nitrocellulose filters, the filters were washed, and the remaining radioactivity was measured by scintillation counting. Stimulation of GTP hydrolysis by ExoT(78-237) or ExoS(78-234) was measured, using a concentration of ExoT or ExoS that stimulated the hydrolysis of less than 25% of the available Rho-GTP. Under these conditions the rate of stimulation of GTP hydrolysis by ExoS was linear for 5 min (4). The intrinsic rates of GTPase activities (in picomoles of GTP per minute per microgram of protein) were greatest for recombinant glutathione S-transferase–RhoA, followed by Rac1 and Cdc42 (data not shown). ExoT(78-237) stimulated the intrinsic GTPase activity of RhoA in a dose-dependent manner and at a rate similar to that stimulated by ExoS(78-234) (Fig. 2). Figure 2 also shows that ExoT(78-237) stimulated the GTPase activity of Rac1 and Cdc42.
FIG. 2.
ExoS and ExoT modulation of intrinsic activity of Rho GTPases. The intrinsic GAP activities of RhoA, Rac1, and Cdc42 were determined by filter-binding assay as described in the text. Nucleotide-free Rho GTPases were loaded with [γ-32P]GTP for 5 min at 37°C. The amount of GTP bound initially (t = 0) was determined, and intrinsic GTPase activity was initiated with the addition of MgCl2. After 5 min, the amount of GTP remaining bound to the Rho GTPase was determined, and the results are presented here as percentages of the amount of GTP bound initially (100%). The amount of GTP bound in the presence of the indicated amount of ExoS(78-234) or ExoT(78-237) was also determined after the addition of MgCl2. After 5 min, the amount of GTP remaining bound to the Rho GTPase was determined, and the results are presented here as percentages of the amount of GTP bound initially (100%). Typical GTP loading of RhoA was 0.6 to 1.0 mol of GTP bound per mole of RhoA. The data are means + standard deviations (error bars).
Rho GTPases regulate numerous eukaryotic cellular functions, including actin cytoskeleton reorganization (5), exocytosis (1), transcription (12), and phagocytosis (2). Like other members of the family of Ras monomeric G proteins, Rho GTPase modulate cell physiology through their ability to cycle between an inactive GDP bound form and an active GTP bound form that is able to interact with downstream effectors. The Rho GTPases possess low intrinsic capacity to perform guanine nucleotide exchange or to catalyze the hydrolysis of GTP to GDP. These two activities are modulated by two types of eukaryotic protein, guanine exchange factors and GAPs. The ability of ExoT to express Rho GAP activity identifies it as a biochemical activity, which may contribute to the pathogenic potential of P. aeruginosa.
Acknowledgments
J.T.B. was supported by AI30162 from the NIH-NIAID. K.A. was supported by the DFG.
We thank Kristin J. Pederson for technical assistance during the study.
ADDENDUM IN PROOF
We acknowledge that Joanne Engel and coworkers have also observed that ExoT is a RhoGAP (B. I. Kazmierczak, T. S. Jou, K. Mostov, and J. N. Engel, Cell. Microbiol., in press).
REFERENCES
- 1.Adamo J E, Rossi G, Brennwald P. The Rho GTPase Rho3 has a direct role in exocytosis that is distinct from its role in actin polarity. Mol Biol Cell. 1999;10:4121–4133. doi: 10.1091/mbc.10.12.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caron E, Hall A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science. 1998;282:1717–1721. doi: 10.1126/science.282.5394.1717. [DOI] [PubMed] [Google Scholar]
- 3.Frank D W. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol. 1997;26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 4.Goehring U-M, Schmidt G, Pederson K J, Aktories K, Barbieri J T. The N-terminal domain of Pseudomonas aeruginosa is a GTPase-activating protein for Rho GTPases. J Biol Chem. 1999;274:36369–36372. doi: 10.1074/jbc.274.51.36369. [DOI] [PubMed] [Google Scholar]
- 5.Hall A. Ras-related GTPases and the cytoskeleton. Mol Biol Cell. 1992;3:475–479. doi: 10.1091/mbc.3.5.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knight D A, Finck-Barbancon V, Kulich S M, Barbieri J T. Functional domains of Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1995;63:3182–3186. doi: 10.1128/iai.63.8.3182-3186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulich S M, Frank D W, Barbieri J T. Purification and characterization of exoenzyme S from Pseudomonas aeruginosa 388. Infect Immun. 1993;61:307–313. doi: 10.1128/iai.61.1.307-313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulich S M, Yahr T L, Mende-Mueller L M, Barbieri J T, Frank D W. Cloning the structural gene for the 49-kDa form of exoenzyme S (exoS) from Pseudomonas aeruginosa strain 388. J Biol Chem. 1994;269:10431–10437. [PubMed] [Google Scholar]
- 9.Liu S, Yahr T L, Frank D W, Barbieri J T. Biochemical relationships between the 53-kilodalton (Exo53) and 49-kilodalton (ExoS) forms of exoenzyme S of Pseudomonas aeruginosa. J Bacteriol. 1997;179:1609–1613. doi: 10.1128/jb.179.5.1609-1613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pederson K J, Barbieri J T. Intracellular expression of the ADP-ribosyltransferase domain of ExoS is cytotoxic for eukaryotic cells. Mol Microbiol. 1998;30:751–760. doi: 10.1046/j.1365-2958.1998.01106.x. [DOI] [PubMed] [Google Scholar]
- 11.Pederson K J, Vallis A J, Aktories K, Frank D W, Barbieri J T. The amino-terminal domain of Pseudomonas aeruginosa ExoS disrupts actin filaments via small-molecular-weight GTP-binding proteins. Mol Microbiol. 1999;32:393–401. doi: 10.1046/j.1365-2958.1999.01359.x. [DOI] [PubMed] [Google Scholar]
- 12.Philips A, Roux P, Coulon V, Bellanger J M, Vie A, Vignais M L, Blanchard J M. Differential effect of Rac and Cdc42 on p38 kinase activity and cell cycle progression of nonadherent primary mouse fibroblasts. J Biol Chem. 2000;275:5911–5917. doi: 10.1074/jbc.275.8.5911. [DOI] [PubMed] [Google Scholar]
- 13.Vallis A J, Finck-Barbancon V, Yahr T L, Frank D W. Biological effects of Pseudomonas aeruginosa type III-secreted proteins on CHO cells. Infect Immun. 1999;67:2040–2044. doi: 10.1128/iai.67.4.2040-2044.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yahr T L, Barbieri J T, Frank D W. Genetic relationship between the 53- and 49-kilodalton forms of exoenzyme S from Pseudomonas aeruginosa. J Bacteriol. 1996;178:1412–1419. doi: 10.1128/jb.178.5.1412-1419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]