ABSTRACT
Evidence has accumulated that gut microbiota and its metabolites, in particular the short-chain fatty acid propionate, are significant contributors to the pathogenesis of a variety of diseases. However, little is known regarding its impact on pediatric bronchial asthma, one of the most common allergic diseases in childhood. This study aimed to elucidate whether, and if so how, intestinal propionate during lactation is involved in the development of bronchial asthma. We found that propionate intake through breast milk during the lactation period resulted in a significant reduction of airway inflammation in the offspring in a murine house dust mite-induced asthma model. Moreover, GPR41 was the propionate receptor involved in suppressing this asthmatic phenotype, likely through the upregulation of Toll-like receptors. In translational studies in a human birth cohort, we found that fecal propionate was decreased one month after birth in the group that later developed bronchial asthma. These findings indicate an important role for propionate in regulating immune function to prevent the pathogenesis of bronchial asthma in childhood.
KEYWORDS: Bronchial asthma, allergic disease, microbiota, Scfas, propionate, lactation period, GPR41, Tlrs, human birth cohort
Introduction
Bronchial asthma (BA), one of the most common allergic diseases in routine clinical practice, is an allergic airway inflammatory disease in which there is an adverse reaction to foreign antigens such as dust mites, pet hair and fungi1,2. BA is divided into two subtypes. Atopic asthma is more common and involves allergic reactions to common environmental allergens with elevated levels of IgE in the blood. The other is non-atopic asthma, which is relatively rare, and allergens are not involved; instead obesity and fragile airways are likely implicated3. It is estimated that more than 300 million people suffer from BA worldwide and there is an urgent need from a public health perspective to understand the pathogenesis of BA to contain the disease1,2.
Maternal lifestyle during the prenatal and lactation periods is known to be an important determinant for the establishment of gut microbiota and the development of BA in their children4. Large human epidemiological studies suggest an association between the onset of BA and childhood circumstances, such as the method of infant feeding5, the method of delivery6 and the presence of pet animals7.
In addition, gut microbiota has been implicated in bronchial asthma, especially pediatric bronchial asthma. Gut microbiota changes most dramatically after birth and during infancy and is influenced by the mode of delivery, nutrition, and the maternal skin and gut microbiota. In addition, there are reports showing that changes in gut microbiota during infancy are involved in allergic diseases, particularly bronchial asthma. For example, Arrietta et al. reported that Lachnospira, Veillonella, Fecalibacterium, and Rothia were decreased, accompanied by reduced acetate, in the feces at 100 days after birth in infants at risk of asthma at one year of age8. Moreover, when these authors transplanted feces from an infant at risk of asthma into GF mice, OVA-induced allergic airway inflammation was exacerbated; the allergic airway inflammation was ameliorated by the addition of the above four bacteria to the feces from the same infant8. On the other hand, another study reported that the diversity of gut bacteria at one and 12 months correlated with serum IgE and allergic conjunctivitis but not with the onset of bronchial asthma9, suggesting the need for further investigations to understand unambiguously the relationship between the gut environment and pediatric asthma.
In recent years, metabolomic analysis has attracted much attention, especially in the intestinal tract, where short-chain fatty acids (SCFAs) and other microbial metabolites affect host immune responses that maintain intestinal homeostasis, and whose dysregulation can lead to disease10. In particular, propionate is a major microbial fermentation metabolite in the gut of many animals including humans, and exerts versatile health-promoting effects systemically beyond the gut11, including the attenuation of cardiac hypertrophy, fibrosis and vascular dysfunction12, as well as the amelioration of colonic inflammation13. These studies suggest that propionate in the gut could modulate the pathogenesis of systemic diseases and gut-specific immune responses. However, little is known about whether intestinal propionate is involved in allergic diseases such as BA.
In the present study, we performed mouse studies to show that propionate intake through breast milk during the lactation period can ameliorate airway inflammation in the offspring in a house dust mite-induced asthma model by suppressing eosinophil function through GPR41 signaling. Moreover, a human birth cohort study with fecal microbiome and water-soluble metabolome analyses suggested that decreased fecal propionate in the first month of life was associate with subsequent BA development.
Results
Propionate suppresses the asthmatic phenotype
To ask whether propionate intake during the lactation period could attenuate allergic airway inflammation later in life, we used a mouse model of allergic airway inflammation by intratracheal administration of house dust mite (HDM). To this end, newborn mice ingested propionate or other SCFAs during the lactation period through the breast milk of their mothers, who had ad libitum access to drinking water containing acetate, propionate or butyrate, or control plain water (Figure 1a). The offspring were then given plain water after weaning at 3 weeks of age and at 6 weeks of age they were sensitized and challenged by intratracheal administration of HDM extract. The numbers of eosinophils, neutrophils and CD4+ T cells in the BALF and lung were then measured (Figure 1a). Among the groups, propionate-fed mice had fewer eosinophil and less CD4+ T cell exudation in the airways compared with mice in the other groups (Figure 1b,c). The infiltration of eosinophils in the lung was also significantly reduced in the propionate-exposed mice (Figure 1d,e). These results were also corroborated in mice in which SCFA administration was started immediately after birth (Supplementary Fig. S1C-F). Moreover, to determine whether these differences in eosinophil counts were due to sex differences, we performed the same experiment in male mice and found that propionate administration decreased eosinophil counts in them as well (Supplementary Fig. S1G-J). Histological analyses also revealed a significant decrease in eosinophil and neutrophil infiltration around the bronchi in the propionate group compared to the other groups of mice (Figure 1f). These results suggest that the administration of propionate during the lactation period attenuated subsequent allergic airway inflammation in mice. Furthermore, HDM-induced production of Th2 cytokines interleukin (IL)-5 and IL-13 by mediastinal lymph node (mLN) cells and bronchoalveolar lavage (BAL) cells was significantly decreased in the propionate group compared with the other three groups; by contrast, a Th17 cytokine IL-17A and a Th1 cytokine IFN-γ were comparable among all four groups (Figure 1g,h). Collectively, these results indicate that the increased intestinal propionate intake in early infancy by administration of propionate during lactation attenuates the development of HDM-induced Th2 responses and thus suppresses the allergic phenotype later in life.
Figure 1.
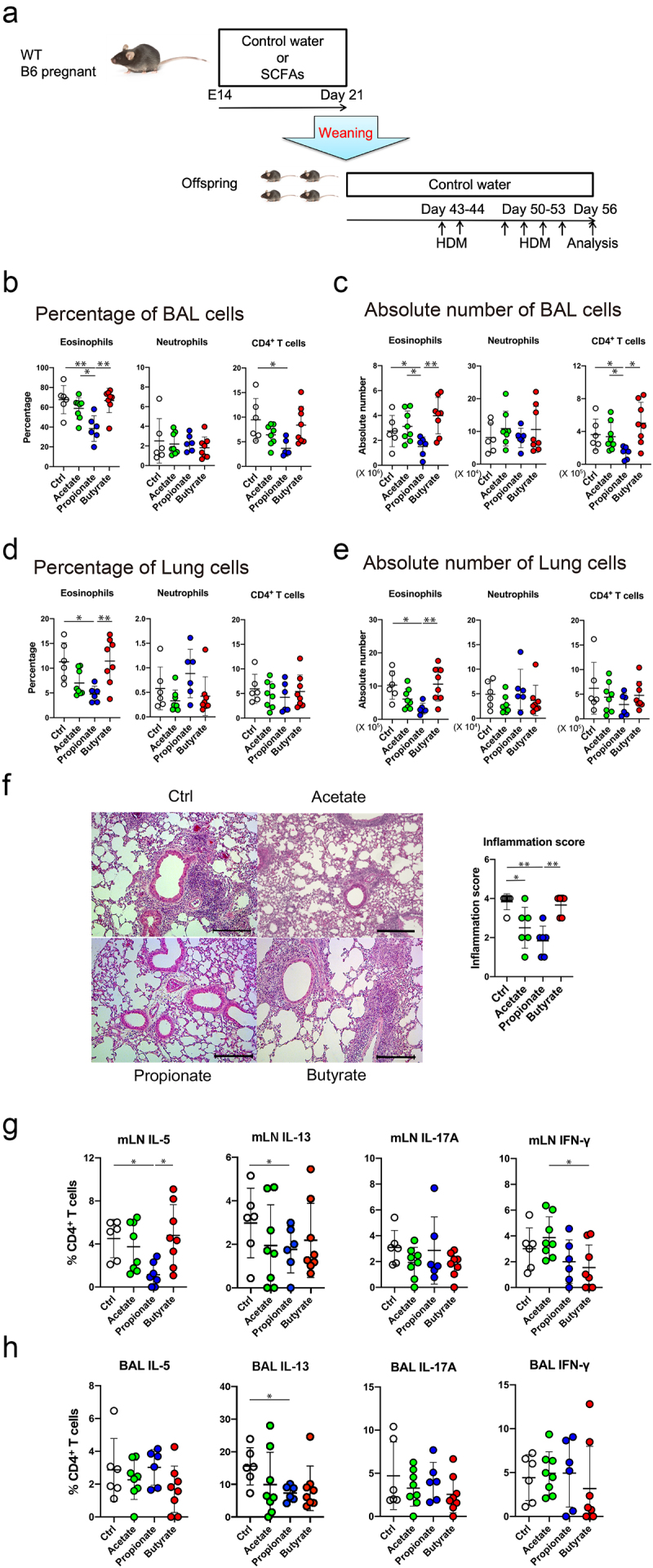
Propionate intake attenuates the eosinophilic airway inflammation and Th2 cytokine production in a murine HDM-induced asthma model. (a) Schematic depiction of the experimental protocol. Details are described in the methods section. (b-e) the percentage (b, d) and absolute number (c, e) of inflammatory cells in the bronchoalveolar lavage (b, c) and lung (d, e) were evaluated 72 h after the last HDM challenge. Data are mean ± SD. *p < 0.05 and **p < 0.01 by one-way ANOVA and Tukey’s test. (f) Representative photomicrographs of lung sections with hematoxylin and eosin staining and histological inflammatory scores. Bars = 50 µm. Data are mean ± SD. *, *p < 0.05 and **p < 0.01 by one-way ANOVA and Tukey’s test. (g, h) the percentage of IL-5-, IL-13, IL-17A, or IFN-γ-producing CD4+ cells in the mediastinal lymph node (mLN) (g) and bronchoalveolar lavage (BAL) (h) by flow cytometry intracellular staining analysis. Data are mean ± SD. *p < 0.05 by one-way ANOVA and Tukey’s test.
GPR41 mediates the suppressive effect of propionate on airway allergy
It is well known that SCFAs, including propionate, exert some of their effects via G protein-coupled receptors (GPCRs) GPR41 and GPR43 expressed by gut epithelium and immune cells, which play a crucial role in maintaining homeostasis in the gut14. Accordingly, we analyzed the involvement of GPR41 and GPR43 in the propionate-mediated suppression of the development of HDM-induced allergic airway inflammation. WT, GPR41 knockout (GPR41−/−) and GPR43 knockout (GPR43−/−) pregnant mice were divided into a propionate water-drinking group and a control water-drinking group. Propionate was added to the drinking water during the 3 weeks of lactation and the offspring mice were sensitized and challenged with HDM by intra-tracheal administration (Figure 2a). The number of inflammatory cells and cytokine production by the lung and BAL cells were then evaluated. Notably, GPR41−/− mice showed comparable numbers of eosinophils and CD4+ T cells in the BALF and lung-infiltrating eosinophils and neutrophils regardless of propionate exposure (Figure 2b-e, Supplementary Figure S2A-D). Moreover, GPR41 deficiency eliminated the propionate-induced suppression of peribronchial and perivascular inflammation (Figure 2f). The reduced HDM-specific IgG1 production induced by propionate treatment was also abolished in GPR41−/− mice (Supplementary Figure S2E). Furthermore, the production of IL-5 and IL-13 by both mLN cells and BAL cells was comparable between the propionate exposed and control GPR41−/− mice (Figure 2g,h). By contrast, in GPR43-knockout mice, as with WT mice, propionate still ameliorated the allergic airway inflammation and suppressed Th2 cytokine production (Figure 2a-d,g,h). These data suggest that GPR41, but not GPR43, acts as a propionate receptor to protect from HDM-induced allergic airway inflammation in mice.
Figure 2.
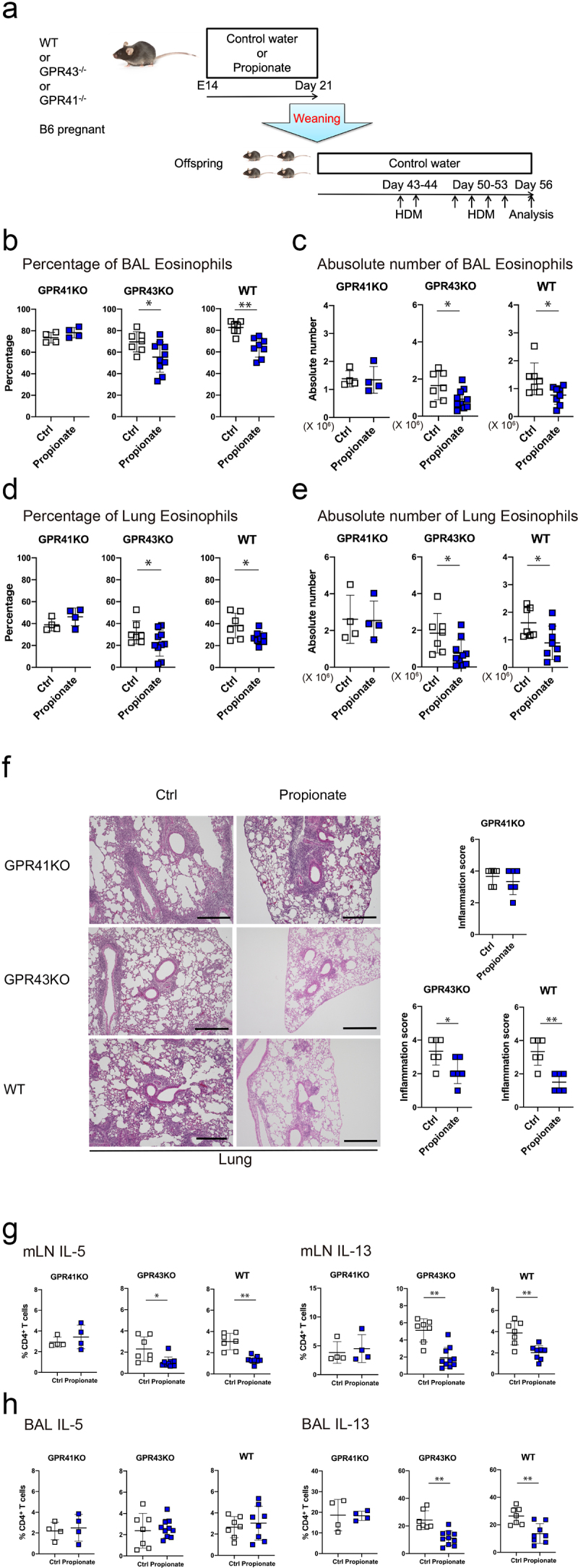
GPR41 deficiency eliminates the inhibitory effect of oral propionate administration in a murine HDM-induced asthma model. (a) Schematic depiction of the experimental protocol. Details are described in the methods section. (b-e) the percentage (b, d) and absolute number (c, e) of eosinophils in the bronchoalveolar lavage (b, d) and lung (c, e) were evaluated 72 h after the last HDM challenge. Data are mean ± SD. *p < 0.05 and **p < 0.01 by student’s t-test. (f) Representative photomicrographs of lung sections with hematoxylin and eosin staining and histological inflammatory scores. Bars = 50 µm. Data are mean ± SD. *, *p < 0.05 and **p < 0.01 by Student’s t-test. (g, h) the percentage of IFN-γ-, IL-4-, IL-5-, IL-13, or IL-17A-producing CD4+ cells in the mediastinal lymph node (mLN) (g) and bronchoalveolar lavage (BAL) (h) cells by flow cytometry intracellular staining analysis. Data are mean ± SD. *p < 0.05 by student’s t-test.
To distinguish whether GPR41 expression in the mother or offspring mice is involved in propionate-induced suppression of HDM-induced allergic airway inflammation, we crossed GPR41-heterozygous females with GPR41-homozygous knockout males, and the resulting GPR41-heterozygous and GPR41-homozygous knockout offspring were used (Figure 3a). We still observed the suppressive effect of propionate on the airway inflammation in GPR41-heterozygous offspring, which was eliminated in GPR41-homozygous knockout offspring (Figure 3b-e), suggesting that this propionate effect depends on GPR41 expression in the offspring themselves and not in the mothers.
Figure 3.
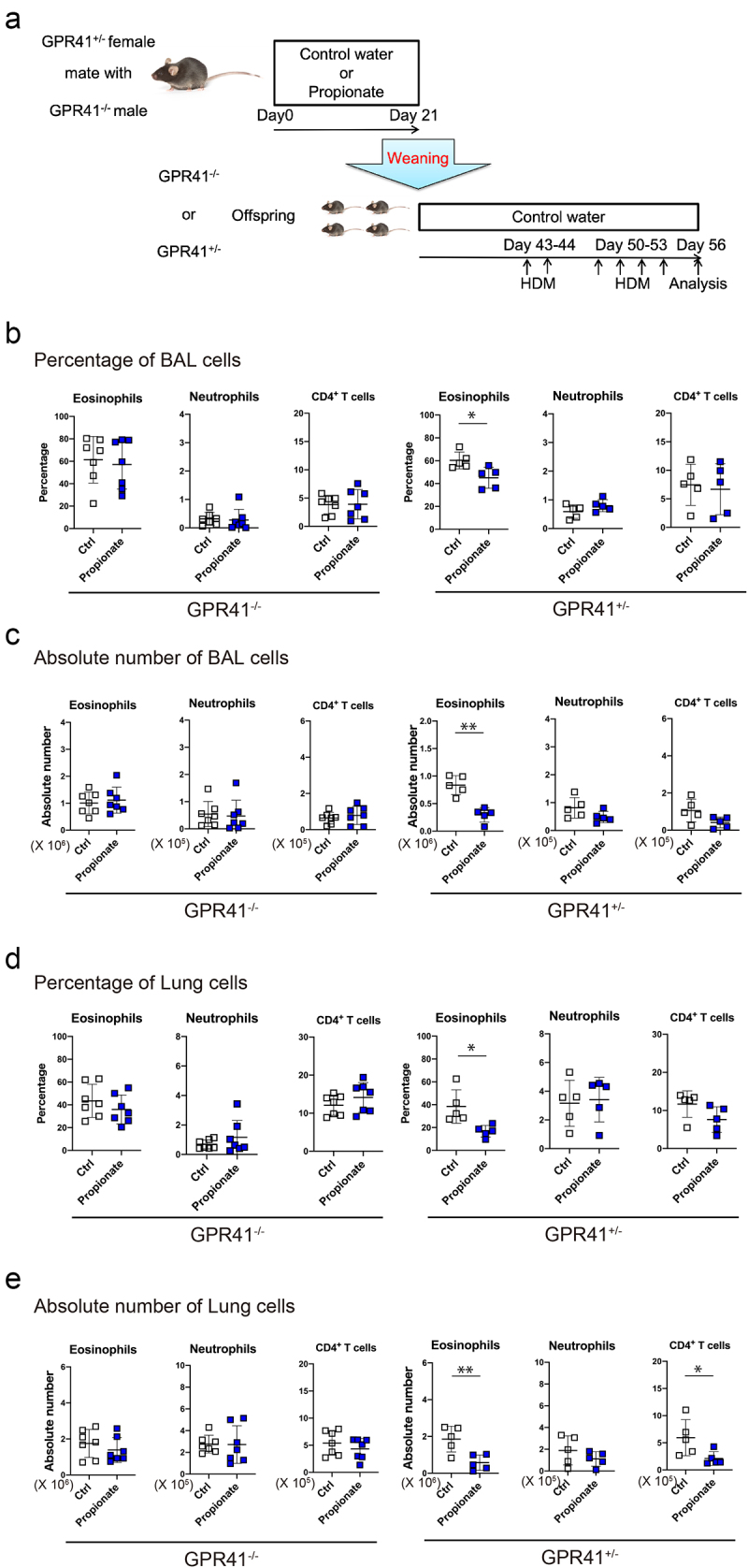
The inhibitory effect of propionate was eliminated despite GPR41 expression by the mother.(a) Schematic depiction of the experimental protocol. (b-e) the percentage (b, d) and absolute number (c, e) of neutrophils and CD4+ T cells in the bronchoalveolar lavage (b, d) and lung (c, e) were evaluated 72 h after the last HDM challenge. Data are mean ± SD. *p < 0.05 and **p < 0.01 by student’s t-test. .
TLR expression is upregulated in eosinophils from propionate-treated mice
To identify the cells in the intestine expressing GRP41, we examined Gpr41 expression by qPCR analysis of cells sorted from the small intestinal lamina propria (SILP) and lungs of wild-type mice. Interestingly, Gpr41 was strongly expressed in eosinophils, from both SILP and lungs, among the various immune cells examined (Figure 4a). This result suggests that GPR41-expressing eosinophils are likely to be the main target of the ingested propionate.
Figure 4.
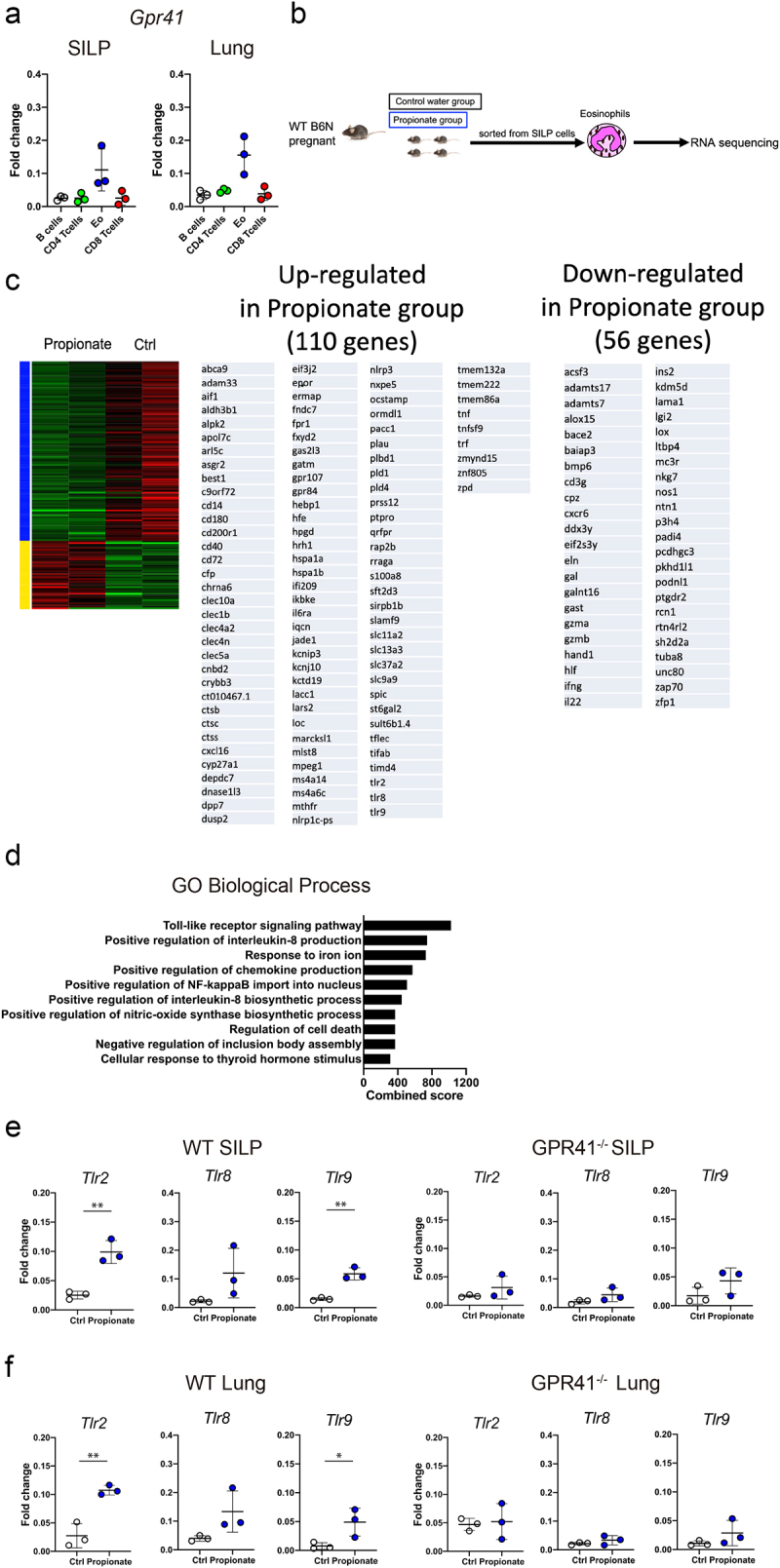
TLR family genes are upregulated in intestinal eosinophils of offspring born from mice fed with propionate (a) Expression levels of Gpr41 mRNA were evaluated by qPCR in eosinophils, B cells, CD4+ T cells, and CD8+ T cells isolated by FACS from small intestinal lamina propria (SILP) and lung cells of wild-type C57BL/6 mice. (b) Schematic depiction of the experimental protocols for isolation of mRnas used in (c-e). (c) Heatmap of differentially expressed genes (DEGs) between the Propionate group and Control water group and a list of the genes upregulated (middle) and downregulated (right) in the Propionate group. (d) Bar plots of the combined score for gene ontology (GO) term enrichment in GO biological processes for DEGs between the propionate group and the Control water group. (e) Expression levels of Tlr2, Tlr8 and Trl9 mRNA in wild-type and GPR41 knockout SILP eosinophils were evaluated by qPCR. Data are mean ± SD. **p < 0.01 by Student’s t-test. (f) Expression levels of Tlr2, Tlr8 and Trl9 mRNA in lung eosinophils were evaluated by qPCR. Data are mean ± SD. *p < 0.05 and **p < 0.01 by Student’s t-test.
In order to perform a bias-free and comprehensive genetic screening of intestinal eosinophils, we next performed RNA sequencing analysis on SILP eosinophils sorted from the offspring of mothers fed with propionate-containing or control water (Figure 4b). One hundred ten genes were upregulated and 56 genes were downregulated in the eosinophils from propionate-fed mice (Figure 4c). Surprisingly, Gene Ontology (GO) revealed that, among the genes upregulated in the propionate-fed group, genes belonging to the toll-like receptor (TLR) signaling pathway in GO biological processes (GO0002224) were the top hit in the combined score ranking (Figure 4d). Consistent with this observation, mRNAs encoding TLR2, TLR8 and TLR9 were highly expressed in SILP eosinophils isolated from the propionate-treated mice compared to the control mice (Figure 4e). By contrast, propionate treatment did not enhance TLR family expression in GPR41 knockout mice (Figure 4e). Finally, analysis of the TLR family in lung eosinophils showed that its expression was also increased in propionate -treated mice (Figure 4f). These results suggest that propionate may facilitate TLR signaling in intestinal eosinophils and also affect lung eosinophils through GPR41.
Propionate is decreased in asthma patients
Finally, we performed fecal metabolome and microbiome analyses on a human birth cohort to address the contribution of propionate in infants to the subsequent development of BA in humans. In this birth cohort, 269 pregnant women, who either themselves and/or the babies’ fathers had a history of allergic diseases, were recruited and agreed to participate15. Fecal samples were collected from these babies/children at one week, one month, one year, and five years of age and breast milk samples were collected from mothers at one week, one month, and six months of ages. At five years of age, 65 patients were unable to continue with the sample collection due to withdrawal of consent or change of hospital. Finally, fecal and breast milk samples from 204 participants were then collected. The participants were classified into two groups based on clinical diagnosis by pediatricians: the BA-onset group (BA group, n = 23) and the non-BA group (NBA group, n = 181) (Supplementary Fig. S3A). When we analyzed the clinical data for the BA and NBA groups, we found a higher proportion of boys, as well as a higher incidence of other allergic diseases such as food allergy and atopic dermatitis, in the BA group compared to the NBA group (Supplementary Fig. S3B), which is consistent with existing clinical studies on asthma in children16–18. On the other hand, there was no difference in gestation period or frequency of antibiotic use between the BA and NBA groups (Supplementary Fig. S3B).
We first performed a comprehensive gas chromatography-mass spectrometry-based targeted water-soluble metabolome analysis of fecal time course samples from the participants. Although statistically not significant, the BA/NBA ratio of fecal propionate at one month of age was lower among the SCFAs (Figure 5a,b), while this decrease was not observed at one week, one year and five years of age (Figure 5c). Moreover, our comprehensive analysis of water-soluble metabolites in addition to SCFAs revealed no significant differences between the BA and NBA groups (Supplementary Fig. S4A).
Figure 5.
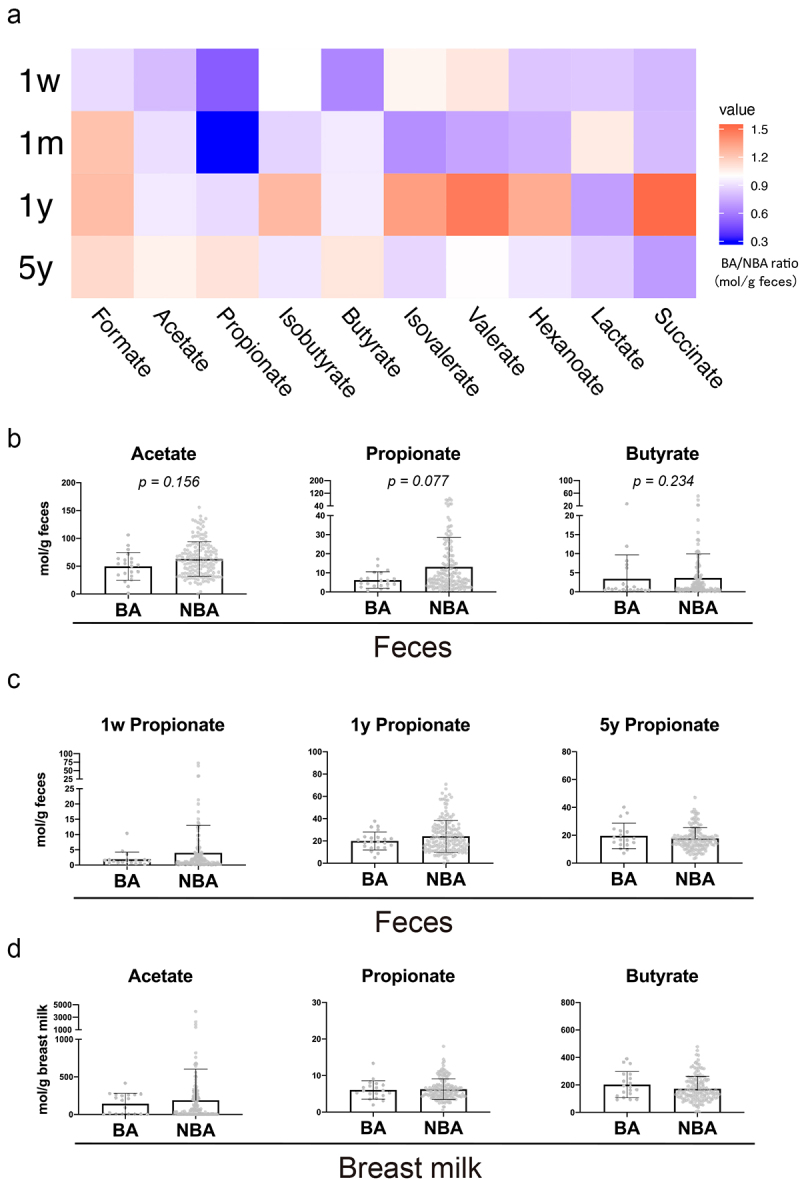
The concentration of fecal propionate was decreased in the human one-month-old BA group compared with the NBA group.Fecal (a-c) and breast milk (d) SCFAs are measured by gas chromatography (a) Heatmap of the ratio of the concentration (mol/g feces) of the ten most abundant fatty acids in feces of the BA group over that of NBA group at 1 week, 1 month, 1 year, and 5 years old. (b) Concentrations of the three most abundant SCFAs in the feces at 1-month-old. Data are mean ± SD. P values based on the Wilcoxon rank-sum test. (c) Concentrations of fecal propionate in the feces at 1 week, 1 year and 5 years old. Data are mean ± SD. P values based on the Wilcoxon rank-sum test. (d) Concentrations of the three most abundant SCFAs in the breast milk when the babies were 1 month old. Data are mean ± SD. P values based on the Wilcoxon rank-sum test.
Since one report has noted a correlation between the concentration of SCFAs in the breast milk and the atopic phenotype19, it was possible that the fecal propionate in the BA group was derived from the breast milk; however, there were no significant differences in SCFA concentrations in the breast milk between BA and NBA groups at one week, one month, and six months (Figure 5d, Supplementary Fig. S4B, C). Taken together with previous studies reporting that fecal SCFAs are mainly produced by the fermentation and metabolism of indigestible dietary fibers by the intestinal microbiota20–22, these observations suggest that the difference in fecal propionate between BA and NBA groups is likely derived from the gut microbiota rather than from breast milk.
To assess bacterial taxa potentially producing SCFA independent of multivariate factors, the level of propionate obtained in a 1-month-old was modeled in the bacterial genus composition using a random forest model23. Interestingly, Bacteroides was the top hit for the propionate production score (Supplementary Fig. S4D). Moreover, we analyzed the correlation between fecal propionate concentration and the intestinal microbiota in the BA groups at one month of age. Three genera, Varibaculum, Bifidobacterium, and Parabacteroides, were positively correlated with fecal propionate in the BA group (Supplementary Fig. S4E). These results suggest that Bacteroides, Varibaculum, Bifidobacterium, and Parabacteroides may contribute to the production of fecal propionate, although further studies are required to test this hypothesis.
Discussion
Here we report that, in the mouse model of HDM-induced allergic airway inflammation, the subsequent eosinophilic airway inflammation was attenuated and Th2 cytokine production was reduced when mice were given propionate during lactation. Moreover, we further discovered that these effects of propionate administration were mediated via GPR41 on eosinophils, likely through upregulation of TLR 2, 8 and/or 9. Finally, in our birth cohort, fecal propionate concentrations were reduced at one month in children who eventually suffered from asthma later in life. Taken together, our observations suggest that the propionate-GPR41 pathway during lactation has a protective role against the later development of HDM-induced asthma in a mouse model. This finding could be extended to human BA inflammation with possible therapeutic benefit.
Some studies in mice have indicated that changes in the gut microbiota affect the pathogenesis of allergic asthma8,24,25. Arrieta et. al. demonstrated that the colonization of germ-free mice with human fecal Lachnospira, Veillonella, Fecalibacterium, and Rothia improves ovalbumin-induced allergic airway inflammation8. However, these studies did not provide detailed mechanistic insight into how the gut microbiota impacts the pathogenesis of BA. Our study goes one step further and provides a hint into how the intestinal environment, including the gut microbiota, contributes to the regulation of BA, by showing that gut microbial-derived propionate can prevent allergic airway inflammation in mice.
The association between propionate and allergic airway inflammation has been reported by Trompette et al26, who showed in adult mice that propionate administration immediately protects from acute airway inflammation in a GPR41-dependent manner. By contrast, our present study suggests a new temporal aspect of the propionate effect in that intestinal propionate in the neonatal period can suppress the later onset of airway inflammation in adult mice, which could also be the case in the prevention of bronchial asthma in human children. Our findings also extend the study by Trompette et al. to show the importance of GPR41 expressed on eosinophils for the prevention of airway inflammation by propionate through TLR expression. In this study, we have added to the list of molecular mechanism of propionate, i.e., prevention of asthmatic airway inflammation. Interestingly, the cardioprotective effects of propionate are suggested to be mediated by Tregs12. Another study has also reported that propionate acts on monocyte-derived dendritic cells to reduce the release of several pro-inflammatory cytokines27. It is therefore possible that the airway protective effect of propionate observed in this study may also involve these anti-inflammatory functions; further studies will clarify the precise underlying mechanisms.
Some functions of SCFAs are transduced via GPCRs, GPR41 and GPR43, on the target cell surfaces. These GPCRs are abundantly expressed in various organs and cells and play a role in some chronic inflammatory diseases such as inflammatory bowel disease, obesity and arthritis28–30. Our present study indicates that propionate-mediated suppression of allergic airway inflammation is dependent on GPR41. GPR41 reportedly has both pro- and anti-inflammatory roles30, with some contradictory findings. Kim et. al. described that GPR41 promotes colitis via ERK and p38 deactivation in intestinal epithelial cells31. By contrast, Niranjana et al. have reported that GPR41−/− mouse aortas exhibited a significant increase in vascular remodeling compared to wild-type mice32. Taken together, these observations may reflect pleiotropic roles of GPR41 in various diseases and a protective role in asthmatic airway inflammation.
We further showed that GPR41 is highly expressed on SILP eosinophils and that propionate upregulated their expression of some TLRs, probably via GPR41. The TLR family plays a guiding role in inducing innate immune responses to microbial pathogens33. Consistent with the fact that the TLR family has both pro-inflammatory and anti-inflammatory effects33, studies focusing on the TLR family in asthma have yielded contradictory results34. Our finding here suggests that the propionate signals through GPR41 play a protective role in BA via upregulating TLR signaling pathways in eosinophils. A previous study has reported that TLR agonists reduce lung eosinophilic infiltration during a virus infection in mice35, although it has not been clear whether the TLR agonists directly act on eosinophils. Taken together with our finding that the expression of some TLR genes in eosinophils was upregulated by propionate, a possible explanation is that the functions of eosinophils could be modulated by propionate via TLR signaling. Further studies are required to corroborate our finding of this protective role and to shed light on the underlying regulatory mechanisms of the propionate-GPR41-TLR axis in BA.
In our study, the human fecal samples were collected over a time period from one week after birth until five years of age. As with the microbiota composition, it is known that fecal SCFA concentrations undergo a drastic change within the first 100 days of life36. Moreover, the relationship between fecal SCFAs and BA has been noted in several large cohort studies of children8,23. For example, Depner et al. have also reported a consistent association between asthma phenotype and fecal butyrate levels, with a trend toward an association between atopic asthma and fecal propionate levels23. However, these studies are limited in that they have analyzed only one timepoint and have not captured the time course of SCFA concentrations in feces. By performing the repetitive analysis of fecal SCFAs over time, our study is novel in that we were able to detect changes in fecal SCFAs that may be associated with prevention of BA.
Although some bacterial genera have been reported to correlate with the development of childhood asthma8,23, few reports have directly analyzed the correlation between SCFAs in feces and intestinal bacterial composition. Our gut microbiota analysis showed a positive correlation between fecal propionate and Varibaculum, Bifidobacterium and Parabacteroides. Therefore, it could be possible that these bacteria are involved in the production of propionate in the neonatal intestine during the lactation period. Further studies are needed to clarify the point.
The suppressive effect of propionate was evaluated in an atopic asthma mouse model (house dust mite-induced allergic airway inflammation) in this study. BA includes another subtype, non-atopic asthma, although it is believed to be a minor component in BA clinical practice; the efficacy of propionate in this subtype should be studied in an appropriate model in the future. Another issue to be resolved is that independent non-Japanese cohort studies have different results from ours8,23. These studies have reported significant differences in butyrate rather than propionate in pediatric bronchial asthma patients. This discrepancy could be due to differences in genotype, diet or gut microbial composition. Future analysis using a large human cohort may lead to a better understanding of the suppressive effect of propionate. Taken together, our study suggests that the propionate-GPR41-TLR axis in eosinophils during lactation may be involved in suppressing the development of BA. Although further studies are required, our results suggest that intestinal propionate could be a promising preventive target for the onset of BA.
Materials and methods
Mice
C57BL/6 mice (CLEA Japan, Inc.), GPR41-knockout mice on a C57BL/6 background28 and GPR43-knockout mice on a C57BL/6 background37 were housed in microisolator cages under specific pathogens-free (SPF) conditions in the animal facility of RIKEN Yokohama Campus. All experiments were performed as approved by the Animal Care and Use Committee of the RIKEN Yokohama Campus (Permission number: AEY2022–008). In some experiments measuring the fecal propionate concentration of weaning mice, mothers and offspring were housed in cages with a wire net flooring through which the fecal would pellets fall down, thus preventing coprophagy of the mothers’ feces by the offspring mice.
SCFA administration
Pregnant wild-type C57BL/6 mice were given drinking water containing 200 mM acetate, 200 mM propionate, 200 mM butyrate, or control plain water, starting from E14 or right after delivery (noted in the Figures and their legends). For GPR41- and GPR43-knockout mice, pregnant mice were given drinking water containing 200 mM propionate or control plain water. Offspring were weaned at day 21 after birth and raised on plain water. Three weeks after weaning, the offspring were subjected to the HDM-induced allergic airway inflammation experiment described below.
HDM-induced allergic airway inflammation
Mice were sensitized and challenged by intratracheal administration of HDM extracts (Greer Laboratories; XPB70D3A25) as described previously with minor modifications38. In brief, mice were sensitized intratracheally with 1 μg HDM in 50 μl PBS two days in a row. Seven days after the last sensitization, the mice were challenged with 10 μg HDM for four consecutive days. Seventy-two hours after the last HDM challenge, the mice were sacrificed and the numbers of eosinophils, neutrophils and lymphocytes in the BAL and lung were evaluated as described elsewhere39.
Cytokine analysis by flow cytometry
Single-cell suspensions were prepared from the mLNs and BAL and stimulated with 20 ng/ml PMA (Calbiochem; 524400) and 1 μg/ml ionomycin (Calbiochem; 407950) for 4 h in the presence of 10 μM brefeldin A (BD Bioscience; 10 μM; 555029). Cells were stained with the indicated antibodies against cell surface molecules together with intracellular cytokines as described previously40.
Histological analysis of the lung
Lung paraffin sections (5 μm thick) were stained with hematoxylin and eosin according to standard protocols. The histological score was evaluated as described elsewhere41,42.
Elisa
The levels of HDM-specific IgG1 were evaluated as described previously41.
qPCR analysis
qPCR was performed using a standard protocol on a Thermal Cycler Dice Real Time System TP800 (TaKaRa) using a TB Green Premix Ex Taq 2 Tli RNaseH Plus (TaKaRa; RR820A).
RNA sequencing analysis
Total RNA was extracted from isolated intestinal eosinophils of propionate-fed and control mice by using the RNeasy Mini kit (QIAGEN; 74104). Library preparation was performed with a TruSeq RNA Library Prep Kit v2 (illumina; RS-122-2001). RNA sequencing was performed on an Illumina Nextseq 2000 in a 50-base single-end mode. mRNA profiles were calculated using R package feature count software and expressed as transcripts per kilobase million. Differentially expressed genes were determined by a weighted mean difference method using the IDEP.9443. GO biological analysis was performed with the Enricher44–46.
Human cohort study design
This study was based on a cohort design to analyze the fecal microbiota of infants enrolled in the CHIBA Study15. This study includes 269 infants born at the Chiba University Hospital (Chiba, Japan) and Seikei-kai Chiba Medical Center (Chiba, Japan). The eligibility criteria of the study were the following: Japanese; infants of any sex, either of whose parents have any allergic disease; and who had obtained consent from a parent. We collected fecal samples at 1 week, 1 month, 1 year, and 5 years of age as much as possible, and mothers’ breast milk when the babies were 1 week, 1 month and 6 months of age. In the end, 65 subjects were lost during follow up periods due to hospital changes and consent withdrawal and the remaining 204 subjects were selected for metabolome analyses and 16S rRNA gene sequencing analysis on the one month fecal sample. Twenty-three of the 204 subjects later diagnosed with BA by a pediatric allergist according to the Japanese Guideline for Childhood Asthma 201447 at 5 years of age were categorized as the BA group, while the rest were in the NBA group. All human experiments have been approved by the research ethics committee of the RIKEN Yokohama Campus and Chiba University (Permission number: H21–17(8)). Written informed consent was obtained from all participants.
Sample collection and storage
The fecal and breast milk samples collected as above were immediately transported on ice to the laboratory. Upon arrival, the samples were immediately frozen in liquid nitrogen and stored at − 80°C until further analyses.
Preparation of fecal samples
Aliquots (5 g) of feces were blended with 30 ml methanol and filtered with a 100 µm mesh filter to remove food residue after vigorous vortexing. The filtrate was centrifuged at 15,000 × g for 10 minutes at 4°C and the supernatant (methanol extract) was used for metabolomics analysis. Fecal microbiome DNA was extracted from the pellet.
Extraction and measurement of SCFAs from breast milk and fecal samples
Extraction and measurement of SCFAs were as previously described48 with some modifications. 10 μl of Milli-Q water containing internal standards (2 mM [1,2-13C2] acetate, 2 mM [2H7]butyrate and 2 mM crotonate) were added to aliquots (90 μl) of breast milk. For fecal samples, aliquots (25 μl) of methanol extract were added to 10 μl of Milli-Q water containing internal standards and then centrifugally dehydrated at 40°C and reconstituted with 100 μl of Milli-Q water. Hydrochloric acid and 200 μl of diethyl ether were added to the solution and mixed well. After centrifugation at 3,000 × g for 10 min, 80 μl of the organic layer was transferred to a glass vial and 16 μl of N-tert-butyldimethylsilyl-N-trifluoroacetamid (Sigma-Aldrich; 394882) was added to derivatize the samples. The vials were incubated at 80°C for 20 min and allowed to stand at RT for 48 h before injection. The analysis was performed using a Shimadzu GCMS-TQ8030 triple quadrupole mass spectrometer with a capillary column (BPX5). The GC oven was programmed as follows: 60°C hold for 3 min, increased to 130°C (8°C/min), increased to 330°C (30°C/min) and finally a 330°C hold for 3 min. The detector and injector temperatures were 230°C and 250°C, respectively. GC was operated in constant linear velocity mode set to 40 cm/sec. Injection volume was set at 1 μl with a split ratio of 1:30. The data were processed and concentrations were calculated by LabSolutions Insight (Shimadzu).
16S microbial community analysis
Fecal DNA was extracted from 10 µg methanol-treated feces as described previously with minor modification49. The 16S rRNA gene analysis of DNA samples was performed as previously described50. Briefly, PCR was performed using 27 fmol 5-AGRGTTTGATYMTGGCTCAG-3′and 338 R 5-TGCTGCCTCCCGTAGGAGT-3′ to amplify the V1–V2 region of the 16S rRNA gene50. The amplified DNA samples (~330 bp) were subsequently purified using AMPure XP (Beckman Coulter; A63882), and quantified using a Quant-iT Picogreen dsDNA assay kit (Invitrogen; P7589) and a TBS-380 Mini-Fluorometer (Turner Biosystems). The 16S sequencing was performed using MiSeq according to the Illumina protocol. The paired-end reads were merged using the fastq-join program based on overlapping sequences. Reads with an average quality value of < 25 and inexact matches to both universal primers were filtered out. Filter-passed reads were used for further analysis after trimming off both primer sequences. For each sample, quality filter-passed reads were arranged in descending order according to the quality value and then clustered into OTUs with a 97% pairwise-identity cutoff using the UCLUST program version 5.2.32 (https://www.drive5.com). Taxonomic assignment of each OTU was made by similarity search against the Ribosomal Database Project (RDP) and the National Center for Biotechnology Information (NCBI) genome database using the GLSEARCH program51. Propionate score was calculated as described elsewhere21
Data analysis
Data are summarized as mean ± SD, unless otherwise indicated. The statistical analysis of the results was performed using the Mann-Whitney U test, independent-samples t test or one-way ANOVA with post-hoc Tukey HSD Test as appropriate.
Supplementary Material
Acknowledgments
We thank T. Kato for analysis support. T.I. is supported by the Research Fellowship for Young Scientists.
Funding Statement
This work was supported in part by grant from Japan Society for the Promotion of Science at https://www.jsps.go.jp/english/e-grants/: Grants-in-Aid for Young Scientists (19K17690 to T.I.), The Japan Agency for Medical Research and Development at https://www.amed.go.jp/en/index.html: AMED (16ek0410029h0001 to N.S.), and The Japan Agency for Medical Research and Development at https://www.amed.go.jp/en/index.html: AMED-CREST (19gm0710009h0006 to H.O.)
Disclosure statement
No potential conflict of interest was reported by the authors.
Authors’ contributions
T.I., and H.O. conceived the study. T.I. designed and performed the experiments and analyses, and cowrote the manuscript. Y.N., R.S., N.S-A., T.N., F.Y., and N.S-I. made the CHIBA Study samples possible and accessible. S.S., and T.J. helped with the mouse experiments. W.S. and M.H. performed and analyzed 16S rRNA sequencing. I.K. provided essential materials and helped to interpret the data. H.O. directed the research and cowrote the manuscript.
Ethics approval and consent to participate
All mouse experiments were performed as approved by the Animal Care and Use Committee of the RIKEN Yokohama Campus (Permission number: AEY2022–008) and carried out according to the committee’s guidelines.
All human experiments have been approved by the research ethics committee of the RIKEN Yokohama Campus and Chiba University. Written informed consent was obtained from all participants.
Consent for publication
Written informed consent was obtained from all participants.
Data availability statement
The 16S rRNA sequence data that support the findings of this study are openly available in DDBJ Sequence Read Archive at https://www.ddbj.nig.ac.jp/ddbj/index.html, reference number DRA014484.
The RNA sequence dat that support the findings of this study are openly available in Gene Expression Omnibus at https://www.ncbi.nlm.nih.gov/geo/, reference number GSE207711.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2023.2206507
References
- 1.Lambrecht BN, Hammad H.. The immunology of asthma. Nat Immunol. 2015;16:45–17. PMID:25521684. doi: 10.1038/ni.3049. [DOI] [PubMed] [Google Scholar]
- 2.Fernando DM, Donata V. Asthma Lancet. 2013;382:1360–1372. PMID: 24041942. doi: 10.1016/S0140-6736(13)61536-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conrad LA, Cabana MD, Rastogi D. Defining pediatric asthma: phenotypes to endotypes and beyond. Pediatr Res. 2021;90:45–51. PMID:33173175. doi: 10.1038/s41390-020-01231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambrecht BN, Hammad H. The immunology of the allergy epidemic and the hygiene hypothesis. Nat Immunol. 2017;18:1076–1083. PMID: 28926539. doi: 10.1038/ni.3829. [DOI] [PubMed] [Google Scholar]
- 5.Loss G, Depner M, Ulfman LH, van Neerven RJ, Hose AJ, Genuneit J, Karvonen AM, Hyvärinen A, Kaulek V, Roduit C, et al. Consumption of unprocessed cow’s milk protects infants from common respiratory infections. J Allergy Clin Immunol. 2015;135:56–62. PMID: 25442645. doi: 10.1016/j.jaci.2014.08.044. [DOI] [PubMed] [Google Scholar]
- 6.Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, Ishikawa E, Kubota H, Swinkels S, Sakai T, Oishi K, et al. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. Plos One. 2016;11:e0158498. PMID: 27362264. doi: 10.1371/journal.pone.0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tun HM, Konya T, Takaro TK, Brook JR, Chari R, Field CJ, Guttman DS, Becker AB, Mandhane PJ, Turvey SE, et al. Exposure to household furry pets influences the gut microbiota of infants at 3–4 months following various birth scenarios. Microbiome. 2017;5:40. PMID: 28381231. doi: 10.1186/s40168-017-0254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152. PMID: 26424567. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 9.Bisgaard H, Li N, Bonnelykke K, Chawes BL, Skov T, Paludan-Müller G, Stokholm J, Smith B, Krogfelt KA. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128:646–652. PMID: 21782228. doi: 10.1016/j.jaci.2011.04.060. [DOI] [PubMed] [Google Scholar]
- 10.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–352. PMID 27231050. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosseini E, Grootaert C, Verstraete W, Van de Wiele T. Propionate as a health-promoting microbial metabolite in the human gut. Nutr Rev. 2011;69:245–258. PMID: 21521227. doi: 10.1111/j.1753-4887.2011.00388.x. [DOI] [PubMed] [Google Scholar]
- 12.Bartolomaeus H, Balogh A, Yakoub M, Homann S, Markó L, Höges S, Tsvetkov D, Krannich A, Wundersitz S, Avery EG, et al. Short-chain fatty acid propionate protects from hypertensive cardiovascular damage. Circulation. 2019;139:1407–1421. PMID 30586752. doi: 10.1161/CIRCULATIONAHA.118.036652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, SCFAs, regulate colonic cc cell homeostasis. Science. 2013;341:569–573. PMID: 23828891. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. PMID: 12496283. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 15.Nakano T, Ochiai S, Suzuki S, Yamaide F, Morita Y, Inoue Y, Arima T, Kojima H, Suzuki H, Nagai K, et al. Breastfeeding promote egg white sensitization in early infancy. Pediatr Allergy Immunol. 2020;31:315–318. PMID:31925979. doi: 10.1111/pai.13208. [DOI] [PubMed] [Google Scholar]
- 16.Biagini Myers JM, Schauberger E, He H, Martin LJ, Kroner J, Hill GM, Ryan PH, LeMasters GK, Bernstein DI, Lockey JE, et al. A pediatric asthma risk score to better predict asthma development in young children. J Allergy Clin Immunol. 2019;143:1803–1810 e2. PMID: 30554722. doi: 10.1016/j.jaci.2018.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teague WG, Phillips BR, Fahy JV, Wenzel SE, Fitzpatrick AM, Moore WC, Hastie AT, Bleecker ER, Meyers DA, Peters SP, et al. Baseline features of the Severe Asthma Research Program (SARP III) cohort: differences with age. J Allergy Clin Immunol Pract. 2018;6:545–554.e4. PMID: 28866107. doi: 10.1016/j.jaip.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McConnell R, Berhane K, Gilliland F, London SJ, Islam T, Gauderman WJ, Avol E, Margolis HG, Peters JM. Asthma in exercising children exposed to ozone: a cohort study. Lancet. 2002;359:386–391. PMID: 11844508. doi: 10.1016/S0140-6736(02)07597-9. [DOI] [PubMed] [Google Scholar]
- 19.Stinson LF, Gay MCL, Koleva PT, Eggesbø M, Johnson CC, Wegienka G, du Toit E, Shimojo N, Munblit D, Campbell DE, et al. Human milk from atopic mothers has lower levels of short chain fatty acids. Front Immunol. 2020;111427. doi: 10.3389/fimmu.2020.01427. PMID: 32903327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gill SK, Rossi M, Bajka B, Whelan K. Dietary fibre in gastrointestinal health and disease. Nat Rev Gastroenterol Hepatol. 2021;18:101–116. PMID: 33208922. doi: 10.1038/s41575-020-00375-4. [DOI] [PubMed] [Google Scholar]
- 21.Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–1345. PMID: 27259147. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 22.Luu M, Monning H, Visekruna A. Exploring the molecular mechanisms underlying the protective effects of microbial SCFAs on intestinal tolerance and food allergy. Front Immunol. 2020;111225. doi: 10.3389/fimmu.2020.01225. PMID: 32612610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Depner M, Taft DH, Kirjavainen PV, Kalanetra KM, Karvonen AM, Peschel S, Schmausser-Hechfellner E, Roduit C, Frei R, Lauener R, et al. Maturation of the gut microbiome during the first year of life contributes to the protective farm effect on childhood asthma. Nat Med. 2020;26:1766–1775. PMID: 33139948. doi: 10.1038/s41591-020-1095-x. [DOI] [PubMed] [Google Scholar]
- 24.Russell SL, Gold MJ, Willing BP, Thorson L, McNagny KM, Finlay BB. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. 2013;4:158–164. PMID: 23333861. doi: 10.4161/gmic.23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. PMID: 22442383. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. PMID: 24390308. [DOI] [PubMed] [Google Scholar]
- 27.Nastasi C, Candela M, Bonefeld CM, Geisler C, Hansen M, Krejsgaard T, Biagi E, Andersen MH, Brigidi P, Ødum N, et al. The effect of short-chain fatty acids on human monocyte-derived dendritic cells. Sci Rep. 2015;5:16148. PMID: 26541096. doi: 10.1038/srep16148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura I, Inoue D, Maeda T, Hara T, Ichimura A, Miyauchi S, Kobayashi M, Hirasawa A, Tsujimoto G. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41). Proc Natl Acad Sci U S A. 2011;108:8030–8035. PMID: 21518883. doi: 10.1073/pnas.1016088108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Souza WN, Douangpanya J, Mu S, Jaeckel P, Zhang M, Maxwell JR, Rottman JB, Labitzke K, Willee A, Beckmann H, et al. Differing roles for SCFAs and GPR43 agonism in the regulation of intestinal barrier function and immune responses. Plos One. 2017;12:e0180190. PMID: 28727837. doi: 10.1371/journal.pone.0180190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ang Z, Ding JL. GPR41 and GPR43 in obesity and inflammation - protective or causative? Front Immunol. 2016;7:28. PMID: 26870043. doi: 10.3389/fimmu.2016.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145:396-406.e1–10. PMID: 23665276. doi: 10.1053/j.gastro.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 32.Natarajan N, Hori D, Flavahan S, Steppan J, Flavahan NA, Berkowitz DE, Pluznick JL. Microbial short chain fatty acid metabolites lower blood pressure via endothelial G protein-coupled receptor 41. Physiol Genomics. 2016;48:826–834. PMID: 27664183. doi: 10.1152/physiolgenomics.00089.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. PMID: 15229469. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 34.Zakeri A, Russo M. Dual role of toll-like receptors in human and experimental asthma models. Front Immunol. 2018;9, 1027. doi: 10.3389/fimmu.2018.01027. PMID: 29867994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwata-Yoshikawa N, Uda A, Suzuki T, Tsunetsugu-Yokota Y, Sato Y, Morikawa S, Tashiro M, Sata T, Hasegawa H, Nagata N, et al. Effects of toll-like receptor stimulation on eosinophilic infiltration in lungs of BALB/c mice immunized with UV-Inactivated severe acute respiratory syndrome-related coronavirus vaccine. J Virol. 2014;88:8597–8614. doi: 10.1128/JVI.00983-14. PMID: 24850731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chow J, Panasevich MR, Alexander D, Vester Boler BM, Rossoni Serao MC, Faber TA, Bauer LL, Fahey GC. Fecal metabolomics of healthy breast-fed versus formula-fed infants before and during in vitro batch culture fermentation. J Proteome Res. 2014;13:2534–2542. PMID: 24628373. doi: 10.1021/pr500011w. [DOI] [PubMed] [Google Scholar]
- 37.Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, Terasawa K, Kashihara D, Hirano K, Tani T, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the SCFA receptor GPR43. Nat Commun. 2013;4, 1829. doi: 10.1038/ncomms2852. PMID: 23652017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plantinga M, Guilliams M, Vanheerswynghels M, Deswarte K, Branco-Madeira F, Toussaint W, Vanhoutte L, Neyt K, Killeen N, Malissen B, et al. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 2013;38:322–335. PMID: 23352232. doi: 10.1016/j.immuni.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 39.Hammad H, Chieppa M, Perros F, Willart MA, Germain RN, Lambrecht BN. House dust mite allergen induces asthma via Toll-like receptor 4 triggering of airway structural cells. Nat Med. 2009;15:410–416. PMID: 19330007. doi: 10.1038/nm.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito T, Hirose K, Norimoto A, Tamachi T, Yokota M, Saku A, Takatori H, Saijo S, Iwakura Y, Nakajima H. Dectin-1 plays an important role in house dust mite-induced allergic airway inflammation through the activation of CD11b+ dendritic cells. J Immunol. 2017;198:61–70. PMID: 27852745. doi: 10.4049/jimmunol.1502393. [DOI] [PubMed] [Google Scholar]
- 41.Ito T, Hirose K, Saku A, Kono K, Takatori H, Tamachi T, Goto Y, Renauld JC, Kiyono H, Nakajima H. IL-22 induces Reg3gamma and inhibits allergic inflammation in house dust mite-induced asthma models. J Exp Med. 2017;214:3037–3050. PMID: 28811323. doi: 10.1084/jem.20162108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaiss MM, Rapin A, Lebon L, Dubey LK, Mosconi I, Sarter K, Piersigilli A, Menin L, Walker AW, Rougemont J, et al. The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity. 2015;43:998–1010. PMID: 26522986. doi: 10.1016/j.immuni.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ge SX, Son EW, Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018;19:534. PMID: 30567491. doi: 10.1186/s12859-018-2486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. PMID: 23586463. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7. PMID: 27141961. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, et al. Gene set knowledge discovery with Enrichr. Curr Protoc. 2021;1:e90. PMID: 33780170. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamasaki Y, Kohno Y, Ebisawa M, Kondo N, Nishima S, Nishimuta T, Morikawa A. Japanese society of allergology; Japanese society of pediatric allergy and clinical immunology. Japanese guideline for childhood asthma 2014. Allergol Int. 2014;63:335–356. PMID: 25178176. doi: 10.2332/allergolint.14-RAI-0767. [DOI] [PubMed] [Google Scholar]
- 48.Sato Y, Sakurai K, Tanabe H, Kato T, Nakanishi Y, Ohno H, Mori C. Maternal gut microbiota is associated with newborn anthropometrics in a sex-specific manner. J Dev Orig Health Dis. 2019;10:659–666. PMID: 31106719. doi: 10.1017/S2040174419000138. [DOI] [PubMed] [Google Scholar]
- 49.Kato T, Fukuda S, Fujiwara A, Suda W, Hattori M, Kikuchi J, Ohno H. Multiple omics uncovers host-gut microbial mutualism during prebiotic fructooligosaccharide supplementation. DNA Res. 2014;21:469–480. PMID: 24848698. doi: 10.1093/dnares/dsu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim SW, Suda W, Kim S, Oshima K, Fukuda S, Ohno H, Morita H, Hattori M. Robustness of gut microbiota of healthy adults in response to probiotic intervention revealed by high-throughput pyrosequencing. DNA Res. 2013;20:241–253. PMID: 23571675. doi: 10.1093/dnares/dst006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Said HS, Suda W, Nakagome S, Chinen H, Oshima K, Kim S, Kimura R, Iraha A, Ishida H, Fujita J, et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res. 2014;21:15–25. PMID: 24013298. doi: 10.1093/dnares/dst037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA sequence data that support the findings of this study are openly available in DDBJ Sequence Read Archive at https://www.ddbj.nig.ac.jp/ddbj/index.html, reference number DRA014484.
The RNA sequence dat that support the findings of this study are openly available in Gene Expression Omnibus at https://www.ncbi.nlm.nih.gov/geo/, reference number GSE207711.


