Abstract
Background
We aimed to estimate the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) seroprevalence among the general population in Conakry, Guinea and Yaounde, Cameroon after the coronavirus disease 2019 Omicron wave.
Methods
We conducted population-based, age-stratified seroprevalence surveys in Conakry and Yaounde (May and June 2022). We collected demographic and epidemiologic information and dried blood spot samples that were tested for SARS-CoV-2 immunoglobulin G (IgG) antibodies using recombinant nucleocapsid and spike proteins with Luminex technology.
Results
Samples were obtained from 1386 and 1425 participants in Guinea and Cameroon, respectively. The overall age-standardized SARS-CoV-2 IgG seroprevalence against spike and nucleocapsid proteins was 71.57% (95% confidence interval [CI], 67.48%–75.33%) in Guinea and 74.71% (95% CI, 71.99%–77.25%) in Cameroon. Seroprevalence increased significantly with age categories. Female participants were more likely than male participants to be seropositive. The seroprevalence in unvaccinated participants was 69.6% (95% CI, 65.5%–73.41%) in Guinea and 74.8% (95% CI, 72.04%–77.38%) in Cameroon. In multivariate analysis, only age, sex, and education were independently associated with seropositivity.
Conclusions
These findings show a high community transmission after the different epidemiological waves including Omicron, especially among people aged >40 years. In addition, our results suggest that the spread of SARS-CoV-2 has been underestimated as a significant proportion of the population has already contracted the virus and that vaccine strategies should focus on vulnerable populations.
Keywords: Cameroon, Guinea, population-based survey, SARS-CoV-2, seroprevalence
The overall age-standardized SARS-CoV-2 IgG seroprevalence against spike and nucleocapsid proteins was 71.57% in Guinea and 74.71% in Cameroon after successive COVID-19 waves, including the Omicron wave from early 2022. Seroprevalence increased with age categories reaching 84% among persons >40 years.
Since the beginning of the coronavirus disease 2019 (COVID-19) pandemic, Africa has experienced 5 distinct waves, with a particularly high number of reported cases in the fourth wave due to the Omicron variant [1].
The Omicron variant has emerged in November 2021 in southern Africa and was associated with a rapid increase of cases [2, 3]. Studies have suggested that the Omicron variant is highly contagious and can evade prior immunity to previous severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants like Delta [3–6]. These characteristics contributed to a wide spread of Omicron, which became the predominant variant worldwide [1]. However, the Omicron variant is associated with milder symptoms and lower risk of severe disease and hospitalization [3, 5, 7, 8].
According to World Health Organization (WHO) data, as of 29 March 2023, 12.5 million COVID-19 cases and 257 664 deaths were confirmed on the African continent, corresponding to 1.6% and 3.7% of the total number of cases and deaths, respectively, reported worldwide [1]. However, several studies on the prevalence of SARS-CoV-2 antibodies showed that COVID-19 has spread widely on the African continent [9–15].
A recent meta-analysis that compiled data from standardized seroprevalence studies conducted in Africa between January 2020 and December 2021 showed that seroprevalence continued to increase from 3.0% (95% confidence interval [CI], 1.0%–9.2%) in the second trimester of 2020 to 65.1% (95% CI, 56.3%–73.0%) in the third trimester of 2021 before the Omicron wave [11]. Conducting repeated seroprevalence studies provides thus better estimates of the extent of the SARS-CoV-2 spread, independent of data on clinical testing. Since the beginning of the COVID-19 pandemic, we conducted 3 independent seroprevalence surveys in Conakry, the capital city of Guinea, and showed that seroprevalence increased from 17.3% in December 2020 just before the second wave, to 28.9% in March/April 2021 during the peak of the second wave, reaching 42.4% in June 2021 [10] after the second wave. Similarly, in Yaounde, the capital city from Cameroon, we showed in 2 independent population-based SARS-CoV-2 serosurveys that seroprevalence increased from 18.6% just before the start of the second wave (January and February 2021) to 51.3% at the decreasing trend of the second wave (April/May 2021) [9].
Since then, eligibility for vaccination against COVID-19 has progressively been expanded in Africa to cover most age groups and especially the most vulnerable populations [16]. In Guinea, the percentage of the population fully vaccinated increased from 0.65% on 1 June 2021 to 18.56% on 1 June 2022. However, in Cameroon, the proportion of the fully vaccinated population rose only from 0.04% to 4.47% for the same period [17]. The massive spread of the Omicron variant also changed the epidemiological curve of COVID-19 in both countries.
From a public health perspective, the availability of up-to-date estimates of antibody seroprevalence is essential to monitor the true extent of the spread of SARS-CoV-2 in the pandemic in order to adapt targeted public health and social measures as well as vaccination strategies to be in line with the local epidemiological context. The aim of this study was to evaluate the impact of the Omicron wave by estimating seroprevalence and risk factors associated with SARS-CoV-2 antibodies in the general population of Conakry and Yaounde, using the same methodology as the previous surveys [9, 10].
METHODS
Study Design and Population
Two population-based surveys were conducted between May and June 2022 in Conakry, Guinea and in May 2022 in Yaounde, Cameroon, after the Omicron wave (Supplementary Figures 1 and 2). The study design was adapted from the WHO population-based age-stratified seroepidemiological investigation protocol for COVID-19 infection [9, 10].
These surveys were done in the same areas that were sampled during our previous seroprevalence studies, which were undertaken before arrival of Omicron variants. The samples were independent from the previous surveys. As for our previous surveys, demographic (sex, age, education, marital status) and epidemiologic (symptoms related to SARS-CoV-2 infection, previous testing, and COVID-19 vaccination) data were collected with the use of an electronic questionnaire with REDCap (Research Electronic Data Capture) platform. Polymerase chain reaction (PCR) testing was offered to all participants with suspicion of COVID-19 infection.
Detection of Antibodies to SARS-CoV-2
Whole blood samples were collected as dried blood spots and tested for SARS-CoV-2 antibodies with a previously validated, highly sensitive and specific Luminex-based multiplex immunoassay (Luminex Corporation, https://www.luminexcorp.com) using recombinant nucleocapsid and spike SARS-CoV-2 proteins [18]. Samples were considered positive for SARS-CoV-2 immunoglobulin G (IgG) antibodies when they reacted simultaneously with nucleocapsid and spike proteins. Samples reacting with only 1 antigen were considered as “indeterminate” because of the difficulty to discriminate between antibody decline or lower specificity of single-antigen reaction, especially with samples from populations in Africa, or with antibodies induced to vaccines based on spike protein only [19–21].
Statistical Analysis
The sample-size justification and the methods for repeated random sampling of households that were used in our previous surveys have been previously published [9, 10]. Seroprevalence estimates were weighted, age-standardized, and stratified for the other characteristics of the study population. To identify risk factors associated with SARS-CoV-2 seropositivity, we used a set of logistic regression models. The effects of the factors were quantified with unadjusted odds ratios (ORs) for univariate models and adjusted OR (aORs) for multivariate models, with their 95% CI. The analyses were done using Stata 16 software (StataCorp, College Station, Texas).
Ethical Considerations
The study was approved by the National Ethics Committee for Health Research of Guinea and Cameroon. Nevertheless, all participants provided written informed consent and were free to decline participation.
RESULTS
Study Population
In Guinea, a total of 1386 samples from 263 households were collected and tested, whereas in Cameroon we obtained samples that were adequate for serostatus evaluation from 1425 participants from 235 households. Demographic and epidemiologic characteristics of the participants are shown in Table 1. In Guinea, about 838 (60%) participants were female, the median age of participants was 18 years (interquartile range [IQR], 8–34 years), 313 (25%) of participants were asymptomatic, and 930 (75%) reported at least 1 symptom. In Cameroon, only 504 (37%) reported at least 1 symptom related to COVID-19 and 857 (63%) did not report any symptom (Table 1). The median age of participants was 26 years (IQR, 15–40 years) and 783 (55%) were female (Table 1). In both countries, a large majority of participants had never been previously tested for SARS-CoV-2 (92% in Guinea vs 82% in Cameroon). The proportion of unvaccinated participants was 63% in Guinea versus 92% in Cameroon.
Table 1.
Weighted and Age-Standardized Seroprevalence of Severe Acute Respiratory Syndrome Coronavirus 2 Antibodies by Sex, Sociodemographic Characteristics, and Medical History in Population-Based Surveys After Omicron Peak—Conakry, Guinea and Yaounde, Cameroon, 2022
| Characteristic | Guinea | Cameroon | ||||||
|---|---|---|---|---|---|---|---|---|
| Participants Tested | Seropositive (IgG Anti-SP+ and IgG Anti-NC+) |
Participants Tested | Seropositive (IgG Anti-SP+ and Anti-NC+) |
|||||
| (n = 1386) | No. (%) | 95% CI | P Value | (n = 1425) | No. (%) | 95% CI | P Value | |
| Age, y | .0000 | .0000 | ||||||
| 0–19 | 740 | 476 (64.32) | 59.51–68.87 | 443 | 297 (67.19) | 62.55–71.52 | ||
| 20–39 | 370 | 290 (78.38) | 73.34–82.69 | 601 | 496 (82.50) | 79.16–85.41 | ||
| ≥40 | 276 | 232 (84.06) | 77.65–88.89 | 381 | 322 (84.47) | 80.33–87.88 | ||
| Sex | 1386 | .0001 | 1425 | .0008 | ||||
| Female | 838 | 631 (74.52) | 70.38–78.26 | 783 | 640 (78.92) | 75.29–82.15 | ||
| Male | 548 | 367 (67.26) | 61.97–72.14 | 642 | 475 (69.98) | 65.81–73.84 | ||
| Marital status | 1383 | .0000 | 1409 | .0000 | ||||
| Single | 939 | 632 (71.08) | 66.65–75.14 | 873 | 648 (73.22) | 69.89–76.30 | ||
| Married/partnership | 398 | 327 (64.50) | 42.10–81.95 | 436 | 369 (74.92) | 38.71–93.39 | ||
| Divorced/separated/widow(er) | 46 | 36 (32.15) | 18.95–48.99 | 100 | 85 (59.53) | 30.04–83.44 | ||
| Education | 1376 | .0000 | 1420 | .0000 | ||||
| Primary school | 548 | 376 (72.49) | 66.79–77.54 | 374 | 280 (75.59) | 70.73–79.87 | ||
| Secondary school | 365 | 286 (78.27) | 70.63–84.36 | 624 | 510 (82.46) | 78.43–85.87 | ||
| University | 138 | 119 (94.15) | 91.38–96.07 | 319 | 276 (86.42) | 65.36–95.55 | ||
| None | 325 | 211 (59.77) | 53.91–65.37 | 103 | 47 (48.69) | 34.97–62.60 | ||
| Household size | 1386 | .0548 | 1418 | .11 | ||||
| ≤6 persons | 244 | 187 (75.67) | 68.53–81.62 | 910 | 726 (74.10) | 70.24–77.61 | ||
| 7-13persons | 384 | 291 (74.9) | 66.51–81.77 | 400 | 293 (72.77) | 68.00–77.06 | ||
| >13 persons | 758 | 520 (68.87) | 64.46–72.97 | 108 | 90 (82.36) | 73.07–88.93 | ||
| No. of symptoms | 1243 | .4582 | 1361 | .8722 | ||||
| No | 313 | 231 (74.28) | 69.26–78.74 | 857 | 663 (74.83) | 71.46–77.91 | ||
| 1–2 symptoms | 458 | 328 (71.88) | 67.07–76.24 | 233 | 189 (74.65) | 66.90–81.09 | ||
| 3–5 symptoms | 351 | 255 (71.49) | 64.43–77.64 | 210 | 158 (71.72) | 63.28–78.86 | ||
| >5 symptoms | 121 | 82 (64.68) | 56.98–71.68 | 61 | 49 (74.43) | 57.67–86.15 | ||
| Previous COVID-19 testing | 1325 | .0008 | 1409 | .003 | ||||
| Never tested | 1217 | 857 (70.62) | 66.54–74.40 | 1159 | 891 (74.01) | 71.10–76.70 | ||
| Tested | 108 | 95 (93.88) | 89.49–96.50 | 250 | 211 (82.13) | 71.59–89.34 | ||
| Vaccination | 1373 | .0000 | 1415 | .1174 | ||||
| Fully vaccinated | 314 | 256 (78.63) | 68.70–86.05 | 100 | 81 (36.52) | 32.36–40.90 | ||
| Partially vaccinated | 191 | 151 (77.63) | 69.99–83.78 | 11 | 11 (46.93) | 46.93–46.93 | ||
| Unvaccinated | 868 | 580 (69.60) | 65.50–73.41 | 1304 | 1015 (74.80) | 72.04–77.38 | ||
| Overall | 1386 | 998 (71.57) | 67.48–75.33 | 1425 | 1115 (74.71) | 71.99–77.25 | ||
Abbreviations: CI, confidence interval; COVID-19, coronavirus disease; IgG, immunoglobulin G; NC, nucleocapsid protein; SP, spike protein.
SARS-CoV-2 Seroprevalence
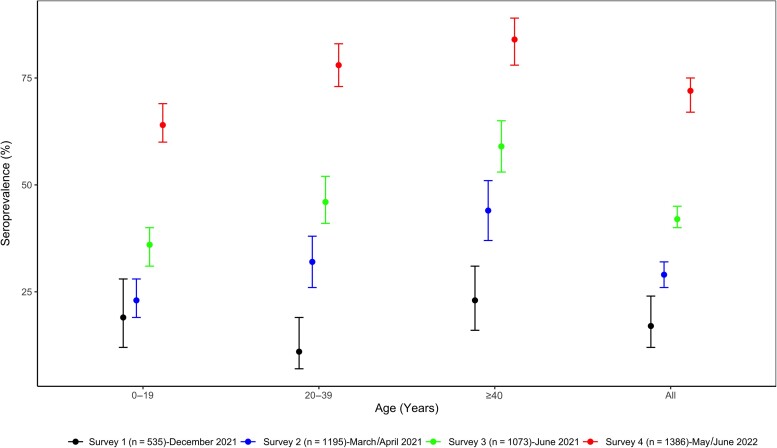
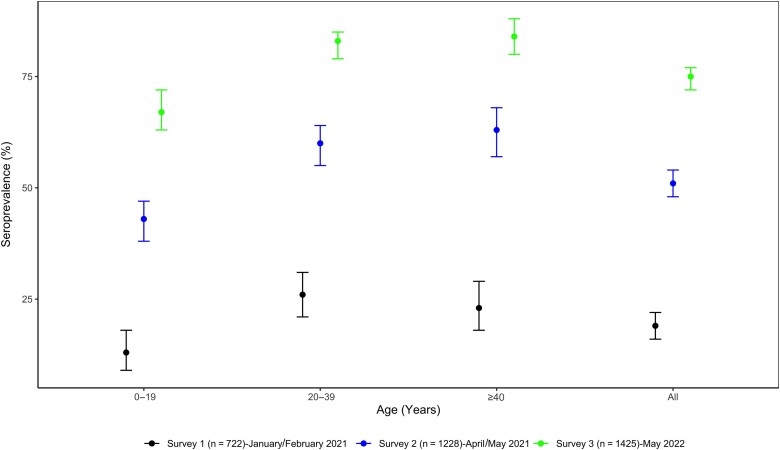
The overall age-standardized SARS-CoV-2 IgG seroprevalence against both spike and nucleocapsid proteins was comparable in both countries: 71.57% (95% CI, 67.48%–75.33%) in Guinea and 74.71% (95% CI, 71.99%–77.25%) in Cameroon (Table 1). In both countries, seroprevalence increased with age categories and was significantly higher among persons aged >40 years, with >8 of 10 participants being seropositive (P = .0000). For all age groups, seroprevalence was significantly higher in this survey than in previous ones in both countries (Figures 1 and 2). Female participants were significantly more likely to be seropositive than male participants (74.52% [95% CI, 70.38%–78.26%] vs 67.26% [95% CI, 61.97%–72.14%]; P = .0001 in Guinea and 78.92% [95% CI, 75.29%–82.15%] vs 69.98% [95% CI, 65.81%–73.84%]; P = .0008 in Cameroon). Seroprevalence was significantly lower among participants who had never been tested for SARS-CoV-2 infection (70.62% [95% CI, 66.54%–74.40%] vs 93.88% [95% CI, 89.49%–96.50%]; P = .0008 in Guinea and 74.01% [95% CI, 71.10%–76.70%] vs 82.13% [95% CI, 71.59%–89.34%]; P = .003 in Cameroon).
Figure 1.
Weighted and age-standardized seroprevalence of severe acute respiratory syndrome coronavirus 2 antibodies in 4 successive population-based surveys, Conakry, Guinea. Dots represent the estimated prevalence and bars represent 95% confidence intervals.
Figure 2.
Weighted and age-standardized seroprevalence of severe acute respiratory syndrome coronavirus 2 antibodies in 3 successive population-based surveys, Yaounde, Cameroon. Dots represent the estimated prevalence and bars represent 95% confidence intervals.
Moreover, the seroprevalence in unvaccinated participants is very high in our survey (69.6% [95% CI, 65.5%–73.41%] in Guinea and 74.8% [95% CI, 72.04%–77.38%] in Cameroon) (Table 1).
The proportion of individuals with spike antibodies only was 16.46% (95% CI, 13.89%–19.39%) in Guinea and 14.20% (95% CI, 12.19%–16.48%) in Cameroon, whereas the proportion of individuals with nucleocapsid antibodies only was 2.98% (95% CI, 2.12%–4.18%) in Guinea and 5.14% (95% CI, 3.81%–6.90%) in Cameroon (Supplementary Table 1). Additionally, the overall age-standardized SARS-CoV-2 seroprevalence against at least spike or nucleocapsid was 91.06% (95% CI, 89.23%–92.61%) in Guinea and 93.26% (95% CI, 91.49%–94.69%) in Cameroon (Supplementary Table 2).
Risk Factors Associated With SARS-CoV-2 Seropositivity
Table 2 gives the unadjusted and adjusted ORs for SARS-CoV-2 seroprevalence. Univariate analysis identified 6 risk factors significantly associated with seropositivity: age, sex, marital status, education, vaccination status, and previous testing for COVID-19. There was no significant association between seropositivity and history of symptoms associated with COVID-19.
Table 2.
Risk Factors Associated With Severe Acute Respiratory Syndrome Coronavirus 2 Antibodies Seropositivity in Population-Based Surveys After Omicron Peak—Yaounde, Cameroon and Conakry, Guinea, 2022: Univariable and Multivariable Binary Logistic Regression Analysis
| Characteristic | Guinea | Cameroon | ||||||
|---|---|---|---|---|---|---|---|---|
| Unadjusted OR (95% CI) | P Value | Adjusted OR (95% CI) | P Value | Unadjusted OR (95% CI) | P Value | Adjusted OR (95% CI) |
P Value | |
| Age, y | 0 | |||||||
| ≤40 | 1 | … | 1 | 1 | ||||
| >40 | 2.37 (1.66–3.38) | .000 | 2.43 (1.61–3.67) | .000 | 1.72 (1.24–2.37) | .002 | 1.55 (1.10–2.17) | .013 |
| Sex | ||||||||
| Male | 1 | 1 | 1 | 1 | ||||
| Female | 1.50 (1.24–1.82) | .000 | 1.62 (1.31–2.01) | .000 | 1.56 (1.21–2.03) | .001 | 1.68 (1.28–2.21) | .001 |
| Marital status | ||||||||
| Single | 1 | … | 1 | … | ||||
| Married/partnership | 2.24 (1.60–3.12) | .000 | … | 1.90 (1.39–2.59) | .000 | … | ||
| Divorced, separated, or widow(er) | 1.75 (.78–3.91) | 0.168 | … | 1.96 (1.09–3.53) | .026 | … | ||
| Education | ||||||||
| None | 1 | 1 | 1 | … | ||||
| Primary school | 1.18 (.87–1.61) | .283 | 1.58 (1.11–2.25) | .011 | 3.59 (2.25–5.73) | .000 | 3.46 (2.17–5.53) | .000 |
| Secondary school | 1.96 (1.29–2.97) | .002 | 2.19 (1.37–3.52) | .002 | 5.31 (3.38–8.33) | .000 | 5.10 (3.24–8.03) | .000 |
| University | 3.38 (2.06–5.57) | .000 | 3.47 (2.09–5.78) | .000 | 7.65 (4.56–12.84) | .000 | 7.95 (4.70–13.45) | .000 |
| Household size | ||||||||
| ≤6 persons | 1 | … | 1 | … | ||||
| 7-13 persons | 0.95 (.56–1.62) | .857 | … | .70 (.53–1.92) | .513 | … | ||
| >13 persons | 0.67 (.44–1.01) | .058 | … | 1.27 (.73–2.19) | .389 | … | ||
| Symptoms | ||||||||
| Asymptomatic | 1 | … | 1 | … | ||||
| Symptomatic | 0.89 (.71–1.12) | .322 | … | 1.07 (.81–1.41) | .642 | … | ||
| Vaccination | ||||||||
| Unvaccinated | 1 | … | 1 | … | ||||
| Vaccinated | 2.06 (1.54–2.75) | .000 | … | 1.36 (.81–2.30) | .203 | … | ||
| Previous COVID-19 testing | ||||||||
| Never tested | 1 | 1 | 1 | … | ||||
| Tested | 3.07 (1.63–5.80) | .001 | 1.97 (.99–3.90) | .053 | 1.62 (1.11–2.36) | .001 | 1.15 (.77–1.71) | .496 |
Abbreviations: CI, confidence interval; COVID-19, coronavirus disease; OR, odds ratio.
In multivariate analysis, only age, sex, and education were independently associated with seropositivity. ORs for female participants were higher compared with males (aOR, 1.62 [95% CI, 1.31–2.01] in Guinea and aOR, 1.68 [95% CI, 1.28–2.21] in Cameroon). Overall, the seroprevalence was significantly higher in patients aged >40 years, resulting in an aOR of 2.43 (95% CI, 1.61–3.67) in Guinea and 1.55 (95% CI, 1.10–2.17) in Cameroon. The OR for participants who had been previously tested for SARS-CoV-2 was higher in Guinea (OR, 3.07; P = .001) and in Cameroon (OR, 1.62; P = .001) compared with that of participants who had never been tested previously, but this relationship was not significant in multivariate models.
The seroprevalence increased significantly according to participants’ level of education. In both countries, ORs almost doubled between participants with a primary education degree and those with a university education degree (1.58 vs 3.47 in Guinea and 3.46 vs 7.95 in Cameroon).
DISCUSSION
In these large serosurveillance studies in Guinea and Cameroon, we estimated IgG seroprevalence among the general population to evaluate the proportion of participants who had been infected with SARS-CoV-2 after the different COVID-19 waves, including the Omicron wave, in the respective capital cities of these 2 countries. We observed that, after the Omicron wave, the overall seroprevalence in the general population was high in both countries, reaching 71.57% in Conakry, Guinea, and 74.71% in Yaounde, Cameroon. The seroprevalence in both countries has risen considerably and significantly over time, from 17.3% in December 2020 in Guinea and from 18.6% in January/February 2021 in Cameroon. To our knowledge, very few population-based seroprevalence studies have been performed in Africa after the Omicron variant became predominant. Nevertheless, our results are consistent with the most recent study performed in Gauteng, South Africa, that was conducted after the Omicron wave [22]. The overall seroprevalence found in this study was 73.1% (95% CI, 72.0%–74.1%). However, even if the Omicron variant has not been associated with severe forms of COVID-19 disease compared to previous variants, these high seroprevalences reflect that today a significant proportion of the general population has anti–SARS-CoV-2 antibodies.
Additionally, like in other surveys [9, 10, 14, 15, 23], seroprevalence increased with age categories and was significantly higher among adults aged >40 years, with a seroprevalence of 84% in both countries for this age group compared to a seroprevalence of <70% among youth aged <19 years. Our previous surveys [9] showed that in Cameroon, seroprevalence was 13.1% (95% CI, 9.3%–18.3%) in the younger age group (<19 years) and 22.9% (95% CI, 17.5%–29.2%) in the older age group (>40 years) at the time of the first survey just before the start of the second wave of COVID-19. In the second survey done at the decreasing trend of the second wave, the seroprevalence in these 2 age groups increased to 42.7% (95% CI, 38.3%–47.3%) and to 62.8% (95% CI, 57.3%–68.0%), respectively. In Guinea, 3 successive surveys have been done [10]. Between the first and third surveys, done just before and after the second wave, respectively, seroprevalence increased from 18.9% (95% CI, 12.1%–28.4%) to 35.5% (95% CI, 31.3%–39.8%) among those <19 years of age and from 22.6% (95% CI, 16.3%–30.6%) to 58.9% (95% CI, 52.6%–64.9%) among those >40 years of age. As reported in these studies and in most seroprevalence studies conducted in Africa, seroprevalence was significantly associated with age [9, 10, 14, 15, 23].
This is an important point to underline since the oldest people are more vulnerable because they are more likely to have severe forms of COVID-19 disease due to comorbidities. For example, in Guinea the prevalence of high blood pressure exceeds 62% among people aged >44 years [24], and we showed that >84% of adults aged >40 years already have antibodies and could therefore already have a protection against the severe forms.
The high level of seroprevalence found in this survey is more likely the effect of past infection than the result of vaccination, as estimated coverage of fully vaccinated individuals at the time of the surveys was very low—18.56% of the general population in Guinea and 4.47% in Cameroon [1, 17]—and in our study 36.78% were fully vaccinated in Guinea and 7.84% in Cameroon. It is known that anti-nucleocapsid antibodies are produced after natural infection and not after vaccination with spike-based vaccines. It is important to note that the increase in vaccination coverage tends to complicate the interpretation of seroprevalence studies, particularly if serological tests detecting only spike are used, because antibody tests that target the spike protein are unable to distinguish between those who have been previously infected and those who have received at least 1 dose of a spike-based SARS-CoV-2 vaccine [20, 21]. In addition, in our study we found that about 70% of unvaccinated participants were seropositive in Guinea and 75% in Cameroon. This finding is slightly lower than the seroprevalence in unvaccinated participants found in our previous survey, which was 82% in Guinea. Although our results indicate that most of the population had developed anti–SARS-CoV-2 antibodies through infection and/or vaccination, further studies are needed to determine how long this acquired immunity lasts and its ability to neutralize other highly circulating variants.
In our multivariate analysis, age, sex, and education were independently associated with seropositivity. Female participants were more likely to be seropositive than male participants, similar to a report from South Africa [22]. Research on gender disparities in COVID-19 infection and clinical outcomes are limited and more studies are needed to fully understand the impact of gender-specific factors, such as health status, biological differences, level and quality of healthcare utilization, profession, and socioeconomic conditions. We found also that the seroprevalence was lowest among participants who had never been tested, as reported elsewhere [22]. This result indicates that even though the PCR test was negative for these previously tested participants, the majority of them developed antibodies and that performing the test was most likely related to an exposure or a suspicion of infection by SARS-CoV-2, except for travelers.
In general, our results on age-standardized SARS-CoV-2 IgG seropositivity are comparable between the 2 countries for all characteristics considered except for vaccination status. The proportion of people vaccinated with at least 1 dose in our sample is very low in Cameroon compared to Guinea (7.84% vs 36.78%) but reflects the national vaccination coverage in the 2 countries (4.46% in Cameroon vs 18.56% in Guinea as of June 2022). In addition, in the univariate analysis, there was a significant difference between vaccination status and seropositivity in Guinea with an OR for vaccinated individuals of 2.06 (95% CI, 1.54–2.75; P = .000). This is not the case for Cameroon (OR, 1.36 [95% CI, .81–2.30]; P = .203).
This study benefits from several strengths, including the large representative sample of the general population of the 2 capital cities, the recent recruitment time-frame post-Omicron waves, the measurement of antibodies, and also the use of spike and nucleocapsid proteins to determine seropositivity; we have taken the most stringent criteria possible because the presence of nucleocapsid or spike alone can be due either to a previous infection (nucleocapsid alone) or to a vaccination (spike alone) or to cross-reactions to other coronaviruses or another artifact. It can also not be excluded that the antigens from the Luminex assay, which are derived from the Wuhan strain, do not optimally detect antibodies against an Omicron infection and that the seroprevalence is underestimated. In addition, sex ratio is relatively similar between the different surveys in both countries. The age structure is slightly different, but we have performed an age standardization to avoid bias. The main limitation of our study relates to the nonrealization of a neutralization assay to identify against which variants one is immunized by the production of antibodies after the Omicron wave.
In conclusion, our testing for SARS-CoV-2 antibodies targeting 2 recombinant nucleocapsid and spike proteins showed that at least three-quarters of the populations of Conakry and Yaounde had been infected with SARS-CoV-2 after the emergence of the Omicron variant. Our results indicate that the seroprevalence of SARS-CoV-2 is high, suggesting higher population exposure to SARS-CoV-2 and potential protection from severe COVID-19 disease than would be indicated by reported cases and vaccine data from routine surveillance systems. More importantly, this massive COVID-19 diffusion in the general population in Africa with humoral immunity suggests implementing a targeted vaccination strategy with priority for the oldest and most vulnerable (eg, people with comorbidities) individuals because currently available vaccines do not prevent the diffusion of the virus but have benefit in reducing the risk of severe outcomes from COVID-19. Our study provided a more accurate understanding of the true extent of SARS-CoV-2 infections in 2 African countries despite low vaccine coverage and can be used alongside other epidemiological information for evidence-based decision making.
Supplementary Material
Contributor Information
Mamadou Saliou Kalifa Diallo, Centre de Recherche et de Formation en Infectiologie de Guinée, Université Gamal Abdel Nasser de Conakry, Conakry, Guinea; TransVIHMI, University of Montpellier, Inserm, Institut de Recherche pour le Developpement, Montpellier, France.
Marie Amougou-Atsama, Centre de Recherche sur les Maladies Émergentes et Re-Emergentes, Yaounde, Cameroon.
Ahidjo Ayouba, TransVIHMI, University of Montpellier, Inserm, Institut de Recherche pour le Developpement, Montpellier, France.
Cece Kpamou, Centre de Recherche et de Formation en Infectiologie de Guinée, Université Gamal Abdel Nasser de Conakry, Conakry, Guinea.
Eric Donald Mimbe Taze, French National Agency for Research on AIDS and Infectious Diseases, Cameroon Site, Central Hospital of Yaounde, Yaounde, Cameroon.
Guillaume Thaurignac, TransVIHMI, University of Montpellier, Inserm, Institut de Recherche pour le Developpement, Montpellier, France.
Haby Diallo, Centre de Recherche et de Formation en Infectiologie de Guinée, Université Gamal Abdel Nasser de Conakry, Conakry, Guinea.
Nadine Boutgam Lamare, Faculty of Medicine and Pharmaceutical Sciences, University of Dschang, Dschang, Cameroon.
Julie Bouillin, TransVIHMI, University of Montpellier, Inserm, Institut de Recherche pour le Developpement, Montpellier, France.
Abdoul Karim Soumah, Centre de Recherche et de Formation en Infectiologie de Guinée, Université Gamal Abdel Nasser de Conakry, Conakry, Guinea.
Sébastien Awono Noah, French National Agency for Research on AIDS and Infectious Diseases, Cameroon Site, Central Hospital of Yaounde, Yaounde, Cameroon.
Emilande Guichet, TransVIHMI, University of Montpellier, Inserm, Institut de Recherche pour le Developpement, Montpellier, France.
Alpha Kabinet Keita, Centre de Recherche et de Formation en Infectiologie de Guinée, Université Gamal Abdel Nasser de Conakry, Conakry, Guinea; TransVIHMI, University of Montpellier, Inserm, Institut de Recherche pour le Developpement, Montpellier, France.
Marie Varloteaux, French National Agency for Research on AIDS and Infectious Diseases, Cameroon Site, Central Hospital of Yaounde, Yaounde, Cameroon.
Martine Peeters, TransVIHMI, University of Montpellier, Inserm, Institut de Recherche pour le Developpement, Montpellier, France.
Anne-Cécile Zoung-Kanyi Bissek, Centre de Recherche sur les Maladies Émergentes et Re-Emergentes, Yaounde, Cameroon; Division of Operational Research in Health, Ministry of Public Health of Cameroon, Yaounde, Cameroon.
Abdoulaye Toure, Centre de Recherche et de Formation en Infectiologie de Guinée, Université Gamal Abdel Nasser de Conakry, Conakry, Guinea.
Eric Delaporte, TransVIHMI, University of Montpellier, Inserm, Institut de Recherche pour le Developpement, Montpellier, France.
Charles Kouanfack, French National Agency for Research on AIDS and Infectious Diseases, Cameroon Site, Central Hospital of Yaounde, Yaounde, Cameroon; Faculty of Medicine and Pharmaceutical Sciences, University of Dschang, Dschang, Cameroon; Central Hospital, Yaounde, Cameroon.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. M. S. K. D., A. T., E. D., and C. K. conceived and designed the study. A. A., G. T., A. K. K., and M. P. developed and did the serological tests. M. S. K. D., M. A.-A., A. A., C. K., E. D. M. T., S. A. N., E. G., and M. V. contributed to the data collection and curation. M. S. K. D., M. P., E. D. M. T., C. K., and A. A. verified the underlying data. M. S. K. D., E. D., A. T., C. K., and M. P. did the data analysis, drafted the first version of the manuscript, and wrote the final version. All authors revised the manuscript and approved the final version. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Acknowledgments. We thank all the study participants, the investigation teams, the community health staff, and logistical support in Guinea and Cameroon. We also thank the national institute of statistics of Guinea, the health department of Conakry, Guinea, the Health Operations Research Division/Ministry of health Cameroon, the French National Agency for Research on AIDS and Infectious Diseases Emerging Infectious Disease Cameroon Site, and Centre de Recherche sur les Maladies Émergentes et Re-Emergentes for assistance in the field. We are also grateful to Caroline Coulon for her support in the execution of this study.
Patient consent. The study was approved by the both National Ethics Committee for Health Research of Guinea and Cameroon. The patient's written consent was obtained and were free to decline participation.
Disclaimer. The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Financial support. This work was supported by the Agence Française de Développement and the Ministère de l’Europe et des Affaires Etrangères, France (Project ARIACOV: Appui à la riposte africaine à l’épidémie de COVID-19, https://www.ariacov.org).
References
- 1. World Health Organization (WHO) . WHO coronavirus disease (COVID-19) dashboard. 2022. Available at:https://covid19.who.int/. Accessed 25 January 2022.
- 2. Viana R, Moyo S, Amoako DG, et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022; 603:679–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tan CW, Chia WN, Zhu F, et al. SARS-CoV-2 Omicron variant emerged under immune selection. Nat Microbiol 2022; 7:1756–61. [DOI] [PubMed] [Google Scholar]
- 4. Mohsin M, Mahmud S. Omicron SARS-CoV-2 variant of concern: a review on its transmissibility, immune evasion, reinfection, and severity. Medicine (Baltimore) 2022; 101:e29165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kimura I, Yamasoba D, Tamura T, et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5. Cell 2022; 185:3992–4007.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tegally H, San JE, Cotten M, et al. The evolving SARS-CoV-2 epidemic in Africa: insights from rapidly expanding genomic surveillance. Science 2022; 378:eabq5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nyberg T, Ferguson NM, Nash SG, et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 Omicron (B.1.1.529) and Delta (B.1.617.2) variants in England: a cohort study. Lancet 2022; 399:1303–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wolter N, Jassat W, Walaza S, et al. Early assessment of the clinical severity of the SARS-CoV-2 Omicron variant in South Africa: a data linkage study. Lancet 2022; 399:437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ndongo FA, Guichet E, Mimbe ED, et al. Rapid increase of community SARS-CoV-2 seroprevalence during second wave of COVID-19, Yaounde, Cameroon. Emerg Infect Dis 2022; 28:1233–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soumah AA, Diallo MSK, Guichet E, et al. High and rapid increase in seroprevalence for SARS-CoV-2 in Conakry, Guinea: results from 3 successive cross-sectional surveys (ANRS COV16-ARIACOV). Open Forum Infect Dis 2022; 9:ofac152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lewis HC, Ware H, Whelan M, et al. SARS-CoV-2 infection in Africa: a systematic review and meta-analysis of standardised seroprevalence studies, from January 2020 to December 2021. BMJ Glob Health 2022; 7:e008793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fryatt A, Simms V, Bandason T, et al. Community SARS-CoV-2 seroprevalence before and after the second wave of SARS-CoV-2 infection in Harare, Zimbabwe. EClinicalMedicine 2021; 41:101172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sagara I, Woodford J, Kone M, et al. Rapidly increasing SARS-CoV-2 seroprevalence and limited clinical disease in three Malian communities: a prospective cohort study. Clin Infect Dis 2021; 74:1030–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nkuba AN, Makiala SM, Guichet E, et al. High prevalence of anti-SARS-CoV-2 antibodies after the first wave of COVID-19 in Kinshasa, Democratic Republic of the Congo: results of a cross-sectional household-based survey. Clin Infect Dis 2022; 74:882–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mulenga LB, Hines JZ, Fwoloshi S, et al. Prevalence of SARS-CoV-2 in six districts in Zambia in July, 2020: a cross-sectional cluster sample survey. Lancet Glob Health 2021; 9:e773–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. World Health Organization (WHO) . WHO SAGE roadmap for prioritizing uses of COVID-19 vaccines. Geneva, Switzerland: WHO; 2022.
- 17. Mathieu E, Ritchie H, Ortiz-Ospina E, et al. A global database of COVID-19 vaccinations. Nat Hum Behav 2021; 5:947–53. [DOI] [PubMed] [Google Scholar]
- 18. Ayouba A, Thaurignac G, Morquin D, et al. Multiplex detection and dynamics of IgG antibodies to SARS-CoV2 and the highly pathogenic human coronaviruses SARS-CoV and MERS-CoV. J Clin Virol 2020; 129:104521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lumley SF, Wei J, O’Donnell D, et al. The duration, dynamics, and determinants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody responses in individual healthcare workers. Clin Infect Dis 2021; 73:e699––709.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nkuba Ndaye A, Hoxha A, Madinga J, et al. Challenges in interpreting SARS-CoV-2 serological results in African countries. Lancet Glob Health 2021; 9:e588––9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ghaffari A, Meurant R, Ardakani A. COVID-19 serological tests: how well do they actually perform? Diagnostics (Basel) 2020; 10:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Madhi SA, Kwatra G, Myers JE, et al. Population immunity and Covid-19 severity with Omicron variant in South Africa. N Engl J Med 2022; 386:1314–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gignoux E, Atthanassiadis F, Garat Yarrow A, et al. Seroprevalence of SARS-CoV-2 antibodies and retrospective mortality in a refugee camp, Dagahaley, Kenya. PLoS One 2021; 16: e0260989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Camara A, Balde NM, Diakite M, et al. High prevalence, low awareness, treatment and control rates of hypertension in Guinea: results from a population-based STEPS survey. J Hum Hypertens 2016; 30:237–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.