Abstract
Over the last century, the number of epidemics caused by RNA viruses has increased and the current SARS-CoV-2 pandemic has taught us about the compelling need for ready-to-use broad-spectrum antivirals. In this scenario, natural products stand out as a major historical source of drugs. We analyzed the antiviral effect of 4 stilbene dimers [1 (trans-δ-viniferin); 2 (11’,13’-di-O-methyl-trans-δ-viniferin), 3 (11,13-di-O-methyl-trans-δ-viniferin); and 4 (11,13,11’,13’-tetra-O-methyl-trans-δ-viniferin)] obtained from plant substrates using chemoenzymatic synthesis against a panel of enveloped viruses. We report that compounds 2 and 3 display a broad-spectrum antiviral activity, being able to effectively inhibit several strains of Influenza Viruses (IV), SARS-CoV-2 Delta and, to some extent, Herpes Simplex Virus 2 (HSV-2). Interestingly, the mechanism of action differs for each virus. We observed both a direct virucidal and a cell-mediated effect against IV, with a high barrier to antiviral resistance; a restricted cell-mediated mechanism of action against SARS-CoV-2 Delta and a direct virustatic activity against HSV-2. Of note, while the effect was lost against IV in tissue culture models of human airway epithelia, the antiviral activity was confirmed in this relevant model for SARS-CoV-2 Delta. Our results suggest that stilbene dimer derivatives are good candidate models for the treatment of enveloped virus infections.
Keywords: Broad-spectrum antiviral, Stilbene dimer derivatives, Influenza virus, SARS-CoV-2, Virucidal
Graphical Abstract
1. Introduction
Among all pathogens responsible for human diseases, viruses are of major public health concern [1]. Due to their diversity and capacity of adaptation, viruses can cause unpredictable epidemics and pandemics. The emergence and spread of viral pandemics has increased dramatically over the past twenty years, due to animal migration, disruption of ecological niches, facilitated cross-species contacts, mundialization and increased travels around the world [2]. These changes have enabled viruses to spread faster and wider, leading to a higher chance to encounter a profitable viral population to emerge and evolve. Viruses are intracellular obligate parasites, which makes hard to eradicate them and forces the development of specific inhibitors targeting viral replication cycle without interfering with the host functions.
The drugs in clinical use today act on key stages of the viral cycle (entry, replication and assembly, release and spread) or interfere with the viral genome [3]. However, despite the numerous assets of the omics technologies, current preventive or therapeutic approaches against viral infections remain limited and viral plasticity can easily reduce the efficacity of virus-specific drugs. The death toll of SARS-CoV-2 pandemic is just a recent example of the flaws of the currently available antiviral therapies.
We need ready-to-use broad-spectrum antivirals to rapidly counteract the spread of new viruses. Ideally, these novel drugs should be affordable, easy to synthesize and should display a high genetic barrier to antiviral resistance. In case of a pandemic outbreak, the massive use of an antiviral would be ineffective in the long term, if the virus can gain resistance just upon a low number of mutations, as for Influenza Virus (IV), where a single amino acid substitution in the neuraminidase gene can confer complete resistance against Oseltamivir [4].
Natural products (NPs) have been one of the main historical sources of drugs [5]. Due to their incredible chemical diversity, NP "scaffolds" inspire the development of a large number of drugs against many diseases [5]. Acyclovir (Zovirax®), used against Herpes viruses, was for example developed based on the nucleoside structures isolated from the Caribbean sponge, Cryptotethya crypta [6].
The classical way to obtain original NPs with unusual structures is to purify extracts from plants, microorganisms or marine organisms, using a bioguided isolation process [7]. Recently, we developed an alternative approach based on the use of the secreted fungal enzymes (secretome) of Botrytis cinerea, to generate chemical complexity from simple substrate NP scaffolds. Using this method, it was possible to obtain libraries of chemo diverse compounds from common NPs such as stilbenes and phenylpropanoids [8], [9], [10], [11], [12]. Some of the resulting compounds, such as trans-δ-viniferin derivatives (TδVD) possess remarkable antimicrobial properties against multidrug resistant strains of Staphylococcus aureus [8], an opportunistic Gram-positive bacterium that colonizes the nose, throat, skin, and gastrointestinal tract of humans [13].
The antimicrobial mechanism of action of TδVD were reported to reliy on their ability to disrupt the bacterial membrane [14]. This prompted us to further investigate whether TδVD could display broad-spectrum antiviral properties also against enveloped viruses, that acquire the membrane of the host cells. We thus tested the antiviral activity of a series of TδVD against a panel of enveloped viruses. We detected antiviral activity against IV, SARS-CoV-2 Delta and Herpes Simplex Virus 2 (HSV-2), with different mechanisms of actions. Importantly, we showed that the barrier to antiviral resistance of these compounds is high, as IV retains its susceptibility even if passed 10 times in presence of suboptimal drug concentrations.
2. Materials and methods
2.1. Cells and tissues
Madin-Darby canine kidney (MDCK), Calu-3, Vero, Vero-E6, Hela and A549 cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured according to the manufacturers’ conditions.
Human airway epithelia (MucilAir™) were purchased from Epithelix (Geneva, Switzerland) [15].
2.2. Viruses
Human IV H1N1 A/Netherlands/602/2009 (A(H1N1)pdm09), B/Wisconsin/01/2010 (Yamagata lineage) and a reassortant strain bearing the HA and NA from A/Vietnam/1203/2004 (H5N1) virus and the backbone of H1N1 A/Puerto Rico/8/1934 (H5N1/PR8) were kindly provided by Prof. Mirco Schmolke (University of Geneva). All IV strains were amplified and titrated in MDCK cells by plaque assay (PA) [4].
SARS-CoV-2 B.1.617.2, Delta variant (hCoV-19/Switzerland/GE-33896105/2021), was amplified in Calu-3 cells and titrated in Vero-E6 cells by PA [16]. For ex vivo experiments, a stock of SARS-CoV-2 Delta (hCoV-19/Switzerland/GE-HUG-35274181/2021) was produced in MucilAir [16]. Information regarding HSV-2 and enterovirus EV-71 can be found in the supplementary Materials and Methods. All experimental work was performed in appropriate biosafety level laboratory.
2.3. Viral infectivity determination
Viral infectivity was determined by at least two independent experiments performed in duplicate. IV and HSV-2 were titrated by PA. SARS-CoV-2 Delta was quantified by RT-qPCR and the infectiousness of EV-A71 was determined by TCID50 [4], [16], [17].
Single replication cycle of SARS-CoV-2 Delta was performed in confluent Calu-3 cells in 96-well plate infected with a MOI of 0.01 PFU/cell. Right after addition of virus on cells, the plates were put at 4 °C for 1 h in order to synchronize the infection. Then, 62.3 µM of compound 3 were added to the cells at 0 hpi or 2 hpi. After 8 h of incubation at 37 °C, the cells were washed with PBS -/- and lysed with a lysis RNA extraction buffer.
2.4. RT-qPCR analysis and viral RNA copies quantification
Viral RNA extracted from Calu-3 supernatants and MucilAir apical washes was quantified by RT-qPCR [18]. For A(H1N1)pdm09, in vitro transcripts of the Influenza A/California/7/2009(H1N1) M gene were used as standard reference [16]. SARS-CoV-2 Delta RNA copies were quantified using as standard reference the transcript of the SARS-CoV-2 E gene [16]. CT values were converted into RNA loads using the slope-intercept form. In all experiments the slope efficiency and R2 ranged between and 0.96–0.99 [15], [19].
2.5. Generation of the TδVD by chemoenzymatic synthesis
The four TδVD (designated as 1–4 in the text) were obtained via chemoenzymatic synthesis [11], starting from resveratrol and pterostilbene, which were dimerized using the enzymatic secretome of Botrytis cinerea [10]. Details about the synthesis and isolation of the compounds can be found in the Supplementary material section.
2.6. Cell viability assay
MDCK (2 x 1E4), Calu-3 (1 x 1E5), Vero (2 x 1E4), Vero-E6 (2 x 1E4), Hela (2x1E4) and A549 (2x1E4) cells were seeded in a 96-well plate one day before the assay. A dose range of each TδVD, spanning from 3.1 μM to 282.1 μM, was added on the cells for 24 h or 48 h. While dose ranges spanning from 24.2 µM to 2.2 mM were added for 1 h. The compounds were dissolved in serum-free DMEM or serum-free DMEM + TPCK-Trypsin (0.2 μg/ml) for MDCK cells; in 2% FBS or serum-free MEM for Calu-3 cells; or in 2% FBS DMEM for Hela, A549, Vero and Vero-E6 cells. After the treatment, MTT reagent (Promega) was added on the cells for 3 h at 37 °C and the absorbance was read at 570 nm [4]. Percentages of viability were calculated by comparing the absorbance in treated wells vs untreated conditions, in two independent biological replicates.
Cell viability in ex vivo tissues was assessed by Resazurin treatment. MucilAir tissues were treated daily on the apical surfaces with different concentrations of compound (from 172.8 µM to 691.3 µM per tissue, in 30 µl). Each day, 250 µl of Resazurin solution (3 µg/ml in MucilAir medium) was added apically for 3 h, before drug administration. The solution was harvested and the apical side of the tissues was washed with PBS+/+ before adding new daily treatments. To determine the toxicity of the treatments compared to an untreated control, the fluorescence levels of resazurin was measured (excitation at 571 nm, emission at 584 nm).
2.7. Influenza Inhibition assay
A dose range of molecules spanning from 0.98 μM to 31.29 μM was pre-incubated with 500 PFU (corresponding to 0.05 MOI) of A(H1N1)pdm09, IBV or H5N1/PR8 for 1 h in serum-free DMEM at 37 °C. The half maximal effective concentration (EC50) values for inhibition curves were determined by immunocytochemistry (ICC) as previously reported [20].
2.8. Dose response assays in cell lines and ex vivo assays (Influenza and SARS-CoV-2 Delta)
Confluent Calu-3 cells in 96-well plate, were infected with a MOI of 0.1 PFU/cell of A(H1N1)pdm09. In the post-treatment experiment, 2.9 or 10.4 μM of molecules 1, 2, 3 or 4 were added or not to the cells after 1 h of infection at 37 °C. The infectious supernatant was harvested at 24 hpi and titrated by plaque assay (PA) in MDCK cells. In the pre-treatment experiment, the cells were pre-incubated or not with 2.9, 10.4 and 22.0 μM of compounds for 1 h at 37 °C. The molecules were then removed and the cells infected as described above. The infectious supernatant was harvested at 24 hpi and titrated by PA in MDCK cells.
For SARS-CoV-2 Delta, confluent Calu-3 cells in 96-well plate were infected with a MOI of 0.1 PFU/cell. In the post-treatment experiment, 2.6–20.7 µM (compound 2) or 7.8–31.1 µM (compound 3) were added or not at 1 hpi. In the pre-treatment experiment, the same amounts of compounds were added or not to confluent Calu-3 cells in 96-well plate, for 1 h at 37 °C, except for the pre-treatment of 3 where the drug concentrations were from 7.8 to 66.3 µM. The cells were then washed and infected with a MOI of 0.1 PFU/cell. For both pre- and post-treatment experiments, at 24 hpi the infectious supernatant was harvested and titrated by RT-qPCR.
MucilAir tissues were infected on their apical side with 1E4 RNA copies/tissue of A(H1N1)pdm09 or SARS-CoV-2 Delta in MucilAir medium (Epithelix), at 37 °C for 5 h. After removal of the inoculum, the apical side was washed three times with PBS + /+ and 345.65 μM of 2 or 3 were administered apically in 30 µl of MucilAir medium. Apical infectious supernatant was harvested daily and 345.65 μM of fresh drug was added. Viral loads were assessed by RT-qPCR and PA.
2.9. Virucidal Assays
A(H1N1)pdm09 (1E6 PFU) and the EC90 (49 µM) concentration of the drugs were incubated for 3 h at 37 °C. The mixture of virus plus molecule was then serially diluted, from 1/9 (5.4 µM) to 1/19000 (2.6 nM), and transferred on MDCK cell, seeded in a 96-well, in serum-free DMEM containing 1% penicillin/streptomycin. After 1 h, the mixture was removed and fresh medium was added. At 16 hpi, viral titers were evaluated by ICC assay and percentages of infection calculated compared to the untreated control, as explained in [20].
SARS-CoV-2 Delta (1E5 PFU) and the maximum non-toxic dose of the molecules (20.7 µM for 2 and 31.1 µM for 3) were incubated for 3 h at 37 °C. The mixture of virus plus molecule was then serially diluted, from 1/10 (2 µM and 3.1 µM) to 1/1E6 (0.02 nM and 0.03 nM), and transferred on Vero-E6 cells. After 1 h, the mixture was removed and the cells were overlaid with DMEM containing 0.8% Avicel (RC-581-NFDR080I, Dupont). After 72 h of incubation at 37 °C, the cells were fixed with 4% formaldehyde solution and stained with 0.1% crystal violet to count the plaques.
2.10. Selection of resistant IAV variants
A(H1N1)pdm09 IV was inoculated with a MOI of 0.1 PFU/cell in Calu-3 cells, seeded in 6-well plate. Compound 3 (2.1–4.2 µM) or the buffer control was administered 1 h after the viral inoculum removal. The cells were incubated in a drug-containing medium for 48 h at 37 °C. Then, the supernatants were collected, centrifuged at 3000 rpm for 5 min to separate the dead cells from the viral suspensions and aliquoted before storage at − 80 °C. Infectious virus yields were determined by PA in MDCK cells.
2.11. TEM analysis
Transmission Electronic Microscopy (TEM) was performed in the Dubochet Center for Imaging (DCI Geneva). Concentrated A(H1N1)pdm09 stock was incubated with control medium, virustatic 1 or virucidal 3 (both at the concentration of 4.4 µM) for 1 h at 37 °C. Following fixation with glutaraldehyde 4% for 24 h at 4 °C, viruses were negatively stained with uranyl acetate on carbon-coated copper grids and analyzed by TEM at 29’000 times magnification with a TEM microscope at 120 kV (Tecnai G2, FEI, Eindhoven).
3. Results
3.1. Chemo-enzymatic generation of TδVD
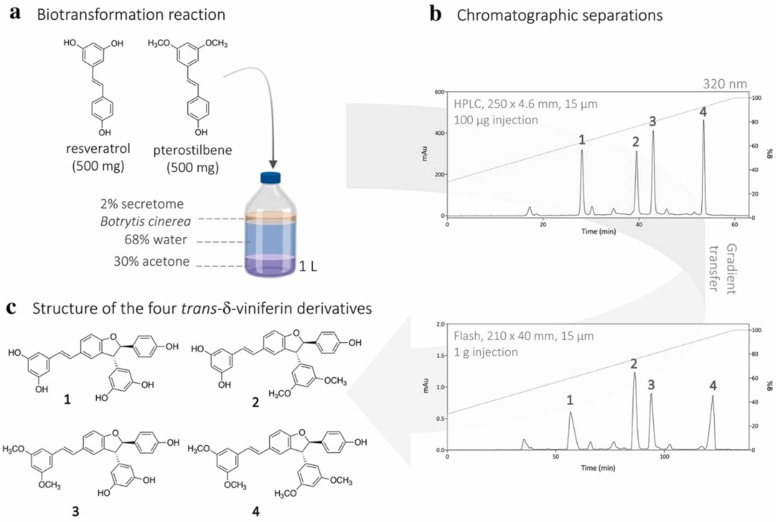
We previously generated 21 compounds, including four TδVD (designated 1, 2, 3 and 4 thereafter) ( Fig. 1), through biotransformation reactions with a mixture of pterostilbene and resveratrol with the enzymatic secretome of B. cinerea [9].
Fig. 1.
Synthesis of the four TδVD derivatives by biotransformation of the resveratrol and pterostilbene. (a) Reaction conditions, (b) analytical HPLC-PDA and preparative Flash-UV chromatograms with UV detection at 320 nm, (c) structure of the four trans-δ-viniferin derivatives isolated (1−4). Purity of the compounds was checked by UHPLC-PDA-ELSD-MS and NMR (Fig. S1 and S2).
To perform the antiviral screening, the protocol was adapted and optimized to obtain high yields of the targeted compounds. The reaction was performed with 500 mg of each substrate. The crude reaction mixture was controlled and purified by state-of-the-art analytical methods. Using this approach, TδVD 1–4 were obtained on the scale of tens of milligram with a purity of > 95%. The purity of the compounds was determined by using both UHPLC-PDA-ELSD-MS and NMR analysis (Fig. S1 and S2). We next tested the antiviral efficacy of these compounds against several enveloped viruses.
3.2. TδVD are non toxic and active against Influenza A virus
The toxicity of each compound was assessed by MTT assay in several cell lines, using the experimental protocol used in the dose-response assay below. All molecules were non toxic with cytotoxic concentrations 50 (CC50) ranging between 345.2 µM and 1201.7 µM upon 1 h of treatment in MDCK cells ( Table 1 and Table S1).
Table 1.
Determination of TδVD selectivity index in MDCK cells. CC50 values were calculated by MTT assay after 24 h of drug incubation (Table S1). The EC50 was calculated by a dose response assay (Fig. S3). N = 2 biological replicates [95% Confidence interval (CI) in parenthesis].
| Compound | CC50 [µM] | Human H1N1 A/Netherlands/602/2009 |
|||
|---|---|---|---|---|---|
| EC50 [µg/ml] (CI) | EC50 [µM] (CI) | SI [µM] | Virucidal activity | ||
| 1 | 183.7 | 1.95 (1.4 – 2.7) | 4.3 (3.1 – 5.9) | 42.7 | - |
| 2 | 74.5 | 1.5 (0.8 – 2.6) | 3.1 (1.7 – 5.4) | 24 | + |
| 3 | 79.5 | 0.9 (0.6 – 1.4) | 1.9 (1.3 – 2.8) | 41.8 | + |
| 4 | 305 | 1.2 (0.95 – 1.6) | 2.4 (1.9 – 3.2) | 127 | - |
The antiviral efficacy against A (H1N1)pdm09 was then assessed and a dose-range of each molecule was incubated for 1 h at 37 °C with a fixed dose of virus (corresponding to 0.05 MOI). MDCK cells were inoculated for 1 h with the mixture of virus and the proportion of infected cells in treated vs non-treated controls was determined by ICC 24 h later. All molecules showed a similar inhibitory profile with an EC50 ranging from 1.9 to 4.3 µM (Table 1 and Fig. S3).
3.3. TδVD have both a direct virucidal effect and an indirect cell-mediated effect on Influenza A (H1N1)pdm09
We then checked whether the molecules display a direct-acting or a cell-mediated antiviral effect.
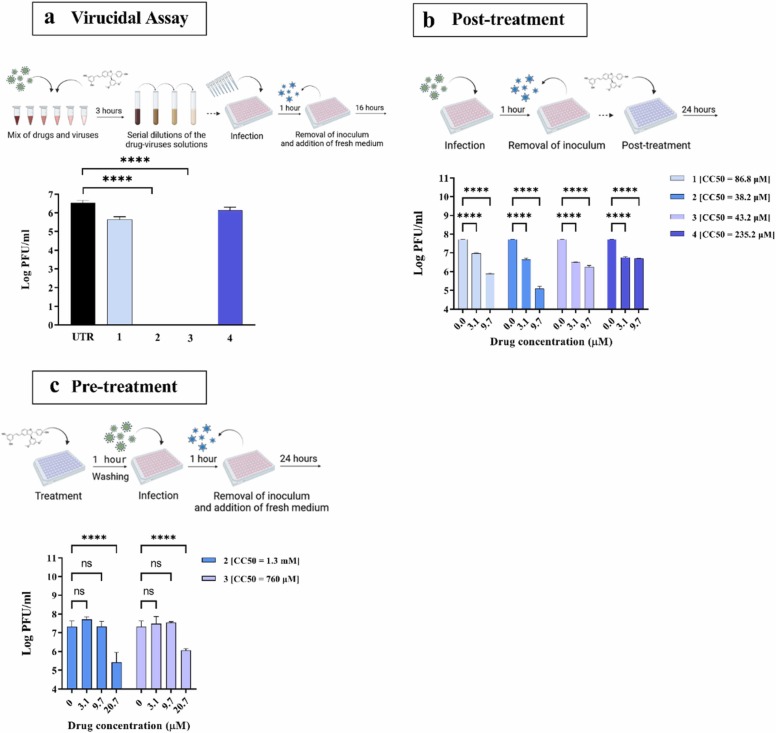
As the dose response in MDCK cells (Table 1 and Fig. S3) suggested a direct interaction between IV and the compounds, we tested the mechanism of action through a virucidal assay [21]. If an antiviral is virucidal, as opposed to virustatic, once it has interacted with the virus, it retains its effect even if the virus/antiviral complex is diluted below the compound’s EC50. Accordingly, the antiviral activity of 1 and 4 was lost upon drug dilution, suggesting a virustatic mode of action ( Fig. 2a). In contrast, both 2 and 3 retained their activity even at dilutions significantly lower than their EC50, indicating that both molecules are virucidal.
Fig. 2.
Mechanism of action of the TδVD 1, 2, 3 and 4 against A(H1N1)pdm09. (a) Virucidal activity assessed in MDCK cells with 1.25E6 PFU of IV and a drug concentration equivalent to the EC90 (49 µM), serially diluted from 1/9 (5.4 µM) to 1/19000 (2.6 nM). The virucidal property of the molecules was assessed at the 1/19000 dilution (2.6 nM). (b) Inhibition of viral replication in Calu-3 cells treated 1 h post-infection with 0.1 MOI of A(H1N1)pdm09. The number of infectious particles was determined at 24 hpi by plaque assay (PA) in MDCK cells. (c) Inhibition of viral replication in Calu-3 cells pre-treated with 2 or 3 for 1 h before infection with 0.1 MOI of A(H1N1)pdm09. Viral titers were determined as in (b). UTR = Untreated condition. CC50 were calculated in the same experimental conditions (Table S1). The results represent the mean and Standard deviation (SD) from two independent experiments performed in duplicate. N = 2 biological replicates. Statistical significance was calculated using Ordinary one-way (2a) and two-way (2b-c) ANOVA analysis: * ** *, p ≤ 0.0001. A schematic representation of the procedure, created with BioRender.com, is shown on top of each panel.
The molecules were then administered after the infection, to mimic therapeutic treatments. For these experiments, we used human bronchial adenocarcinoma Calu-3 cells, that better recapitulate respiratory infections in vivo and are commonly used as a model for IV infection [22], [23], [24]. We first confirmed the absence of toxicity in this cell line (Table S1). Cells were infected for 1 h, the inoculum washed out and drugs were added for 24 h. In these experimental conditions, the viral titers were significantly reduced by the lowest dose of each compound, indicating that the molecules are effective when administered after infection (Fig. 2b). Finally, we assessed whether the 2 virucidal compounds (2 and 3) were also acting via an indirect cell-mediated effect by adding them to the cells for 1 h at 37 °C prior infection with A(H1N1)pdm09 (Fig. 2c). In these conditions, the molecules also exerted an antiviral effect, suggesting that the compounds act through both a direct and an indirect cell-mediated effect against the virus.
As 3 showed a strong virucidal activity (Fig. 2a) and a good selectivity index (Table 1), we further characterized its mechanism of action and the genetic barrier to antiviral resistance.
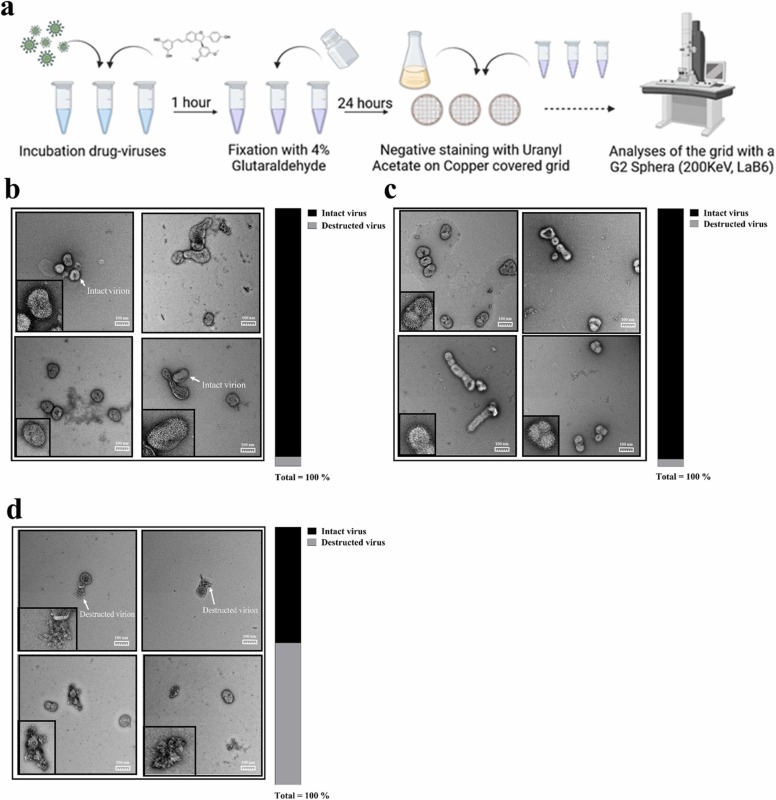
To confirm the virucidal activity of 3, we visualized treated viral particles integrity by TEM, using 1 as a virustatic control. Concentrated A(H1N1)pdm09 viruses were incubated for 1 h at 37 °C with the EC50 of 1 and 3. Negative staining showed that the viruses incubated either with control medium or in presence of 1 ( Fig. 3a-b) retained their intact structures, while the ones pre-incubated with 3 were disrupted (Fig. 3c). Compound 3 also reduced the total number of visible particles: 40 particles for 3 (18 intact +22 destructed) compared to 86 for UTR (83 intact + 3 destructed) and 75 (73 intact + 2 destructed) for 1.
Fig. 3.
Effect of virustatic or virucidal TδVD on viral particles. (a) Schematic representation of the procedure, created with BioRender.com. Concentrated A(H1N1)pdm09 stock was incubated with the drugs at a concentration ∼EC50. The solution was then fixed with glutaraldehyde 4% for 24 h at 4 °C. Viruses were negatively stained with uranyl acetate on carbon-coated copper grids, and analyzed by TEM at 29’000 times magnification. (b-d) Representative images of the virus incubated with control medium (b), virustatic 1 (c) or virucidal 3 (d). The percentage of intact and destructed viral particles are shown for each condition. The total numbers of viruses per condition were: UTR = 86 total (83 intact + 3 destructed); 1 = 75 total (73 intact + 2 destructed); 3 = 40 total (18 intact +22 destructed). N = 2 biological replicates.
To further investigate the mechanism behind the cell-mediated antiviral activity, we tested the activity of 3 in A549 cells, a cell-line which has been shown to be easily transfected with H1N1/PR8 luciferase minireplicon vectors [25], [26]. In this system, we did not observe any decrease in luciferase activity in presence of the antiviral treatment (Fig. S4), suggesting that 3 does not target directly intracellular viral replication, but another step of the viral life cycle.
3.4. TδVD 3 presents a high barrier to antiviral resistance
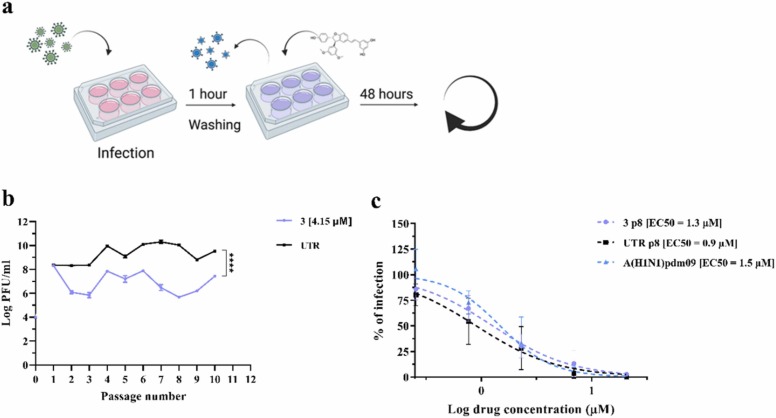
As a high genetic barrier to resistance is an essential prerequisite for a novel antiviral, we assessed it for 3 against IV. We serially passaged A(H1N1)pdm09 (generated in MDCK cells) 10 times in Calu-3 cells, with 2.1 µM (EC50) of 3 at passage 1 (p1) and doubled it to 4.2 µM (a dose which reduced significantly the viral titer without toxicity) up to p10 ( Fig. 4a). In parallel, the virus was passaged in absence of drug, to monitor the appearance of adaptation mutations to Calu-3 cells. At each passage, supernatants were titrated and the MOI of 0.1 was kept constant for all the passages and conditions (Fig. 4b) as previously done to test antiviral resistance against oseltamivir [4].
Fig. 4.
Assessment of the genetic barrier to resistance of A(H1N1)pdm09 against 3. (a) A schematic representation of the resistance assay, created with BioRender.com. (b) Comparison of viral titers from supernatants collected from Calu-3 cells infected with A(H1N1)pdm09 for 48 h in the presence of 2.1 µM of 3 in the first passage and 4.2 µM from the second and follow up passages. The titer (plaque assay performed in MDCK cells) is expressed in plaque-forming units per milliliter (log PFU/ml). The results represent the mean and SD of the Area Under the Curve (AUC) from two independent titrations performed in duplicate. Statistical significance was calculated using a two-way ANOVA analysis: * ** *, p ≤ 0.0001. (c) Antiviral effect of 3 against A(H1N1)pdm09 variants grown for eight passages in Calu-3 cells and measured by a dose-response assay in MDCK cells. Mean EC50 values and SD were calculated from two independent experiments performed in duplicate. 3 p8 = virus passaged eight times in the presence of 3; UTR p8 = virus passaged eight times in absence of drug; A(H1N1)pdm09 Stock = virus produced in MDCK cells and never passaged in Calu-3.
From p2 to p10, the extent of viral inhibition stayed constant (∼ 2 log reduction) and no emergence of antiviral resistance was observed. This result was confirmed by comparing the EC50s against the variants passaged 8 times in presence of drug, the variant passaged in the presence of control medium or against the viral stock. No difference in EC50 was observed across the variants, confirming the absence of resistance emergence after 8 passages (Fig. 4c).
3.5. TδVD 2 and 3 display antiviral activity against other Influenza strains, Herpes Simplex Virus 2 and SARS-CoV-2 Delta
We tested the activity of 2 and 3 against human IBV/Wisconsin/01/2010 (Yamagata lineage, IBV), and against a lab reassortant variant (H5N1/PR8) bearing the HA and NA from A/Vietnam/1203/2004 (H5N1) virus and the backbone of H1N1 A/Puerto Rico/8/1934. Both strains were sensitive to the two compounds, with EC50 of respectively 10.0 µM (CI = 8.41–12.1 µM) and 11 µM (CI = 7.6–15.7 µM) for 2 and 5.7 µM (CI = 4.4–7.4 µM) and 7.5 µM (CI = 5.9–9.2 µM) for 3 (Fig. S5).
We next measured the antiviral activity of 3 against HSV-2. This compound was effective only when pre-incubated for 2 h with the virus and then left on cells (Fig. S6b). Even in these conditions the EC50 was low (59.4 µM) and the activity was only virustatic (Fig. S6c). We thus did not investigate further the mechanism of action of the molecule against HSV-2.
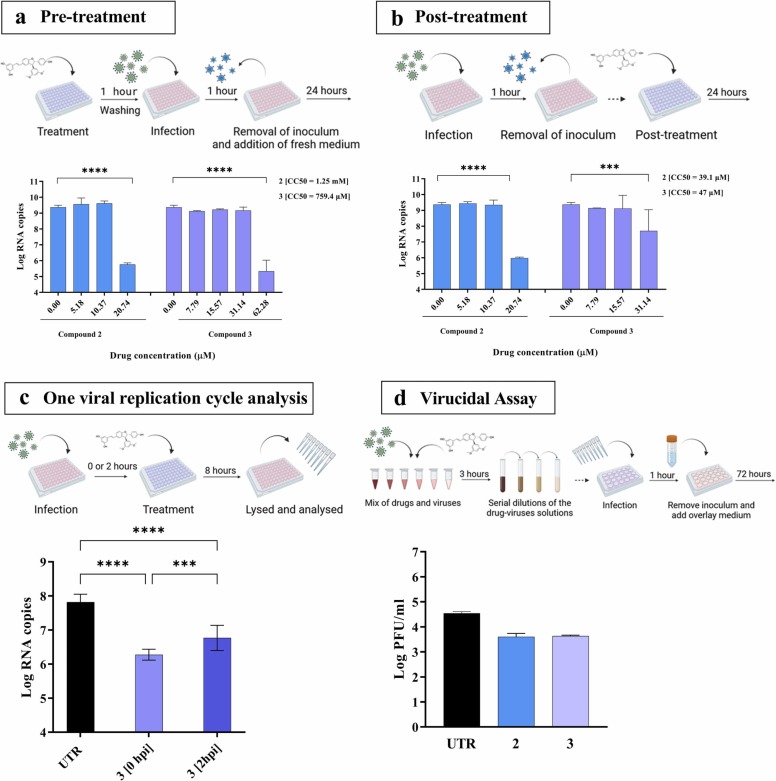
Finally, we tested 2 and 3 in dose-response against SARS-CoV-2 Delta variant ( Fig. 5). We observed a significant reduction of viral replication by pre-treating Calu-3 cells with 20.7 or 62.3 µM of compounds 2 and 3 respectively, corresponding to a Selectivity Index (SI) = 60.2 or = 12.2 respectively (Fig. 5a-b). Similarly, the compounds were active when added 1 h after infection, at 20.7 µM for compound 2, with a SI of 1.8 or 31.1 µM for 3, with a SI of 1.4 (Fig. 5a-b). To better dissect the mechanism of action against SARS-CoV-2 Delta, we administered 3 at 0 or at 2 hpi and the measured viral transcripts 8 h post infection, and therefore within the first cycle of viral replication (Fig. 5c). A significant reduction of RNA copies occurred at both times of treatment compared to the untreated control, suggesting that the molecules hinder the very early steps of viral replication (Fig. 5c). Finally, no virucidal activity was observed (Fig. 5d). These data suggest that the antiviral potency is higher against IV compared to SARS-CoV-2 Delta and is restricted to a cell-mediated effect in the latter virus.
Fig. 5.
Mechanism of action of 2 and 3 against SARS-CoV-2 Delta variant. (a) Inhibition of viral replication in Calu-3 cells, pre-treated with 2 or 3 1 h before infection with 0.1 MOI of SARS-CoV-2 Delta. (b) Inhibition of viral replication in Calu-3 cells, post-treated 1 h after infection as in (a). Viral loads were determined at 24 hpi by RT-qPCR. (c) Inhibition of viral replication in Calu-3 cells, infected with 0.1 MOI of SARS-CoV-2 Delta and treated with 3 (62.3 µM) at 0 and 2 hpi. Viral loads were determined at 8 hpi by RT-qPCR. (d) Virucidal activity assessed in Vero E6 cells with 1E5 PFU and the maximum non-toxic dose of each drugs (20.7 µM for 2 and 31.1 µM for 3) were incubated for 3 h at 37 °C. The mixture of virus plus molecule was then serially diluted, from 1/10 (2 µM and 3.1 µM) to 1/1E6 (0.02 nM and 0.03 nM) and transferred on cells. Mean values and SD were calculated from two independent experiments performed in duplicate. Statistical significance was calculated using Ordinary one-way (2c) and two-way (2a-b) ANOVA analysis: * ** , p ≤ 0.001; * ** *, p ≤ 0.0001. A schematic representation of the procedure created with BioRender.com is shown on top of each panel.
We also tested the effect of compounds 1–4 against the non-enveloped Enterovirus A71(EV-A71), but did not observe any activity when applied both before and after infection (Table S2).
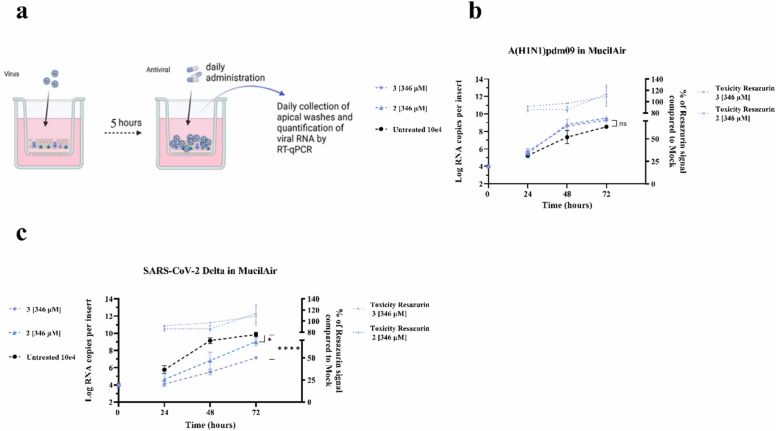
3.6. Antiviral effect of TδVD 2 and 3 against A(H1N1)pdm09 and SARS-CoV-2 Delta in ex vivo human airway epithelia
Human airway epithelia cultured at an air-liquid interface nicely reproduce the pseudostratified architecture and defense mechanism of the human respiratory mucosa. We infected the tissues with A(H1N1)pdm09 or SARS-CoV-2 Delta (10E4 RNA copies) and administered 2 or 3 on either their apical side (to mimic in vivo nasal administration, Fig. 6) or their basal side (to mirror oral or intravenous treatment, Fig. S7), daily for 3 days starting from 5 hpi. To our surprise, the compounds administered apically or basally did not display any antiviral efficacy against IV in this ex vivo model (Fig. 6b and S7a). However, a strong antiviral efficacy against SARS-CoV-2 Delta was observed when the compounds were administered apically (Fig. 6c). Interestingly, we did not observe any reduction of viral replication of SARS-CoV-2 when tissues were treated basally, suggesting that the compound is not able to cross the tissue and reach SARS-CoV-2 infected cells to interfere with replication. Of note, Imdevimab or Casirivimab, two monoclonal Abs administered intravenously in patients and used as positive controls show an antiviral effect upon basal administration. The lack of effect against IV confirms that the molecules have different mechanisms of action against different viruses and suggests that the efficacy against IV may be reduced due to the presence of mucus or mucociliary clearance in HAE. To test this hypothesis, we performed an experiment in MDCK cells infected with IV pre-incubated with the drugs in presence or absence of mucus harvested from ex vivo tissues (10% final concentration). Fig. S8 shows that the direct effect of 3 against IV is lost in presence of mucus, suggesting that it may protect the virus from the drugs direct antiviral effect.
Fig. 6.
Antiviral efficacy of 2 and 3 against A(H1N1)pdm09 or SARS-CoV-2 Delta in ex vivo human airway epithelia reconstituted at the air–liquid interface. (a) Schematic of ex vivo infection in MucilAir tissues. (b-c) Tissues were infected apically with A(H1N1)pdm09 (b) or SARS-CoV-2 Delta (c) (1E4 viral RNA copies per tissue at time 0). Daily administration of 346 µM of 2 and 3 were performed to the apical side of the tissues starting 5 hpi. Viral loads, expressed in RNA copies/tissue, were quantified by RT-qPCR from daily collected apical washes. The results represent the mean and SD from two independent experiments. Statistics were obtained by analyzing the AUC for b and c. Statistical significance was calculated using a two-way ANOVA analysis: * , p ≤ 0.1; * ** *, p ≤ 0.0001.
4. Discussion
Despite the outstanding progress in medical science of the last decades and the advances in preventing and controlling infectious diseases, the burden induced by viral infections remains enormous. The global society in which we live favors the spread of viral infections, which take advantage also from unhealthy lifestyle and sanitary habits. Moreover, rapidly acquired viral genetic variability has made rational drug and vaccine design against viruses extremely hard. Drug resistance often results from changes in a very small number of amino acid residues in the targeted viral protein. To rapidly counteract the spread of new viruses, we need to develop broad-spectrum antivirals with high genetic barrier to antiviral resistance. NP appear as a major historical source of drugs. Thanks to their remarkable chemical diversity, NP are used as "scaffolds" for the development of a large number of compounds, against both infectious and non-infectious diseases [5]. We previously demonstrated that the secretome of B. cinerea could be exploited to generate trans-δ-viniferin derivatives from stilbenes [8], [9], [10], [11], [12]. Trans-δ-viniferin derivatives turned out to possess remarkable antimicrobial properties against multidrug resistant strains of Staphylococcus aureus, by destabilizing their membrane [8], [14]. Currently, little is known about their pharmacokinetic properties. A single study in rats treated with compound 1 has demonstrated poor oral availability and rapid metabolization [27]. A recent in silico docking study suggests that the resveratrol dimer trans-δ-viniferin possesses a high affinity for several targets, among them the active site of SARS-CoV-2 MPro [28]. In this context, the present work aimed to evaluate the broad-spectrum antiviral properties of four TδVD (1, 2, 3 and 4). When pre-incubated with the virus, all compounds are able to reduce the infectivity of A(H1N1)pdm09 to the same extent. However, only 2 and 3 display virucidal activity. This phenomenon might be attributable to the O-methylation pattern, as shown previously with S. Aureus [8], [14], as virustatic compounds 1 and 4 have no or both sides with O-methyl groups, respectively, unlike virucidal compounds 2 and 3, which have O-methyl groups on one side and OH groups on the other. 1, 2 3 and 4 also displayed an indirect cell-mediated inhibition of IV. To further dissect the virucidal mechanism of action of TδVD, we performed TEM on IV treated with 3 and discovered that this molecule directly destabilizes the viral envelope. As the latter is inherited from the host cell and is not likely to undergo drug selective pressure, 3 also displays a high genetic barrier to antiviral resistance, differing from Oseltamivir for instance, the most common antiviral used to treat IV infections [4]. Despite no obvious differences in structure or lipid composition between cellular and viral membranes, compound 3 could disrupt IV envelope at a dose that is not cytotoxic. Such results have already been described for other drugs targeting viral envelope [29]. One hypothesis to explain this observation could be that cellular membranes are fluid and able to self-repair, while viral envelopes are more static and therefore more sensitive to the drugs.
In line with these findings, 2 and 3 could also inhibit the replication of IBV/Wisconsin/01/2010 (Yamagata lineage) and a lab reassortant bearing the HA and NA from A/Viet Nam/1203/2004 (H5N1) virus and the backbone of H1N1 A/Puerto Rico/8/1934 (H5N1/PR8). With regard to SARS-CoV-2 Delta, we pinpointed a cell-mediated antiviral effect with higher EC50 compared to IV. The mechanism behind this cell-mediated antiviral action may be linked to an action on cellular membranes interfering with the membranous webs needed for of the early replication step of the viral cycle and is currently under investigation. As both IV and SARS-CoV-2 Delta are respiratory viruses of major public health concern, we assessed the antiviral efficacy of 2 and 3 in a more relevant model of airway epithelia reconstituted in vitro. To our surprise, in this model, while the antiviral activity of 2 and 3 against SARS-CoV-2 Delta was confirmed, the molecules were not effective against IV. One possible explanation for this difference of effect between both viruses could be, that, in the case of IV, the presence of mucus interferes with the main direct effect of the molecule, and the contribution of the intracellular effect is not sufficient to inhibit significantly viral growth. Additional investigations are needed to dissect the cell mediated action of the drug against the two viruses. Moreover, the absence of efficacy against SARS-CoV-2 upon basal compound administration suggests that these molecules should be administered topically in vivo, more specifically intranasally rather than orally or by intravenous injection. Finally, additional experiments showed that only 3 is able to reduce HSV-2 replication in pre-incubation conditions and with a virustatic mechanism of action. The presence and absence of virucidal activity against H1N1 and HSV2, respectively, is probably related to the structural differences between these two enveloped viruses. The large amount of viral proteins inserted in the envelope of HSV-2 or the underlying proteinaceous tegument layer may render it more resistant to TδVD disruption. Finally, we did not find any activity against EV-A71, a non-enveloped virus, confirming the probability of a membrane-mediated antiviral effect. Additional in vitro and in vivo studies will be needed to better dissect the precise antiviral mechanism of action of our trans-δ-viniferins. Also, an optimized formulation or new trans-δ-viniferins derivatives may help to reduce molecule toxicity, increase its selectivity index and reach inhibition against IV in presence of mucus and ciliated cells. The production of a given compound can also be optimized in some cases. For example, compounds 1 and 4 could be obtained in very high yields by performing the enzymatic reaction with a single stilbenoid, resveratrol and pterostilbene respectively. Optimizing the enzymatic reaction to obtain compounds 2 and 3 is more difficult as both stilbenes must be present. Mass production should probably be approached by chemical synthesis, as it has been already published for compound 1 [30]. Nonetheless, our results open the door to future clinical application of viniferin derivatives as broad-spectrum antivirals against enveloped viruses.
Funding Source
All sources of funding should be acknowledged, and you should declare any extra funding you have received for academic research of this work. If there are none state ‘there are none’.
CRediT authorship contribution statement
Arnaud Charles-Antoine Zwygart, Conceptualization, Methodology, Investigation, Writing - Original Draft, Writing - Review & Editing, Visualization, Chiara Medaglia, Conceptualization, Methodology, Investigation, Writing - Original Draft, Writing - Review & Editing, Robin Huber, Conceptualization, Methodology, Investigation, Writing - Original Draft, Writing - Review & Editing, Visualization, Romain Poli, Investigation, Writing - Review & Editing, Laurence Marcourt, Writing - Review & Editing, Sylvain Schnee, Writing - Review & Editing, Emilie Michellod, Writing - Review & Editing. Beryl Mazel-Sanchez, Investigation, Writing - Review & Editing. Samuel Constant, Resources, Writing - Review & Editing. Song Huang, Resources, Writing - Review & Editing, Meriem Bekliz, Methodology, Resources, Writing - Review & Editing. Katia Gindro, Conceptualization, Methodology, Writing - Review & Editing. Sophie Clément, Writing - Review & Editing, Emerson Ferreira Queiroz, Conceptualization, Resources, Writing - Review & Editing, Supervision, Project administration, Funding acquisition. Caroline Tapparel, Conceptualization, Resources, Writing - Review & Editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The study was funded thanks to the financial support of the “Fondation privée des HUG”, the Carigest Foundation and the faculty of Medicine of the University of Geneva to CT. Swiss National Science Foundation for (grant 205321_182438/1 E.F.Q. and K.G.). We thank Pre. Isabella Eckerle for providing the SARS-CoV-2 strains.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.biopha.2023.114825.
Appendix A. Supplementary material
Supplementary material
.
Data Availability
Data will be made available on request.
References
- 1.Gelderblom H.R. Structure and Classification of Viruses. In: th, Baron S., editors. Medical Microbiology. Galveston (TX); 1996. [PubMed]
- 2.Da S. Prioritizing the continuing global challenges to emerging and reemerging viral infections. Front. Virol. 2021 doi: 10.3389/fviro.2021.701054. [DOI] [Google Scholar]
- 3.Majumder J., Minko T. Recent developments on therapeutic and diagnostic approaches for COVID-19. AAPS J. 2021;23(1):14. doi: 10.1208/s12248-020-00532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medaglia C., Zwygart A.C., Silva P.J., Constant S., Huang S., Stellacci F., et al. Interferon lambda delays the emergence of influenza virus resistance to oseltamivir. Microorganisms. 2021;9(6) doi: 10.3390/microorganisms9061196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newman D.J., Cragg G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020;83(3):770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 6.Sagar S., Kaur M., Minneman K.P. Antiviral lead compounds from marine sponges. Mar. Drugs. 2010;8(10):2619–2638. doi: 10.3390/md8102619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J.W.H., Vederas J.C. Drug discovery and natural products: end of an era or an endless frontier? Science. 2009;325(5937):161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 8.Righi D., Huber R., Koval A., Marcourt L., Schnee S., Le Floch A., et al. Generation of stilbene antimicrobials against multiresistant strains of Staphylococcus aureus through biotransformation by the enzymatic secretome of Botrytis cinerea. J. Nat. Prod. 2020;83(8):2347–2356. doi: 10.1021/acs.jnatprod.0c00071. [DOI] [PubMed] [Google Scholar]
- 9.Gindro K., Schnee S., Righi D., Marcourt L., Nejad Ebrahimi S., Codina J.M., et al. Generation of antifungal stilbenes using the enzymatic secretome of Botrytis cinerea. J. Nat. Prod. 2017;80(4):887–898. doi: 10.1021/acs.jnatprod.6b00760. [DOI] [PubMed] [Google Scholar]
- 10.Huber R., Koval A., Marcourt L., Heritier M., Schnee S., Michellod E., et al. Chemoenzymatic synthesis of original stilbene dimers possessing wnt inhibition activity in triple-negative breast cancer cells using the enzymatic secretome of Botrytis cinerea Pers. Front Chem. 2022;10 doi: 10.3389/fchem.2022.881298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber R., Marcourt L., Koval A., Schnee S., Righi D., Michellod E., et al. Chemoenzymatic synthesis of complex phenylpropanoid derivatives by the Botrytis cinerea secretome and evaluation of their wnt inhibition activity. Front Plant Sci. 2021;12 doi: 10.3389/fpls.2021.805610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber R., Marcourt L., Quiros-Gurerrero L.M., Luscher A., Schnee S., Michellod E., et al. Chiral separation of stilbene dimers generated by biotransformation for absolute configuration determination and antibacterial evaluation. Front Chem. 2022 doi: 10.3389/fchem.2022.912396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong S.Y., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G., Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol Rev. 2015;28(3):603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattio L.M., Dallavalle S., Musso L., Filardi R., Franzetti L., Pellegrino L., et al. Antimicrobial activity of resveratrol-derived monomers and dimers against foodborne pathogens. Sci. Rep. 2019;9(1):19525. doi: 10.1038/s41598-019-55975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Essaidi-Laziosi M., Brito F., Benaoudia S., Royston L., Cagno V., Fernandes-Rocha M., et al. Propagation of respiratory viruses in human airway epithelia reveals persistent virus-specific signatures. J. Allergy Clin. Immunol. 2018;141(6):2074–2084. doi: 10.1016/j.jaci.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cagno V., Medaglia C., Cerny A., Cerny T., Zwygart A.C., Cerny E., et al. Methylene Blue has a potent antiviral activity against SARS-CoV-2 and H1N1 influenza virus in the absence of UV-activation in vitro. Sci. Rep. 2021;11(1):14295. doi: 10.1038/s41598-021-92481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tseligka E.D., Sobo K., Stoppini L., Cagno V., Abdul F., Piuz I., et al. A VP1 mutation acquired during an enterovirus 71 disseminated infection confers heparan sulfate binding ability and modulates ex vivo tropism. PLoS Pathog. 2018;14(8) doi: 10.1371/journal.ppat.1007190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medaglia C., Kolpakov I., Zwygart A.C., Zhu Y., Constant S., Huang S., et al. An anti-influenza combined therapy assessed by single cell RNA-sequencing. Commun. Biol. 2022;5(1):1075. doi: 10.1038/s42003-022-04013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Essaidi-Laziosi M., Geiser J., Huang S., Constant S., Kaiser L., Tapparel C. Author correction: interferon-dependent and respiratory virus-specific interference in dual infections of airway epithelia. Sci. Rep. 2020;10(1):12523. doi: 10.1038/s41598-020-69600-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kocabiyik O., Cagno V., Silva P.J., Zhu Y., Sedano L., Bhide Y., et al. Non-Toxic Virucidal Macromolecules Show High Efficacy Against Influenza Virus Ex Vivo and In Vivo. Adv. Sci. (Weinh. ) 2021;8(3):2001012. doi: 10.1002/advs.202001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cagno V., Andreozzi P., D'Alicarnasso M., Jacob Silva P., Mueller M., Galloux M., et al. Broad-spectrum non-toxic antiviral nanoparticles with a virucidal inhibition mechanism. Nat. Mater. 2018;17(2):195–203. doi: 10.1038/nmat5053. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y., Chidekel A., Shaffer T.H. Cultured human airway epithelial cells (calu-3): a model of human respiratory function, structure, and inflammatory responses. Crit. Care Res Pr. 2010:2010. doi: 10.1155/2010/394578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ong H.X., Traini D., Young P.M. Pharmaceutical applications of the Calu-3 lung epithelia cell line. Expert Opin. Drug Deliv. 2013;10(9):1287–1302. doi: 10.1517/17425247.2013.805743. [DOI] [PubMed] [Google Scholar]
- 24.Zeng H., Goldsmith C., Thawatsupha P., Chittaganpitch M., Waicharoen S., Zaki S., et al. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type i interferon response in polarized human bronchial epithelial cells. J. Virol. 2007;81(22):12439–12449. doi: 10.1128/JVI.01134-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazel-Sanchez B., Boal-Carvalho I., Silva F., Dijkman R., Schmolke M. H5N1 Influenza A Virus PB1-F2 Relieves HAX-1-mediated restriction of avian virus polymerase PA in human lung cells. J. Virol. 2018;92(11) doi: 10.1128/JVI.00425-18. doi: ARTN e00425-1810.1128/JVI.00425-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pleschka S., Jaskunas S.R., Engelhardt O.G., Zurcher T., Palese P., GarciaSastre A. A plasmid-based reverse genetics system for influenza A virus. J. Virol. 1996;70(6):4188–4192. doi: 10.1128/jvi.70.6.4188-4192.1996. doi: Doi 10.1128/Jvi.70.6.4188-4192.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao P., Lei Y., Zhang T., Ma C., Jin B., Li T. Pharmacokinetics, bioavailability, metabolism and excretion of delta-viniferin in rats. Acta Pharm. Sin. B. 2016;6(3):243–252. doi: 10.1016/j.apsb.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joshi R.S., Jagdale S.S., Bansode S.B., Shankar S.S., Tellis M.B., Pandya V.K., et al. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J. Biomol. Struct. Dyn. 2021;39(9):3099–3114. doi: 10.1080/07391102.2020.1760137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song J.M., Seong B.L. Viral membranes: an emerging antiviral target for enveloped viruses. Expert Rev. Anti Infect. Ther. 2010;8(6):635–638. doi: 10.1586/eri.10.51. [DOI] [PubMed] [Google Scholar]
- 30.Natori Y., Ito M., Anada M., Nambu H., Hashimoto S. Catalytic asymmetric synthesis of (−)-E-δ-viniferin via an intramolecular C–H insertion of diaryldiazomethane using Rh2(S-TFPTTL)4. Tetrahedron Lett. 2015;56:4324–4327. doi: 10.1016/j.tetlet.2015.05.072. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.