Abstract
The effects of polymyxin B (PMB) on the Escherichia coli outer (OM) and cytoplasmic membrane (CM) permeabilities were studied by monitoring the fluxes of tetraphenylphosphonium, phenyldicarbaundecaborane, and K+ and H+ ions. At concentrations between 2 and 20 μg/ml, PMB increased the OM permeability to lipophilic compounds and induced a leakage of K+ from the cytosol and an accumulation of lipophilic anions in the cellular membranes but did not cause the depolarization of the CM. At higher concentrations, PMB depolarized the CM, forming ion-permeable pores in the cell envelope. The permeability characteristics of PMB-induced pores mimic those of bacteriophage- and/or bacteriocin-induced channels. However, the bactericidal effect of PMB took place at concentrations below 20 μg/ml, indicating that this effect is not caused by pore formation. Under conditions of increased ionic strength, PMB made the OM permeable to lipophilic compounds and decreased the K+ gradient but was not able to depolarize the cells. The OM-permeabilizing effect of PMB can be diminished by increasing the concentration of Mg2+. The major new findings of this work are as follows: (i) the OM-permeabilizing action of PMB was dissected from its depolarizing effect on the CM, (ii) the PMB-induced ion-permeable pores in bacterial envelope were registered, and (iii) the pore formation and depolarization of the CM are not obligatory for the bactericidal action of PMB and dissipation of the K+ gradient on the CM.
Lipid bilayers usually are quite permeable to lipophilic compounds (15, 41, 44). However, the outer membrane (OM) of gram-negative bacteria forms a rather effective permeability barrier against various lipophilic substances, including antibiotics. At least 10- to 100-fold slower rates of lipophilic compound permeation through the OM bilayer compared to those through the cytoplasmic membrane (CM) are observed because of the highly charged lipopolysaccharide (LPS)-formed outer monolayer and the stabilization of this layer by divalent cations (20, 39). These compounds also cannot traverse the porins, the narrow channels for inorganic ions and small hydrophilic nutrients (19, 38).
Polymyxin B (PMB) is a decapeptide antibiotic characterized by a heptapeptide ring containing four 2,4-diaminobutyric acids. An additional peptide chain covalently bound to the γ-amino group carries an aliphatic chain attached to the peptide through an amide bond. The molecule carries five positively charged residues of diaminobutyric acid (52). Due to its molecular mass (about 1,200 Da), charge, and amphiphilicity, PMB should be excluded by the OM. However, several polycationic compounds, including PMB, are known to penetrate the OM using pathways other than porins. Although the detailed mechanism of the OM permeability barrier disruption remains undetermined, complex formation by PMB with LPS is expected to be the first stage (26, 50, 56, 60). Being bulkier than the inorganic divalent cations that it displaces, PMB changes the packing order of LPS and increases the permeability of the OM to a variety of molecules, inducing also its own uptake (“self-promoted” uptake). Aminoglycosides and cationic antimicrobial peptides, such as insect cecropins and mammalian neutrophil defensins, might also access their targets by this pathway (21, 22).
PMB is bactericidal to almost all gram-negative bacteria at rather low concentrations (52). However, severe side effects (45, 48) prevent the intensive use of PMB in medicine. Despite the absence of PMB resistance in clinical isolates or the possibility of induction of a genetically stable resistance against PMB by mutagenesis (43), there are species and strains which either are a priori resistant or have obtained resistance to PMB (23, 33, 42, 59). The main reason for the resistance is decreased PMB binding due to the reduced anionicity of LPS because of esterification of phosphate groups in the core region and/or lipid A by amino compounds (18a, 23, 42, 59). Although a variety of analogues of PMB have been produced in order to clarify the relationship between its structure and the biological activity (31, 52, 57, 60), the mechanisms of bactericidal and cytotoxic actions are still under discussion (32, 43, 62). The different CM functions such as active transport and respiration were found to be affected by PMB (for a review, see reference (52)). Therefore, it is considered that the bactericidal action of PMB depends on its interaction with the CM. The lethal action of PMB is proposed to be the result of the interaction of PMB with the acidic phospholipids exposed on the CM (52, 55).
However, more than 20 years ago La Porte et al. (29) demonstrated that the interaction of immobilized PMB with the OM alone is sufficient to block the respiration and growth of Escherichia coli. Later it was shown that immobilized cationic immune protein attacin (8) as well as the aminoglycoside antibiotic gentamicin conjugated to bovine serum albumin (24) are also able to inhibit cell growth.
In order to obtain more information on the self-promoted uptake and understand the mechanism of bactericidal action of PMB, its effects on the OM and the CM were studied. We monitored the fluxes of tetraphenylphosphonium (TPP+), phenyldicarbaundecaborane (PCB−), and K+ and H+ ions with selective electrodes. Simultaneous measurements of ion fluxes through the bacterial envelope and the sensitivities of the ion gradients to ionophoric antibiotics gave us valuable information about the permeabilities and functions of both the CM and the OM. We were able to dissect the OM-disorganizing action of PMB from the damaging action of PMB on the CM. We observed that, at high concentrations, PMB induced ion-permeable pores in the envelope and depolarized the CM, but the bactericidal effect of PMB was expressed at lower concentrations and was not dependent on depolarization of the CM.
MATERIALS AND METHODS
Materials.
TPP+ chloride and the potassium salt of PCB− were obtained from Biocell Products, Helsinki, Finland. Polymyxin B sulfate (PMB; 7730 U of PMB base/mg), polymyxin B nonapeptide (PMBN), and gramicidin D (GD) were purchased from Sigma.
Bacteria, phage, and growth conditions.
Propagation of E. coli AN180 (F− argE3 thi mtl xyl str-704), with a wild-type cell envelope, and E. coli KO1489, a sodium dodecyl sulfate-sensitive derivative of MC4100 [araD-139 Δ(argF-lac)U-169 rps-L150 relA-7 deoC-1 ptsF-25 rbsR thi supF Z1a:tn 10sds-16] was carried out as described previously (10, 11). The strains were grown in Luria-Bertani medium (46) at 37°C with aeration. The final cell batch was grown from a diluted (2 × 108 cells/ml) overnight culture. Cells were harvested at 1.1 × 109 cells/ml, concentrated 100-fold by centrifugation, and resuspended in 100 mM Tris-HCl (pH 7.0) to obtain 2 × 1011 to 3 × 1011 cells/ml. For pH measurements the cells were washed once and resuspended in 0.5 mM MOPS (morpholinepropanesulfonic acid)-Tris in 100 mM NaCl (pH 7.0). The concentrated cells were kept on ice until used (maximally, 5 h). Tris-EDTA-treated cells were prepared as described previously (25). The bacteria were incubated in 100 mM Tris containing 10 mM EDTA (pH 7.0) at 37°C for 10 min. Bacteriophage T4 was grown, and the number of infectious particles was determined as described previously (18, 25). The phage was concentrated from cell lysates and was purified with a two-phase system (polyethylene glycol 6000–dextran sulfate), also as described previously (18).
For the viability measurements the cells (3 × 109 cells/ml) were preincubated for 10 min at 37°C in 100 mM sodium phosphate or Tris-HCl buffers, treated with PMB or PMBN for an additional 10 min, diluted with 0.9% NaCl, and dispersed on agar plates prepared with 1% peptone, 0.5% yeast extract, 1% NaCl, and 1.5% agar (pH 7.0) for determination of the number of CFU.
Measurements of ion fluxes and determination of membrane voltage.
For the ion flux experiments, 50 to 80 μl of the concentrated cell suspension was added to an appropriate buffer in a 5-ml reaction vessel. The vessel was thermostated, and the cell suspension was aerated by magnetic stirring. The concentrations of TPP+, PCB−, and K+ and H+ ions in the medium were monitored with selective electrodes connected to 520A pH/ISE meters (Orion Research Inc.). The K+- and H+-selective electrodes were from Orion Research Inc. (models 93-19 and 81-02, respectively). Ag/AgCl reference electrodes (model 9001 or 9002; Orion Research Inc.) were indirectly connected to the measuring vessels through an agar salt bridge. The characteristics of the TPP+- and PCB−-selective electrodes have been described previously (10, 17). The electrodes are available through Biocell Products.
The internal TPP+ concentration was calculated from the external one, assuming that 200 Klett units (A540) correspond to 1 × 109 cells/ml, 1.25 × 109 cells correspond to 1 mg of dry mass, and the intracellular water volume of E. coli is 1.1 μl/mg of dry mass (3). The means of preparation of the cells and the methods used to study the respiration-driven H+ transport have been described previously (18). The changes in membrane voltage (ΔΨ) were calculated by a modified Nernst equation, also as described previously (12, 25).
Measurement of ion fluxes was carried out simultaneously in three reaction vessels, and a typical registration course of ion movements is presented in the figures.
RESULTS
The OM porins freely exchange inorganic ions but form a permeability barrier to lipophilic compounds like ionophoric antibiotics (nigericin, GD) or lipophilic ions like TPP+ (for reviews, see references 20 and 39). On the other hand, the CM is rather impenetrable to inorganic ions such as K+ but does not prevent membrane voltage (ΔΨ; negative inside)-driven accumulation of TPP+ in the bacterial cytosol. Therefore, analysis of the interaction of lipophilic cations with bacterial cells is a simple but informative way to estimate both the OM and the CM permeabilities (for details, see reference 11).
The OM-permeabilizing action of PMB can be dissected from its depolarizing effect on the CM.
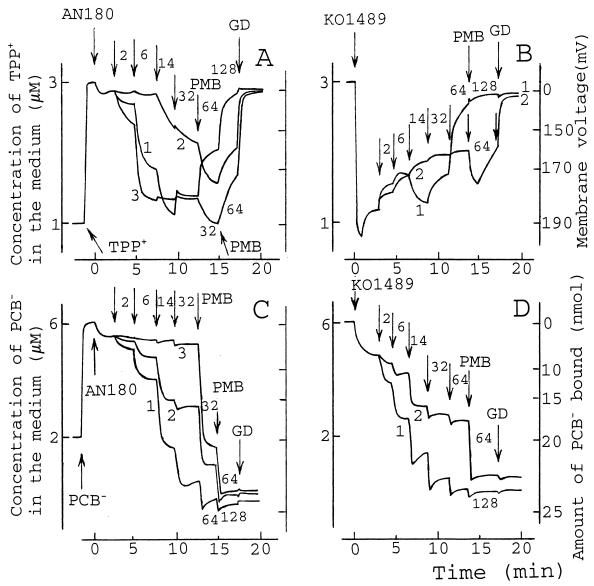
Several PMB-induced stages in the accumulation of TPP+ by AN180 cells with wild-type OM could be defined. Low concentrations of PMB (2 to 14 μg/ml) induced a decrease in the TPP+ concentration in the medium because of accumulation of this lipophilic cation in the cells as a result of a PMB-induced increase in the OM permeability (Fig. 1A, curve 1). However, at concentrations over 32 μg/ml, PMB induced a leakage of initially accumulated TPP+, indicative of depolarization of the CM.
FIG. 1.
Effects of PMB, PMBN, and EDTA on the accumulation of TPP+ (A and B) and PCB− (C and D) by E. coli AN180 (A and C) and KO1489 (B and D) cells. The experiments were performed at 37°C in 100 mM Tris-HCl (pH 8.0). The cell concentration was 3 × 109 cells/ml. Arrows, if not stated otherwise, indicate the addition of PMB (curve 1), PMBN (curve 2), or EDTA (curve 3). The number next to the arrow indicates the final concentration (in micrograms per milliliter) of PMB or PMBN after the last addition. EDTA was added to 0.03, 0.12, 1, and 5 mM (A and C), and GD was added to 5 μg/ml. The results shown are representative of three independent experiments.
Both PMB and EDTA remove divalent cations from the LPS layer, increasing the permeability of the OM to lipophilic compounds (2, 20, 41, 60). In Tris buffer, EDTA at concentrations exceeding 20 μM induced the uptake of TPP+ by AN180 cells, and the maximal amount of TPP+ was accumulated when the concentration of EDTA achieved 120 μM. A further increase in the EDTA concentration (up to 5 mM) had no considerable effect on the accumulation of TPP+ (Fig. 1A, curve 3). Low concentrations of PMB added to the suspension of AN180 cells containing EDTA induced the release of a small amount of accumulated TPP+ (data not shown), but 32 μg of PMB per ml stimulated an additional uptake of TPP+. At higher concentrations, PMB induced the depolarization of the cells. In the case of the deacylated PMB derivative PMBN, the accumulation of TPP+ started at concentrations above 14 μg/ml (Fig. 1A, curve 2) and reached the maximal level at 32 μg/ml. However, subsequent addition 32 μg of PMB per ml led to an additional uptake of TPP+, followed by its release if higher concentrations of PMB were used.
In the case of lipophilic compound-permeable KO1489 cells, low concentrations of PMB (2 to 6 μg/ml) induced the release of a small amount of accumulated TPP+ (Fig. 1B, curve 1). Further additions of PMB stimulated an additional uptake of TPP+. At concentrations above 32 μg/ml, PMB induced leakage of accumulated TPP+. Under these conditions PMBN induced the release of only a small amount of accumulated TPP+, but subsequent PMB addition induced supplementary accumulation of the lipophilic cation (Fig. 1B, curve 2).
On the basis of additional experiments with different titration steps (data not shown), we can conclude that PMB at concentrations of 2 to 20 μg/ml permeabilizes the OM to lipophilic compounds but does not depolarize the CM. Approximately 30 μg of PMB per ml is needed to induce the depolarizing effect. There was no strong correlation between the OM permeability to lipophilic compounds and the depolarizing efficiency of PMB. The permeable KO1489 and AN180 cells with the wild-type OM were depolarized by approximately the same concentration of PMB.
PMB induces an effective binding of lipophilic anions to cell membranes.
It has been shown (4, 15, 44) that lipophilic anions bind to artificial membranes several orders of magnitude more strongly and translocate across bilayers several orders of magnitude more rapidly than structurally similar cations. However, the CMs of intact microbial cells accumulate very small amounts of lipophilic anions (10, 17), and the association-exclusion mechanism is mostly based on an electrostatic repulsion caused by the negatively charged phospholipids (cardiolipin, phosphatidylglycerol) that form the outer layer of the CM (10).
The uptake of PCB− by AN180 cells was induced by PMB at concentrations exceeding 2 μg/ml, and the amount of PCB− bound gradually increased with an increase in the concentration of PMB (Fig. 1C, curve 1). PMB-induced accumulation of PCB− became biphasic (the initial binding immediately after the addition of PMB was followed by a slow release) when the concentration of PMB exceeded 32 μg/ml.
Although EDTA (in Tris buffer) permeabilized AN180 cells to TPP+ rather effectively, this chelator did not considerably increase the level of accumulation of PCB−. PMB additions were needed to induce PCB− binding (Fig. 1C, curve 3). PMBN also stimulated the binding of PCB− to the cell membranes, and the saturation was achieved at a concentration of 14 to 32 μg/ml, but addition of PMB induced an additional accumulation of PCB− (Fig. 1C, curve 2).
In the absence of PMB, KO1489 cells accumulated about three times larger amounts of PCB− than AN180 cells (Fig. 1D). However, the final amount of PCB− bound as well as the dependence of the amount bound on the concentration of PMB in the medium was rather similar to that observed for AN180 cells (Fig. 1D, curve 1). PMBN increased the amount of PCB− bound to KO1489 cells, but not as effectively as PMB did (Fig. 1D, curve 2). The addition of EDTA to Tris buffer had no considerable effect on the accumulation of PCB− by KO1489 cells (data not shown). Despite the low efficiency of the effect of EDTA on PCB− binding (Fig. 1C, curve 3), Tris-EDTA-treated AN180 cells (see Materials and Methods) accumulated larger amounts of PCB, close to the amount bound by KO1489 cells (10).
The CM depolarizing and the OM permeabilizing effects of PMB have different ionic strength dependencies.
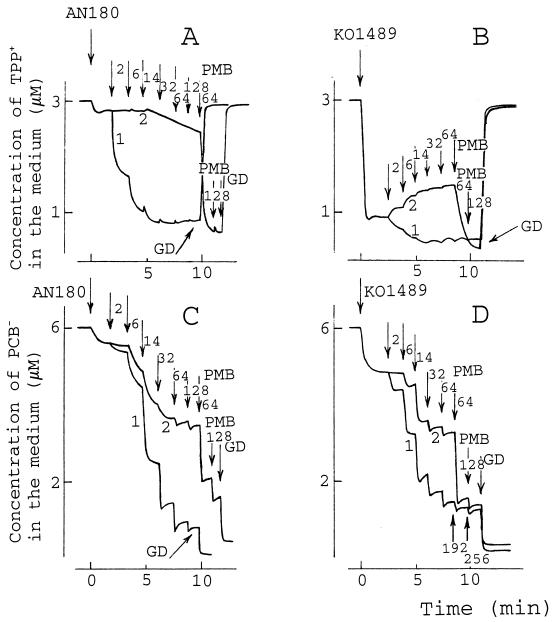
When the phosphate (sodium or potassium salt) concentration in the medium was increased from 25 to 100 mM, the CM-depolarizing effect of PMB disappeared and only an OM-permeabilizing effect was observed (data not shown). At low PMB concentrations (up to 14 μg/ml in 100 mM phosphate), TPP+ accumulation was also a PMB concentration-dependent process. However, the subsequent increase in the concentration of PMB (exceeding 120 μg/ml) had no depolarizing effect and only the addition of GD induced the leakage of TPP+ (Fig. 2A, curve 1). Under these conditions PMBN had considerably lower OM-permeabilizing activity than in Tris buffer (compare Fig. 1A, curve 2, and Fig. 2A, curve 2). In the case of permeable KO1489 cells, PMB induced an additional accumulation of TPP+ starting from the lowest concentrations (2 to 6 μg/ml) (Fig. 2B, curve 1), but no depolarizing effect was observed even at high concentrations. PMBN induced the release of accumulated TPP+, but a subsequent PMB addition stimulated additional accumulation (Fig. 2B, curve 2).
FIG. 2.
Effects of PMB and PMBN on accumulation of TPP+ (A and B) and PCB− (C and D) by E. coli AN180 (A and C) and KO1489 (B and D) cells. The initial conditions of the experiments were as described in the legend to Fig. 1, but the experiments were performed in 100 mM sodium phosphate (pH 8.0). Arrows, if not stated otherwise, indicate the addition of PMB (curve 1) and PMBN (curve 2). A number next to the arrow indicates the final concentration (in micrograms per milliliter) of PMB or PMBN after the last addition. GD was added to a concentration of 5 μg/ml. The results shown are representative of three independent experiments.
In 100 mM phosphate buffer the maximal binding of PCB− was not achieved by PMB alone but needed the addition of GD. The same result was observed with both AN180 and KO1489 cells, indicating that the maximal amount of PCB− can be bound only after depolarization of the CM (Fig. 2C and D). On the other hand, it is clear that the different levels of PCB− binding caused by PMB and PMBN cannot be explained only by the ability of PMB to depolarize the CM (compare Fig. 1 and 2).
The PMB-caused bactericidal effect is not dependent on depolarization of the cells.
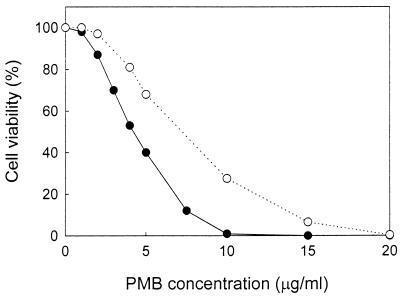
More than 99% of the cells (of both strains) were not able to form colonies after 10 min of incubation in sodium phosphate buffer containing 20 μg of PMB per ml (Fig. 3). KO1489 cells lost their viabilities in the presence of concentrations of PMB lower than those required for the loss of AN180 cell viability, indicating that the OM permeability to lipophilic compounds is one of the factors controlling cell sensitivity to PMB. However, in both cases the ability of the cells to form colonies was lost under conditions in which PMB was not able to depolarize the CM (Fig. 2A and B). These results indicate that the depolarization of the CM is not obligatory for the bactericidal action of PMB.
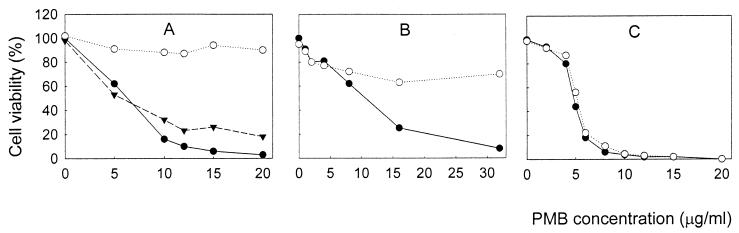
FIG. 3.
Effect of PMB on the viabilities of E. coli AN180 (○) and KO1489 (●) cells. The viabilities of the cells were measured as described in Materials and Methods, using 100 mM sodium phosphate (pH 8.0) as the incubation medium. Each datum point represents the mean of values from three independent experiments. The standard errors of the means were all less than 10%.
Mg2+ abolishes the bactericidal action of PMB by stabilizing the OM structure.
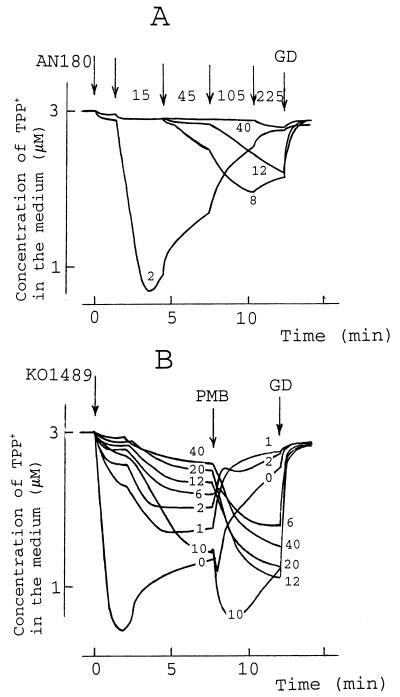
Mg2+ is known to inhibit the actions of many OM permeabilizers that act by either chelating or replacing cations in the OM (60) and antagonize the bactericidal action of PMB (37). PMB-induced permeabilization of the OM, as measured by determination of the level of TPP+ uptake, was also dependent on the Mg2+ concentration (Fig. 4A). At concentrations up to 2 mM no considerable effect on cell sensitivity to PMB was detected. In the presence of higher Mg2+ concentrations increased amounts of PMB were necessary to make the OM permeable to TPP+ and the depolarizing activity of PMB became considerably weaker. Further increases in the concentration of Mg2+ (over 12 mM) abolished the depolarizing activity of PMB, and at concentrations over 40 mM the OM-permeabilizing activity was totally blocked (Fig. 4A).
FIG. 4.
Effect of Mg2+ on PMB-induced TPP+ uptake by E. coli AN180 (A) or KO1489 (B) cells. The experiments were performed at 37°C in 100 mM Tris-HCl (pH 8.0), and MgCl2 was added to the concentrations (in millimolar) indicated in the figure. The cells were added to a final concentration of 3 × 109 cells/ml, and GD was added to a final concentration of 5 μg/ml. (A) Arrows, if not stated otherwise, indicate the addition of PMB, and a number next to the arrow indicates the final concentration (in micrograms per milliliter) of PMB after the last addition. (B) PMB was added to a concentration 80 μg/ml. The results shown are representative of three independent experiments.
In the case of KO1489 cells, a 1 mM concentration of Mg2+ had already reduced the amount of TPP+ accumulated, and a two-step mode of TPP+ uptake became apparent (Fig. 4B). However, this Mg2+ concentration had no considerable effect on the depolarizing activity of PMB. In general, an increase in the concentration of Mg2+ considerably suppressed the initial accumulation of TPP+, and PMB treatment did not result in the release of TPP+ but increased the amount of TPP+ accumulated. However, an anomaly was registered at Mg2+ concentrations close to 10 mM. The additional increase in the concentration of Mg2+ reduced the rate of TPP+ accumulation as well as caused a reduction in the initial level of uptake. However, the TPP+ uptake stimulated by PMB and the depolarizing effect of GD were well expressed even with Mg2+ at a concentration of 40 mM.
It is known (37) that an Mg2+ concentration of 40 to 50 mM abolishes the antibacterial effects of PMB. Figure 5 shows that Mg2+ at a concentration of 40 mM effectively rescued AN180 cells, including EDTA-treated ones. The rescue was effective only when the dilution solution also contained 40 mM Mg2+, indicating that Mg2+ did not prevent the adsorption of PMB onto the cells. Experiments with the permeable KO1489 cells indicated that Mg2+ ions did not inhibit the bactericidal effect of PMB, addressing the importance of the role of OM in the rescue. However, the Tris-EDTA-treated cells were even less sensitive to PMB than the cells with an intact OM. This could be due to the lower level of adsorption of PMB onto the treated cells. It is known (2) that cells lose LPS during the Tris-EDTA treatment. It is also evident that cells in Tris buffer were more sensitive to PMB than cells in 100 mM phosphate (compare Fig. 3 and Fig. 5).
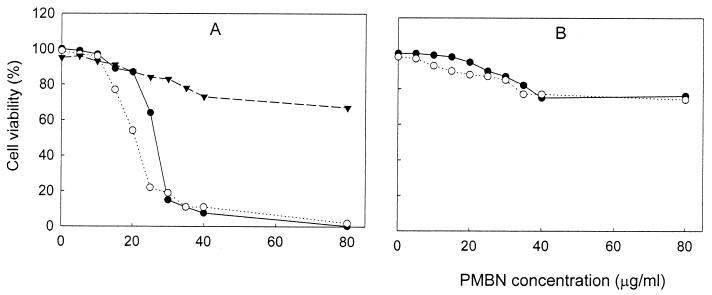
FIG. 5.
Effect of Mg2+ on the viability of PMB-treated cells. The viability of the cells was measured as indicated in Materials and Methods, using 100 mM Tris-HCl (pH 8.0) as the incubation medium. ●, experiments carried out in the absence of Mg2+; ○, experiments carried out in the presence of 40 mM Mg2+; ▾, experiments carried out in the presence of 40 mM Mg2+ but Mg2+ was not included in the 0.9% NaCl solutions used for the dilutions before plating. (A) AN180 cells; (B) Tris-EDTA-treated AN180 cells; (C) KO1489 cells. Each data point represents the mean of values from three independent experiments. The standard errors of the means were all less than 10%.
PMB-induced efflux of K+ ions is not caused by depolarization of the CM.
The K+ content of the cells is considered to reflect the integrity of the CM (14), and depolarization of the CM is expected to cause K+ leakage. Irrespective of the cells (AN180 or KO1489) or medium (100 mM Tris or 100 mM phosphate) used, 2 to 15 μg of PMB per ml induced K+ efflux (Fig. 6). In 100 mM phosphate buffer the maximal rate of efflux was achieved at 14 μg/ml PMB and the following increase in the concentration of PMB had no further effect. GD induced an additional release of K+ ions (Fig. 6A, curve 1).
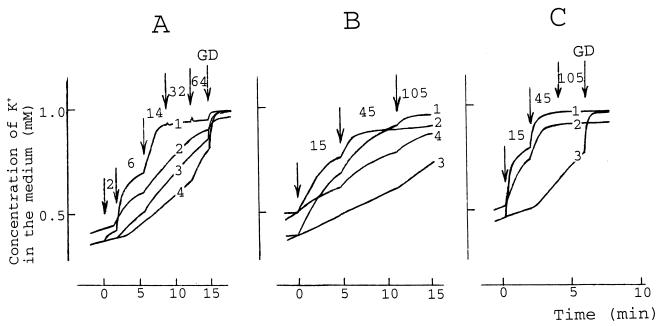
FIG. 6.
Effects of PMB and PMBN on the efflux of K+ from E. coli cells. The experiments were performed at 37°C (A and C) or 25°C (B) in buffers consisting of 100 mM sodium phosphate at pH 8.0 (A, curves 1 to 3) or pH 7.0 (A, curve 4) or in 100 mM Tris-HCl at pH 8.0 (B and C). In panel B, curves 3 and 4, and panel C, curve 3, the incubation medium contained 8 mM Mg2+. The cell concentration was 3 × 109 cells/ml, and GD (A and C) was added to 5 μg/ml. Arrows, if not stated otherwise, indicate the addition of PMBN (A, curves 3 and 4) or PMB. The number next to the arrow indicates the final concentration (in micrograms per milliliter) of PMB or PMBN after the last addition. Curve 2 in each panel and curve 4 in panel B are data from experiments with KO1489 cells; the rest of the experiments were carried out with AN180 cells. The results shown are representative of three independent experiments.
The PMB titration-induced “steps” of K+ leakage were also expressed in Tris buffer. The maximal K+ leakage was observed at a PMB concentration of 45 μg/ml without GD (Fig. 6C, curves 1 and 2). This indicates that PMB-induced depolarization of the CM is involved in the dissipation of the K+ gradient but is not the principal cause of the PMB-induced K+ leakage.
In the presence of 8 mM Mg2+ PMB had a very weak effect on the K+ gradient in AN180 cells at 25°C (Fig. 6B, curve 3), and considerably higher concentrations of PMB were necessary to dissipate the gradient at 37°C (Fig. 6C; compare curves 1 and 3). The influence of Mg2+ was weaker in the case of KO1489 cells, but a higher concentration of PMB was necessary to induce K+ efflux in the presence of Mg2+ (Fig. 6B; compare curves 2 and 4). These results indicate that the release of intracellular K+ is one of the earliest consequences of the PMB action and that most of the K+ ions are released in the presence of nondepolarizing concentrations of PMB and/or when the depolarization of the CM does not occur due to the medium conditions (Fig. 2A and B).
PMBN-induced dissipation of the K+ gradient does not cause cell death.
It is known (36, 60, 61) that PMBN is much less toxic to gram-negative bacteria than PMB. Under the conditions of our experiments PMBN had a rather small bactericidal effect in the case of AN180 cells (Fig. 7). However, at concentrations above 30 μg/ml it effectively killed KO1489 cells in Tris (but not 100 mM phosphate) medium. At pH 8.0, PMBN abolished the capability of the cells to form colonies at lower concentrations compared to the capability at pH 7.0. At concentrations over 6 μg/ml, PMBN induced K+ leakage from AN180 cells suspended in 100 mM sodium phosphate buffer, and the maximal rate of the efflux was achieved at a PMBN concentration of 14 μg/ml (Fig. 6A, curve 3). However, the same concentrations of PMBN at pH 7.0 (Fig. 6A, curve 4) induced a weaker efflux of K+ than the efflux induced by PMBN at pH 8.0, even though the net charge of PMB does not vary from pH 3 to 8 (62).
FIG. 7.
KO1489 (A) and AN180 (B) cell sensitivity to PMBN. The viabilities of the cells were measured as indicated in Materials and Methods. The incubation media were 100 mM Tris-HCl (pH 8.0) (in panel A, ○; in panel B, ○ and ●, 100 mM Tris-HCl (pH 7.0) (in panel A, ●), and 100 mM sodium phosphate (pH 8.0) (in panel A, ▾). (B) ○, Tris-EDTA-treated cells; ●, nontreated cells. Each datum point represents the mean of values from three independent experiments. The standard errors of the means were all less than 10%.
Such pH dependence was not observed with PMB (data not shown). It is evident that PMB interacts with the cells more strongly than PMBN and that the dissipation of the K+ gradient by PMB or PMBN is not enough to block the capability of the cells to form colonies.
PMB-treated cells are able to maintain a considerable ΔpH.
The CM depolarization by PMB can be caused by (i) inhibition of ΔΨ-generating systems and/or (ii) an increase in the permeability of the CM. Due to the stringent coupling between H+ and other ion gradients with ΔΨ, H+ flux is one of the most sensitive indicators of the energy state of the CM. Under aerobic conditions the addition of PMB induced an acidification of the cell suspension (Fig. 8). PMB-induced acidification occurred in two stages: an initial fast stage and a subsequent slow one. At PMB concentrations up to 30 μg/ml, the amount of H+ released during the first stage of acidification correlated well with the amount of PMB added. Therefore, the rapid acidification, as well as the release of TPP+ in the presence of low concentrations of PMB (Fig. 1B), might be a result of replacement of the cell surface-bound cations by PMB. At 40 μg/ml the phase of slow acidification also became considerably weaker. PMB induced only a slow alkalinization of the bacterial suspension when the final concentration of PMB exceeded 60 μg/ml. However, GD-induced alkalinization in the presence of PMB indicated that PMB-treated cells are able to keep a considerable pH gradient (ΔpH) on their CMs.
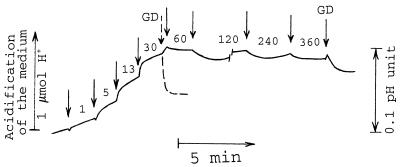
FIG. 8.
Effect of PMB on the pH of the bacterial suspension. The experiment was performed at 32°C. The incubation medium contained 0.5 mM MOPS in 100 mM NaCl (pH adjusted by Tris to 6.75) and 3 × 109 AN180 cells/ml. Arrows, if not stated otherwise, indicate the addition of PMB, and a number next to the arrow indicates the final concentration (in micrograms per milliliter) of PMB after the last addition. GD was added to a final concentration of 5 μg/ml. The results shown are representative of three independent experiments.
PMB induces ion-permeable pores in the cellular envelope.
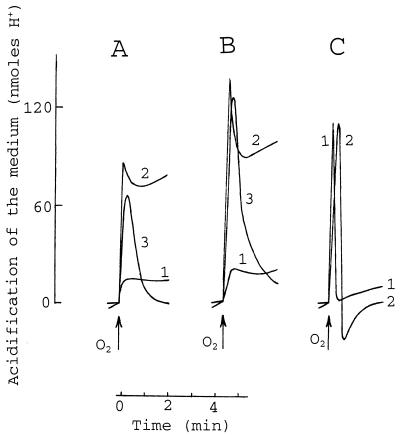
Discrete PMB-induced channel-like events in ionic conductance were detected when planar bilayers made of LPS and phospholipid monolayers were studied (47, 62). The formation of channels in the bacterial CM can be studied by analyzing the mode and amplitude of the oxygen-induced acidification of anaerobic bacterial suspension (16, 18, 35). The addition of PMB to an anaerobic suspension of AN180 cells induced a rapid acidification of the medium, and no alkalinization was observed even at the highest concentrations of PMB used (over 250 μg/ml). Alkalinization of the medium was observed only after the addition of GD (data not shown). The introduction of small amounts of oxygen into the anaerobic suspension of PMB-treated cells caused an extrusion of H+ ions (Fig. 9A, curve 2) that was more substantial than that in the absence of PMB (Fig. 9A, curve 1). The ratio of amount of H+ ions extruded/amount of oxygen atoms added (H+/O ratio) started to increase at a PMB concentration of 30 μg/ml and reached the maximum at 80 μg/ml. Further increases in the PMB concentration (up to 500 μg/ml) only decreased the H+/O ratio. This stimulatory effect was also registered at 10°C (Fig. 9A, curve 3), when the cell membranes are in a “frozen” gel state and ion carriers, such as valinomycin, are not effective (18). In the presence of GD, the respiration-driven acidification of the incubation medium was completely reversible at both temperatures (Fig. 9C), but it was reversible only at the low temperature in the presence of PMB (Fig. 9A) or phage T4 (Fig. 9B). PMB-induced pores mimicked the phage-induced channels but not the GD-formed channels in respect to the dissipation of the oxygen pulse-created proton gradient. However, the maximal H+/O ratio for PMB-treated cells was lower than that for phage T4-infected ones. In addition, the amplitude of the oxygen-induced acidification had a tendency to decrease, and after 30 to 45 min the PMB-treated cells did not respond to oxygen.
FIG. 9.
Effects of PMB, phage T4, and GD on the E. coli respiration-driven proton pump. The anaerobic incubation medium (pH 6.75) contained 0.5 mM MOPS, 100 mM NaCl (see Fig. 8), 3 × 109 AN180 cells/ml, and 100 μg of PMB per ml (A), 12 × 109/ml infectious particles of phage T4 (B), or 10 μg of GD per ml (C). Oxygen pulse (added as air-saturated H2O) contained 8.25 nmol of O2. The temperature of the medium was 32°C (A and B, curves 1 and 2, and C, curve 1) or 10°C (A and B, curve 3, and C, curve 2). The experiment was repeated three times with similar results, and the data from one representative experiment are shown.
DISCUSSION
Recently, two models were presented explaining, at the molecular level, the interaction of PMB with bacterial membranes as well as the bactericidal action of this antibiotic. According to the detergent-like mechanism (62), PMB alone or together with lipid molecules of the membrane matrix forms transient water-filled membrane lesions when PMB is present above a certain threshold concentration. However, the main conclusion drawn from this model is based on experiments with artificial membranes. According to the periplasmic contact formation model (32, 43), PMB forms contacts between the two phospholipid interfaces that enclose the periplasmic space and triggers the metabolic changes that lead to bacterial stasis in the early growth phase. Rapid intermembrane exchange of phospholipids without fusion was shown to occur across the stable vesicle-vesicle contacts formed by stoichiometric amounts of PMB (5, 6), and PMB-induced mixing of phospholipids between the outer layer of the CM and the inner layer of the OM could also be possible. The results presented here indicate that both the proposed PMB-induced processes, pore formation and membrane contact, can occur in vivo and could consist of different stages of the antimicrobial action of PMB.
Interaction of PMB with the OM.
The OM permeability to lipophilic compounds is one of the factors controlling cell sensitivity to PMB. The OM-permeable KO1489 cells are killed at lower PMB concentration than AN180 cells with the wild-type membrane, although the depolarizing concentration of PMB does not considerably depend on the OM permeability. It is known (50) that hydrophobic interactions are the main driving force for the association of PMB with LPS and that the positive charges help only with the correct positioning of the molecule at the surface of the LPS. The interaction of the nonpolar region of the PMB molecule with LPS is independent of the pH and the salt concentration (57). This explains why higher concentrations of PMBN are necessary to increase the OM permeability and to induce the efflux of K+ and why these effects are considerably more dependent on medium composition and pH compared to the concentrations of PMB needed to achieve these results.
Our experiments confirmed the cell-protecting role of Mg2+ and indicated that it is connected to Mg2+-dependent stabilization of the LPS layer. It appears that high concentrations of Mg2+ block the self-promoted entry of PMB into the periplasm but not the binding of PMB to the OM surface (Fig. 5A). In the case of strain KO1489, Mg2+ does not protect the cells because their high degree of OM permeability is not caused by the depletion of divalent cations.
PMB-induced pores.
The increase in ionic strength inhibited the depolarizing action of PMB more than it affected the influence on OM permeability and K+ gradient or the bactericidal activity. A threshold concentration of PMB is required for the formation of the depolarizing pores (62). An electrostatic interaction concentrates PMB locally and enhances the insertion of PMB clusters into the CM. It is known (34, 49) that binding of PMB to the acidic phospholipids is sensitive to the charge-screening effect of the high ionic strength. K+ leakage and the failure to form colonies can, probably, be induced by lower local concentrations of PMB, and therefore, these processes are not blocked by surface charge screening in 100 mM phosphate buffer (Fig. 3 and Fig. 6A).
PMB-induced ion-permeable pores are the main reason for the depolarization of the CM. K+ ions are directly involved in energy metabolism in bacteria (14), but the PMB-induced leakage of this cation did not affect ΔΨ in 100 mM phosphate medium (Fig. 2). PMB-induced pores mimic the phage-induced channels in respect to their mode of dissipation of the proton gradient. It has been shown (1, 53, 54) that the entry of phage T4 DNA into the cytosol occurs through the sites of fusion between the OM and the CM. In such a case the phage-induced pores connect the cytosol directly to the cell exterior. We suggest that PMB also induces the fusion of the CM with the OM and forms envelope-crossing pores. The channel-forming colicins also induce pores with similar characteristics, depolarizing the cells but not dissipating the ΔpH (16, 58), and it is known (9, 30) that they form stable contacts between cellular membranes. GD forms ion-permeable channels in the CM and links the cytosol to the intermembrane space (periplasm), where an acidic pH at the outer surface of the CM is expected (27, 28, 51). The phage or PMB-induced pores should be permeable to protons, but the level of proton flux through the pores is low because of the low H+ concentration in the medium. This probably is the reason for a considerable ΔpH on the CM in the presence of high concentrations of PMB.
Multidirectional bactericidal action of PMB.
Despite the insignificant effect on ΔpH, PMB-induced pores like bacteriocin or bacteriophage-induced channels depolarize the CM and should be bactericidal. However, it was demonstrated that the pore formation in the CM is not a prerequisite for the antimicrobial effect of PMB and probably only guarantees the killing. The capability of the cells to form colonies is lost in the presence of nondepolarizing concentrations of PMB. It should be mentioned that in our experiments the bactericidal concentration of PMB was rather high because of the high concentration of cells (3 × 109 ml) used.
It was shown (Fig. 6) that PMB-induced destruction of the cellular barrier to K+ develops in several stages, with only the last one being the leakage of K+ through the PMB-induced ion-permeable pores. However, the PMB-induced destruction of the osmotic barrier and the release of the intracellular K+ cannot be considered the primary causes of the bactericidal effect. PMBN is able to destroy the potassium gradient without deleterious effects on the cells (Fig. 6 and 7).
The local fusion of the OM and the CM is a prerequisite for envelope-crossing pore formation, and a direct PMB-mediated contact between the OM and the CM must be made at early stages of this process. This would lead to a late growth phase like stasis (32, 43). The exchange of phospholipids between the inner surface of the OM and the outer surface of the CM could also be possible at the later stages of pore formation. This would lead to an increase in the pH at the surface of the CM because of the decreased surface charge and the inhibition of cell growth (7).
“Dilution” of the negatively charged phospholipids in the outer layer of the CM by phosphatidylethanolamine originating from the inner layer of the OM could be the reason for the decrease in the barrier of the CM to lipophilic anions. PCB− binds very poorly to the membranes made of negatively charged phospholipids (10). PMBN stimulated the binding of PCB− to the cell membranes, increasing the permeability of the OM to lipophilic compounds and neutralizing the negative charge at the outer surfaces of the membranes. However, PMB considerably more strongly increases the level of binding, even if no depolarization of the CM occurs. The induction of the phospholipid exchange through the intermembrane contacts would also explain the bactericidal action of the immobilized derivatives of PMB (13, 29). It is possible, however, that not only PMB-induced phospholipid exchange but also direct neutralization of the negative charge at the outer surface of the CM is harmful to the cells (7). PMB (but not PMBN) induces an additional accumulation of TPP+ by cells with the permeable OM (Fig. 1A and B and Fig. 2B). The cause of this phenomenon could be (i) the inactivation of multidrug efflux pumps (40, 63) and/or (ii) the increase in membrane voltage because of inactivation of some ΔΨ-consuming systems. In both cases serious PMB-induced destruction of cell functions would occur without apparent destruction of the CM permeability barrier.
PMBN is bactericidal only to KO1489 cells and only in Tris buffer. In the case of cells with a wild-type or slightly modified OM (like that found after Tris-EDTA treatment), PMBN is able to increase the permeability of CM to K+ but does not induce more severe consequences. Bactericidal amounts of PMBN access the surface of CM only through the highly permeable OM of KO1489 cells and when the negative surface charge of the CM is not screened.
It is obvious that PMB has several cell-damaging effects: (i) the disturbance of surface charges, lipid compositions, and structures of the membranes; (ii) dissipation of the K+ gradient; and (iii) depletion of the membrane voltage. This is probably the reason for the slow emergence of resistance to PMB. Several genetic changes are simultaneously necessary to alter the cell in a way in which all these steps are compensated for.
ACKNOWLEDGMENTS
We are indebted to Marja-Leena Perälä and Paulius Slavinskas for technical assistance.
This investigation was supported by grants 62993 and 37725 from the Finnish Academy of Sciences (to D.H.B.).
REFERENCES
- 1.Bayer M E. Adsorption of bacteriophages to adhesions between wall and membrane of Escherichia coli. J Virol. 1968;2:346–356. doi: 10.1128/jvi.2.4.346-356.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayer M E, Leive L. Effect of ethylenediaminetetraacetate upon the surface of Escherichia coli. J Bacteriol. 1977;130:1364–1381. doi: 10.1128/jb.130.3.1364-1381.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boulanger B, Letellier L. Characterization of ion channels involved in the penetration of phage T4 DNA into Escherichia coli cells. J Biol Chem. 1988;263:9767–9775. [PubMed] [Google Scholar]
- 4.Bühler R, Stürmer W, Apell H-J, Läuger P. Charge translocation by the Na, K-pump. I. Kinetics of local field changes studies by time-resolved fluorescence measurement. J Membr Biol. 1991;121:141–161. doi: 10.1007/BF01870529. [DOI] [PubMed] [Google Scholar]
- 5.Cajal Y, Rogers J, Berg O G, Jain M K. Intermembrane molecular contacts by polymyxin B mediate exchange of phospholipids. Biochemistry. 1996;35:299–308. doi: 10.1021/bi9512408. [DOI] [PubMed] [Google Scholar]
- 6.Cajal Y, Ghanta J, Easwaran K, Surolia A, Jain M K. Specificity for the exchange of phospholipids through polymyxin B mediated intermembrane molecular contacts. Biochemistry. 1996;35:5684–5695. doi: 10.1021/bi952703c. [DOI] [PubMed] [Google Scholar]
- 7.Card G L, Trautman J K. Role of anionic lipids in bacterial membranes. Biochim Biophys Acta. 1990;1047:77–82. doi: 10.1016/0005-2760(90)90263-w. [DOI] [PubMed] [Google Scholar]
- 8.Carlsson A, Nyström T, de Cock H, Bennich H. Attacin—an insect immune protein—binds LPS and triggers the specific inhibition of bacterial outer-membrane protein synthesis. Microbiology. 1998;144:2179–2188. doi: 10.1099/00221287-144-8-2179. [DOI] [PubMed] [Google Scholar]
- 9.Cramer W A, Heymann J B, Schendel S L, Deriy B N, Cohen F S, Elkins P A, Stauddacher C V. Structure-function of the channel-forming colicins. Annu Rev Biophys Biomol Struct. 1995;24:611–641. doi: 10.1146/annurev.bb.24.060195.003143. [DOI] [PubMed] [Google Scholar]
- 10.Daugelavičius R, Bakienė E, Beržinskienė J, Bamford D H. Binding of lipophilic anions to microbial cells. Bioelectrochem Bioenerget. 1997;42:263–274. [Google Scholar]
- 11.Daugelavičius R, Bamford J K H, Grahn A M, Lanka E, Bamford D H. The IncP plasmid-encoded cell envelope-associated DNA transfer complex increases cell permeability. J Bacteriol. 1997;179:5195–5202. doi: 10.1128/jb.179.16.5195-5202.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daugelavičius R, Bamford J K H, Bamford D H. Changes in host cell energetics in response to bacteriophage PRD1 DNA entry. J Bacteriol. 1997;179:5203–5210. doi: 10.1128/jb.179.16.5203-5210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drabick J J, Bhattacharjee A K, Hoover D L, Siber G E, Morales V E, Young L D, Brown S L, Cross A S. Covalent polymyxin B conjugate with human immunoglobulin G as an antiendotoxin reagent. Antimicrob Agents Chemother. 1998;42:583–588. doi: 10.1128/aac.42.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epstein W. Osmoregulation by potassium transport in Escherichia coli. FEMS Microbiol Rev. 1986;39:73–78. [Google Scholar]
- 15.Flewelling R F, Hubbell W L. The membrane dipole potential in a total membrane potential model. Applications to hydrophobic ion interactions with membranes. Biophys J. 1986;49:541–552. doi: 10.1016/S0006-3495(86)83664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gould J M, Cramer W A. Studies on the depolarization of the Escherichia coli cells membrane by colicin E1. J Biol Chem. 1977;252:5491–5497. [PubMed] [Google Scholar]
- 17.Grinius L, Daugelavičius R, Alkimavičius G. Studies of the membrane potential of Bacillus subtilis and Escherichia coli cells by the method of penetrating ions. Biochimya. 1981;45:1222–1230. . (English translation.) [Google Scholar]
- 18.Grinius L, Daugelavičius R. Depolarization of Escherichia coli cytoplasmic membrane by bacteriophage T4 and lambda: evidence for induction of ion-permeable channels. Bioelectrochem Bioenerget. 1988;19:235–245. [Google Scholar]
- 18a.Gunn J S, Lim K B, Krueger J, Kim K, Guo L, Hackett M, Miller S I. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 19.Hancock R E W. Role of porins in outer membrane permeability. J Bacteriol. 1987;169:929–933. doi: 10.1128/jb.169.3.929-933.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hancock R E W. The bacterial outer membrane as a drug barrier. Trends Microbiol. 1997;5:37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 21.Hancock R E W, Chapple D. Peptide antibiotics. Antimicrob Agents Chemother. 1999;43:1317–1323. doi: 10.1128/aac.43.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock R E W, Falla T, Brown M. Cationic bactericidal peptides. Adv Microb Physiol. 1995;37:135–175. doi: 10.1016/s0065-2911(08)60145-9. [DOI] [PubMed] [Google Scholar]
- 23.Helander I M, Kato Y, Kilpeläinen I, Kostiainen R, Lindner B, Nummila K, Sugiyama T, Yokochi T. Characterization of lipopolysaccharides of polymyxin-resistant and polymyxin-sensitive Klebsiella pneumoniae O3. Eur J Biochem. 1996;237:272–278. doi: 10.1111/j.1432-1033.1996.0272n.x. [DOI] [PubMed] [Google Scholar]
- 24.Kadurugamuwa J, Clarke A J, Beveridge T J. Surface action of gentamicin on Pseudomonas aeruginosa. J Bacteriol. 1993;175:5798–5805. doi: 10.1128/jb.175.18.5798-5805.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalasauskaitė E V, Kadišaitė D L, Daugelavičius R J, Grinius L L, Jasaitis A A. Studies on energy supply for genetic processes. Requirement for membrane potential in Escherichia coli infection by phage T4. Eur J Biochem. 1983;130:123–130. [PubMed] [Google Scholar]
- 26.Katsu T, Yoshimura S, Tsuchiya T, Fujita Y. Temperature dependence of action of polymyxin B on Escherichia coli. J Biochem. 1984;95:1645–1653. doi: 10.1093/oxfordjournals.jbchem.a134777. [DOI] [PubMed] [Google Scholar]
- 27.Kemper M A, Urrutia M M, Beveridge T J, Koch A L, Doyle R J. Proton motive force may regulate cell wall-associated enzymes of Bacillus subtilis. J Bacteriol. 1993;175:5690–5696. doi: 10.1128/jb.175.17.5690-5696.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch A L. The pH in the neighborhood of membranes generating a protonmotive force. J Theor Biol. 1986;120:73–84. doi: 10.1016/s0022-5193(86)80018-2. [DOI] [PubMed] [Google Scholar]
- 29.La Porte D C, Rosenthal K S, Storm D R. Inhibition of Escherichia coli growth and respiration by polymyxin B covalently attached to agarose beads. Biochemistry. 1977;16:1642–1648. doi: 10.1021/bi00627a019. [DOI] [PubMed] [Google Scholar]
- 30.Lazdunski C L. Colicin import and pore formation: a system for studying protein transport across membranes? Mol Microbiol. 1995;16:1059–1066. doi: 10.1111/j.1365-2958.1995.tb02331.x. [DOI] [PubMed] [Google Scholar]
- 31.Li C, Budge L P, Driscoll C D, Willardson B M, Allman G W, Savage P B. Incremental conversion of outer-membrane permeabilizers into potent antibiotics for gram-negative bacteria. J Am Chem Soc. 1999;121:931–940. [Google Scholar]
- 32.Liechty A, Chen J, Jain M K. Origin of antibacterial stasis by polymyxin B in Escherichia coli. Biochim Biophys Acta. 2000;1463:55–64. doi: 10.1016/s0005-2736(99)00178-9. [DOI] [PubMed] [Google Scholar]
- 33.McLeod G I, Spector M P. Starvation- and stationary-phase-induced resistance to the antimicrobial peptide polymyxin B in Salmonella typhimurium is RpoS (ςs) independent and occurs through both phoP-dependent and -independent pathways. J Bacteriol. 1996;178:3683–3688. doi: 10.1128/jb.178.13.3683-3688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller I R, Bach D, Teuber M. Effect of polymyxin B on the structure and the stability of lipid layers. J Membr Biol. 1978;39:49–56. doi: 10.1007/BF01872754. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell P, Moyle J, Mitchell R. Measurement of H+/O in mitochondria and submitochondrial vesicles. Methods Enzymol. 1979;55:627–640. doi: 10.1016/0076-6879(79)55071-x. [DOI] [PubMed] [Google Scholar]
- 36.Morris C M, George A, Wilson W W, Chaplin F R. Effect of polymyxin B nonapeptide on daptomycin permeability and cell surface properties in Pseudomonas aeruginosa, Escherichia coli, and Pasteurella multocida. J Antibiot. 1995;48:67–72. doi: 10.7164/antibiotics.48.67. [DOI] [PubMed] [Google Scholar]
- 37.Newton B A. The properties and mode of action of the polymyxins. Bacteriol Rev. 1956;20:14–27. doi: 10.1128/br.20.1.14-27.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikaido H. Porins and specific diffusion channels in bacterial outer membranes. J Biol Chem. 1994;269:3905–3908. [PubMed] [Google Scholar]
- 39.Nikaido H. Outer membrane. In: Neidhardt F C, Curtiss III R, Ingraham J L, Linn E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 29–47. [Google Scholar]
- 40.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikaido H, Vaara T. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nummila K, Kilpeläinen I, Zähringer U, Vaara M, Helander I M. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol Microbiol. 1995;16:271–278. doi: 10.1111/j.1365-2958.1995.tb02299.x. [DOI] [PubMed] [Google Scholar]
- 43.Oh J-T, Cajal Y, Skowronska E M, Belkin S, Chen J, Van Dyk T K, Sasser M, Jain M K. Cationic peptide antimicrobials induce selective transcription of micF and osmY in Escherichia coli. Biochim Biophys Acta. 2000;1463:43–54. doi: 10.1016/s0005-2736(99)00177-7. [DOI] [PubMed] [Google Scholar]
- 44.Pickar A D, Benz R. Transport of oppositely charged lipophilic probe ions in lipid bilayer membranes having various structures. J Membrane Biol. 1978;44:353–376. [Google Scholar]
- 45.Rifkind D J. Prevention by polymyxin B of endotoxin lethality in mice. J Bacteriol. 1967;93:1463–1464. doi: 10.1128/jb.93.4.1463-1464.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Schröder G, Branderburg K, Seydel U. Polymyxin B induces transient permeability fluctuations in asymmetric planar lipopolysaccharide/phospholipid bilayer. Biochemistry. 1992;31:631–638. doi: 10.1021/bi00118a001. [DOI] [PubMed] [Google Scholar]
- 48.Seale T W, Rennert O M. Mechanisms of antibiotic-induced nephrotoxicity. Ann Clin Lab Sci. 1992;12:1–9. [PubMed] [Google Scholar]
- 49.Sixl F, Galla H M. Polymyxin interaction with negatively charged lipid bilayer membranes and the competitive effect of Ca++ Biochem Biophys Acta. 1981;643:626–635. doi: 10.1016/0005-2736(81)90358-8. [DOI] [PubMed] [Google Scholar]
- 50.Srimal S, Surolia N, Balasubramanian S, Surolia A. Titration calorimetric studies to elucidate the specificity of the interactions of polymyxin B with lipopolysaccharide and lipid A. Biochem J. 1996;315:679–686. doi: 10.1042/bj3150679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stock J B, Rauch B, Rosemen S. Periplasmic space in Salmonella typhimurium and Escherichia coli. J Biol Chem. 1977;252:7850–7861. [PubMed] [Google Scholar]
- 52.Storm D R, Rosenthal K S, Swanson P E. Polymyxin and related peptide antibiotics. Annu Rev Biochem. 1977;46:723–763. doi: 10.1146/annurev.bi.46.070177.003451. [DOI] [PubMed] [Google Scholar]
- 53.Tarahovsky Y S, Khusainov A A, Deev A A, Kim Y V. Membrane fusion during infection of Escherichia coli cells by phage T4. FEBS Lett. 1991;289:18–22. doi: 10.1016/0014-5793(91)80899-e. [DOI] [PubMed] [Google Scholar]
- 54.Tarahovsky Y S, Khusainov A A, Daugelavičius R, Bakienė E. Structural changes in Escherichia coli membranes induced by bacteriophage T4 at different temperatures. Biophys J. 1995;68:157–163. doi: 10.1016/S0006-3495(95)80170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teuber M, Bader J. Action of polymyxin on bacterial membranes. Binding capacities of polymyxin B of inner and outer membranes isolated from Salmonella typhimurium G30. Arch Microbiol. 1976;109:51–58. doi: 10.1007/BF00425112. [DOI] [PubMed] [Google Scholar]
- 56.Thomas C J, Gangadhar B P, Surolia N, Surolia A. Kinetics and mechanism of the recognition of endotoxin by polymyxin B. J Am Chem Soc. 1998;120:12428–12434. [Google Scholar]
- 57.Thomas C J, Surolia A. Kinetics of the interaction of endotoxin with polymyxin B and its analogs: a surface plasmon resonance analysis. FEBS Lett. 1999;445:420–424. doi: 10.1016/s0014-5793(99)00150-7. [DOI] [PubMed] [Google Scholar]
- 58.Tokuda H, Konisky J. Mode of action of colicin Ia: effect of colicin on the Escherichia coli proton electrochemical gradient. Proc Natl Acad Sci USA. 1978;75:2579–2583. doi: 10.1073/pnas.75.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaara M. Effect of ionic strength on polymyxin resistance of pmrA mutant of Salmonella. FEMS Microbiol Lett. 1981;11:321–326. [Google Scholar]
- 60.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vaara M, Vaara T. Sensitization of gram-negative bacteria to antibiotics and complement by a nontoxic oligopeptide. Nature. 1983;303:526–528. doi: 10.1038/303526a0. [DOI] [PubMed] [Google Scholar]
- 62.Wiese A, Münstermann M, Gutsmann T, Linder B, Kawahara K, Zähringer U, Seydel U. Molecular mechanisms of polymyxin B-membrane interactions: direct correlation between surface charge density and self-promoted transport. J Membr Biol. 1998;162:127–138. doi: 10.1007/s002329900350. [DOI] [PubMed] [Google Scholar]
- 63.Zheleznova E E, Markham P, Edgar R, Bibi E, Neyfakh A A, Brennan R G. A structure-based mechanism for drug binding by multidrug transporters. Trends Biochem Sci. 2000;25:39–43. doi: 10.1016/s0968-0004(99)01514-5. [DOI] [PubMed] [Google Scholar]