Abstract
Background
ALKBH5 is aberrantly activated and exerts critical roles in facilitating the development of glioblastoma. However, the underlying activation mechanism by which ALKBH5 protein is increased in glioblastoma is not completely understood. Our study aimed to elucidate the signaling pathways involved in mediating ALKBH5 protein stability.
Methods
The contribution of deubiquitinating enzymes (DUB) to the fluctuation of ALKBH5 protein expression was globally profiled with western blot analysis. Mass spectrometry and immunoprecipitation were performed to identify the USP36 and ALKBH5 interaction. The effects of USP36 on the stability of ALKBH5 were detected with in vivo and in vitro ubiquitination assays. Cell proliferation assays, neurosphere formation, limited dilution assay, and intracranial tumor growth assays were implemented to assess the collaborative capacities of USP36 and ALKBH5 in tumorigenesis.
Results
Ubiquitin-specific peptidase 36 (USP36), as a potential ALKBH5-activating DUB, played an essential role in stabilization of ALKBH5 and regulation of ALKBH5-mediated gene expression in glioblastoma. The depletion of USP36 drastically impaired cell proliferation deteriorated the self-renewal of GSCs and sensitized GSCs to temozolomide (TMZ) treatment. Furthermore, the deletion of USP36 substantially decreased the in vivo tumor growth when monitored by bioluminescence imaging. Our findings indicate that USP36 regulates the protein degradation and expression of ALKBH5, and the USP36-ALKBH5 axis orchestrates glioma tumorigenesis.
Conclusion
Our findings identify USP36 as a DUB of ALKBH5 and its role in glioblastoma progression, which may serve as a potential therapeutic target for glioblastoma treatment.
Keywords: ALKBH5, deubiquitination, glioblastoma stem cells, USP36
Key Points.
USP36 expression is increased in GSCs and clinically correlated with ALKBH5 level
USP36 stabilizes ALKBH5 expression through its deubiquitinase activity
USP36 depletion impairs the growth of glioblastoma cells and xenograft tumors.
Importance of the Study.
Glioblastoma is one of the deadliest types of brain tumors, and exhibits cellular heterogeneity, with a subpopulation of undifferentiated and self-renewing glioma stem cells (GSCs) at the peak of the hierarchy. The hard-to-treat GSCs are responsible for cancer formation and chemotherapy resistance, and development of new strategies to eradicate GSC is an unmet need. Herein, we examined the DUB of ALKBH5 and found that USP36, as a deubiquitinating enzyme of ALKBH5, increased ALKBH5 protein expression by inhibiting its ubiquitin-proteasome proteolytic pathway. USP36 was remarkably increased in GSCs and associated with poor patient survival. The depletion of USP36 drastically impaired cell proliferation and self-renewal capacities of GSCs sensitized GSCs to temozolomide (TMZ) treatment, and deteriorated the in vivo tumor growth. These results open up a new landscape of signaling pathways and provide vital information for designing strategies to optimize glioblastoma treatment.
Glioblastoma (GBM) is the most common and aggressive primary malignant brain tumor, which manifests remarkable inter- and intratumoral heterogeneity driven by genetic and epigenetic landscapes as well as transcriptional circuits.1,2 Of those, N6-methyladenosine (m6A) RNA methylation is one of the most prevalent reversible epitranscriptomic modifications of mRNA.3,4 A growing body of evidence has shown that m6A modifications in mRNA could determine the RNA fate and functions,5,6 and are crucial for multiple fundamental biological processes including tumor metastasis,7 circadian rhythm,8 tissue development,9 cell differentiation,10 and tumorigenesis.11 The formation of the m6A methylation landscape is governed by the multicomponents as RNA methyltransferase complex, RNA demethylase, and RNA readers.12 ALKBH5, as a pivotal m6A demethylase, plays an important role in various cancers and attracts considerable pharmacological interest from a broad range of researchers.13–15 In our previous study,16 the expression of ALKBH5 was increased in glioblastoma stem cells, further promoting the proliferation of GSCs. Although accumulating studies focus on the regulatory roles of ALKBH5 in many types of tumors, its own regulatory mechanism of ALKBH5 expression, especially the mechanism of overexpression of ALKBH5 in GBM, remains largely unknown.
Ubiquitination, one of the most universal forms of protein post-translational modification,17 plays an essential role in controlling a myriad aspects of cellular functions. The ubiquitination process is reversible and can be catalyzed by a series of deubiquitylating enzymes termed deubiquitinases (DUB).18,19 The DUB may promote protein stability and alter signaling pathways, which deregulate certain protein functions leading to profound impacts on multiple malignant cancers including GBM.20–22 USP36 is a deubiquitinating enzyme belonging to ubiquitin-specific protease (USP) family. Emerging studies showed USP36 was a key modulator to regulate multiple cellular biological processes by deubiquitination leading to decreased proteasomal degradation of proteins involved in tumorigenesis,23,24 immune reactions,25 cell cycle progression,26 and autophagy.27
In our study, we reveal a mechanism involving post-translational modification of m6A demethylase-ALKBH5, in which the deubiquitinase-USP36 promotes the protein stability of ALKBH5 by directly cleaving its polyubiquitin chains. Conversely, ablation of USP36 significantly decreased ALKBH5 expression and consequently inhibited tumor growth in glioblastoma xenograft models. Furthermore, we report high USP36 expression predicts poor prognosis of GBM patients and a direct correlation of elevated expression of USP36 with decreased survival of patients with GBM. We therefore have sufficient reasons to believe that the USP36-ALKBH5 axis is essential for gliomagenesis and our results may provide vital information for designing strategies to overcome drug resistance, thereby achieving optimized glioblastoma treatment.
Materials and Methods
Plasmids and Reagents
All the USP expression plasmids were kindly provided by Dr. Jianhua Yang (Texas Children’s Cancer Center, Houston, TX). USP36 and ALKBH5 cDNA were further cloned into pcDNA3 and/or pLVX-AcGFP1-N1 vectors. Mutant USP36 plasmids were also introduced by the QuikChange site-directed mutagenesis Kit (Agilent Technologies). The USP36 siRNAs (SASI_Hs01_00194136 and SASI_Hs01_00194135), CRIPSR/Cas9 gRNA, Temozolomide (TMZ), cycloheximide (CHX), MG132, puromycin and DAPI were purchased from Sigma.
Cell Culture, Transfection, and Treatment
NHA-E6/E7/hTERT (immortalized normal human astrocytes), SW1783, HS683, U-251MG, U87MG, LN229, and 293T cells were cultured in DMEM medium supplemented with 10% bovine calf serum (HyClone). Human GSCs were maintained in a DF12 medium (Gibco) with B27, epidermal growth factor (EGF, 10 ng/mL), and basic fibroblast growth factor (bFGF, 10 ng/mL). Cell lines were authenticated by short tandem repeat profiling and were routinely tested for mycoplasma con N229 and 293
tamination in every 6 months.
The plasmid and siRNAs transfection were performed using Turbofect (#R0531, ThermoFisher Scientific) and X-tremeGENE siRNA transfection reagents (#04476093001, Roche Diagnostics), respectively. For lentiviral production, packing (psPAX2) and envelope (pMD2.G) plasmids were cotransfected into 293T cells. Stable clones were selected by culturing cells in a medium with 2 mg/ml puromycin for 2 weeks.
Real-time PCR and Western Blot
Real-time PCR and western blot assay were performed as described previously.22 The primers and antibodies used for PCR and western blot analysis are listed in the Supplementary Table.
In vivo and in vitro Deubiquitination
In vivo and in vitro deubiquitylation of ALKBH5 by USP36 was performed as described previously.22 For in vivo deubiquitination, cells were lysed using RIPA lysis buffer and cell lysates were immunoprecipitated using the indicated magnetic beads or antibodies; for in vitro ubiquitination, the ALKBH5 included ubiquitylated His-ALKBH5 was purified with HisPur Ni-NTA magnetic beads. The ubiquitylated ALKBH5 protein was incubated with recombinant USP36 proteins (R&D systems Company, #E-628) in a deubiquitylation buffer at 37 °C for 2 h.
Neurosphere Formation Assay
For each cell line, three 35-mm culture dishes were seeded with 100 cells per dish in 2.5 ml of neural stem cell media and cultured for 10 days. Then, the numbers of neurospheres were counted with a microscope.
In vitro limiting dilution assay was performed as described previously.16 Briefly, GSCs were dissociated into single cells and then plated in 96-well plates at a cell number of 5, 10, 50, 100, or 200 cells per well. Wells with no neurosphere were counted for each group after 10 d. Extreme limiting dilution assays were analyzed using software available at https://bioinf.wehi.edu.au/software/elda/.
Immunohistochemical (IHC) and Immunofluorescence (IF) Analysis
Tissue array slides (US Biomax, INC) were deparaffinized, rehydrated, and then followed by antigen retrieval. Next, the slides were blocked and then incubated with primary antibodies at 4 °C overnight. IHC staining was done with horseradish peroxidase followed by DAB Substrate Kit (Vector laboratories, #SK-4100).
For immunofluorescence analysis, cells were fixed with 4% paraformaldehyde and incubated with indicated primary antibody at 4 °C overnight, followed by incubation with the second antibody for 1 h at room temperature. Coverslips were mounted on slides using an antifade mounting medium with DAPI. Immunofluorescence images were analyzed on a confocal microscope (Zeiss).
Cell Proliferation Assay
GSC cells were seeded in a 96-well plate (1000 cells/well), and cell proliferation was measured from day 1 to day 4 by CCK-8 assay. Also, the GSC cells were seeded in a 96-well plate (500/well) and treated with vehicle or TMZ for 3 days. Cell proliferation was measured by CCK-8 assay.
Mice and Animal Housing
All male and female mice used in the experiments were athymic nude mice at 6–8 weeks of age. The mice were purchased from the Charles River Laboratories. The animals were maintained in VCU animal facilities in accordance with the current regulations and standards of the U.S. Department of Agriculture and the Department of Health and Human Services. All mouse experiments were reviewed and approved by Institutional Animal Care and Use Committees of the Virginia Commonwealth University.
Intracranial Tumor Assay
For the tumor growth experiment, mice were anesthetized and injected intracranially with GSC cells as described previously.11 For bioluminescence imaging (BLI), mice were given the substrate D-luciferin (150 mg/kg) by subcutaneous injection and then subjected to IVIS Spectrum instrument (PerkinElmer) to monitor the tumor growth of tumor cells. Brain tumor burden was measured by the photon flux of the BLI signal and Ex vivo BLI was subsequently performed on removed organs to define the precise tumor size of the glioma. For the studies of measuring mice survival, mice were euthanized when reaching moribund condition or experimental endpoint, and the data were analyzed by Kaplan–Meier plot.
Analysis of USP36 mRNA Expression in TCGA and REMBRANDT Datasets
USP36 mRNA expression data were retrieved from cBioportal for Cacner Genomics (https://www.cbioportal.org/), Betastasis (www.betastasis.com), and R2 (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). USP36 mRNA expressions were stratified by quartiles of all expressions: <25% quartile = low expression, 25–75% quartiles = medium expression, and >75% quartile = high expression.
Quantification and Statistical Analyses
Quantitative data were presented as the mean ± SEM or SD. Statistical analysis between the two groups was accessed by a two-tailed t-test. Correlation experiments were conducted with the Pearson correlation test and survival analysis was conducted using the Kaplan–Meier model with a two-sided log-rank test. All graphs and statistical analyses were completed with GraphPad Prism 8.0. Statistical significance was defined as a P < .05.
Results
Identification of USP36 as a Novel Mediator of ALKBH5 Signaling
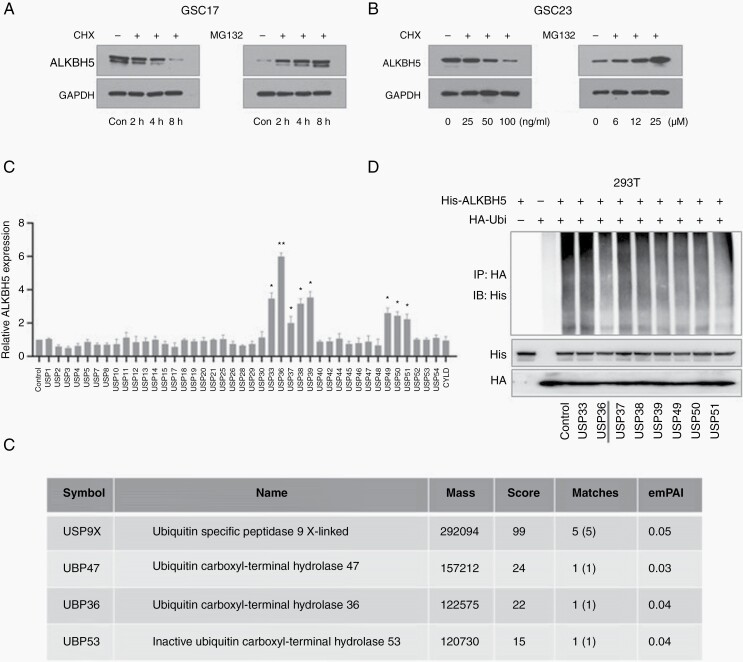
Our previous study has revealed that the signaling pathway of ALKBH5 plays a pivotal role in sustaining glioma stem cell properties and glioma tumorigenesis.16 However, virtually nothing is known about the specific mechanism of ALKBH5 protein regulation in tumorigenesis. In an attempt to determine whether the ALKBH5 protein degradation is mediated through the ubiquitin-proteasome proteolytic pathway, we studied the ALKBH5 protein alteration in GSC11, GSC17, and GSC23 cells with CHX- or MG132-over time with a variety of concentrations. We found the ALKBH5 protein expression in GSC17 cells and GSC11 cells were decreased in a time-dependent manner after treatment with CHX. On the contrary, the protein levels of ALKBH5 were gradually increased with the longer MG132 incubation time (Figure 1A and Supplemental Figure S1A). In addition, we found the ALKBH5 protein levels were decreased or increased in a concentration-dependent manner in GSC23 cells upon CHX or MG132 treatment (Figure 1B). Furthermore, we assessed the ubiquitylation of ALKBH5 in 293T cells and found the degradation of ALKBH5 was significantly increased together with the ubiquitin ligase (Supplemental Figure S1B). Collectively, our data indicate that the turnover of ALKBH5 is mainly regulated by a degradation process through the ubiquitin-proteasome proteolytic pathway.
Figure 1.
USP36 is a novel mediator of ALKBH5. (A) The ALKBH5 expressions were detected in GSC17 cells treated with cycloheximide (CHX) or MG132 for the indicated time. (B) The ALKBH5 expressions were detected in GSC23 cells treated with CHX or MG132 for the indicated concentration. (C) LN229 cells were transfected with different deubiquitinases plasmids. The band intensity of ALKBH5 was quantified and normalized to the internal control (data are mean ± SD for triplicate biological replicates; Student t-test; *P < .05; **P < .01). (D) 293T cells were transfected with indicated plasmids. Cells were treated with 25 μM MG132 for 6 h and cell lysates were immunoprecipitated with an anti-HA antibody. The immunoprecipitates were analyzed by immunoblotting with anti-His antibodies. (E) The 293T cells were transfected with His-ALKBH5. His-ALKBH5 protein was harvested with HisPur Ni-NTA magnetic beads and subjected to mass spectrometry. The table shows that four DUB proteins with P < .05 were identified by the mass spectrometry analysis. ).
To identify the specific DUBs involved with ALKBH5 regulation, we first applied a gain-of-function screen by monitoring ALKBH5 expression. We screened 42 DUBs in LN229 glioblastoma cells, which usually have effects on tumorigenesis or exhibit high expression in the central nervous system. Of the 42 DUBs we detected, USP33, USP36, USP37, USP38, USP39, USP49, USP50, and USP51 could substantially increase the protein expression of ALKBH5 in LN229 cells under CHX treatment (Figure 1C and Supplemental Figure S1C). However, USP36 had the most obvious effects on ALKBH5 levels. Then, we examined the deubiquitination effects of these candidates and found that USP36 could significantly inhibit the ubiquitination of ALKBH5 (Figure 1D).
To further verify the molecular mechanism underlying the ALKBH5 proteasome degradation system, we next identify protein interaction partners of ALKBH5 with mass spectrometry. We identified 4 DUBs with a cutoff of P < .05 (Figure 1E), and combined with our aforementioned results, we confirmed that USP36 is the deubiquitylating enzyme of ALKBH5. Together, the above results suggest that USP36 is a specific deubiquitinating enzyme of ALKBH5 and increases ALKBH5 protein expression by inhibiting its ubiquitin-proteasome proteolytic pathway.
USP36 Stabilize ALKBH5 Through its Deubiquitinase Activity
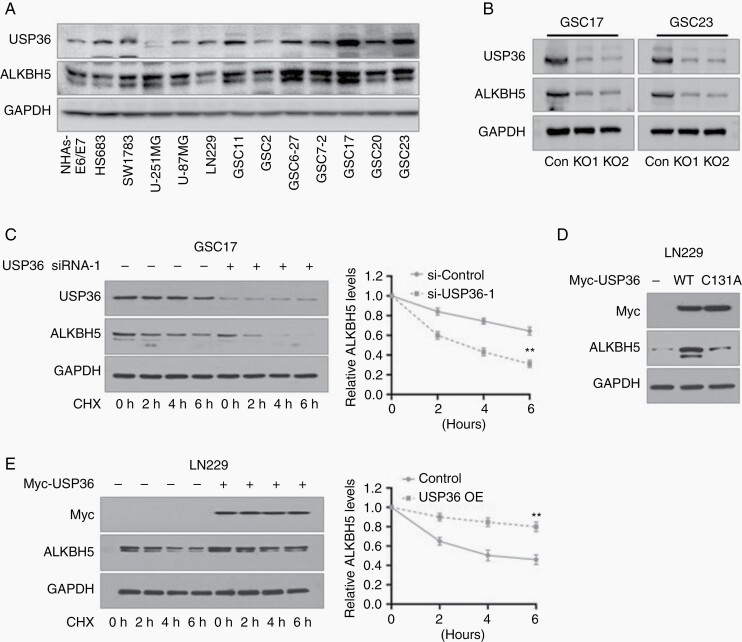
A growing body of studies has shown that USP36 is associated with malignancy in various tumors.23,24 We first detected the expression of USP36 in glioblastoma cell lines to assess the protein correlation of USP36 and ALKBH5. As anticipated, the protein expression of USP36 was significantly increased in GSCs. When non-GSC glioma cell lines were compared with NHA cells, the expression levels of USP36 were directly correlated with that of ALKBH5 (Figure 2A).
Figure 2.
USP36 stabilizes ALKBH5 in glioblastoma cells through its deubiquitinase activity. (A) Immunoblotting analysis of USP36 and ALKBH5 in glioma stem cells (GSCs), glioblastoma cell lines, and non-GSC glioma cell lines. (B) Immunoblotting analysis of ALKBH5 and USP36 protein expression in GSC17 and GSC23 cells after depleted USP36 expressions. (C) The ALKBH5 expressions were analyzed by immunoblotting in GSC17 cells under the treatment with USP36 siRNA-1 and CHX. Western blotting band intensity of ALKBH5 was quantified and normalized to the internal control (data are mean ± SD for triplicate biological replicates; Student t-test; **, P < .01). (D) LN229 cells were transfected with USP36 wild-type and mutant plasmids. The expression of ALKBH5 was analyzed with immunoblotting. (E) The ALKBH5 expressions were analyzed in USP36 overexpressed LN229 cells under treatment with 25 μM MG132. Western blotting band intensity of ALKBH5 was quantified and normalized to the internal control (data are mean ± SD for triplicate biological replicates; Student t-test; **, P < .01).
To further confirm the regulatory effects of USP36 on ALKBH5 expression, we used CRIPSR/Cas9 to knockdown USP36 expressions in GSC17 and GSC23 cells. As expected, USP36 depletion significantly inhibited the turnover of ALKBH5 (Figure 2B). Meanwhile, to confirm whether the protein fluctuation of ALKBH5 was due to the transcriptional discrepancy, we validated the mRNA expression of ALKBH5 and found the USP36 deletion had no effects on the mRNA expression of ALKBH5 (Supplemental Figure S2A and S2B). Moreover, the depletion of USP36 using two specific siRNAs enhanced the degradation of ALKBH5 and this effect became more apparent with increasing CHX incubation time (Figure 2C and Supplemental Figure S2C–D).
Previous studies indicate that the C131 residue in the N-terminal USP domain is a key residue for USP36 and contributes to its catalytic activity.25 Thus, we mutated the residue C131 to alanine to generate a USP36 catalytically inactive mutant, and then ectopically expressed wild-type USP36 or the mutant plasmids in LN229 cells. We found that the elevation of USP36 markedly increased the ALKBH5 expression; however, the catalytically inactive C131A mutant of USP36 failed to increase ALKBH5 levels (Figure 2D). Furthermore, the overexpressed USP36 dramatically prolonged ALKBH5 stability with the treatment of CHX (Figure 2E). Collectively, these results indicate that USP36 directly stabilizes ALKBH5 protein expression in glioblastoma cells in a deubiquitinase activity-dependent manner.
USP36 Directly Interacts with ALKBH5
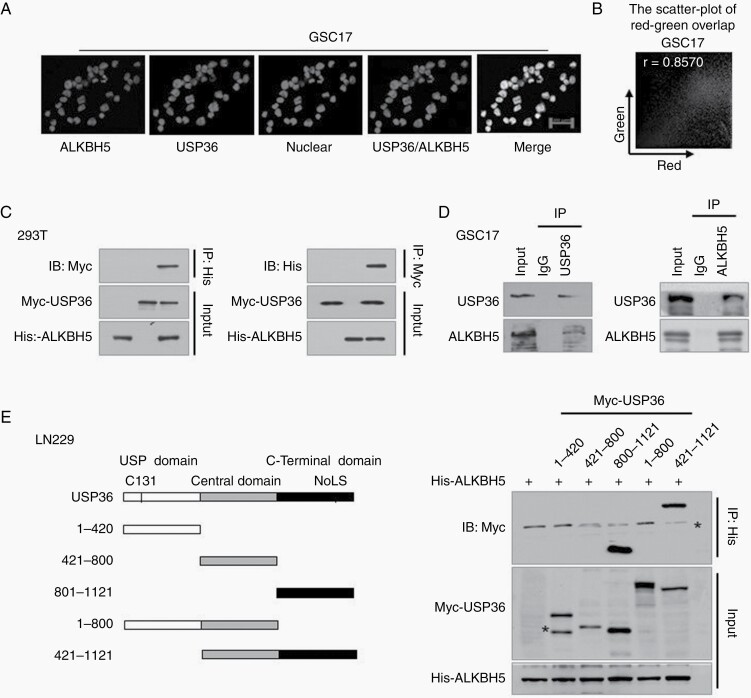
We then investigated the direct interaction between USP36 and ALKBH5. First, we wanted to determine whether the USP36 could co-localize with ALKBH5 in GSCs. Immunofluorescence assay in GSC17 and GSC23 cells showed that the USP36 co-localized with ALKBH5, primarily in the nuclei (Figure 3A and Supplemental Figure S3A). The quantification of the overlap between USP36 and ALKBH5 was determined with the Image Pro Plus software. The results showed that there is a linear relationship between the USP36 and ALKBH5 signal intensities, as the Pearson’s correlation between ALKBH5 and USP36 in GSC17 and GSC23 cells are 0.8570 and 0.8009, respectively (Supplemental Figure S3B). Moreover, the results showed that USP36 colocalizes with ALKBH5 because the overlap coefficient of USP36 and ALKBH5 signals in GSC17 and GSC23 cells are 0.8687 and 0.8144 respectively (Figure 3B and Supplemental Figure S3B).
Figure 3.
USP36 interacts with ALKBH5 directly. (A) Immunofluorescence assays of USP36 and ALKBH5 were performed in GSC17 cells. Scale bar, 20 μm. (B) The quantification of overlap between USP36 and ALKBH5 in (A) was determined with the Image Pro Plus software. The representative red-green scatter-plot of USP36 and ALKBH5 images are shown. (C) 293T cells were transfected with His-ALKBH5 and Myc-USP36, and cell lysates were immunoprecipitated with His-tag or Myc-tag antibody followed by immunoblotting analysis using the indicated antibodies. (D) GSC17 cell lysates were immunoprecipitated with an anti-USP36 or ALKBH5 antibody and the immunoprecipitates were analyzed by immunoblotting using the indicated antibodies. (E) A series of USP36-deletion constructs were generated and co-transfected with His-ALKBH5 into 293T cells. The cell lysates were immunoprecipitated with an anti-His antibody and then subjected to immunoblotting analysis with indicated antibodies. Stars indicated non-specific western bands in this immunoprecipitation result.
Next, we examined the exogenous interaction in 293T cells ectopically expressing both proteins. As shown in Figure 3C, USP36 was indeed physically co-immunoprecipitated with ALKBH5, and vice versa. Furthermore, we assessed the endogenous interaction between USP36 and ALKBH5 in GSC17 cells. Consistently, the reciprocal co-immunoprecipitation experiment showed the endogenous USP36 and ALKBH5 interacted with each other (Figure 3D). In line with this observation, endogenous USP36 protein could directly interact with ALKBH5 in GSC23 cells, and the alternative is also true (Supplemental Figure S3C and D).
To determine which domain of USP36 interacts with ALKBH5, we constructed a panel of Myc-tagged USP36 deletion mutants, including the N-terminal USP domain (1–420), central domain (421–800), the C-terminal nucleolar localization signal (NoLS)-containing domain (801–1121), and two combined domains (1–800 and 401–1121) (Figure 3E). The results showed that C-termianl USP domain-USP36800-1121 could special co-immunoprecipitate with ALKBH5, but not the deletion mutant lacking this domain (Figure 3E and Supplemental Figure S3E). Taken together, these results indicate that USP36 directly interacts with ALKBH5 through its C-terminal domain.
ALKBH5 is Deubiquitinated Directly by USP36
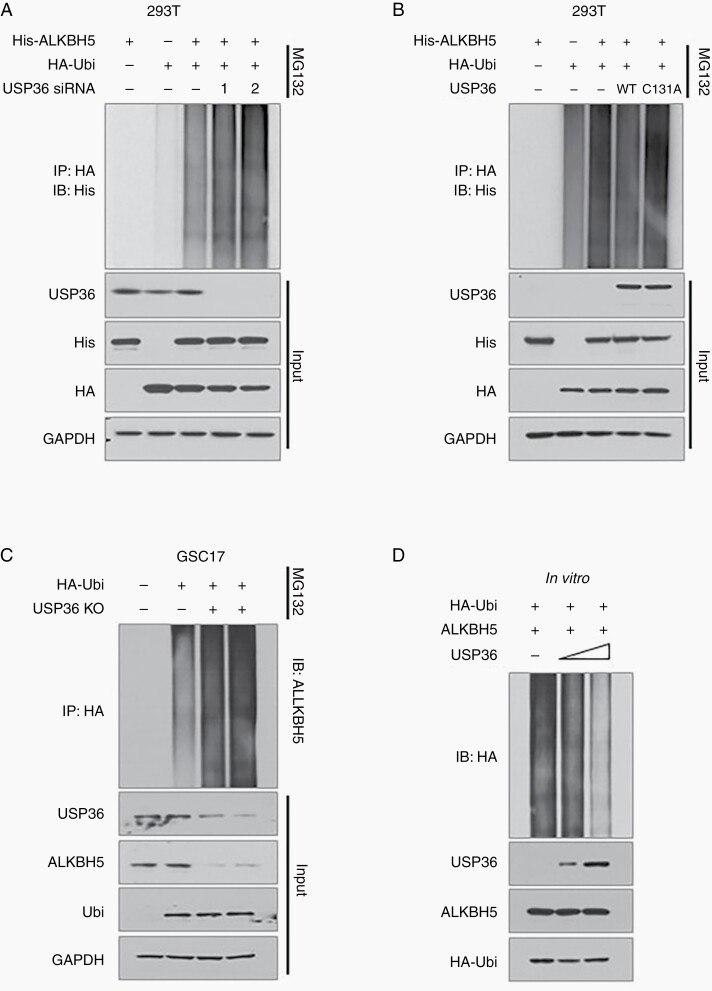
To test whether the deubiquitinase activity of USP36 is required for ALKBH5 regulation, we further evaluated the effect of USP36 on ALKBH5 ubiquitination. First, we found the downregulation of USP36 promoted the ubiquitination of exogenous ALKBH5 in 293T cells (Figure 4A). Then we ectopically expressed USP36 and catalytic inactive mutant into 293T cells and found the wild-type USP36 remarkably decreased the ubiquitination of ALKBH5 (Figure 4B, lane4). However, USP36-C131A did not alter ALKBH5 ubiquitination (Figure 4B, lane 5). Conversely, we examined the ubiquitination of endogenous ALKBH5 in USP36-depleting GSC17 and GSC23 cells and found that USP36 deletion led to a significant accumulation of endogenous ubiquitinated ALKBH5 (Figure 4C, Supplemental Figure S4). Subsequent in vitro deubiquitination assays with purified His-ALKBH5, HA-Ubi, and different amounts of Myc-USP36, showed that USP36 remarkably diminished ALKBH5 ubiquitination, and the efficiency was reinforced with the quantitation of USP36 proteins that were appended to the reactions (Figure 4D). These results strongly indicate that USP36 interacts with and deubiquitinates ALKBH5 directly.
Figure 4.
USP36 stabilizes ALKBH5 by inhibiting its ubiquitination. (A) 293T cells were transfected with indicated plasmids or specific siRNA. Cell lysates were immunoprecipitated with anti-HA antibody and then analyzed by immunoblotting. (B) 293T cells were transfected with indicated plasmids, and followed by treatment with MG132 for 6 hours before cell harvest. Cell lysates were immunoprecipitated with anti-HA antibody and then analyzed by immunoblotting using the indicated antibodies. (C) The endogenous ubiquitination of ALKBH5 was analyzed in USP36-knockout GSC17 cells after transfected with HA-Ubi. Cell lysates were immunoprecipitated with anti-HA antibody and then analyzed by immunoblotting using the indicated antibodies. (D) 293T cells were transfected with His-ALKBH5 and HA-Ubi and then treated with MG132 for 6 hours. ALKBH5 protein was purified using the magnetic bead. The ubiquitinated ALKBH5 was then incubated with 50 ng or 500 ng USP36 in a deubiquitination buffer. The resulting reactions were subjected to immunoblotting analysis.
USP36 Maintain the Stemness of GSCs Through ALKBH5 and USP36-ALKBH5 Axis Promotes Glioma Tumorigenicity
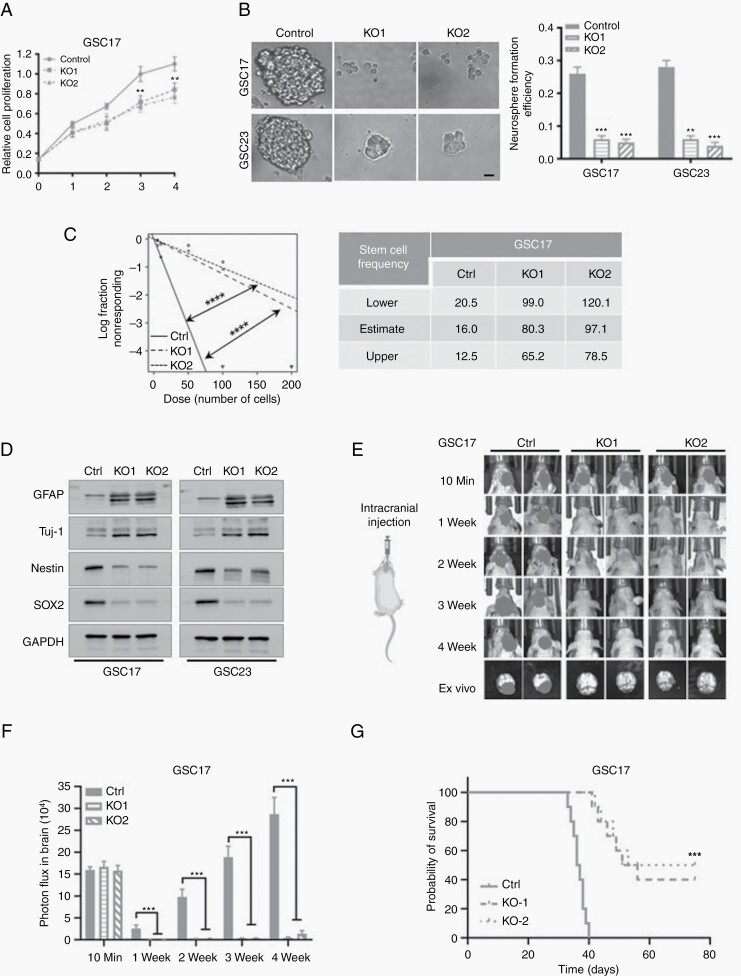
Previous studies have suggested that cell proliferation is required for self-renewal of GSC. Therefore, we investigated the roles of the USP36-ALKBH5 axis in glioma cell proliferation and stemness. Depletion of USP36 significantly decreased the viable cell number and inhibited cell proliferation (Figure 5A and Supplemental Figure S5A). Then we examined the stemness of the GSC cells with a neurosphere formation assay. The results indicated that the knockout of USP36 substantially declined the sphere sizes and neurosphere formation efficiency in comparison with control cells (Figure 5B). An extreme limiting dilution analysis (ELDA) was performed to check the GSC frequency and self-renewal capacity. We found the USP36 deletion significantly reduced the tumorsphere formation frequency (Figure 5C and Supplemental Figure S5B). Next, we explored the relationship between USP36 and specific stem cell markers and found the USP36 expression was significantly decreased after inducing differentiation of GSC cells (Supplemental Figure S5C). Additionally, USP36 knockout substantially inhibited the expression of stem cell markers, Nestin and SOX2, but upregulated the differentiation markers, GFAP and Tuj1, in GSCs (Figure 5D). Therefore, the data indicate that USP36 knockout suppresses the stemness of GSC cells.
Figure 5.
Knockout of USP36 impaired the stemness and tumorigenesis of glioblastoma. (A) The cell proliferation of GSC17 cells after ablation of USP36 was assessed by the CCK8 assay. Data are mean ± SEM, for triplicate biological replicates with three technical replicates in each biological one. Student’s t-test; **P < .01. (B) The neurosphere formation efficiency (spheres/cells plated) of the GSC17 and GSC23 cells were detected after the depletion of USP36. The significance was determined by Student’s t-test. Data are mean ± SEM for triplicate samples. **P < .01; ***P < .001. Scale bar, 100 μm. (C) Extreme limiting dilution assay showed the frequencies of neurosphere formation in USP36-depleted GSC17 cells. The significance of the difference was determined by Chi-square test. ****P < .0001 (n = 3 independent experiments). (D) The protein expressions of downstream targets of ALKBH5 in USP36 knockout or control GSC17 and GSC23 cells were analyzed by western blot analysis. (E) Representative BLI images of the GSC17 cell are shown after intracranial injection. (F) The value of bioluminescence was quantitated by measuring photon flux. Values are the mean ± SD, n = 8, Two-sided Mann–Whitney test, ***P < .001. (G) Kaplan–Meier survival analyses for mice injected with control or USP36 sgRNA GSC17 cells (mean ± SD, n = 10; log-rank test, ***P < .001).
To investigate the effects of the USP36-ALKBH5 axis on tumor formation, we performed in vivo intracranial tumor assay with luciferase-labeled GSC cells and bioluminescence imaging (BLI) to track the tumor growth. Accord to the BLI, all mice injected with control cells developed tumors with characteristic glioblastoma features. In contrast, depletion of USP36 significantly inhibited tumor formation compared with the control group. Furthermore, the ex vivo imaging of mice brains confirmed that USP36 knockdown remarkably diminished the tumor growth (Figure 5E–F, Supplemental Figure S5D-E). We next determined the effect of USP36-ALKBH5 axis on the survival of glioma-bearing mice. Knockdown of USP36 in GSC11 and GSC17 cells extended the survival time of the implanted mice compared with the control (Figure 5G, Supplemental Figure S5F). Furthermore, the protein expressions of USP36 and ALKBH5 were down-regulated in brain tumors of USP36-deleted GSC17 or GSC11 cells as compared with control cells. In addition, we examined the cell proliferation marker Ki-67 expression in the brain tumors and found the expressions of Ki-67 were decreased in tumors formed by USP36-depleted GSC17 or GSC11 cells (Supplemental Figure S5G).
Next, to ascertain whether ALKBH5 is a major contributor to the functions of USP36 in GSC proliferation and tumor growth, we asked whether adding back ALKBH5 could reverse the effects of ALKBH5 inhibition. Firstly, we re-expressed ALKBH5 in USP36-depleted GSC17 and GSC11 cells by using lentiviral transduction of ALKBH5 to establish stable cell lines. As shown in the Supplemental Figure S5H, the protein levels of ALKBH5 were increased after re-expression of ALKBH5. However, the protein levels of USP36 were not changed by the re-expression of ALKBH5 in the USP36-depleted GSC17 and GSC11 cells. Secondly, we examined the effects of re-expression of ALKBH5 after USP36 deletion on the cell proliferation, neurosphere formation, and self-renewal capacities. We found the deterioration of cell proliferation, neurosphere formation, and self-renewal capacities induced by USP36 deletion were notably suppressed by the ALKBH5 re-expression (Supplemental Figure S5I-L). Furthermore, we determined the effects of the re-expression of ALKBH5 on tumor growth by using the in vivo intracranial tumor assay. We found that the re-expression of ALKBH5 in the USP36-depleted GSC17 and GSC11 cells largely abolished tumor growth inhibition by USP36 knockout (Supplemental Figure S5M). Collectively, these results consistently demonstrate that the USP36-ALKBH5 axis plays a critical role in GSC cell proliferation and tumorigenesis.
USP36 is Increased in Glioma Patient Samples and Correlated with ALKBH5, and USP36 Knockout Sensitized GSCs to TMZ Treatment
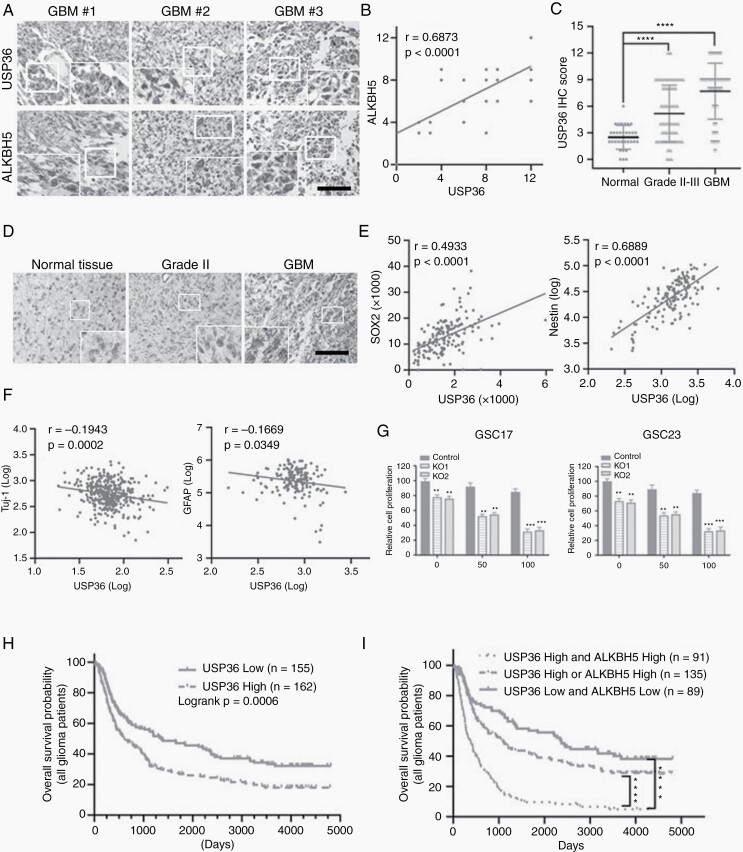
To better understand the potential clinical correlation between USP36 and ALKBH5, we next examined the expressions of USP36 and ALKBH5 in 43 paired human glioblastoma specimens. The results of immunochemistry assays showed that the expression levels of USP36 in GBM samples were directly correlated with ALKBH5 levels (Figure 6A). The quantification of immunochemistry staining and statistical analysis disclosed the significant correlations between the protein expression of USP36 and ALKBH5 (Figure 6B). We then inspected whether the protein levels of USP36 correlated with the grade of glioma malignancy in 34 normal brain tissues, 87 grade II-III astrocytomas, and 51 glioblastomas. We found that USP36 expression was strongly expressed in gliomas and the expression level was dramatically higher in glioblastoma than in lower-grade astrocytoma, which can be positively correlated to increasing glioma grades (Figure 6C and D). To further verify the correlation between USP36 and ALKBH5 protein expression, we excavated the USP36 mRNA expression in TCGA and REMBRANDT database. In the TCGA and REMBRANDT datasets, we found there was a slightly higher mRNA expression of USP36 observed in glioma; however, the mRNA expression levels were not found to correlate with glioma grade (Supplemental Figure 6A). Notably, the protein expression of USP36 is significantly increased in GBM and correlates with tumor grade (Supplemental Figure 6B). The expression of SOX2, Nestin, Vimentin, and CD44 exhibited a positive correlation with USP36 expression, and Tuj-1, GFAP, MOG, and OLIG1 exhibited a negative correlation with USP36 expression (Figure 6E-F, Supplemental Figure S6C). Collectively, these data further support the critical role of USP36 in controlling ALKBH5 protein expression in human glioblastomas.
Figure 6.
USP36 is increased in glioma specimens and correlates with ALKBH5 protein level, and USP36 knockout sensitized GSCs to TMZ treatment. (A) Immunohistochemical staining detected the expression of USP36 and ALKBH5 in 43 glioblastoma specimens. Scale bar, 200 μm. (B) Correlation between ALKBH5 and USP36 protein expressions in human specimens was analyzed by the Pearson correlation test. IHC staining of USP36 and ALKBH5 was scored as 1–12 according to the percentage of positive cells and staining intensity in a sample. Note that the scores of some samples overlap. (C) The expression levels of active USP36 in 34 normal, 87 grade II and III astrocytomas were compared with those in 50 glioblastoma specimens. The significance was determined by statistical analysis t-test. ****P < .0001. (D) IHC staining of USP36 was carried out on normal, grade II astrocytoma and glioblastoma specimens. Scale bars, 200 μm. (E/F) Correlation between USP36 and indicated gene expressions in the TCGA dataset was analyzed by Pearson correlation test. (G) The USP36 knockout or control GSC17 and GSC23 cells were treated with vehicle (0.05% DMSO) or TMZ (50 or 100 μM in the vehicle) for 3 days. The cell proliferation was assessed by the CCK8 assay. Student’s t-test, **P < .01, ***P < .001. (H) Kaplan–Meier survival curves were used to assess the overall survival of primary and recurrent glioma patients (all patients) in the CGGA database. The median was the cutoff value used to classify USP36 mRNA expression as high or low. Log-rank test, P = .0006. (I) Kaplan–Meier survival curves were used to assess the overall survival of primary glioma patients and recurrent in the CGGA database based on the expression levels of USP36 and/or ALKBH5 expressions. Log-rank test, ****P < .0001.
Previous studies reported that the stemness of GSCs made them resistant to TMZ; therefore, USP36 knockout that inhibits GSC stemness might sensitize the cells to TMZ. The USP36 knockout or control GSC17 and GSC23 cells were treated with TMZ (0, 50 μM or 100 μM) and cell proliferation was compared at the end of three days. The results revealed that, in both GSC17 and GSC23 cells, the USP36 knockout significantly sensitized the cells to TMZ treatment (Figure 6G). Therefore, this data indicates that loss of USP36 increased the sensitivity of GSCs to TMZ treatment.
Next, we sought to confirm the clinical significance of USP36 which may result in poor clinical outcomes in GBM patients by using the CCGA database (http://www.cgga.org.cn/) analysis. We found that elevated USP36 expression conferred a poor prognosis in both primary and recurrent glioma patients (Figure 6H and Supplemental Figure S6D). Furthermore, to analyze the compound signatures of USP36 and ALKBH5, we have also downloaded ALKBH5 data of the glioma patients from the dataset. We then divided all of the glioma patients (including both primary and recurrent glioma patients) into three groups according to the expressions of USP36 and ALKBH5: 1) both ALKBH5 and USP6 expressions are high and 2) both ALKBH5 and USP36 expressions are low, and only one of the proteins is high (ALKBH5 or USP36 expression is high). Following the analysis of the data of the three groups, we found that the survival rate of USP36-high/ALKBH5-high group is significantly decreased compared with the survival rate of USP36-low/ALKBH5-low group. Also, the survival rate of USP36-high/ALKBH5-high group is significantly decreased compared with the survival rate of the group with only one of the proteins is high (Figure 6I). Taken together, these data not only validate our hypothesis that USP36 promotes glioma tumorigenesis through stabilizing ALKBH5, but also identify USP36 as a potential therapeutic target for glioma therapy.
Discussion
In this study, we have revealed an important role for USP36 in ALKBH5 accumulation via deubiquitination, which leads to the proliferation and self-renewal of GSC in glioma tumorigenesis (Supplemental Figure S6E). Our molecular profiling results, together with clinical findings, show that the USP36-ALKBH5 axis is essential for the glioblastoma process, and pursuing this axis for targeted therapy may lead to the discovery of pharmacotherapeutic agents to expand the menu of available strategies for glioblastoma treatment.
ALKBH5 is an influential m6A demethylase (also reported m6A eraser), which has generated remarkable biological interest among various researchers.28 Abnormal expression of ALKBH5 participates in tumorigenesis of multiple cancers such as glioblastoma,16 pancreatic cancer,29 breast cancer,30 gastric cancer,31 lung cancer,32 and ovarian cancer.33 Moreover, emerging studies have demonstrated that ALKBH5 affects many biological processes, including embryonic stem cell cardiac commitment, brain development, post-ischemic angiogenesis, ROS-induced DNA damage response and immune response, and chemotherapy resistance.32,34 However, little is known about the dysregulation of ALKBH5 in cancers, especially the high expression in glioblastoma. Herein, we showed that the ALKBH5 is regulated by the ubiquitin-proteasome pathway, and the deubiquitination process is responsible for the high expression in glioblastoma.
Chemotherapy is one of the main therapeutic strategies, and temozolomide (TMZ), an oral alkylating agent, is the first-line chemotherapy after surgical excision for glioma patients.35 However, the chemoresistance of TMZ restrain its usage in the clinic.36 The drug resistance of TMZ is likely secondary to a variety of mechanisms including the m6A modification. Ding and colleagues identified that the abundance of circ_0072083 was increased in TMZ-resistant glioma tissues and cells. The circ_0072083 promoted TMZ resistance via enhancing NANOG level induced by ALKBH5-mediated demethylation in TMZ-resistant glioma cells.37 Meanwhile, Liu et al. found the LncRNA SOX2OT was increased in TMZ-resistant cells and recurrent GBM patient samples. The LncRNA SOX2OT recruited ALKBH5, which binds with SOX2, demethylating the SOX2 transcript, leading to enhanced SOX2 expression in TMZ-resistant glioma cells.38
Deubiquitination induced by DUBs, the reversed course of ubiquitin-proteasome system-mediated proteolysis, has been demonstrated to impose a significant influence on the modulation of a broad range of cellular mechanisms including malignant transformation.39 Furthermore, growing evidence indicates that DUBs, including USP36, stabilize several CSC (cancer stem cell)-associated transcriptional factors and mediate the CSC self-renewal, proliferation, and differentiation.40 Previous studies have revealed overexpression of USP36 in other various types of human cancers, including breast and lung cancer.26 In breast cancer cells, USP36 interacted with and deubiquitinated c-Myc. Knockdown of USP36 in the cells by shRNA reduced the levels of c-Myc and suppressed cell proliferation.26 Meanwhile, in neuroblastoma, the expression of USP36 was increased. USP36 KD led to CDH7 destabilization and promoted neuronal differentiation.25 These results are in concordance with our study that USP36 play a critical role in proliferation and self-renewal of GSCs. Moreover, our study identified that expression of USP36 was highly increased in GSCs, which is clinically correlated with ALKBH5 levels in glioblastoma specimen samples and inversely correlated with glioblastoma patients’ survival rate. Furthermore, we showed that knockout of USP36 can significantly promote the drug sensitivities of GSCs to TMZ treatment.
In our study, we additionally uncovered that USP36 interacts with and stabilizes ALKBH5, leading to the upregulation of ALKBH5-target genes. Moreover, segmentation of USP36-ALKBH5 axis inhibits cell proliferation and self-renewal of GSCs and drug sensitivities of GSCs. Considering the striking functional roles of ALKBH5 in the tumorigenesis of glioblastoma, we suggest that USP36-induced activation of ALKBH5 may be a starting point for the development of more effective therapies for glioblastoma.
Supplementary material
Supplementary material is available online at Neuro-Oncology (http://neuro-oncology.oxfordjournals.org/).
Acknowledgements
We thank Dr Jianhua Yang (Texas Children’s Cancer Center, Houston, TX) for providing DUB-expressing plasmids. We give thanks for the services of the VCU MCC Mouse Model and Microscopy Core Facilities, which are supported, in part, by NCI P30 CA016059 grant.
Conflict of interest statement: None declared.
Contributor Information
Guoqiang Chang, Department of Human and Molecular Genetics, Institute of Molecular Medicine, VCU Massey Cancer Center, Virginia Commonwealth University, School of Medicine, Richmond, Virginia 23298, USA.
Gloria S Xie, Department of Human and Molecular Genetics, Institute of Molecular Medicine, VCU Massey Cancer Center, Virginia Commonwealth University, School of Medicine, Richmond, Virginia 23298, USA.
Li Ma, Department of Neurosurgery, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.
Linlin Li, Department of Human and Molecular Genetics, Institute of Molecular Medicine, VCU Massey Cancer Center, Virginia Commonwealth University, School of Medicine, Richmond, Virginia 23298, USA.
Hope T Richard, Department of Pathology, Virginia Commonwealth University, School of Medicine, Richmond, Virginia 23298, USA.
Author contributions
Conception and design: G. Chang, H. Richard. Development of methodology: G. Chang, G. Xie, L. Ma. Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): G. Chang, G. Xie, L. Ma, P. Li, L. Li. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): G. Chang, G. Xie, H. Richard. Writing, review, and/or revision of the manuscript: G. Chang, G. Xie, H. Richard. Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): L. Ma, P. Li, L. Li. Study supervision: G. Chang, H. Richard.
References
- 1. Qazi MA, Vora P, Venugopal C, et al. Intratumoral heterogeneity: pathways to treatment resistance and relapse in human glioblastoma. Ann Oncol. 2017;28(7):1448–1456. [DOI] [PubMed] [Google Scholar]
- 2. Perrin SL, Samuel MS, Koszyca B, et al. Glioblastoma heterogeneity and the tumour microenvironment: implications for preclinical research and development of new treatments. Biochem Soc Trans. 2019; 47(2):625–638. [DOI] [PubMed] [Google Scholar]
- 3. Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science 2018; 361(6409):1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012; 485(7397):201–206. [DOI] [PubMed] [Google Scholar]
- 5. Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014; 505(7481):117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012; 149(7):1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang G, Shi L, Ye Y, et al. YTHDF3 induces the translation of m(6)A-enriched gene transcripts to promote breast cancer brain metastasis. Cancer Cell 2020; 38(6):857–871.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fustin JM, Doi M, Yamaguchi Y, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013; 155(4):793–806. [DOI] [PubMed] [Google Scholar]
- 9. Wang Y, Li Y, Toth JI, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014; 16(2):191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weng H, Huang H, Wu H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell 2018; 22(2):191–205.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fang R, Chen X, Zhang S, et al. EGFR/SRC/ERK-stabilized YTHDF2 promotes cholesterol dysregulation and invasive growth of glioblastoma. Nat Commun. 2021; 12(1):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deng X, Su R, Weng H, et al. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018; 28(5):507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shen C, Sheng Y, Zhu AC, et al. RNA demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell 2020; 27(1):64–80.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu F, Wei J, Cui X, et al. Post-translational modification of RNA m6A demethylase ALKBH5 regulates ROS-induced DNA damage response. Nucleic Acids Res. 2021; 49(10):5779–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang S, Zhao BS, Zhou A, et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 2017;31(4):591–606.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem. 2017; 86:129–157. [DOI] [PubMed] [Google Scholar]
- 18. Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009; 78:363–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mevissen TET, Komander D. Mechanisms of deubiquitinase Specificity and Regulation. Annu Rev Biochem. 2017; 86:159–192. [DOI] [PubMed] [Google Scholar]
- 20. Bonacci T, Emanuele MJ. Dissenting degradation: deubiquitinases in cell cycle and cancer. Semin Cancer Biol. 2020; 67(Pt 2):145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mennerich D, Kubaichuk K, Kietzmann T. DUBs, hypoxia, and cancer. Trends Cancer 2019; 5(10):632–653. [DOI] [PubMed] [Google Scholar]
- 22. Ma L, Lin K, Chang G, et al. Aberrant activation of beta-catenin signaling drives glioma tumorigenesis via USP1-mediated stabilization of EZH2. Cancer Res. 2019; 79(1):72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang J, Ren P, Xu D, et al. Human UTP14a promotes colorectal cancer progression by forming a positive regulation loop with c-Myc. Cancer Lett. 2019; 440-441:106–115. [DOI] [PubMed] [Google Scholar]
- 24. Mondal T, Juvvuna PK, Kirkeby A, et al. Sense-antisense lncRNA pair encoded by locus 6p22.3 determines neuroblastoma susceptibility via the USP36-CHD7-SOX9 regulatory axis. Cancer Cell 2018; 33(3):417–434.e7. [DOI] [PubMed] [Google Scholar]
- 25. Sun XX, He X, Yin L, et al. The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc Natl Acad Sci USA. 2015; 112(12):3734–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fraile JM, Campos-Iglesias D, Rodriguez F, et al. Loss of the deubiquitinase USP36 destabilizes the RNA helicase DHX33 and causes preimplantation lethality in mice. J Biol Chem. 2018; 293(6):2183–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Taillebourg E, Gregoire I, Viargues P, et al. The deubiquitinating enzyme USP36 controls selective autophagy activation by ubiquitinated proteins. Autophagy 2012; 8(5):767–779. [DOI] [PubMed] [Google Scholar]
- 28. Lan Q, Liu PY, Haase J, et al. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 2019;79 (7):1285–1292. [DOI] [PubMed] [Google Scholar]
- 29. Tang B, Yang Y, Kang M, et al. m(6)A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol Cancer. 2020; 19(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang C, Samanta D, Lu H, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016; 113(14):E2047–E2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yue B, Cui R, Zheng R, et al. Essential role of ALKBH5-mediated RNA demethylation modification in bile acid-induced gastric intestinal metaplasia. Mol Ther Nucleic Acids. 2021; 26:458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jin D, Guo J, Wu Y, et al. m(6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol Cancer. 2020; 19(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fukumoto T, Zhu H, Nacarelli T, et al. N(6)-Methylation of adenosine of FZD10 mRNA contributes to PARP inhibitor resistance. Cancer Res. 2019; 79(11):2812–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li XC, Jin F, Wang BY, et al. The m6A demethylase ALKBH5 controls trophoblast invasion at the maternal–fetal interface by regulating the stability of CYR61 mRNA. Theranostics 2019; 9(13):3853–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lang F, Liu Y, Chou FJ, Yang C. Genotoxic therapy and resistance mechanism in gliomas. Pharmacol Ther. 2021; 228:107922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li F, Chen S, Yu J, et al. Interplay of m(6) A and histone modifications contributes to temozolomide resistance in glioblastoma. Clin Transl Med. 2021; 11(9):e553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ding C, Yi X, Chen X, et al. Warburg effect-promoted exosomal circ_0072083 releasing up-regulates NANGO expression through multiple pathways and enhances temozolomide resistance in glioma. J Exp Clin Cancer Res. 2021; 40(1):164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu B, Zhou J, Wang C, et al. LncRNA SOX2OT promotes temozolomide resistance by elevating SOX2 expression via ALKBH5-mediated epigenetic regulation in glioblastoma. Cell Death Dis. 2020; 11(5):384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lange SM, Armstrong LA, Kulathu Y. Deubiquitinases: from mechanisms to their inhibition by small molecules. Mol Cell. 2022; 82(1):15–29. [DOI] [PubMed] [Google Scholar]
- 40. Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018; 17(1):57–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.