Abstract
Hypertrophic scar is a problem for numerous patients, especially after burns, and is characterized by increased fibroblast proliferation and collagen deposition. Increasing evidence demonstrates that lncRNAs contribute to the development and progression of various diseases. However, the function of lncRNAs in hypertrophic scar formation remains poorly characterized. In this study, a novel fibroblast proliferation-associated lncRNA, named lncRNA FPASL (MSTRG.389905.1), which is mainly localized in the cytoplasm, is found to be downregulated in hypertrophic scar, as detected by lncRNA microarray and qRT-PCR. The full-length FPASL is characterized and further investigation confirms that it has no protein-coding potential. FPASL knockdown in fibroblasts triggers fibroblast proliferation, whereas overexpression of FPASL directly attenuates the proliferation of fibroblasts. Furthermore, target genes of the differentially expressed lncRNAs in hypertrophic scars and the matched adjacent normal tissues are enriched in fibroblast proliferation signaling pathways, including the PI3K/AKT and MAPK signaling pathways, as determined by GO annotation and KEGG enrichment analysis. We also demonstrate that knockdown of FPASL activates the PI3K/AKT and MAPK signaling pathways, and specific inhibitors of the PI3K/AKT and MAPK signaling pathways can reverse the proliferation of fibroblasts promoted by FPASL knockdown. Our findings contribute to a better understanding of the role of lncRNAs in hypertrophic scar and suggest that FPASL may act as a potential novel therapeutic target for hypertrophic scar.
Keywords: fibroblast, hypertrophic scar, lncRNA FPASL, MAPK, PI3K/AKT
Introduction
A hypertrophic scar occurs after the damage of deep dermis by burns or trauma [1]. These hypertrophic scars can cause symptoms such as itching, pigmentation and burning. In severe cases, they can lead to limited mobility due to lack of elasticity or contracture, which results in psychological and cosmetic disturbance to the patients [ 2, 3] . Although many treatment options for hypertrophic scar are available [ 4, 5] , unfortunately, these treatments fail to effectively improve the overall prognosis of hypertrophic scar patients, which is mainly caused by the complex pathogenesis of hypertrophic scar. Thus, it is important to identify the key molecules that promote or inhibit hypertrophic scar formation. Hypertrophic scar is caused by the excessive accumulation of extracellular matrix (ECM) components partly owing to abnormal fibroblast proliferation and differentiation [6]. Growing evidence indicates that proliferative and activated fibroblasts contribute to the formation and development of hypertrophic scar [ 7, 8] . However, the defined molecular changes in fibroblasts from hypertrophic scar tissues remain to be determined.
Among the ncRNAs, long noncoding RNAs (lncRNAs) comprise a heterogeneous family of RNA molecules longer than 200 nucleotides with no or limited protein-coding potential [9]. Similar to miRNAs [10], lncRNAs are a significant class of RNA transcripts that regulate gene expression and signal transduction [ 11, 12] . Clarifying the role and mechanisms of lncRNAs in various diseases may deepen our understanding of lncRNAs. Compelling evidence has suggested that lncRNAs play crucial roles in a variety of cellular and disease processes, including cell proliferation [ 13, 14] , differentiation [15], autophagy [ 16, 17] and apoptosis [ 18, 19] . Some specific lncRNAs are correlated with skin disorders, including melanoma [20], systemic lupus erythematosus [21], keloid [22], and hypertrophic scar [23]. For example, lncRNA PAPPA-AS1 can induce the development of hypertrophic scar through TLR4 [24]. However, the current studies on the regulation of hypertrophic scar by lncRNAs are rather limited. LncRNAs that have potential regulatory roles in hypertrophic scar remain to be discovered.
In the present study, we screened the expression profiles of lncRNAs from normal tissues and hypertrophic scar tissues, and a novel lncRNA, FPASL, was identified and confirmed to be significantly decreased in hypertrophic scar. We first found that lncRNA FPASL is associated with fibroblast proliferation. Further studies demonstrated that the PI3K/AKT and MAPK signaling pathways are activated in hypertrophic scar. We preliminary demonstrated that FPASL might regulate fibroblast proliferation through the PI3K/AKT and MAPK signaling pathways.
Materials and Methods
Ethics statement
This study was approved by the Medical Ethics Committee of the Affiliated Hospital of Ningxia Medical University (No. 2018-087). Patients admitted to our hospital for scar removal were provided with information about the purpose of the study, and written informed consent was obtained from each participant.
Tissue samples
Hypertrophic scar tissues and their corresponding adjacent normal tissues were obtained from eighteen different patients who were admitted to the Affiliated Hospital of Ningxia Medical University for scar removal. The diagnosis of hypertrophic scar was confirmed by routine pathological examination.
Cell isolation and culture
Hypertrophic scar tissues and their corresponding adjacent normal tissues were taken from five patients who had not been treated for burns before surgery. The samples were pruned to remove excess adipose tissue and rinsed three times with phosphate buffer saline (PBS) containing gentamicin (Biotopped, Beijing, China) and penicillin-streptomycin solution (Solarbio, Beijing, China), and then incubated in DPBS (HyClone, Logan, USA) containing 0.125% Trypsin-EDTA solution (Solarbio) at 4°C overnight. The next day, the solution was removed and the tissues were incubated in DMEM-F12 medium (HyClone) containing 0.1% collagenase type I (Solarbio) at 37°C for 6–8 h, and filtered through a 200-mesh screen. The isolated cells were cultured in DMEM-F12 medium containing 10% fetal calf serum (Biological Industries, Beit-Haemek, Israel) and 1% penicillin-streptomycin solution at 37°C in an atmosphere of 5% CO 2. The cell strains were maintained and analyzed from passage 3 to passage 6.
Microarray-based lncRNA profiling
Total RNA was extracted from 5 paired hypertrophic scar tissues and adjacent normal tissues using Trizol reagent (TIANGEN, Beijing, China). Microarray hybridization was performed using the microarray provided by Beijing Biomarker Technologies Corporation (Beijing, China) according to the manufacturer′s standard protocols. The acquired raw data and array images were extracted with Agilent Feature Extraction software (version 11; Agilent, Santa Clara, USA). GeneSpring GX v12.0 (Agilent) was used for RNA quantification and subsequent data processing. The differentially expressed LncRNAs were identified through filtering on fold change (≥ 2) and P value ( P<0.05).
Gene Ontology (GO) and KEGG pathway analysis
To better understand the biological functions and potential mechanisms of ncRNAs and mRNAs in the mechanism of lncRNA acting on hypertrophic scarHS, we performed GO enrichment and KEGG pathway analyses on predicted target genes of differentially expressed lncRNAs (DElncRNAs). GO enrichment analysis of the differentially expressed genes (DEGs) was implemented using the clusterProfiler R packages. Enrichment analysis uses hypergeometric testing to find GO entries that are significantly enriched compared to the entire genome background. Briefly, GO analysis (www. geneontology.org) consists of three components: biological process (BP), cellular component (CC), and molecular function (MF). KEGG is a database resource for understanding the high-level functions and utilities of biological systems, such as cells, organisms and ecosystems, from molecular level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies ( http://www.genome.jp/kegg/). ClusterProfiler R packages were used to find KEGG pathways that are significantly enriched compared to the entire genome background.
Fluorescence in situ hybridization (FISH)
LncRNA FISH Probe Mix (RiboBio, Guangzhou, China) was used to perform FISH assay. Briefly, fibroblasts were fixed with 4% paraformaldehyde (Solarbio) and treated with protease K, glycine, and acetylation reagents. After pre-hybridization at 37°C for 30 min, fibroblasts were hybridized with Cy3-labelled FPASL probes at 37°C overnight. The nucleus was stained with DAPI. The 18S and U6 were used as internal controls, as the 18S was distributed in the cytoplasm and U6 in the nucleus.
RNA isolation from nuclear and cytoplasmic fractions
The Ambion® PARIS™ system (Thermo Scientific, Waltham, USA) was used for the isolation of RNA from the nucleus and cytoplasm of fibroblasts. Cultured cells were first homogenized in ice-cold cell disruption buffer to prepare a total cell lysate. Since the homogenization was performed quickly on ice and in the presence of detergent, both protein and RNA were purified directly from this lysate. For RNA isolation, a part of the total cell lysate was immediately mixed with an equal volume of lysis/binding solution. Total RNA was then purified from the mixture using an RNA binding glass fiber filter. After three rapid washing steps, high-quality RNA was eluted in a concentrated form. Then, total RNAs from the nucleus and cytoplasm were used for qRT-PCR.
Quantitative reverse transcription PCR
Total RNA was extracted from human scar tissues and scar-derived fibroblasts using Trizol reagent (TIANGEN). Isolated RNA was reverse transcribed to cDNA using PrimeScript RT Master Mix (TaKaRa, Dalian, China). Quantitative PCR was performed using TB Green qPCR Master Mix (TaKaRa) on the Analytik Jena Real-Time PCR detection system (Frechen, Germany). A total volume of 20 μL was subjected to the following PCR conditions: 95°C, 30 s for initial denaturation, 35 cycles of 95°C for 5 s and TM for 34 s. Relative expression was calculated using the 2 –ΔΔCt method, and normalized to that of the housekeeping gene GAPDH. Primers are shown in Supplementary Table S1.
Rapid amplification of cDNA ends (RACE)
The full-length cDNA sequence of FPASL was obtained from HSFB RNA using the SMARTer RACE 5′/3′ kit (TaKaRa). Nested 5′ and 3′ RACE products were obtained using a GeneRacerTM kit (Invitrogen, Carlsbad, USA). The sequences of the designed gene-specific primers for the PCR step of the RACE procedure are presented in Supplementary Table S2. PCR products were extracted using a SanPrep Column DNA Gel Extraction kit (Sangon Biotech, Shanghai, China), cloned into the pGM-T vector, and analyzed by Sanger sequencing.
Western blot analysis
To obtain total protein, cells were lysed in lysis buffer containing protease inhibitor, phosphatase inhibitor and PMSF. Equal amounts of protein (30 μg) were separated by 10% SDS-PAGE and then electroblotted onto PVDF membranes (Millipore, Billerica, USA). After being blocked with 5% BSA in Tris-buffered saline with 0.1% Tween 20 (TBST) buffer for 4 h at 4°C and washed with DPBS three times, the membranes were incubated overnight at 4°C with indicated primary antibodies. The membranes were washed again and incubated with the corresponding HRP-conjugated secondary antibody. Finally, the protein bands were visualized with the ChemiDoc™ XRS+System. Detailed information of the primary antibodies used is shown in Supplementary Table S3.
Cell Counting Kit-8 (CCK-8) assay
Fibroblasts were inoculated into a 96-well plate at a density of 3000 cells/well. After cell culture for 0, 24, 48, 72, 96, and 120 h respectively, 10 μL CCK8 reagent (APExBIO, Houston, USA) was added into each well. After incubation at 37°C for 2 h, the OD value was detected at 450 nm with a microplate reader, and the data were recorded for analysis.
EdU assay
Cell proliferation was determined by EdU assay using the 5-ethynyl-2′-deoxyuridine labelling/detection kit (RiboBio). A total of 200 μL diluent A was added to each confocal dish inoculated with cells and incubated in the cell incubator for 2 h. Cells were washed with PBS buffer, fixed with 4% paraformaldehyde for 30 min, and decolorized with 200 μL of 2 mg/mL glycine for 5 min on a shaker at room temperature. Then, 200 μL of 0.5% Triton X-100 was added to each well. After being washed once with PBS buffer, each well was incubated with 200 μL of 1× Apollo stain in the dark for 30 min. Then, 0.5% Triton X-100 was added to each well and incubated for 10 min. Each well was stained with 200 μL diluent F and then washed twice with PBS buffer. After staining, the cells were observed under a laser confocal microscope (ZEISS, Oberkochen, Germany).
Lentivirus infection
Lentivirus-expressing lncRNA-FPASL and negative control were transfected into fibroblasts. First, fibroblasts were seeded into 6-well plates at 1×10 5 cells per well and incubated for 24 h at 37°C, and then transfected with 1×10 8 transducing units (TUs)/mL of lentivirus (10 μL). Then, the medium was refreshed, and incubation was continued for 48 h. Subsequently, RT-qPCR was performed to detect overexpression efficiency.
Locked nucleic acid (LNA) GapmeRs
The previously characterized human fibroblasts were transfected with either LNAs targeting lncRNA-FPASL (20 nM) or with LNA control (20 nM) (Qiagen, Hilden, Germany). GapmeR transfections were performed using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s protocols. Following 24 h of transfection, the cells were used for subsequent experiments. The GapmeR sequences of FPASL are listed in Supplementary Table S4.
Statistical analysis
Data are presented as the mean±standard deviation. The data were analyzed using GraphPad Prism 6. Statistical significance was tested by two-tailed Student’s t test for comparisons between two groups and one-way analysis of variance (ANOVA) with post hoc analysis contrasts for multigroup comparisons. A P value<0.05 was considered statistically significant.
Results
lncRNA expression profiles are altered in hypertrophic scar
It has widely been accepted that the excessive proliferation of fibroblasts is the hallmark of hypertrophic scar. Human primary fibroblasts from hypertrophic scar tissues and the matched adjacent normal tissues were successfully cultured and identified in this study ( Supplementary Figure S1A,B). Moreover, we evaluated the levels of cell proliferation in fibroblasts from hypertrophic scar tissues (HSFBs) and matched adjacent normal tissues (NSFBs). CCK8 assay and EdU staining assay showed that the cell viability of HSFBs was significantly higher than that of NSFBs. qRT-PCR and western blot analysis confirmed the increased PCNA expression and decreased p27 expression in HSFBs, which are associated with cell proliferation ( Supplementary Figure S1C–F). Taken together, these results demonstrate that there is a hyperproliferation of fibroblasts in hypertrophic scar.
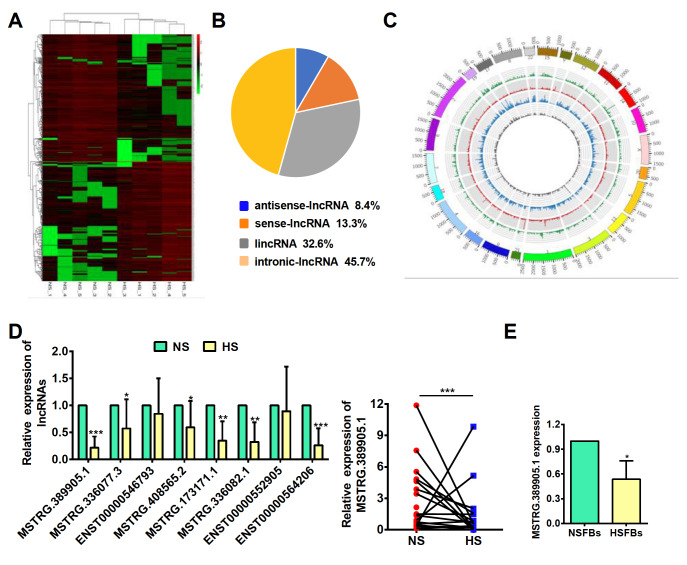
We obtained the differentially expressed lncRNAs in hypertrophic scar tissues and adjacent normal tissues by analysing the lncRNA microarray data. As expected, a variety of lncRNAs were identified to be aberrantly expressed in hypertrophic scar tissues. From 5 paired samples, we found that there are a total of 1739 lncRNAs that are significantly differentially expressed (log 2FC>1 and P<0.05). Among them, 891 lncRNAs are upregulated, while 848 are downregulated in hypertrophic scar tissues ( Figure 1A). As shown in Figure 1B, the composition of these significantly differentially expressed lncRNAs in terms of genomic origin is different, including lincRNAs, antisense lncRNAs, intronic lncRNAs and sense lncRNAs. Meanwhile, the circos plot shows the distribution and expression of detected and significantly differentially expressed lncRNAs on human chromosomes ( Figure 1C). Subsequently, we selected the 8 most downregulated lncRNAs with log 2FC>2, and P<0.01, an expression level greater than 10, and an RPKM greater than 1 in at least one group, and confirmed their expressions by qRT-PCR in 18 paired hypertrophic scar tissues and adjacent normal tissues. The results showed that 6 of them were significantly downregulated in hypertrophic scar tissues, of which lncRNA MSTRG.389905.1, a lincRNA, was the one with the lowest expression level, and was therefore selected for further study ( Figure 1D). Additionally, lncRNA MSTRG.389905.1 was obviously decreased in HSFBs ( Figure 1E), suggesting that this lncRNA may play an essential role in the proliferation of fibroblasts. Therefore, we named it fibroblast proliferation-associated lncRNA in hypertrophic scar (FPASL).
Figure 1 .
lncRNA FPASL is a fibroblasts-enriched lncRNA downregulated in hypertrophic scar
(A) A cluster heatmap presenting the significantly dysregulated lncRNAs in hypertrophic scar (HS) tissues relative to the matched adjacent normal tissues (NS). (B) Pie charts of the lncRNA classification, including lincRNAs, antisense lncRNAs, intronic lncRNAs and sense lncRNAs. (C) Circos plot displaying the distribution and expression of lncRNAs on human chromosomes. The outermost layer was a chromosome map of the human genome. The inner circles from outside to inside correspond to the distribution and expression of detected circRNAs on the chromosomes; green indicates sense lncRNAs, red indicates lincRNAs, gray indicates antisense lncRNAs, and blue indicates intronic lncRNAs. (D) The expressions of eight significantly deregulated lncRNAs, including lncRNA MSTRG.389905.1, in 18 paired NS and HS samples were measured by qRT-PCR. (E) qRT-PCR analysis of FPASL (lncRNA MSTRG.389905.1) expression in primary fibroblasts from hypertrophic scar tissues (HSFBs) and matched adjacent normal tissues (NSFBs). * P<0.05, ** P<0.01, and *** P<0.001.
Characterization and distribution of lncRNA FPASL
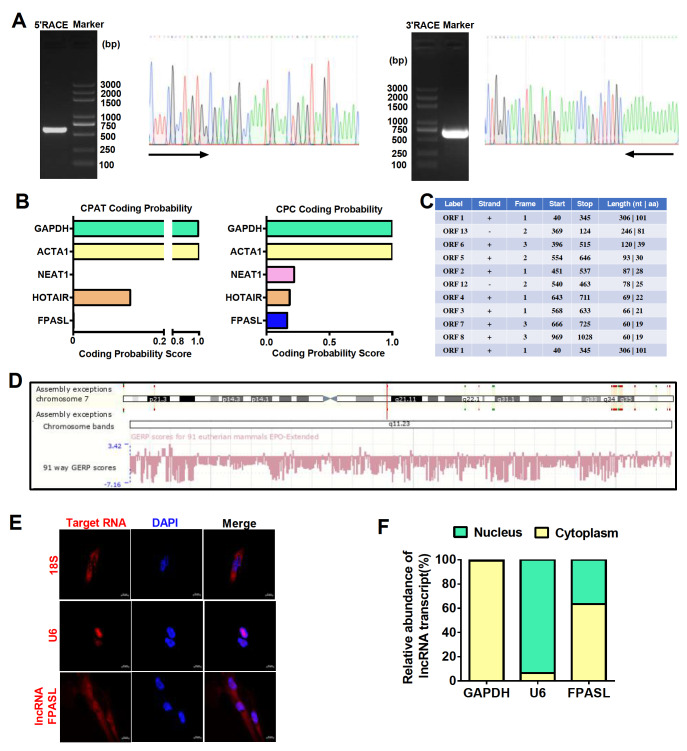
As a novel lncRNA, according to the microarray results, the full sequence of FPASL needs to be determined. Therefore, 5′ and 3′RACE was performed. Both the 5′ and 3′ ends of the representative FPASL were extended in different lengths: 539 nt at the 5′ end and 624 nt at the 3′ end ( Figure 2A and Supplementary Figure S2A). Some lncRNAs have the ability to encode micropetides. Thus, we evaluated whether the FPASL transcript is a true noncoding RNA using the Coding Potential Assessment Tool (CPAT) and Contrastive Predictive Coding (CPC). As expected, both tools revealed that the FPASL transcript has a very low coding-potential score ( Figure 2B). Additionally, open reading frame (ORF) analysis showed 13 ORFs in the full sequence of FPASL. For all ORFs longer than 30 nt in the FPASL sequence, no ORF was found to match any protein sequence in the Swiss-Prot database ( Figure 2C). These data indicated that FPASL is indeed a long noncoding RNA (lncRNA). Furthermore, we found that FPASL is located on chromosome 7 (q11,23) and is well conserved across different species ( Figure 2D). The predicted FPASL secondary structure based on the minimum free energy (MFE) and partition function was assessed using the RegRNA Server. The analyses revealed that FPASL has stem-loop structures with relatively high base-pairing potential ( Supplementary Figure S2B). To investigate its possible function, we explored the cellular location of FPASL. FISH results indicated that FPASL is expressed in both the cytoplasm and nucleus ( Figure 2E). Moreover, cytoplasmic and nuclear RNA extraction experiments were performed, and FPASL expression was detected by qRT-PCR. The results showed that FPASL was predominantly located in the cytoplasm of fibroblasts ( Figure 2F). These results prompt us to study the function of FPASL in the fibroblast cytoplasm of hypertrophic scar.
Figure 2 .
The novel lncRNA FPASL is mainly expressed in the cytoplasm of fibroblasts
(A) 5′- and 3′-rapid amplification of cDNA ends (RACE) assays in fibroblasts were used to detect the whole sequence of FPASL. Left: an image of PCR products from the 5′-RACE and 3′-RACE assays separated by gel electrophoresis. Right: sequencing of PCR products indicated the boundary between the universal anchor primer and FPASL sequences. (B) The protein-coding ability of FPASL was predicted by the Coding Potential Assessment Tool (CPAT) and Contrastive Predictive Coding (CPC). GAPDH, ACTA1 (mRNA, highly coded protein), NEAT1, and HOTAIR (lncRNA, lowly coded protein) were included as controls. (C) ORF finder was used to analyze the protein-coding ability of lncRNA FPASL. (D) Ensemble Genome Browser (Human GRCh38/p13 (GCA_000001405.28)) was used to search the location and conservativeness of FPASL in different species. (E) A FISH assay was performed to observe FPASL expression in fibroblasts. Mixed 18S RNA (exclusively expressed in the cytoplasm) and U6 (exclusively expressed in the nucleus) were included as controls. One of the representative results is presented. Scale bar: 10 μm. (F) Cytoplasmic and nuclear RNAs were separated and reverse transcribed to generate cDNA. PCR was conducted to detect the expression of GAPDH, U6, and FPASL in both cytoplasmic and nuclear cDNAs.
lncRNA FPASL inhibits fibroblast proliferation in hypertrophic scar
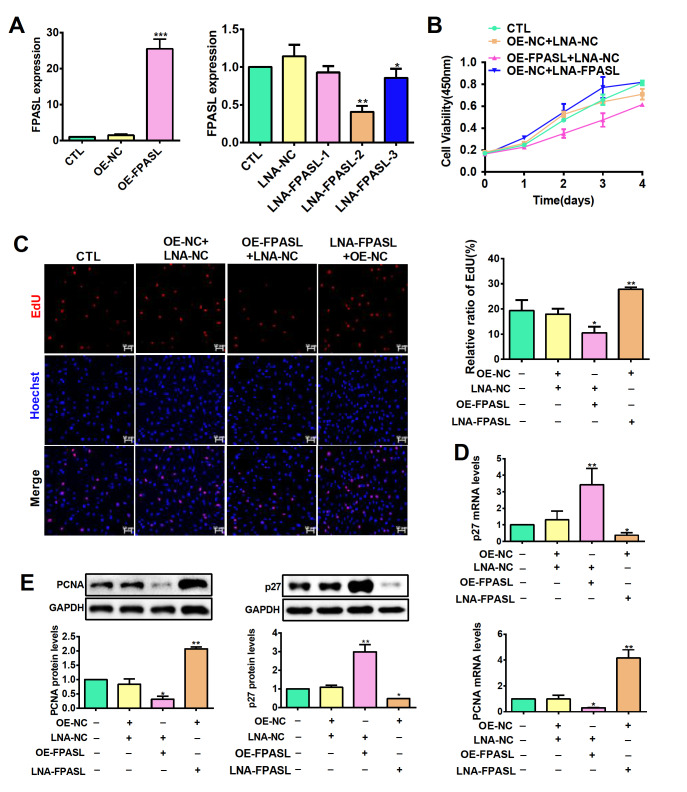
To identify the potential mechanisms by which FPASL functions in hypertrophic scar, we constructed primary fibroblasts stably overexpressing FPASL by lentivirus infection and used locked nucleic acid (LNA) GapmeRs to knockdown the expression of FPASL in fibroblasts. The knockdown or overexpression efficiencies were verified by RT-PCR ( Figure 3A). Next, the proliferation of fibroblast was tested. CCK8 assay showed that knockdown of FPASL significantly increased fibroblast growth, whereas forced FPASL expression had the opposite effect in fibroblasts ( Figure 3B). These results were validated by EdU staining assay, which showed that the proportion of EdU-positive cells in FPASL-overexpressing fibroblasts was significantly lower than that in the control fibroblasts ( Figure 3C). In contrast, FPASL knockdown increased the viability and proliferation of fibroblasts ( Figure 3B,C). Meanwhile, overexpression of FPASL significantly suppressed the mRNA and protein expression levels of PCNA, while p27 expression was increased. The mRNA and protein expression levels of PCNA were enhanced, and p27 expression was decreased by knockdown of FPASL ( Figure 3D,E). These results strongly support the notion that FPASL plays an important role in fibroblast proliferation.
Figure 3 .
lncRNA FPASL suppresses fibroblast proliferation
(A) FPASL expression was measured by qRT-PCR in fibroblasts infected with OE-FPASL or transfected with LNA-FPASL. (B) Cell viability of fibroblasts detected by CCK8 assay after infection with OE-FPASL or transfection with LNA-FPASL. (C) EdU staining assays were applied to detect the proliferation of fibroblasts infected with OE-FPASL or transfected with LNA-FPASL. Scale bar: 50 μm. (D) The mRNA expressions of PCNA and p27 were measured by qRT-PCR after fibroblasts were infected with OE-FPASL or transfected with LNA-FPASL. (E) The protein expressions of PCNA and p27 were measured by western blot analysis after fibroblasts were infected with OE-FPASL or transfected with LNA-FPASL. * P<0.05, ** P<0.01, and *** P<0.001.
lncRNA FPASL regulates the MAPK and PI3K/AKT signaling pathways in hypertrophic scar
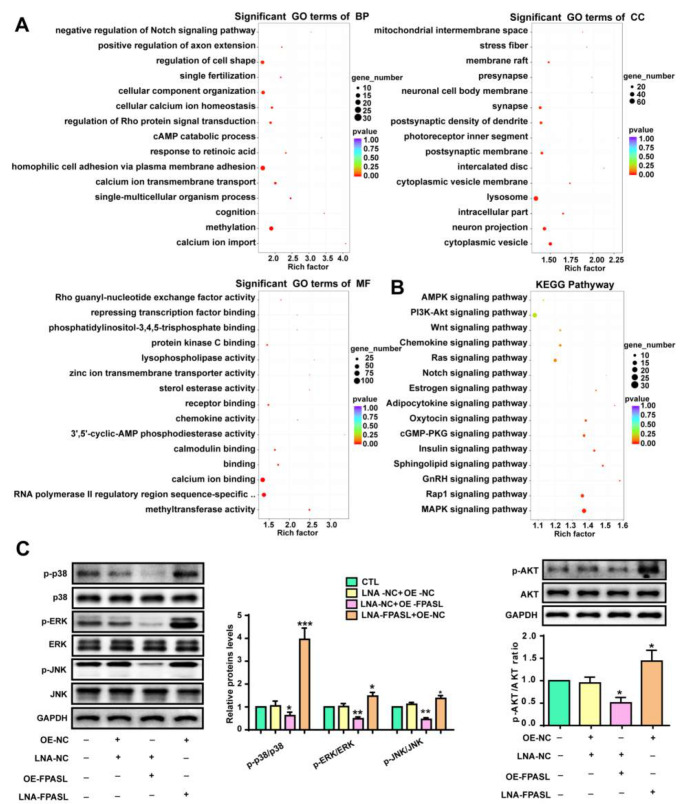
It has been reported that different lncRNAs trigger the activation of multiple signal transduction pathways. GO annotation and KEGG enrichment analysis were performed with the target genes of the differentially expressed lncRNAs to better understand the functional significance of these lncRNAs. GO analysis results revealed that lncRNA target genes were categorized according to biological process (BP), cellular component (CC), and molecular function (MF). BP was significantly enriched for calcium ion import, methylation and single-multicellular organism process. Significant CC terms revealed enrichment in cytoplasmic vesicles, neuron projections and intracellular parts. Finally, the top three MFs identified were methyltransferase activity, RNA polymerase II regulatory region sequence-specific DNA binding and calcium ion binding. These enriched terms confirmed that aberrant lncRNA expression profiles play an important role in the progression of hypertrophic scar ( Figure 4A). We then specifically analyzed and identified KEGG pathways related to cell proliferation. Among them, the most significantly enriched pathway is the PI3K/AKT signaling pathway, and the MAPK signaling pathway shows the greatest upregulation ( Figure 4B), suggesting that the two signaling pathways may be related to the proliferation of fibroblasts in hypertrophic scar. Thus, we measured p38, ERK, JNK and AKT activation in HSFBs after overexpression of FPASL or transfection with LNA-FPASL. We found dramatic decreases in AKT, ERK, p38 and JNK phosphorylation in fibroblasts with FPASL overexpression, while their phosphorylation levels were increased in fibroblasts transfected with LNA-FPASL ( Figure 4C). Altogether, these data indicate that the MAPK and PI3K/AKT signaling pathways are activated in hypertrophic scar and FPASL suppresses their phosphorylation.
Figure 4 .
lncRNA FPASL knockdown activates the MAPK and PI3K/AKT signaling pathways
(A) The top 15 enriched biological process, cellular component and molecular function terms determined by GO analysis in hypertrophic scar. (B) The top 15 upregulated signaling pathways related to cell proliferation were found to be enriched by KEGG analysis in hypertrophic scar. (C) Western blot analysis was performed to detect p-AKT, AKT, p-ERK, ERK, p-p38, p38, p-JNK and JNK levels in fibroblasts with lncRNA FPASL knockdown or overexpression. * P<0.05, ** P<0.01, and *** P<0.001.
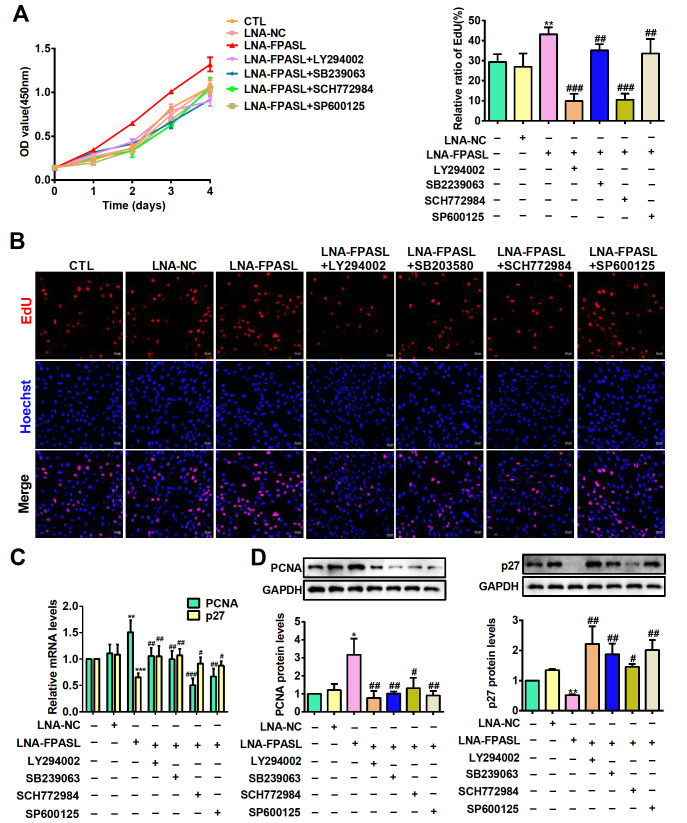
lncRNA FPASL inhibits fibroblast proliferation by suppressing MAPK and PI3K/AKT signaling pathway activation
As FPASL can regulate the PI3K/AKT and MAPK signaling pathways in hypertrophic scar, we assume that FPASL may regulate fibroblast proliferation via these two pathways. Thus, specific inhibitors of PI3K (LY294002), p38 (SB239063), ERK (SCH772984) and JNK (SP600125) were used to determine whether the PI3K/AKT and MAPK signaling pathways are involved in FPASL-regulated fibroblast proliferation. Fibroblasts were stimulated with LY294002, SB239063, SCH772984 and SP600125 at concentrations of 0, 10, 20 and 50 μM for 24 h, and then the cell viability was measured by CCK-8 assay. The results showed that the inhibition ratio was prominently increased by these inhibitors at concentrations of 0, 10, 20 and 50 μM, indicating that these inhibitors suppressed cell viability in a dose-dependent manner ( Supplementary Figure S3 and Supplementary Table S5). Subsequently, 50 μM LY294002 and 20 μM SB239063, SCH772984 and SP600125 were selected to treat fibroblasts. The results of cell proliferation ( Figure 5A,B) showed that the inhibitors LY294002, SCH772984, SB239063 and SP600125 recovered the proliferation of fibroblasts transfected with LNA-FPASL. Correspondingly, the mRNA and protein levels of PCNA and p27 were also analyzed by qRT-PCR and western blot analysis, respectively. As shown in Figure 5C,D, the inhibitors LY294002, SB239063, SCH772984 and SP600125 obviously decreased the expression of PCNA in fibroblasts transfected with LNA-FPASL but increased the expression of p27. Taken together, the above results suggest that knockdown of FPASL promotes fibroblast proliferation by activating the PI3K/AKT and MAPK signaling pathways.
Figure 5 .
lncRNA FPASL affects fibroblast proliferation by suppressing MAPK and PI3K/AKT signaling pathway activation
(A) CCK8 assay was performed to detect the effects of LY294002 (a specific inhibitor of the PI3K/AKT signaling pathway), SB239063 (a specific inhibitor of the p38 signaling pathway), SP600125 (a specific inhibitor of the JNK signaling pathway), and SCH772984 (a specific inhibitor of the ERK signaling pathway) on fibroblast viability induced by LNA-FPASL. (B) The effects of LY294002, SB239063, SCH772984 or SP600125 on fibroblast proliferation induced by LNA-FPASL measured by EdU staining. Scale bar: 50 μm. (C) The mRNA expressions of PCNA and p27 were evaluated by qRT-PCR in fibroblasts treated with LY294002, SB239063, SCH772984 or SP600125 after transfection with LNA-FPASL. (D) Western blot analysis was used to measure the protein expressions of PCNA and p27 in fibroblasts treated with LY294002, SB239063, SCH772984 or SP600125 after transfection with LNA-FPASL. * P<0.05, ** P<0.01 vs LNA-NC; # P<0.05, ## P<0.01, ### P<0.001 vs LNA-FPASL.
Discussion
Hypertrophic scar is a pathologically significant fibrotic skin disease, and the major characteristic of hypertrophic scar is the overproliferation of dermal fibroblasts and the subsequent metabolic disorder of collagen-based ECM proteins [25]. Thus, we cultured primary fibroblasts from hypertrophic scar tissues and adjacent normal tissues, and further demonstrated that there is a hyperproliferation of fibroblasts in hypertrophic scar. A better understanding of the underlying molecular mechanisms responsible for fibroblast proliferation could help us to design or identify novel therapeutic methods for the treatment hypertrophic scar.
lncRNAs are important regulators of multiple cellular processes, biomarkers for disease prognosis and possible therapeutic targets [26]. For example, lncRNA NR2F1-AS1 induces breast cancer lung metastasis [27]. However, the lncRNAs with aberrant expression in hypertrophic scar remain unknown. In this study, we screened the altered expression profiles of lncRNAs using lncRNA microarray analysis of clinical tissues. Many lncRNAs were found to be significantly dysregulated, implying that lncRNAs participate in hypertrophic scar formation. Herein, we focused on FPASL, which exhibited relatively low expression in hypertrophic scar. To better understand the functional or molecular role of FPASL, we characterized the full-length of FPASL through RACE and revealed that FPASL is mainly located in the cytoplasm of fibroblasts. The result provides a basis for further functional and mechanistic studies because the function of lncRNAs is closely related to their cellular localization.
LncRNAs participate in several aspects of the hypertrophic scar and regulate the expression of key components of related pathways [ 28, 29] . In the present study, we found that FPASL is a mediator of fibroblast proliferation. lncRNAs, residing in the cytoplasm, can act by activating specific signaling pathways, which is one of the mechanisms [30]. For example, LINC00941 promotes colorectal cancer metastasis by activating the TGF-β/SMAD2/3 signaling pathway [31]. AC067945.2 decreases collagen through the VEGF and Wnt signaling pathways in hypertrophic scar [32]. Here, GO annotation and KEGG enrichment analysis of differentially expressed lncRNAs were performed. The PI3K/AKT signaling pathway had the highest enrichment, and the MAPK signaling pathway showed the greatest upregulation, suggesting that the two signaling pathways may play important roles in hypertrophic scar formation. Moreover, we found that downregulation of FPASL expression activated the PI3K/AKT, ERK, JNK and p38 signaling pathways. It is well established that the PI3K/AKT and MAPK signaling pathways can increase cell proliferation. This could explain why FPASL knockdown led to fibroblast proliferation. By using specific inhibitors of these two signaling pathways, we further confirmed that FPASL regulates fibroblast proliferation through the PI3K/AKT and MAPK signaling pathways.
In summary, we identified a novel lncRNA, FPASL, in hypertrophic scar and revealed that FPASL regulates fibroblast proliferation through the PI3K/AKT and MAPK signaling pathways in the progression of hypertrophic scar. This study forms a new theoretical basis for hypertrophic scar progression and provides a possible approach for targeted therapy of hypertrophic scar.
Supplementary Data
Supplementary data is available at Acta Biochimica et Biophysica Sinica online.
Supporting information
Acknowledgments
The authors gratefully thank the patients from the Affiliated Hospital of Ningxia Medical University for providing the hypertrophic scar and the matched normal skin tissues. The authors also thank the NHC Key Laboratory of Metabolic Cardiovascular Diseases Research and Ningxia Key Laboratory of Vascular Injury and Repair Research for the assistance in using the instruments.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the National Natural Science Foundation of China (Nos. 82060110 and 81860555), the Basic Scientific Research Operating Expenses from the Public Welfare Research Institutes at the Central Level of the Chinese Academy of Medical Sciences (No. 2019PT330002), the Key Research and Development Projects in Ningxia Hui Autonomous Region (Nos. 2020BFH02003, 2020BEG03005, 2020BFH02001, 2021BEG02033, and 2021BEG02028), and the Natural Science Foundation of Ningxia Hui Autonomous Region (No. 2021AAC03337).
References
- 1.El9 Ayadi A, Jay JW, Prasai A. Current approaches targeting the wound healing phases to attenuate fibrosis and scarring. Int J Mol Sci. . 2020;21:1105. doi: 10.3390/ijms21031105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida IR, Gonçalves AC, Corrêa FB, Castro JC, Guirro EC, Junior JA, Coltro PS. Evaluation of clinical and biomechanical features of scars resulting from the treatment of burn contractures comparing acellular dermal matrices. Ann Surg. . 2023;277:198–205. doi: 10.1097/SLA.0000000000005371. [DOI] [PubMed] [Google Scholar]
- 3.Lv K, Liu H, Xu H, Wang C, Zhu S, Lou X, Luo P, et al. Ablative fractional CO2 laser surgery improving sleep quality, pain and pruritus in adult hypertrophic scar patients: a prospective cohort study. Burns Trauma. . 2021;9:40. doi: 10.1093/burnst/tkab023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shibuya Y, Hokugo A, Okawa H, Kondo T, Khalil D, Wang L, Roca Y, et al. Therapeutic downregulation of neuronal PAS domain 2 (Npas2) promotes surgical skin wound healing. eLife. . 2022;11:e71074. doi: 10.7554/eLife.71074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Hou K, Wu Y, Liu Z. Use of adipose stem cells against hypertrophic scarring or keloid. Front Cell Dev Biol. . 2021;9:823694. doi: 10.3389/fcell.2021.823694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang N, Xue L, Younas A, Liu F, Sun J, Dong Z, Zhao Y. Co-delivery of triamcinolone acetonide and verapamil for synergistic treatment of hypertrophic scars via carboxymethyl chitosan and Bletilla striata polysaccharide-based microneedles. Carbohydrate Polyms. . 2022;284:119219. doi: 10.1016/j.carbpol.2022.119219. [DOI] [PubMed] [Google Scholar]
- 7.Tu L, Lin Z, Huang Q, Liu D. USP15 enhances the proliferation, migration, and collagen deposition of hypertrophic scar–derived fibroblasts by deubiquitinating tgf-βr1 in vitro. Plast Reconstr Surg. . 2021;148:1040–1051. doi: 10.1097/PRS.0000000000008488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang X, Chai B, Duan R, Zhou Y, Huang X, Li Q. Inhibition of fkbp10 attenuates hypertrophic scarring through suppressing fibroblast activity and extracellular matrix deposition. J Investig Dermatol. . 2017;137:2326–2335. doi: 10.1016/j.jid.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 9.Bridges M, Daulagala A, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021, 220: e202009045 . [DOI] [PMC free article] [PubMed]
- 10.Ding N, Xie L, Ma F, Ma S, Xiong J, Lu G, Zhang H, et al. miR-30a-5p promotes glomerular podocyte apoptosis via DNMT1-mediated hypermethylation under hyperhomocysteinemia. Acta Biochim Biophys Sin. . 2022;54:126–136. doi: 10.3724/abbs.2021005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Q, Gong J, Bi J, Yang X, Zhang L, Lu C, Li L, et al. KGF-2 regulates stap-2–mediated signal transducer and activator of transcription 3 signaling and reduces skin scar formation. J Investig Dermatol. . 2022;142:2003–2013. doi: 10.1016/j.jid.2021.12.018. [DOI] [PubMed] [Google Scholar]
- 12.Lovell CD, Anguera MC. Long noncoding rnas that function in nutrition: lnc-ing nutritional cues to metabolic pathways. Annu Rev Nutr. . 2022;42:251–274. doi: 10.1146/annurev-nutr-062220-030244. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Gan Y, Liu J, Li J, Zhou Z, Tian R, Sun R, et al. Downregulation of MEIS1 mediated by ELFN1-AS1/EZH2/DNMT3a axis promotes tumorigenesis and oxaliplatin resistance in colorectal cancer. Sig Transduct Target Ther. . 2022;7:87. doi: 10.1038/s41392-022-00902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue ST, Zheng B, Cao SQ, Ding JC, Hu GS, Liu W, Chen C. Long non-coding RNA LINC00680 functions as a ceRNA to promote esophageal squamous cell carcinoma progression through the miR-423-5p/PAK6 axis. Mol Cancer. . 2022;21:69. doi: 10.1186/s12943-022-01539-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S, Sun G, Wu P, Chen C, Kuang Y, Liu L, Zheng Z, et al. WDR82-binding long noncoding RNA lncEry controls mouse erythroid differentiation and maturation . J Exp Med. . 2022;219 doi: 10.1084/jem.20211688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, He M, Zhang M, Sun Q, Zeng S, Zhao H, Yang H, et al. The role of non-coding RNAs in colorectal cancer, with a focus on its autophagy. Pharmacol Ther. . 2021;226:107868. doi: 10.1016/j.pharmthera.2021.107868. [DOI] [PubMed] [Google Scholar]
- 17.Zhou C, Yi C, Yi Y, Qin W, Yan Y, Dong X, Zhang X, et al. LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/β-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol Cancer. . 2020;19:118. doi: 10.1186/s12943-020-01237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meza-Sosa KF, Miao R, Navarro F, Zhang Z, Zhang Y, Hu JJ, Hartford CC, et al. SPARCLE, a p53-induced lncRNA, controls apoptosis after genotoxic stress by promoting PARP-1 cleavage. Mol Cell. . 2022;82:785–802.e10. doi: 10.1016/j.molcel.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY, Liu Q, Zhao G, et al. Correction to: The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol Cancer. . 2021;20:120. doi: 10.1186/s12943-021-01387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D, Chen J, Li B, Jiang Q, Liu L, Xia Z, Zheng Q, et al. A noncoding regulatory RNA Gm31932 induces cell cycle arrest and differentiation in melanoma via the miR-344d-3-5p/Prc1 (and Nuf2) axis. Cell Death Dis. . 2022;13:314. doi: 10.1038/s41419-022-04736-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J, Deng LJ, Xia YR, Leng RX, Fan YG, Pan HF, Ye DQ. Involvement of N6-methyladenosine modifications of long noncoding RNAs in systemic lupus erythematosus. Mol Immunol. . 2022;143:77–84. doi: 10.1016/j.molimm.2022.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Deng Y, Xu Y, Xu S, Zhang Y, Han B, Liu Z, Liu X, et al. Secondary data mining of GEO database for long non-coding RNA and Competing endogenous RNA network in keloid-prone individuals. Aging. . 2020;12:25076–25089. doi: 10.18632/aging.104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang Y, Zhou R, Fu X, Wang C, Wang D. HOXA5 counteracts the function of pathological scar-derived fibroblasts by partially activating p53 signaling. Cell Death Dis. . 2021;12:40. doi: 10.1038/s41419-020-03323-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan P, Wang Y, Li J, Fang M. lncRNA PAPPA-AS1 induces the development of hypertrophic scar by upregulating TLR4 through interacting with TAF15. Mediators Inflamm. . 2021;2021:1–23. doi: 10.1155/2021/3170261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ning X, Wiraja C, Chew WTS, Fan C, Xu C. Transdermal delivery of Chinese herbal medicine extract using dissolvable microneedles for hypertrophic scar treatment. Acta Pharmaceutica Sin B. . 2021;11:2937–2944. doi: 10.1016/j.apsb.2021.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen BQ, Dragomir MP, Yang C, Li Q, Horst D, Calin GA. Targeting non-coding RNAs to overcome cancer therapy resistance. Sig Transduct Target Ther. . 2022;7:121. doi: 10.1038/s41392-022-00975-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Zhang P, Wu Q, Fang H, Wang Y, Xiao Y, Cong M, et al. Long non-coding RNA NR2F1-AS1 induces breast cancer lung metastatic dormancy by regulating NR2F1 and ΔNp63. Nat Commun. . 2021;12:5232. doi: 10.1038/s41467-021-25552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu N, Kataoka M, Wang Y, Pu L, Dong X, Fu X, Zhang F, et al. LncRNA LncHrt preserves cardiac metabolic homeostasis and heart function by modulating the LKB1-AMPK signaling pathway. Basic Res Cardiol. . 2021;116:48. doi: 10.1007/s00395-021-00887-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmad S, Abbas M, Ullah MF, Aziz MH, Beylerli O, Alam MA, Syed MA, et al. Long non-coding RNAs regulated NF-κB signaling in cancer metastasis: micromanaging by not so small non-coding RNAs. Semin Cancer Biol. . 2022;85:155–163. doi: 10.1016/j.semcancer.2021.07.015. [DOI] [PubMed] [Google Scholar]
- 30.Jia Z, Qian Z, Tang Y, Li X, Shi Y, Xin H, Fan Y, et al. LncRNA WEE2-AS1 knockdown inhibits the proliferation, migration and 3 invasion of glioma cells via regulating miR-29b-2-5p/TPM3 axis. Oncol Res. . 2022;doi: 10.3727/096504022X16493267465621 doi: 10.3727/096504022X16493267465621. [DOI] [PubMed] [Google Scholar]
- 31.Wu N, Jiang M, Liu H, Chu Y, Wang D, Cao J, Wang Z, et al. LINC00941 promotes CRC metastasis through preventing SMAD4 protein degradation and activating the TGF-β/SMAD2/3 signaling pathway. Cell Death Differ. . 2021;28:219–232. doi: 10.1038/s41418-020-0596-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Li J, Li Q, Li X, Gao Y, Hua X, Zhou B, et al. Overexpression of lncrna ac067945.2 down-regulates collagen expression in skin fibroblasts and possibly correlates with the vegf and wnt signalling pathways. Cell Physiol Biochem. . 2018;45:761–771. doi: 10.1159/000487167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.