Abstract
Intron retention is a type of alternative splicing where one or more introns remain unspliced in a polyadenylated transcript. Although many viral systems are known to translate proteins from mRNAs with retained introns, restriction mechanisms generally prevent export and translation of incompletely spliced mRNAs. Here, we provide evidence that the human nuclear poly(A)-binding protein, PABPN1, functions in such restrictions. Using a reporter construct in which nuclear export of an incompletely spliced mRNA is enhanced by a viral constitutive transport element (CTE), we show that PABPN1 depletion results in a significant increase in export and translation from the unspliced CTE-containing transcript. Unexpectedly, we find that inactivation of poly(A)-tail exosome targeting by depletion of PAXT components had no effect on export and translation of the unspliced reporter mRNA, suggesting a mechanism largely independent of nuclear RNA decay. Interestingly, a PABPN1 mutant selectively defective in stimulating poly(A) polymerase elongation strongly enhanced the expression of the unspliced, but not of intronless, reporter transcripts. Analysis of RNA-seq data also revealed that PABPN1 controls the expression of many human genes via intron retention. Notably, PABPN1-dependent intron retention events mostly affected 3′-terminal introns and were insensitive to PAXT and NEXT deficiencies. Our findings thus disclose a role for PABPN1 in restricting nuclear export of intron-retained transcripts and reinforce the interdependence between terminal intron splicing, 3′ end processing, and polyadenylation.
Keywords: constitutive transport element, PABPN1, RNA export, intron retention, polyadenylation
INTRODUCTION
In contrast to prokaryotes where translation is generally cotranscriptional, eukaryotic cells possess a nuclear envelope that physically separates translating ribosomes in the cytoplasm from nascent transcripts in the nucleus. Accordingly, the export of RNAs outside of the nucleus is a critical step of eukaryotic gene expression and must undergo key checkpoints to ensure translation of appropriately matured mRNAs in the cytoplasm. For most mRNAs, the process of RNA export begins during transcription by the cotranscriptional association of various RNA-binding proteins, most notably the Aly/REF, THO, and UAP56 components of the conserved transcription-export (TREX) complex, which recruits the main export receptor complex consisting of NXF1 and NXT1 to assemble into an export-competent mRNP (Stewart 2019; Xie and Ren 2019). The binding between the NXF1/NXT1 complex and FG-repeat nucleoporins allows translocation through the nuclear pore complex (NPC) (Fribourg et al. 2001; Grant et al. 2002), after which NXF1/NXT1 heterodimers are displaced from the mRNP at the cytoplasmic face of the NPC.
The export of RNAs into the cytoplasm is generally a point of no return for the nuclear phase of gene expression, and therefore, it is critical that export-competent mRNPs are only released when splicing and polyadenylation steps are fully completed. Although the underlying mechanisms that implement quality control steps for mRNP export are not totally understood, studies in yeast and mammalian cells indicate that the nuclear basket of the NPC contributes to prevent premature export of intron-containing transcripts. Specifically, depletion of the nuclear basket component Mlp1 in yeast and its TPR homolog in human cells causes leakage of incompletely spliced transcripts into the cytoplasm (Galy et al. 2004; Coyle et al. 2011; Rajanala and Nandicoori 2012). Interestingly, the quality control mediated by the NPC nuclear basket is likely to be coordinated with poly(A) tail status of the mRNP, as yeast Mlp1 interacts with the nuclear poly(A)-binding protein (PABP) Nab2 (Fasken et al. 2008), whose depletion results in the accumulation of unspliced pre-mRNAs in the nucleus (Schmid et al. 2012; Soucek et al. 2016), thus suggesting a role for Nab2 in pre-mRNA decay at the NPC. Although the human ortholog of yeast Nab2, ZC3H14, was shown to associate with the splicing machinery (Soucek et al. 2016), a role for ZC3H14 in nuclear RNA degradation remains unclear. Conversely, the main nuclear PABP in human cells, PABPN1, is functionally connected to the RNA exosome complex of 3′–5′ exonucleases via associations with a dimer consisting of the zinc-finger protein ZFC3H1 and MTR4, which together with RBM26/27 and ZC3H3, form the poly(A) RNA exosome targeting (PAXT) connection (Meola et al. 2016; Ogami et al. 2017; Silla et al. 2020). Accordingly, depletion of PABPN1 and other PAXT components in human cells results in the stabilization of selected polyadenylated transcripts, most of which are noncoding RNAs (Beaulieu et al. 2012; Bresson et al. 2015; Meola et al. 2016; Ogami et al. 2017). Interestingly, the PAXT component ZFC3H1 was shown to compete with the RNA export factor Aly/REF and restrict the export of selected noncoding RNAs (Ogami et al. 2017; Silla et al. 2018; Lee et al. 2022), suggesting that ZFC3H1-mediated nuclear retention targets RNAs for polyadenylation-dependent decay. However, whether ZFC3H1-mediated nuclear retention also targets incompletely processed mRNAs, such as intron-containing polyadenylated transcripts, for PAXT-dependent RNA surveillance remains unknown.
Due to the existence of cellular quality control pathways that prevent export of intron-retained transcripts, several viruses have developed mechanisms that allow nuclear export of unspliced RNAs whose intronic sequences are translated to produce essential viral proteins (Rekosh and Hammarskjold 2018). One well-characterized mechanism involves the human immunodeficiency virus (HIV) Rev protein, which recognizes a structured cis-acting sequence, the Rev response element (RRE), that is not present in fully spliced HIV mRNAs but included in unspliced or incompletely spliced viral transcripts (Hammarskjold et al. 1989; Malim et al. 1989). Rev promotes the export of unspliced and incompletely spliced HIV RNAs by directly interacting with Exportin-1 (CRM1), a key karyopherin that docks cellular cargos, in this case a Rev-RNA complex, to the NPC for nuclear export (Fischer et al. 1995; Fornerod et al. 1997). Other retroviruses, such as the Mason–Pfizer monkey virus (MPMV) and Rous sarcoma virus (RSV), have evolved mechanisms that are independent of viral proteins to export unspliced or incompletely spliced viral transcripts (Gales et al. 2020). MPMV and RSV use a stem–loop RNA motif referred to as the constitutive transport element (CTE) (Ernst et al. 1997b), which mediates nuclear export of unspliced viral genomic RNA by directly recruiting the cellular NXF1/NXT1 export receptor complex (Teplova et al. 2011) to allow translocation through the NPC.
Interestingly, endogenous transcripts with retained introns have been detected in the cytoplasm of human cells (Brugiolo et al. 2017). Furthermore, translation of intronic sequences to increase proteome diversity was shown in yeast (Moldon et al. 2008) and appears to be exploited by human cells as revealed by recent proteogenomic analysis (Laumont et al. 2016; Zhu et al. 2018). How nuclear export and translation of these intron-containing transcripts escape nuclear and cytoplasmic RNA surveillance mechanisms remains elusive. Given the now recognized widespread nature of intron retention in mammalian cells (Braunschweig et al. 2014; Boutz et al. 2015), how incompletely spliced polyadenylated RNAs escape nuclear decay and how are they distinguished from mature mRNAs that are destined for nuclear export remain poorly understood.
Given the apparently contrasting roles of PABPN1 in stimulating polyadenylation, protecting nascent poly(A) tails, and RNA export (Wigington et al. 2014) versus its function in PAXT-dependent exosome-mediated RNA decay (Tudek et al. 2018), PABPN1 is a likely candidate involved in key checkpoints in the pathway leading to the generation of export-competent mRNPs. As the difference between export-competent and nuclear-degraded transcripts potentially resides in the differential composition of RNA-binding proteins, we investigated the protein interaction network of PABPN1 in living human cells using proximal-dependent biotinylation assays. We report that PABPN1 is physically connected to the nuclear basket nucleoporins, TPR, NUP153, and NUP50. We demonstrate that PABPN1 loss-of-function leads to increased export and translation of an intron-containing reporter mRNA exported in a CTE-dependent manner. RNA-seq data analysis also identified more than a hundred introns that showed increased retention in PABPN1-deficient cells, but not in cells depleted for PAXT, NEXT, and exosome components. Notably, close to 90% of these PABPN1-sensitive intron retention events affected 3′-terminal introns. Collectively, our findings disclose important PAXT-independent roles for PABPN1 in the post-transcriptional control of terminal intron splicing and the nuclear retention of intron-retained transcripts.
RESULTS
Proximity-dependent biotinylation map of human PABPN1
To elucidate how PABPN1 selectively targets transcripts in the nucleus, we searched for proteins acting in the vicinity of PABPN1 in living human cells by proximity-dependent biotinylation assays (BioID). For the BioID analysis, we generated a stable HEK293T cell line that conditionally expresses PABPN1 fused to the BirA* biotin ligase. Specific PABPN1-proximal proteins were determined by a quantitative proteomics approach (SILAC) that classifies interactions on the basis of specificity (ratio of peptide intensities between the streptavidin pull-down from cells that express BirA–PABPN1 versus control cells) and protein abundance, as estimated by the sum of peptide signal intensities of a given protein normalized to its molecular mass (Ong et al. 2002). In total, 70 PABPN1-proximal proteins were reproducibly identified from two independent BioID assays with a SILAC enrichment ratio >2.0, a minimum of two unique peptides, and an amino acid sequence coverage >10% (Fig. 1A; Supplemental Table S6). Gratifyingly, several components of the PAXT connection were recovered: ZFC3H1, RBM26, and RBM27 (Fig. 1A), confirming the validity of our BioID approach. Furthermore, a link between ZC3H14, a nuclear poly(A)-binding protein that associates with PAXT component ZFC3H1 (Meola et al. 2016), and PABPN1 was also identified (Fig. 1A). Gene ontology (GO) analysis showed the enrichment of factors involved in nuclear RNA processing, including RNA splicing and export (Fig. 1B,C). Notably, auxiliary components of the exon junction complex (EJC), such as Pinin and ACIN1, as well as spliceosome components SF3B2, SF1, and the SFPQ-NONO heteromer, were identified proximal to PABPN1 (Fig. 1C, RNA processing). Several factors with a connection to mRNA export were also identified in the PABPN1 BioID analysis. This includes POLDIP3 (SKAR) and ZC3H11A, two proteins associated with the human TREX complex (Dufu et al. 2010; Folco et al. 2012) as well as TPR, NUP153, and NUP50, which are components of the basket of the nuclear pore complex (Fig. 1A,C, RNA export). This unbiased proteomics approach therefore identified several proteins proximal to PABPN1 in the nucleus, including components of PAXT as well as multiple factors involved in pre-mRNA processing and nuclear export.
FIGURE 1.
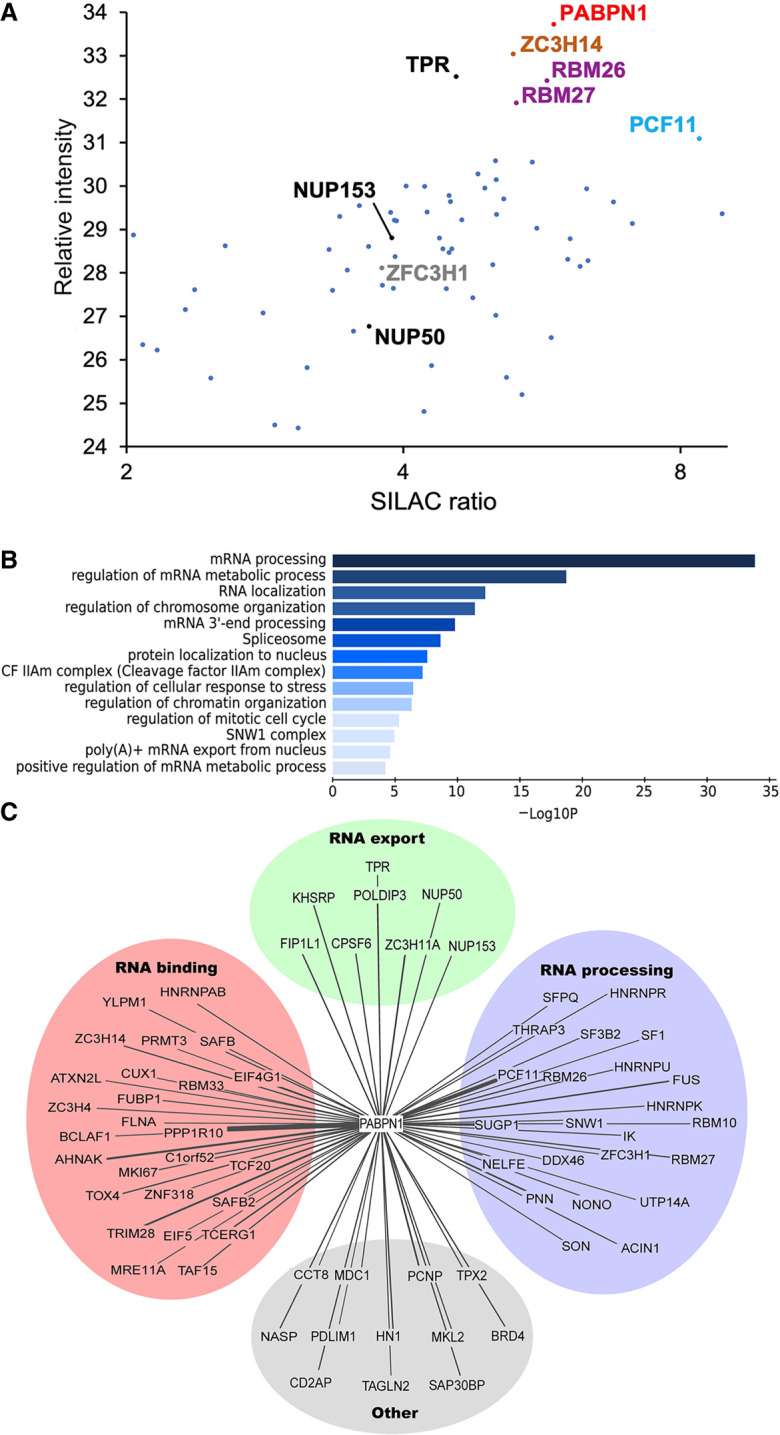
Identification of PABPN1 association network by proximity-dependent biotinylation (PDB). (A) Scatterplot showing results from PDB assays using BirA–PABPN1 coupled to mass spectrometry quantifications plotted by SILAC ratio on the x-axis (peptide intensity originating from the BirA–PABPN1 BioID assay versus control), which reflects specificity; and relative peptide intensity up the y-axis (total peptide intensity for each protein), reflecting relative abundance of each protein in the streptavidin pull-down. (B) Gene ontology (GO) enrichment analysis of the proximity interactors of PABPN1 based on their biological processes. The x-axis represents the log-transformed P-value (Fisher's exact test) of GO terms. (C) Enrichment network visualization for results of the PABPN1 PDB-MS assays. The related and redundant GO terms associated with the 70 PABPN1-proximal proteins were manually clustered into one of four “meta” GO terms: RNA export, RNA processing, RNA binding, and other. Lines denote PABPN1-protein associations, with line thickness being indicative of evidence strength (based on SILAC ratio) for a predicted interaction.
PABPN1 is required for the negative regulation of a CTE-containing reporter mRNA with a retained intron
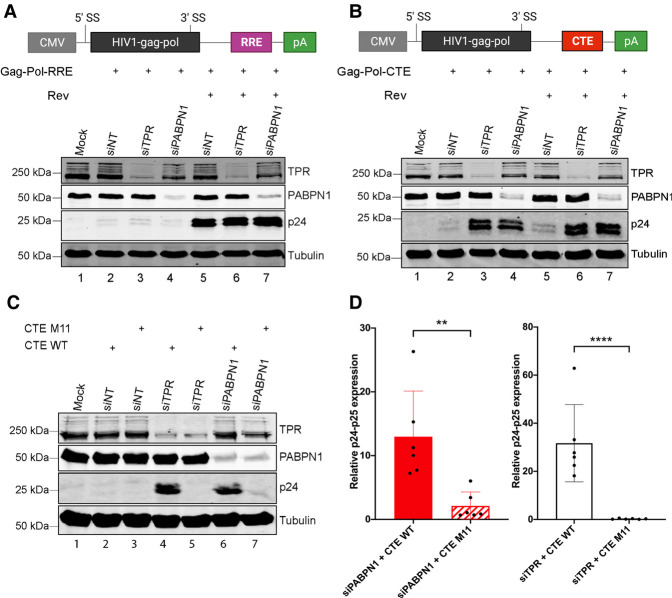
The proximal association of PABPN1 with basket nucleoporins of the nuclear pore complex (NPC) caught our attention, as the TPR–NUP153–NUP50 complex is part of a NPC-based surveillance mechanism that prevents export of incompletely spliced mRNAs into the cytoplasm (Palazzo and Lee 2018). Specifically, both yeast Mlp1/Mlp2 and human TPR have been shown to function as a gatekeeper in the NPC to prevent premature export of partially spliced pre-mRNAs (Galy et al. 2004; Vinciguerra et al. 2005; Coyle et al. 2011; Rajanala and Nandicoori 2012). This role of human TPR has been previously characterized using a reporter construct (see Fig. 2A) that expresses HIV mRNA sequences with a single constitutive RNA transport element (CTE) from the Mason–Pfizer monkey virus (MPMV), revealing that a TPR knockdown results in a significant increase in the export of unspliced gag-pol mRNA (Coyle et al. 2011). We therefore transfected the Gag-Pol-CTE reporter construct into HEK293T cells that were previously treated with specific siRNAs and measured HIV p24gag expression, which is only expressed from unspliced gag-pol mRNA. As shown in Figure 2B, only small levels of p24gag protein were detected in cells transfected with a control nontarget siRNA (compare lanes 1,2), as most of the unspliced mRNA is retained in the nucleus (Coyle et al. 2011). Depletion of TPR and NUP153 resulted in a significant increase of p24gag levels (Fig. 2B, compare lanes 3, 5 to lane 2; Fig. 2C for quantifications), consistent with previous reports indicating that the human TPR–NUP153 complex prevents the export and expression of mRNAs that retain introns (Coyle et al. 2011; Rajanala and Nandicoori 2012). Notably, knockdown of PABPN1 in HEK293T cells also caused a significant increase of p24gag protein levels compared to control siRNAs (Fig. 2B, lane 4; Fig. 2C). As a control for plasmid copy number variation, the HIV Gal-Pol-CTE reporter construct was cotransfected with a plasmid expressing an intronless mRNA. While p24gag levels increased in PABPN1-depleted cells, protein expression from the intronless mRNA was unaffected (Supplemental Fig. S1). Unexpectedly, the effect of TPR and NUP153 depletions on p24gag expression were small compared to the knockdown of PABPN1 in HeLa cells (Fig. 2B, compare lane 9 to lanes 8 and 10; Fig. 2D for quantifications), suggesting that the NPC-based mechanism of RNA surveillance may be more important in specific cell types. The maintenance of negative regulation in HeLa cells after depletion of TPR and NUP153, but not after PABPN1 knockdown, also suggested that PABPN1 and the NPC-based mechanism of RNA surveillance function independently. To test this idea, we examined the effect of codepleting PABPN1 and TPR in HEK293T cells with the rationale that if these proteins function in the same regulatory pathway, codepletion should result in a similar effect as single depletions. As shown in Figure 2E,F, codepletion of PABPN1 and TPR resulted in a significant increase in p24gag levels relative to either single depletion (compare lane 5 to lanes 3-4), consistent with the view that PABPN1 and TPR function in independent pathways. Taken together, these results disclose a central role for PABPN1 in a regulatory pathway that prevents expression of a transcript with a retained intron.
FIGURE 2.
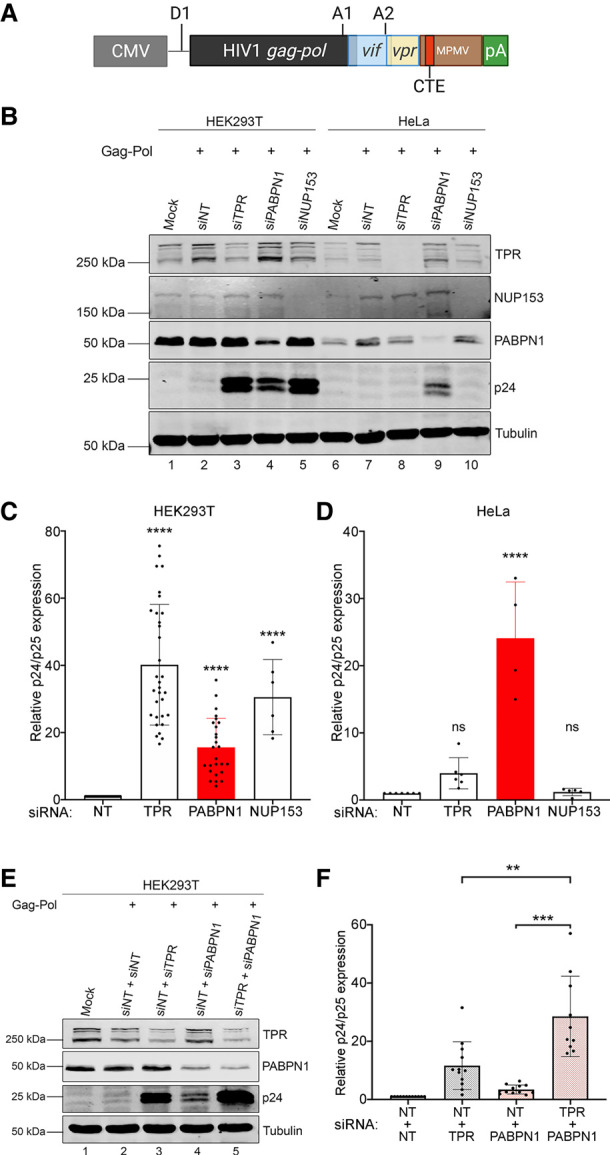
PABPN1 prevents the expression of an unspliced viral RNA in human cells. (A) Schematic of the HIV Gag-Pol-CTE reporter plasmid showing the major splice donor site (D1) and splice acceptor sites A1 and A2, which result in the expression of vif and vpr isoforms, respectively, whereas the unspliced mRNA allows Gag-Pol expression. The CTE element from MPMV is shown as a red box. (B) Western blot analysis of the indicated proteins using total extracts prepared from HEK293T (lanes 1–5) and HeLa (lanes 6–10) cells that were previously cotransfected with the Gag-Pol-CTE construct and the indicated siRNAs (lanes 2–5,7–10) or mock transfected (lane 1,6). siNT, nontarget siRNA. (C,D) Levels of p24/p25gag were normalized to tubulin levels and were expressed relative to values for HEK293T (C) and HeLa (D) cells treated with nontarget (siNT) siRNAs. Data and error bars represent the means and standard deviations, respectively, from independent experiments. ns, P-value >0.05; (****) P-value <0.0001. (E) Western blot analysis of the indicated proteins using total extracts prepared from HEK293T cells that were previously cotransfected with the Gag-Pol-CTE construct and the indicated mixture of siRNAs (lanes 2–5) or mock transfected (lane 1). (F) Levels of p24/p25gag were normalized to tubulin levels and were expressed relative to values for cells treated with nontarget (siNT + siNT) siRNAs. Data and error bars represent the means and standard deviations, respectively, from independent experiments. (**) P-value <0.01; (***) P-value <0.001.
The NXF1/NXT1 RNA export pathway is required for expression of CTE-containing mRNAs with retained introns in PABPN1-deficient cells
During HIV infection, the viral protein Rev is responsible for the export of incompletely spliced and unspliced HIV mRNAs via recognition of the Rev response element (RRE) (Nekhai and Jeang 2006) and the activity of cellular helicases (Yedavalli et al. 2004). This Rev-dependent mRNA export pathway is dependent on Exportin-1, contrasting with the NXF1/NXT1 pathway that promotes nuclear export of incompletely spliced transcripts containing CTE-like RNA elements (Rekosh and Hammarskjold 2018). To assess whether PABPN1 deficiency affected Rev/RRE-dependent mRNA export, we transfected the HIV Gag-Pol reporter that included the RRE (Fig. 3A, top), in the presence and absence of Rev, in PABPN1-depleted cells. As expected, Rev stimulated p24gag expression from the unspliced gag-pol mRNA in the presence of the RRE (Fig. 3A, compare lane 5 to lane 2), but not when the CTE was used (Fig. 3B, compare lane 5 to lane 2). Depletion of PABPN1 did not affect p24gag expression from the RRE-containing construct, neither in the absence nor in the presence of Rev (Fig. 3A, lane 7 vs. 5 and lane 4 vs. 2), whereas a PABPN1 deficiency caused a robust increase in p24gag expression when the unspliced gag-pol mRNA included the MPMV CTE (Fig. 3B, lane 4 vs. 2). The positive effect of depleting PABPN1 on the expression of the CTE-containing unspliced mRNA was not stimulated further by the addition of Rev (Fig. 3B, lane 7 vs. lane 4). The effect of TPR depletion on unspliced gag-pol mRNAs was also restricted to CTE-containing transcripts (Fig. 3A,B), consistent with previous results (Coyle et al. 2011). We also introduced previously described mutations in the MPMV CTE (M11, see Supplemental Fig. S2) that abolish interaction with NXF1 (Ernst et al. 1997a; Teplova et al. 2011) and found that the depletion of PABPN1 had no stimulatory effect on p24gag expression when the CTE was mutated (Fig. 3C, compare lanes 6,7; quantifications in Fig. 3D). Collectively, these results indicate that a functional CTE is required to allow export of the unspliced gag-pol mRNA in PABPN1-depleted cells.
FIGURE 3.
PABPN1 depletion enhances CTE-mediated, but not Rev/RRE-mediated, expression of unspliced transcript. (A,B) (Top) Schematic of the HIV Gag-Pol reporter construct with HIV RRE (A) and MPMV CTE (B) RNA transport elements. (Bottom) Western blot analysis of the indicated proteins using total extracts prepared from HEK293T cells that were previously cotransfected with either the Gag-Pol-RRE (A) or the Gag-Pol-CTE (B) constructs and the indicated siRNAs (lanes 2–7). The absence (lanes 2–4) or the presence (lanes 5–7) of the HIV Rev protein was also analyzed. (C) Western blot analysis of the indicated proteins using total extracts prepared from HEK293T cells that were previously cotransfected with either the wild-type Gag-Pol-CTE (lanes 2,4,6) or a Gag-Pol-CTE construct with mutations in the CTE element (M11; lanes 3,5,7) and the indicated siRNAs (lanes 2–7). (D) Levels of p24/p25gag were normalized to tubulin levels and were expressed relative to values for cells treated with nontarget (siNT) siRNAs. Data and error bars represent the means and standard deviations, respectively, from independent experiments. (**) P-value <0.01; (****) P-value <0.0001.
ZC3H14 is not required for the negative regulation of the intron-retained CTE-containing mRNA
In addition to PABPN1, human cells also express additional poly(A)-binding proteins in the nucleus, including the ubiquitously expressed zinc finger RNA-binding protein ZC3H14 (Wigington et al. 2014). Interestingly, the budding yeast homolog of human ZC3H14, Nab2, has previously been shown to directly interact with Mlp1/2 (Green et al. 2003), which is the yeast homolog of human TPR. The interaction between yeast Mlp1 and the Nab2 nuclear PABP has been proposed to promote nuclear export of poly(A) RNA (Fasken et al. 2008) as well as contribute to the quality control of RNA nuclear export (Bonnet et al. 2015). Given the functional significance of the Nab2–Mlp1 association in quality control of RNA export in yeast and recent evidence demonstrating that human ZC3H14 prevents premature export of specific intron-containing mRNA in neuronal cells (Wigington et al. 2014; Morris and Corbett 2018), we examined whether ZC3H14 functions with PABPN1 to negatively control the expression of the CTE-containing reporter RNA. We therefore transfected the HIV Gag-Pol-CTE construct in HEK293T cells that were previously treated with PABPN1- or ZC3H14-specific siRNAs. As shown in Supplemental Figure S3A,B, knockdown of ZC3H14 in both HEK293T and HeLa cells did not result in the accumulation of p24gag protein relative to cells treated with control siRNAs (compare lane 4 to lane 2; quantifications in Supplemental Fig. S3C,D). This contrasts with the knockdown of PABPN1 that stimulated p24gag expression in both cell lines (Supplemental Fig. S3). We thus conclude that the poly(A)-binding activity of ZC3H14 is not required to restrict the expression of unspliced gag-pol transcripts that are exported via a CTE.
PABPN1-dependent surveillance of an incompletely spliced mRNA functions independently of PAXT-mediated RNA decay
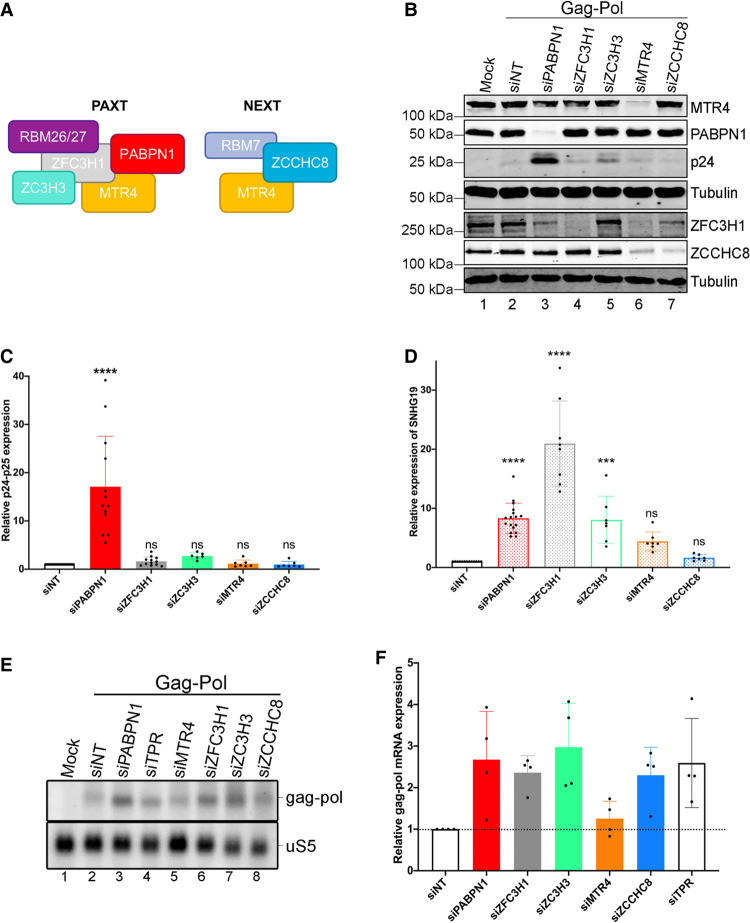
PABPN1 functions in the degradation of nuclear polyadenylated noncoding RNAs via a connection with PAXT (see Fig. 4A), which includes MTR4, ZFC3H1, ZC3H3, and RBM26/RBM27 proteins (Meola et al. 2016; Silla et al. 2020). To determine whether the function of PABPN1 in restricting the expression of an mRNA with retained introns is mediated via its connection with PAXT-dependent RNA decay, we assessed the outcome of Gag-Pol-CTE expression in HEK293T cells deficient for PAXT components ZFC3H1, ZC3H3, and MTR4 (Fig. 4B; Supplemental Fig. S4). Strikingly, apart from PABPN1, knocking down the expression of any of the PAXT component had no significant effect on the expression of the unspliced gag-pol mRNA as measured by p24gag protein levels (Fig. 4B,C). We also tested the depletion of ZCCHC8 (Fig. 4A,B), a component of the NEXT connection (Lubas et al. 2011), which also showed no significant increase in p24gag expression compared to the control siRNA (Fig. 4C). As a control, we confirmed that depletion of PAXT components PABPN1, ZFC3H1, ZC3H3, and MTR4 resulted in the accumulation of a polyadenylated lncRNA produced from a snoRNA host gene, a known target of PAXT (Beaulieu et al. 2012; Meola et al. 2016), but not of the NEXT connection (Fig. 4D).
FIGURE 4.
The role of PABPN1 in repressing the expression of unspliced transcripts is independent of PAXT. (A) Schematic representation of key components of PAXT and NEXT. (B) Western blot analysis of the indicated proteins using total extracts prepared from HEK293T cells that were previously cotransfected with the Gag-Pol-CTE construct and the indicated siRNAs (lanes 2–7) or mock transfected (lane 1). (C) Levels of p24/p25gag were normalized to tubulin levels and were expressed relative to values for cells treated with nontarget (siNT) siRNAs. Data and error bars represent the means and standard deviations, respectively, from independent experiments. ns, P-value >0.05; (****) P-value <0.0001. (D) RT-qPCR analysis of PAXT substrate SNHG19 using total RNA harvested from HEK293T cells transfected with the indicated siRNAs. Data and error bars represent the means and standard deviations, respectively, from independent experiments. ns, P-value >0.05; (***) P-value <0.001; (****) P-value <0.0001. (E) Northern blot analysis of total RNA prepared from HEK293T cells previously cotransfected with the Gag-Pol-CTE construct and the indicated siRNAs. The unspliced gag-pol transcript is shown (top) and the uS5 mRNA was used as a loading control (bottom). (F) Levels of unspliced gag-pol transcript were normalized to the levels of uS5 mRNA and expressed relative to values for cells treated with nontarget (siNT) siRNAs. Data and error bars represent the means and standard deviations, respectively, from independent experiments.
We next analyzed the levels of the 6.8-Kb unspliced gag-pol mRNA in cells depleted for PABPN1 as well as for RNA decay (PAXT and NEXT) and nuclear retention (TPR and ZFC3H1) factors to determine the extent to which p24gag protein levels (Fig. 4B,C) correlated with unspliced gag-pol mRNA. Unexpectedly, we did not find a significant difference in unspliced gag-pol mRNA levels between PABPN1-deficient cells and cells knocked down for other PAXT components (Fig. 4E,F). Rather, Northern blot analysis of total RNA prepared from cells depleted for PAXT or NEXT components generally showed a 2.5-fold increase in unspliced gag-pol mRNA levels compared to cells treated with the control siRNA (Fig. 4E, lanes 3–8 vs. lane 2; quantifications in Fig. 4F). Although it appears surprising that the unspliced gag-pol transcript showed increased levels in NEXT depletion, unadenylated gag-pol transcription products could be targeted by NEXT. Knockdown of TPR expression also resulted in similar accumulation of unspliced gag-pol mRNA (Fig. 4E, lane 4 vs. lane 2; Fig. 4F). Similar results were obtained using RT-qPCR analysis (Supplemental Fig. S5). We conclude that the contribution of PABPN1 in preventing the expression of the intron-retained reporter mRNA is largely independent of its role in PAXT-mediated RNA decay.
Unspliced gag-pol transcripts accumulate in the cytoplasm of PABPN1-deficient cells
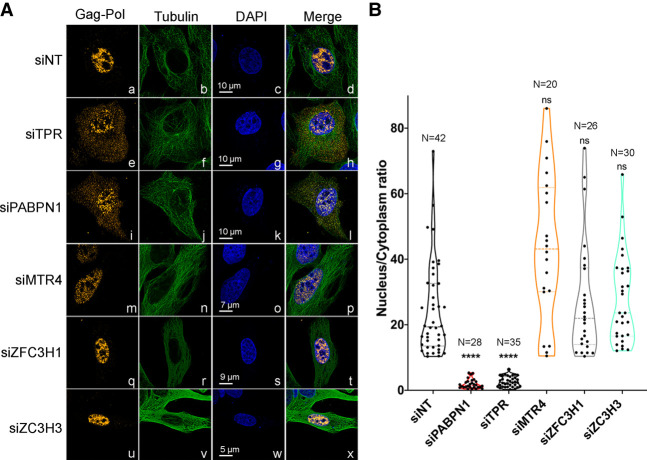
Our data indicate that although depletion of PAXT components resulted in a marginal increase in unspliced gag-pol RNA, only PABPN1 and TPR knockdown led to significant accumulation of p24gag protein (Fig. 4). Accordingly, this suggests that PABPN1 could be involved in retaining the unspliced reporter mRNA in the nucleus independently of its role in PAXT-dependent RNA decay. To directly address this possibility, we performed RNA fluorescent in situ hybridization (FISH) analysis using probes spanning gag-pol intronic sequences to visualize the unspliced RNA in human cells (Supplemental Fig. S6A,B). We used U2OS cells for the FISH analysis, as their large nucleus and cytoplasm facilitate quantification of nucleus-to-cytoplasm signal ratios. Importantly, we confirmed that depletion of PABPN1 and TPR in U2OS cells results in p24gag accumulation (Supplemental Fig. S6C, lanes 2,3). Consistent with previous biochemical results (Coyle et al. 2011), depletion of TPR resulted in the detection of unspliced gag-pol mRNA in both the nucleus and the cytoplasm (Fig. 5A, panels e–h), whereas the unspliced gag-pol mRNA was primarily nuclear in control cells (Fig. 5A, panels a–d). Notably, unspliced gag-pol mRNA was clearly detected in the cytoplasm of PABPN1-deficient U2OS (Fig. 5A, panels i–l) and HeLa cells (Supplemental Fig. S6D), consistent with the accumulation of p24gag protein in cells depleted of PABPN1 (Fig. 2; Supplemental Fig. S6C). In contrast, the unspliced gag-pol mRNA was mainly nuclear in cells depleted for PAXT components MTR4, ZFC3H1, and ZC3H3 (Fig. 5A, panels m–x). Importantly, quantification analysis of nuclear-to-cytoplasmic ratio of FISH fluorescence intensity from several individual cells showed a significant change in the subcellular distribution of the gag-pol unspliced mRNA in PABPN1- and TPR-deficient cells (Fig. 5B). We conclude that PABPN1 restricts p24gag expression by preventing the nuclear export of intron-containing gag-pol mRNA.
FIGURE 5.
Unspliced gag-pol transcripts accumulate in the cytoplasm of PABPN1-deficient cells. (A) Deconvoluted images of U2OS cells that were previously cotransfected with the wild-type Gag-Pol-CTE construct (a–x) and control (a–d), TPR-specific (e–h), PABPN1-specific (i–l), MTR4-specific (m–p), ZFC3H1-specific (q–t), and ZC3H3-specific (u–x) siRNAs were simultaneously analyzed by FISH using Cy3-labeled probes for gag-pol sequences (a, e, i, m, q, and u) and immunostaining for the cytosolic tubulin (b, f, j, n, r, and v). DNA stained with DAPI shows the nucleus of each cell (c, g, k, o, s, and w). Scale bar sizes are indicated. (B) Quantification of nucleus-to-cytoplasmic ratios of unspliced gag-pol transcripts shows a marked decrease in TPR and PABPN1-expressing cells when compared to control (siNT) cells. The number of cells analyzed is indicated for each condition, with at least two independent RNA FISH staining experiments. ns, P-value >0.05; (****) P-value <0.0001.
mRNA 3′ end processing and polyadenylation are required for PABPN1-dependent surveillance of intron-retained gag-pol transcripts
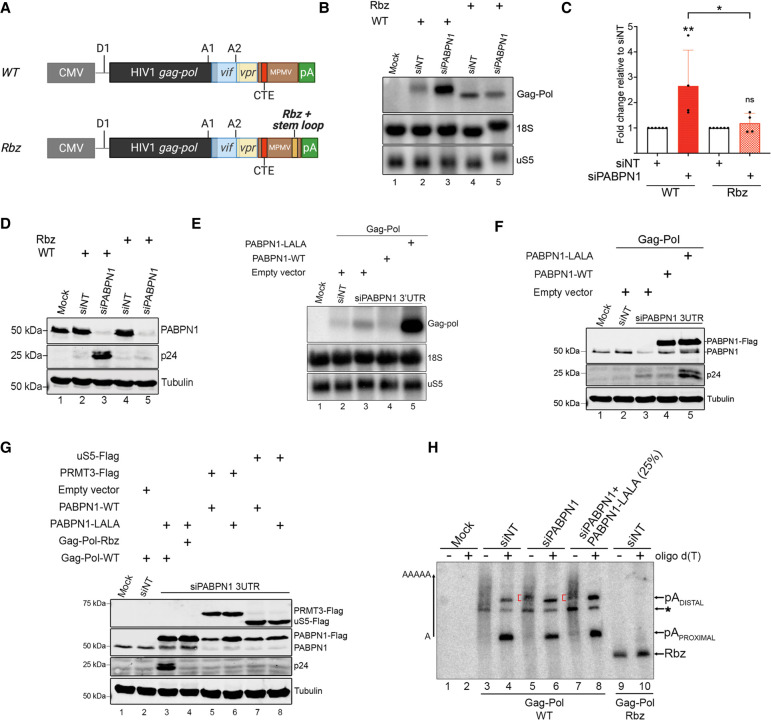
The restriction of unspliced gag-pol mRNA export by PABPN1 suggests a mechanism dependent on RNA 3′ end processing and polyadenylation. To address whether cleavage and polyadenylation are required for the negative control of the intron-containing reporter mRNA by PABPN1, we generated a gag-pol construct in which a variant of the hepatitis Delta ribozyme was inserted upstream of the natural MPMV polyadenylation sites (Fig. 6A, Rbz construct). We chose the hepatitis Delta ribozyme because we and others have previously shown that it cleaves efficiently in human cells (Bird et al. 2005; Beaulieu et al. 2012; Bergeron et al. 2015). A hairpin loop structure derived from the 3′ end of human histone mRNAs was also inserted at the 5′ end of the ribozyme cleavage site to stabilize the non-polyadenylated gag-pol transcripts (Bird et al. 2005; Bergeron et al. 2015). Wild-type and ribozyme constructs were transfected into HEK293T cells that were concomitantly treated with control and PABPN1-specific siRNAs, and the expression of the unspliced gag-pol mRNA was analyzed by Northern blotting. As shown in Figure 6B, the shorter, non-polyadenylated unspliced gag-pol mRNA expressed from the ribozyme construct was insensitive to PABPN1 deficiency (compare lanes 4,5 and quantifications in Fig. 6C), whereas the wild-type unspliced gag-pol transcript showed increased levels in PABPN1-deficient cells (Fig. 6B, lanes 2,3; Fig. 6C), consistent with our previous results. Western blot analysis confirmed the accumulation of p24gag protein expressed from the wild-type reporter in PABPN1-depleted cells, but not from the non-polyadenylated version of the unspliced gag-pol mRNA (Fig. 6D, compare lanes 3,5). These data argue for a mechanism of PABPN1 surveillance that requires the presence of a 3′ poly(A) tail.
FIGURE 6.
RNA 3′ end processing and polyadenylation is required for PABPN1-dependent repression of unspliced mRNA expression. (A) Schematic of the wild-type (WT) and ribozyme (Rbz) HIV Gag-Pol-CTE reporter contructs showing the insertion of hepatitis delta ribozyme and histone mRNA stabilizing stem–loop (pale orange box) upstream of the MPMV polyadenylation (pA) sites. (B) Northern blot analysis of total RNA prepared from HEK293T cells previously cotransfected with either the wild-type (lanes 2,3) or the ribozyme (lanes 4,5) Gag-Pol-CTE construct and the indicated nontarget (lanes 2,4) and PABPN1-specific (lanes 3,5) siRNAs. The unspliced gag-pol transcript is shown (top). The uS5 mRNA and 18S rRNA were used as loading controls. (C) Levels of unspliced gag-pol transcript were normalized to the levels of uS5 mRNA and expressed relative to values for cells treated with nontarget (siNT) siRNAs. Data and error bars represent the means and standard deviations, respectively, from independent experiments. ns, P-value >0.05; (*) P-value <0.05; (**) P-value <0.01. (D) Western blot analysis of the indicated proteins using extracts prepared from HEK293T cells that were previously cotransfected with either the wild-type (lanes 2,3) or the ribozyme (lanes 4,5) Gag-Pol-CTE construct and the indicated nontarget (lanes 2,4) and PABPN1-specific (lanes 3,5) siRNAs. (E,F) Northern blot (E) and western blot (F) analysis using extracts of HEK293T cells previously cotransfected with the wild-type Gag-Pol-CTE construct (lanes 2–5) and either nontarget (lane 2) or PABPN1-specific (lanes 3–5) siRNAs. Transfections also included constructs expressing either wild-type (lane 4) or LALA (lane 5) versions of PABPN1, as well as an empty vector control (lanes 2,3). (G) Western blot analysis using extracts of HEK293T cells previously cotransfected with nontarget (lane 2) or PABPN1-specific (lanes 3–8) siRNAs and the indicated plasmid constructs. (H) RNase H cleavage assay using total RNA prepared from HEK293T cells transfected with PABPN1-specific (lanes 5–8) and control (lanes 3,4,9,10) siRNAs together with the PABPN1 LALA mutant (lanes 7,8) along with either wild-type (lanes 3–8) or ribozyme (lanes 9,10) Gag-Pol CTE constructs. RNase H assays were performed in the presence (+) and absence (−) of oligo d(T). Due to the significant accumulation of gag-pol transcripts in PABPN1 LALA-expressing cells (part E, lane 5), only 25% of the RNase H assay for the LALA mutant (lanes 7,8) was loaded on gel to compare poly(A) tail distribution with other samples. Red brackets indicate the position of the cluster of PABPN1-dependent 3′ fragments. Transcripts cleaved at the proximal and distal poly(A) sites are indicated on the right. Transcripts cleaved at an unidentified poly(A) site are indicated by an asterisk.
Besides the absence of a 3′ poly(A) tail, the gag-pol ribozyme construct is unlikely to be cleaved by the canonical mRNA 3′ end processing machinery. To get further evidence that polyadenylation is important for PABPN1-dependent control of the unspliced gag-pol transcript, we used a dominant-negative version of PABPN1 with alanine substitutions at residues L119 and L136 (LALA mutant) that binds RNA but does not stimulate poly(A) polymerase (PAP) activity (Kuhn et al. 2009; Bresson and Conrad 2013). HEK293T were therefore depleted of endogenous PABPN1 using siRNAs that target the 3′-UTR of the PABPN1 mRNA and cotransfected with cDNA constructs expressing either wild-type or LALA mutant versions of PABPN1. Consistent with our data using exon-specific siRNAs, depletion of PABPN1 using 3′-UTR-specific siRNAs resulted in increased levels of unspliced gag-pol transcript compared to nontarget siRNAs (Fig. 6E, compare lane 3 to lane 2). Remarkably, the expression of the PABPN1 LALA mutant resulted in a substantial increase in unspliced gag-pol RNA compared to PABPN1-deficient cells (Fig. 6E, compare lane 5 to lane 3), whereas the wild-type version of PABPN1 rescued the accumulation of gag-pol RNA observed after PABPN1 knockdown (compare lane 4 to lane 3). Consistent with these RNA analyses, Western blot revealed a marked accumulation of p24gag protein in cells that expressed the PABPN1 LALA mutant compared to PABPN1-deficient cells (Fig. 6F, compare lane 5 to lane 3). The expression of the PABPN1 LALA mutant had no stimulatory effect on the expression of the gag-pol transcripts produced from the ribozyme construct (Fig. 6G, compare lanes 3,4) or on intronless polyadenylated mRNAs expressed from cDNAs (Fig. 6G, compare lanes 5–8). The stimulatory effect of the PABPN1 LALA mutant suggests that RNA polyadenylation is important in preventing premature export of the intron-containing reporter mRNA by PABPN1.
We next specifically examined the effect of a PABPN1 deficiency and of the LALA mutant on the 3′ end of the gag-pol transcripts using RNase H cleavage assays. RNase H treatment of total RNA in the presence of a DNA oligonucleotide complementary to a region located in the 3′-UTR MPMV sequence of the reporter mRNA will release heterogeneous 3′ fragments because of transcripts with different poly(A) tail lengths. As this approach uses a DNA oligonucleotide positioned at the 3′ end of the reporter RNA to generate 3′ fragments, the total population of gag-pol transcripts (spliced and unspliced) were analyzed in the RNase H assay. As shown in Figure 6H, the depletion of PABPN1 (lane 5) and the overexpression of the PABPN1 LALA mutant (lane 7) resulted in the accumulation of a cluster of 3′ fragments that was not detected in the RNase H assay using RNA from control cells (lane 3). We also analyzed 3′ end cleavage sites by adding oligo d(T) to the RNase H reaction, which causes the heterogenous population of polyadenylated 3′ fragments to collapse into discrete unadenylated products, indicating poly(A) site positions. Two major poly(A) sites were detected (Fig. 6H, see lanes 4,6,8), which were confirmed and mapped by 3′ RACE analysis (Supplemental Fig. S7). As controls for our RNase H assays, 3′ fragments were not detected using total RNA from untransfected cells (Fig. 6H, lanes 1,2), whereas cells transfected with the ribozyme construct produced a single oligo d(T)-insensitive 231-nt-long RNA fragment resulting from non-polyadenylated reporter transcripts (lanes 9,10). Together, these results indicate that polyadenylation of the gag-pol reporter transcript is affected by PABPN1.
Increased intron retention in PABPN1-deficient human cells
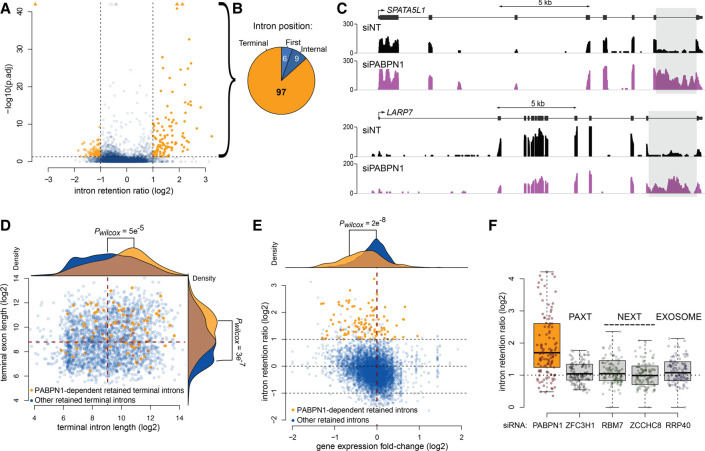
The use of the CTE-containing mRNA reporter allowed for a detailed characterization of the mechanism underlying the increased protein expression from an intron-containing transcript in PABPN1-deficient cells. Accordingly, we next asked whether PABPN1 loss-of-function affects the expression of endogenous genes via intron retention. We therefore analyzed RNA-seq data from PABPN1-depleted and control HeLa cells (Beaulieu et al. 2012) to examine for differentially retained introns using intron retention analysis and detector (iREAD) (Li et al. 2020). Briefly, iREAD counts all reads that overlap intronic regions. We then normalized these counts on gene expression to identify differentially retained introns in a condition-specific manner. Because the RNA-seq data of PABPN1-deficient cells was obtained using poly(A)-selected RNA (Beaulieu et al. 2012), the intron retention analysis mostly focused on post-transcriptional splicing events. As shown in Figure 7A, iREAD analysis identified 112 introns with significantly increased retention in PABPN1-deficient cells relative to control cells (Supplemental Table S7), whereas decreased intron retention was only marginal in the absence of PABPN1. Notably, nearly 87% of PABPN1-dependent intron retention events concerned the 3′-terminal intron of the affected gene (Fig. 7B), as illustrated by the increase in RNA-seq coverage in the 3′-terminal introns of SPATA5L1 and LARP7 genes in PABPN1-depleted cells (Fig. 7C). Interestingly, the genes sensitive to PABPN1-dependent retention of the 3′-terminal intron were enriched for genes with longer terminal introns and longer 3′-terminal exons (Fig. 7D). Furthermore, 86% and 68% of the differentially retained introns are within genes with transcripts isoforms annotated as “retained introns” (Fisher's exact test P-value <2.2 × 10−16, 7.5 odds ratio) and “NMD targets” (Fisher's exact test P-value = 9.475 × 10−13, 4 odds ratio), respectively, suggesting that those genes normally express inefficiently spliced transcripts. We also found that the mRNAs expressed from genes sensitive to PABPN1-dependent intron retention were generally down-regulated in PABPN1-deficient cells (Fig. 7E).
FIGURE 7.
Increased intron retention in PABPN1-deficient cells. (A) Volcano plot of the change in intron retention ratio (intron reads normalized on exon reads) in cells depleted for PABPN1 relative to control cells. Each point represents a detectable retained intron event (see Materials and Methods) in at least one condition (n = 6739). Intron significantly less or more retained in PABPN1-depleted cells relative to control with adjusted P-value (p.adj) < 0.05 and absolute log2 intron retention ratio >1 are colored in orange. Data points with p.adj coordinates that fall outside of the axes range are represented as triangles. (B) Position within their parent transcript of the 112 introns that are significantly more retained in cells depleted for PABPN1. (C) Read coverage over the SPATA5L1 and LARP7 genes. The gray area highlights their respective last intron, which are more retained in cells depleted for PABPN1 (purple) compared to control cells (black). (D) Log2 nucleotide length for all detectable retained terminal introns in at least one condition against the length of their following terminal exon (n = 2414). The points representing the 97 terminal introns significantly more retained in cells depleted for PABPN1 are colored in orange whereas retained terminal introns insensitive to PABPN1 are colored in blue. The density distribution of the log2 nucleotide length is drawn on top of the panel for the intron and on the right of the panel for the corresponding following exons. (E) Log2 fold change in intron retention against the log2 fold change of gene expression in cells depleted for PABPN1 relative to control (n = 6739 introns from 2721 different genes). The 112 introns significantly more retained in cells depleted for PABPN1 and their 101 associated genes are represented by orange points whereas retained introns not significantly affected by PABPN1 are colored in blue. The density distribution of the log2 fold change of gene expression is drawn on top of the panel in orange for those 101 genes or in blue for the other genes. (F) Boxplot of the change in intron retention ratio in cells depleted for PABPN1, the PAXT component ZFC3H1, the NEXT components RBM7 and ZCCHC8, and the exosome component RRP40 (Meola et al. 2016) for the 97 terminal introns identified as more retained in PABPN1-depleted cells in our own study. The distribution of the fold change is significantly shifted from 1 in PABPN1-depleted cells (pWilcoxon = 2 × 10−13).
Our results indicated that PABPN1 largely functions independently of its role in PAXT-mediated RNA decay in preventing the expression from the unspliced gag-pol reporter mRNA (Figs. 4, 5). To assess whether PABPN1-sensitive 3′-terminal introns are retained after inactivation of PAXT, NEXT, and RNA exosome complexes, we calculated intron retention ratios using an independent RNA-seq data set where PABPN1, ZFC3H1, RBM7, ZCCHC8, and RRP40 were knockdown (Meola et al. 2016). Gratifyingly, our set of PABPN1-sensitive 3′-terminal introns showed a significant increase in the mean intron retention ratio using this independent RNA-seq data set prepared from PABPN1 depletions (Fig. 7F). In contrast, the mean intron retention ratio for the same set of PABPN1-sensitive introns was not significantly changed in cells deficient for PAXT (ZFC3H1), NEXT (RBM7 and ZCCHC8), and exosome (RRP40) functions (Fig. 7F). These observations were confirmed by quantitative RT-PCR analysis from independent depletion experiments using primer sets spanning exon–intron regions that were located in either an internal or the 3′-terminal intron of SPATA5L1 and LARP7, as well as in the control HPRT gene that did not show PABPN1-dependent intron retention (Supplemental Fig. S8). These RT-qPCR results reinforce the conclusion that the PABPN1-dependent accumulation of intron-retained pre-mRNAs is not the consequence of a deficiency in PAXT-mediated RNA decay. Collectively, we conclude that PABPN1 controls the expression of many human genes via intron retention.
DISCUSSION
While a variety of sequencing and imaging approaches have established that many introns are spliced cotranscriptionally in most organisms, evidence of cleaved and polyadenylated transcripts with one or more remaining introns has been mounting in mammalian cells in the past decade (Braunschweig et al. 2014; Boutz et al. 2015; Gordon et al. 2021). A recent study in fact suggests that a large fraction of human genes splice their 3′-terminal intron after cleavage and polyadenylation (Drexler et al. 2020). These incompletely spliced transcripts, often referred to as retained intron RNAs, are usually found in the nucleus and can be spliced post-transcriptionally (Boutz et al. 2015). Yet, how these transcripts with a retained intron are distinguished from mature polyadenylated mRNAs that are destined for nuclear export remains poorly understood. In the current study, we provide data suggesting that the state of the 3′ end poly(A) tail is key to surveillance mechanisms that prevent premature export of incompletely spliced transcripts. Specifically, our findings unveil an important role for PABPN1 in such a nuclear surveillance process.
Our study reports the protein association network of human PABPN1 as determined by proximity-dependent biotinylation (PDB) assays. In addition to being more sensitive at detecting transient protein–protein interactions, PDB-MS also identifies proximal proteins in the context of living cells (Gingras et al. 2019). Consequently, PBD-MS also includes proximal connections that engage indirectly via RNA interactions. Our PDB-MS analysis validated PABPN1-associated proteins previously identified by affinity capture coupled to MS, including PAXT components RBM26, RBM27, and ZFC3H1 (Meola et al. 2016; Silla et al. 2020), the splicing regulator SNW1 (Lleres et al. 2010), as well as the chromatin reader protein BRD4 (Erber et al. 2019; Shu et al. 2020). Beyond confirming previous associations, our proximity-dependent biotinylation map of PABPN1 unveiled a connection with the NPC nuclear basket, via PABPN1-dependent biotinylation of TPR, NUP153, and NUP50. We suspect that the connection between PABPN1 and the nuclear basket is transient and/or low affinity since we were unsuccessful at capturing a stable complex between PABPN1 and TPR by affinity purification approaches. Nevertheless, the presence of PABPN1 at the nuclear periphery is consistent with previous electron microscopy analysis of the Chironomus tentans salivary gland Balbiani ring mRNA (Bear et al. 2003) and the fact that PABPN1 shuttles between nucleus and cytoplasm in yeast and human cells (Calado et al. 2000; Lemieux and Bachand 2009).
Given the established role of the nuclear basket in retaining incompletely spliced RNAs from premature nuclear export (Galy et al. 2004; Palancade et al. 2005; Coyle et al. 2011; Rajanala and Nandicoori 2012), the proximal association between PABPN1 and key nuclear basket proteins led us to examine whether PABPN1 functions in restricting the nuclear export of intron-containing transcripts using a well-characterized reporter mRNA (Coyle et al. 2011; Rajanala and Nandicoori 2012). Collectively, our data showed that PABPN1 is required for the nuclear retention of the unspliced gag-pol mRNA reporter in three different human cell lines: HEK293T, HeLa, and U2OS. Accordingly, the unspliced mRNA leaked into the cytoplasm of PABPN1- and TPR-deficient cells, resulting in the accumulation of p24gag protein. Our data also indicated that PABPN1 and TPR act independently in the nuclear retention of the unspliced gag-pol mRNA (Fig. 2), suggesting that certain cell types may have redundant pathways with different mechanisms to prevent the untimely export of incompletely spliced transcripts. It should be noted, however, that the role of TPR in nuclear retention of incompletely spliced RNAs has recently been questioned in human cells, as cytoplasmic accumulation of intron-containing transcripts was not found to increase after siRNA-mediated depletion of TPR (Lee et al. 2020; Zuckerman et al. 2020). One possibility is that nuclear basket-dependent nuclear retention of intron-containing pre-mRNAs is cell type-specific, as suggested by our results in HeLa cells where depletion of TPR and NUP153 showed negligible accumulation of p24gag compared to PABPN1 depletion (Fig. 2B–D). These studies (Lee et al. 2020; Zuckerman et al. 2020) also did not inactivate nonsense-mediated RNA decay (NMD), which could conceal the cytoplasmic accumulation of intron-containing RNAs despite TPR depletion.
Based on previous findings revealing that the fission yeast homolog of human PABPN1, Pab2, functions in a polyadenylation-dependent pre-mRNA degradation pathway that targets intron-containing genes (Lemieux et al. 2011), we anticipated a similar mechanism based on PABPN1-dependent pre-mRNA decay that would result in leakage of unspliced gag-pol transcripts into the cytoplasm of PABPN1-deficient human cells. Unexpectedly, our data indicated that the function of PABPN1 in the nuclear retention of the unspliced gag-pol mRNA is independent of PAXT, arguing against a mechanism that solely relies on RNA turnover. Accordingly, the increased levels of gag-pol mRNA in cells deficient for ZFC3H1 and ZC3H3 (Fig. 4E,F) was not sufficient to escape nuclear retention (Figs. 4B,C, 5), whereas similar RNA accumulation leaked into the cytoplasm of PABPN1-deficient cells. Notably, it was surprising to find that knockdown of ZFC3H1 did not result in the cytoplasmic accumulation of unspliced gag-pol transcripts, as this PAXT component was shown to function in the nuclear retention of selected polyadenylated noncoding RNAs destined for nuclear decay by the RNA exosome (Ogami et al. 2017; Silla et al. 2018). It remains unclear, however, whether ZFC3H1 and PAXT target polyadenylated pre-mRNAs for nuclear degradation.
The role of PABPN1 in RNA nuclear retention described in this study was likely highlighted by the presence of the MPMV constitutive transport element (CTE) in the reporter construct. Accordingly, mutations that impair the secondary structure of the MPMV CTE and perturb its recognition by the NXF1/NXT1 complex (Ernst et al. 1997a; Teplova et al. 2011) significantly affected the expression of Gag and Gag-Pol polypeptides when PABPN1 was depleted (Fig. 3C,D). During viral infection, CTE-like cis-acting nuclear export elements can override nuclear retention of intron-containing transcripts to serve both as unspliced viral mRNAs encoding for structural proteins as well as genomic RNAs packaged into virions (Rekosh and Hammarskjold 2018). Our findings therefore raise the possibility that CTE-containing viruses may need to interfere with PABPN1 function and/or expression to promote efficient export of unspliced viral RNAs during the viral life cycle. Indeed, PABPN1-associated functions have previously been demonstrated to be targeted by specific viral proteins during infection. The NS1 protein of influenza A virus was shown to bind and interfere with PABPN1 ability to stimulate processive poly(A) tail elongation of cellular mRNAs (Chen et al. 1999). Kaposi's sarcoma-associated herpesvirus viral proteins also influence the polyadenylation-dependent RNA decay function of PABPN1 to either protect viral RNAs from nuclear decay (Ruiz et al. 2019) or promote turnover of cellular transcripts (Lee and Glaunsinger 2009). As such, it may not be surprising that CTE-containing retroviruses target PABPN1 to promote the nuclear export of intron-containing viral transcripts.
Previous studies established that poly(A) tails are generally longer in the nucleus compared to the cytoplasm (Sheiness and Darnell 1973; Palatnik et al. 1979), a finding corroborated by more recent analyses indicating that newly synthesized mRNAs have long poly(A) tails relative to steady state transcripts (Eisen et al. 2020). Interestingly, poly(A) tail length analysis by long read sequencing revealed that intron-containing transcripts have longer poly(A) tails than the bulk of nuclear RNAs in HeLa cells (Alles et al. 2021), suggesting the existence of a nuclear deadenylation process that is coordinated with RNA splicing before nuclear export. Consistent with this view, several lines of evidence indicate that polyadenylation plays a critical role in PABPN1-dependent nuclear retention of the unspliced gag-pol reporter RNA. First, ribozyme-cleaved unspliced transcripts that bypassed 3′ end polyadenylation were no longer sensitive to PABPN1 depletion. Second, a PABPN1 mutant (LALA) selectively defective in stimulating poly(A) polymerase elongation, but retaining poly(A) binding activity, strongly enhanced expression of the unspliced gag-pol transcript. Third, analysis of poly(A) tail length distribution by RNase H cleavage assays revealed that the absence of PABPN1 and the PABPN1 LALA mutant affected the steady state length distribution of poly(A) tails of the reporter mRNA. Together, these data support a direct polyadenylation-dependent role of PABPN1 in the control of the unspliced gag-pol transcript, although we cannot totally exclude the possibility of an indirect effect. Based on our data and the aforementioned studies, we propose that PABPN1 can coordinate splicing completion with nuclear export via poly(A) tail length control. Specifically, PABPN1 would stimulate long poly(A) tails to intron-containing transcripts, decreasing nuclear export rates and favoring nuclear retention, while completion of splicing may limit PABPN1-dependent poly(A) tail extension and promote nuclear export. Recent RIP-seq analysis in fact supports this view, showing that PABPN1-enriched transcripts are often incompletely spliced and decorated by long poly(A) tails (Nicholson-Shaw et al. 2022), findings that are consistent with observations that many human introns are spliced after cleavage and polyadenylation (Drexler et al. 2020; Coté et al. 2021). In the absence of PABPN1, or in cells overexpressing the PABPN1 LALA mutant, incompletely spliced transcripts may not gain sufficiently long poly(A) tails to slow down export kinetics, with the potential of escaping other RNA nuclear retention and decay pathways, an event that was likely enhanced by the presence of the strong MPMV RNA export element in the viral reporter transcript.
Using a tool to detect intron retention events from RNA-seq data, we have identified more than a hundred introns that showed increase retention in PABPN1-deficient cells, but not in cells depleted for PAXT, NEXT, and exosome components. Remarkably, close to 90% of these PABPN1-sensitive intron retention events affected the 3′-terminal intron. These observations are in fact consistent with previous RT-qPCR analysis demonstrating roles for PAP and PABPN1 in the removal of 3′-terminal introns of selected candidate genes (Muniz et al. 2015). Furthermore, as our intron retention analysis was performed on poly(A)-selected RNA-seq data (Beaulieu et al. 2012), these findings suggest that PABPN1 post-transcriptionally controls the splicing of the 3′-terminal intron of many human genes, which echoes recent evidence indicating that a large fraction of genes splice their 3′-terminal intron after cleavage and polyadenylation (Drexler et al. 2020). Our findings are therefore compatible with multiple lines of evidence supporting that terminal intron removal, 3′ end processing, and polyadenylation are interdependent (Niwa and Berget 1991; Vagner et al. 2000; Millevoi et al. 2006; Rigo and Martinson 2008; Muniz et al. 2015); still, the mechanisms underlying the coupling between these RNA processing steps remain poorly understood. Our RNA-seq data also revealed that a significant fraction of genes affected by PABPN1-dependent intron retention were down-regulated. Intron retention can lead to reduced mRNA levels by triggering nonsense-mediated RNA decay (NMD) in the cytoplasm (Yap et al. 2012; Wong et al. 2013). Accordingly, further analyses are required to establish whether the transcripts sensitive to PABPN1-dependent intron retention show reduced levels in PABPN1-deficient cells because they are targeted by NMD as a result of premature export of intron-containing transcripts.
In summary, our study discloses important PAXT-independent roles for PABPN1 in the post-transcriptional control of terminal intron splicing and the nuclear retention of intron retained transcripts, thereby building on several recent observations that link poly(A) tail length status to splicing and nuclear export. More generally, these findings suggest that the status of the 3′ end poly(A) tail bound by PABPN1 could serve as a signal that can licence terminal intron splicing and nuclear export rates.
MATERIALS AND METHODS
Human cell culture
HeLa, HEK293T, HEK293-FT and U2OS-FT cells were grown in Dulbecco's Modified Eagle's Medium supplemented with 10% tetracycline-free fetal bovine serum (FBS) in a humidified CO2 incubator (37°C, 5% CO2). Inducible expression of BirA–PABPN1 was achieved by flippase-mediated recombination in HEK293-FT cells, as previously described (Bergeron et al. 2015). Activation of BirA-tagged proteins was achieved with 50 µm biotin for 24 h. Cells were transfected with siRNAs at 25 nM using Lipofectamine 2000 (Life Technologies), except for siPABPN1 3UTR (10 nM each) and maintained for 72 h. Sequences of siRNAs used in this study are listed in Supplemental Table S1.
SILAC-coupled BioID assays
For SILAC and BioID experiments, proteins were metabolically labeled with stable isotopes of arginine and lysine in cell culture, as previously described (Dionne et al. 2019; Landry-Voyer et al. 2020). Briefly, HEK293-FT cells expressing BirA–PABPN1 were grown in media containing labeled amino acids. Twenty-four to 48 hours after induction with doxycycline and biotin, cells were collected in lysis buffer and incubated at 4°C for 20 min. Lysates were centrifuged for 10 min at 13,000 rpm at 4°C and equal amounts of proteins were incubated with Streptavidin Sepharose beads (Sigma) for 3 h at 4°C. Beads were then washed extensively and subjected to on-bead digestion with trypsin. LC-MS/MS and analysis of SILAC ratios were performed as described previously (Dionne et al. 2019; Landry-Voyer et al. 2020).
Protein analyses and antibodies
Preparation of total cell extracts was previously described (Bergeron et al. 2015; Dionne et al. 2019; Landry-Voyer et al. 2020). A total of 50–100 µg of proteins was separated by SDS-PAGE, transferred to nitrocellulose membranes, and analyzed by immunoblotting using primary antibodies (see Supplemental Table S2). Membranes were then probed with either a donkey anti-rabbit antibody conjugated to IRDye 800CW (926-32213; 1:15,000; Li-Cor) or a goat anti-mouse antibody conjugated to Alexa Fluor 680 (A-21057; 1:15,000; Life Technologies). Protein detection was performed using an Odyssey infrared imaging system (Li-Cor).
RNA analyses
Total RNA was extracted using TRIzol RNA isolation reagents (Thermo Fisher) and analyzed by northern blotting and RT-qPCR as described previously (Bergeron et al. 2015; Landry-Voyer et al. 2020) with the following modifications: An amount of 10 µg/well of total RNA was mixed with 0.6 volume of loading dye (75% formamide, 45 mM tricine, 45 mM triethanolamine, 0.75 mM EDTA, 0.03% bromophenol blue, 0.6 M formaldehyde) and loaded onto a 1% denaturing agarose gel (1% agarose, 30 mM tricine, 30 mM triethanolamine, 0.4 M formaldehyde) in running buffer (30 mM tricine, 30 mM triethanolamine) at 200 V. RNAs were then transferred onto nylon membranes and hybridized using DNA probes listed in Supplemental Table S3. Signals were detected using a Typhoon Trio instrument and quantified by Quantity One 1-D analysis software (Bio-Rad Laboratories).
RT-qPCR analyses were performed as previously described (Yague-Sanz et al. 2021). Briefly, 5 µg of total RNA was treated with 1 unit of RNase-free DNase RQ1 (Promega, M6101) for 30 min at 37°C and inactivated with 1 µL of 25 mM EDTA for 10 min at 65°C. Reverse transcription reactions were in a volume of 20 µL using random hexamers or oligo d(T) and 2 units of M-MLV RT for 60 min at 42°C and inactivated for 20 min at 65°C. qPCR reactions were performed in triplicates on a LightCycler 96 system (Roche) in a final volume of 15 µL using 6 µL from a 1:20 dilution of each cDNA, 0.15 µM of forward and reverse primers, and 7.5 µL of the 2× PerfeCTa SYBR Green SuperMix from Quantabio. Analysis of gene expression changes were calculated relative to the appropriate control samples and were measured with the ΔΔCT method using the gene GAPDH as an internal reference. The oligonucleotides used in the qPCR experiments are listed in Supplemental Table S4.
Single molecule fluorescence in situ hybridization (smFISH)
The list of the 24 primary DNA probes complementary to the HIV-1 gag-pol sequence as well as the FLAP (fluorescent secondary probe) used for smFISH are listed in Supplemental Table S5. Probe sequences were selected using Oligostan software (Tsanov et al. 2016) keeping only filters 1, 2, and 4 and synthesized by Integrated DNA Technologies (IDT). The secondary FLAP probe was conjugated to two cyanine-3 at the 5′ and 3′ ends. smFISH coupled to immunofluorescence analysis experiments were as described previously (Nguyen et al. 2015) with the following modifications: cells were permeabilized in 70% ethanol at 4°C overnight and washed with 1× SSC/20% formamide for 15 min at RT. For hybridization, each coverslip was incubated with 80 µL hybridization mix (mix 1: 2× SSC, 0.7 mg/mL yeast tRNA, 40% formamide; 4 µM Flap-structured duplexes; Mix 2: 0.4 mg/mL BSA, 4 mM Ribonucleoside Vanadyl Complexes; 21% dextran sulfate) at 37°C overnight in the dark. After hybridization, cells were washed twice with 1× SSC/20% formamide for 30 min at 37°C.
Microscopy
Visualization of the images was carried out on a ZEISS LSM880 microscope equipped with a Plan-Apochromat 40×/1.4 oil immersion objective with a 1.6× zoom. Eight Z-sections of 1.54 µm were acquired for each condition and parameters acquisition times have been defined to avoid pixel saturation to ensure appropriate visualization and intensity quantification. These conditions have been adjusted in the same way between conditions. For display purposes, image sections have been deconvolved using the ZEN 2.6 iterative algorithm. A macro made with the software ImageJ (Schneider et al. 2012) was used to cluster five Z-sections to form a single image that is presented in the results section. The five Z-sections selected were the same for each of the channels. Cropping and changes of 300 dpi resolution were made in Photoshop for data presentation. Image quantifications were performed using CellProfiler 4.1.3 software (Carpenter et al. 2006). Briefly, the nuclei were identified as the primary objects, the whole cell, visualized using tubulin, was identified as the secondary object; the cytoplasm was identified as a tertiary object (by subtracting the nucleus from the whole cell). A nucleus/cytoplasm ratio was then measured based on signal intensity, and the measurements were exported to GraphPad Prism version 9 (GraphPad Software) for statistical analyses.
Quantifications and statistical analysis
All statistical analyses were performed using GraphPad Prism version 9. Data were analyzed by a one-way ANOVA test (multiple independent groups) followed by a post hoc Dunnett test (when P < 0.05). In the case of non-normal distributions, a Kruskal–Wallis test followed by a post-hoc Dunn test (when P < 0.05), respectively, were used. Statistical details of each experiment (test used, value of n, replicates, etc.) can be found in the Results and legends to figures.
3′ rapid amplification of cDNA ends (3′ RACE)
A total of 2 µg of RNA was previously treated with 1 unit of RNase-free DNase RQ1 (Promega, M6101) for 15 min at room temperature and inactivated with 1 µL of 25 mM EDTA for 10 min at 65°C. The DNase-treated RNA was then incubated with 50 µM oligo(dT)-anchor primer (5′-GGCCACGCGTCGACTAGTACGATCTTTTTTTTTTTTTTTTTTTTTTTT-3′) at 37°C for 1 h, heated at 95°C for 5 min, and quickly chilled on ice before the addition of 1 µL of 10 µM of reverse PCR anchor-primer, 1 µL of 10 mM dNTPs, 1.5 µL of 50 mM MgCl2, 5 µL of Taq buffer, 1 µL of Taq polymerase, 5 µL of cDNA and 1 µL of forward primer containing MPMV-specific sequences (before CTE : 5′-TAATACGACTCACTATAGGTGTCGAGGGATCTCGAGA-3′ or after CTE : 5′-TAATACGACTCACTATAGGCGGATGATGTCTTGGCCTCT-3′). PCR reactions were incubated with the following program: 98°C for 3 min, 35 cycles of 98°C for 30 sec, 63°C–68°C for 30 sec, 72°C for 2 min, and final extension at 72°C for 8 min. PCR products were separated on a 1.2% agarose gel, gel purified (Qiagen), and analyzed by Sanger sequencing.
RNase H cleavage assay
Total RNA (20 µg) was incubated with one antisense DNA oligonucleotides (5′-ATGTCACCCTTTTTAATTATA-3′) complementary to sequences upstream of the MPMV cleavage sites (as illustrated in Supplemental Fig. S7) and with or without oligo(dT). The MPMV RNA:DNA hybrids were then cleaved by RNase H (37°C for 1 h). The reaction was stopped by adding 15 µL of 2× Formamide loading dye followed by heating at 65°C for 5 min and gel analysis. The samples were analyzed by standard Northern blot protocol on 4.5% polyacrylamide/8 M urea gels (Lemay et al. 2010; Beaulieu et al. 2012). To detect the RNA signals, three DNA probes (listed in Supplemental Table S3) were designed and labeled with 32P.
Analysis of intron retention from RNA-seq data
Our own siNT and siPABPN1 RNA-seq data (Beaulieu et al. 2012) are available under the sequence read archive (SRA) number SRP015926. Raw reads were filtered with Trimmomatic (Bolger et al. 2014) with options “ILLUMINACLIP:${adapters}:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36” and were mapped on the human genome (hg38) using HISAT2 (Kim et al. 2019) with options “–rna-strandness RF –min-intronlen 20 –no-discordant –no-mixed”.
FeatureCounts (Liao et al. 2014) with options “-p -s 0 -J -P -B -d 0 -D 500000 – C” was used to count fragments on exons aggregated at the gene level and on exon–exon junctions based on the GencodeV25 release of gene annotation downloaded from Ensembl (Howe et al. 2021). Based on the same GencodeV25 annotation, count of fragments on introns or exon–intron junctions, as well as the Shannon entropy in intron coverage were quantified with the iREAD tool using default parameters (Li et al. 2020).
Differentially retained introns were defined based on the three following criteria:
The intron had to be considered as retained in at least one condition based on IRead default filters; that is, that the intron is independent of (= not overlapping) any other annotated transcript, that it is supported by at least 20 intronic reads and at least one exon–intron junction read accounting together for at least one FPKM, and that the intron coverage is uniformly distributed across the region (Shannon entropy score ≥0.9).
The intronic fragment count normalized on the gene expression (i.e., the exonic fragment counts) had to be twice higher in one condition compared to the other (absolute log2 fold-change of normalized intron retention ratio >1).
To assess statistical significance of differential intron retention, a binomial regression model was fit on the data at the individual intron level, explaining the change caused by PABPN1 knockdown on the proportion of spliced fragments out of all fragments traversing the exon–intron junctions. To consider an intron as significantly differentially retained, a threshold on the P-value adjusted for multiple testing (false discovery rate) of a Wald test on the fitted siPABPN1 effect (condition coefficient) was set at smaller or equal to 5%.
To assess how the PABPN1-dependant introns are retained in other conditions, we exported the coordinates of the differentially retained intron (and associated exons) in .bed format and converted them from the hg38 to the hg19 genome space using the UCSC liftOver tool (Navarro Gonzalez et al. 2021). Using these converted coordinates, we calculated intronic and exonic coverage from the publicly available coverage files (.bigwig) from the GSE84172 study where RNA has been sequenced after depletion of components of the Exosome, PAXT, and NEXT complexes (Meola et al. 2016).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Marie-Louise Hammarskjold for the HIV Gag-Pol reporter constructs, Stephen Goff and Yosef Sabo for fruitful discussions, and Poulami Choudhuri for critical reading of the manuscript. M.L. and C.Y-S. were supported by awards from the Fonds de recherche du Québec—Santé (FRQS). This work was supported by the Canadian Institutes of Health Research Grant (to F.B.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079294.122.
MEET THE FIRST AUTHOR
Lauren Kwiatek.

Meet the First Author(s) is an editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Lauren Kwiatek is the first author of this paper, “PABPN1 prevents the nuclear export of an unspliced RNA with a constitutive transport element and controls human gene expression via intron retention.” Lauren is a research assistant in the Pavillon de Recherche Appliquée sur le Cancer at the University of Sherbrooke, where she completed her second master's degree under the supervision of Professor François Bachand. Previously, Lauren obtained a master's degree in proteomics and biochemistry. Her main research objective is to better understand the functions of PABPN1 in mammalian cells in order to discover new roles.
What are the major results described in your paper and how do they impact this branch of the field?
The main results of this study demonstrate a role for PABPN1 in restricting nuclear export of intron-retained transcripts. Furthermore, our data also reinforce the interdependence between terminal intron splicing, 3′ end processing, and polyadenylation. These findings bring new insights into how RNAs are processed according to the state of PABPN1-bound poly(A) tails, which could serve as a signal authorizing the splicing of terminal introns and affect nuclear export rates.
What led you to study RNA or this aspect of RNA science?
It was somewhat by chance that I embarked on RNA research. Indeed, I studied proteomics during my first master's degree. But an internship in the laboratory of François Bachand allowed me to discover RNA and its many facets! Indeed, we still know very little about the full potential of RNA. We are only at the tip of the iceberg.
What are some of the landmark moments that provoked your interest in science or your development as a scientist?
Having inspiring teachers during my graduate studies in proteomics and biochemistry made me want to dive into the study of gene regulation.
If you were able to give one piece of advice to your younger self, what would that be?
Have more confidence in yourself and you will see that you can go very far!
Are there specific individuals or groups who have influenced your philosophy or approach to science?
First of all, I would say my director, François Bachand, is incredible and helped me a lot during my master's degree and also to advance the PABPN1 project. He is always open to new experiences and discussions. Then, my mentor, Benoît Laurent, who helped me keep my positivity in science and also gave me a lot of advice during my master's degree.
REFERENCES
- Alles J, Legnini I, Pacelli M, Rajewsky N. 2021. Rapid nuclear deadenylation of mammalian messenger RNA. iScience 26: 105878. 10.1016/j.isci.2022.105878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear DG, Fomproix N, Soop T, Bjorkroth B, Masich S, Daneholt B. 2003. Nuclear poly(A)-binding protein PABPN1 is associated with RNA polymerase II during transcription and accompanies the released transcript to the nuclear pore. Exp Cell Res 286: 332–344. 10.1016/S0014-4827(03)00123-X [DOI] [PubMed] [Google Scholar]
- Beaulieu YB, Kleinman CL, Landry-Voyer AM, Majewski J, Bachand F. 2012. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet 8: e1003078. 10.1371/journal.pgen.1003078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron D, Pal G, Beaulieu YB, Chabot B, Bachand F. 2015. Regulated intron retention and nuclear pre-mRNA decay contribute to PABPN1 autoregulation. Mol Cell Biol 35: 2503–2517. 10.1128/MCB.00070-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird G, Fong N, Gatlin JC, Farabaugh S, Bentley DL. 2005. Ribozyme cleavage reveals connections between mRNA release from the site of transcription and pre-mRNA processing. Mol Cell 20: 747–758. 10.1016/j.molcel.2005.11.009 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet A, Bretes H, Palancade B. 2015. Nuclear pore components affect distinct stages of intron-containing gene expression. Nucleic Acids Res 43: 4249–4261. 10.1093/nar/gkv280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Bhutkar A, Sharp PA. 2015. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev 29: 63–80. 10.1101/gad.247361.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, Frey B, Irimia M, Blencowe BJ. 2014. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res 24: 1774–1786. 10.1101/gr.177790.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresson SM, Conrad NK. 2013. The human nuclear poly(a)-binding protein promotes RNA hyperadenylation and decay. PLoS Genet 9: e1003893. 10.1371/journal.pgen.1003893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresson SM, Hunter OV, Hunter AC, Conrad NK. 2015. Canonical poly(A) polymerase activity promotes the decay of a wide variety of mammalian nuclear RNAs. PLoS Genet 11: e1005610. 10.1371/journal.pgen.1005610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugiolo M, Botti V, Liu N, Muller-McNicoll M, Neugebauer KM. 2017. Fractionation iCLIP detects persistent SR protein binding to conserved, retained introns in chromatin, nucleoplasm and cytoplasm. Nucleic Acids Res 45: 10452–10465. 10.1093/nar/gkx671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A, Kutay U, Kuhn U, Wahle E, Carmo-Fonseca M. 2000. Deciphering the cellular pathway for transport of poly(A)-binding protein II. RNA 6: 245–256. 10.1017/S1355838200991908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7: R100. 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Li Y, Krug RM. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J 18: 2273–2283. 10.1093/emboj/18.8.2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coté A, Coté C, Bayatpour S, Drexler HL, Alexander KA, Chen F, Wassie AT, Boyden ES, Berger S, Churchman LS, et al. 2021. Pre-mRNA spatial distributions suggest that splicing can occur post-transcriptionally. bioRxiv 10.1101/2020.04.06.028092 [DOI] [Google Scholar]
- Coyle JH, Bor YC, Rekosh D, Hammarskjold ML. 2011. The Tpr protein regulates export of mRNAs with retained introns that traffic through the Nxf1 pathway. RNA 17: 1344–1356. 10.1261/rna.2616111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne KL, Bergeron D, Landry-Voyer AM, Bachand F. 2019. The 40S ribosomal protein uS5 (RPS2) assembles into an extra-ribosomal complex with human ZNF277 that competes with the PRMT3-uS5 interaction. J Biol Chem 294: 1944–1955. 10.1074/jbc.RA118.004928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler HL, Choquet K, Churchman LS. 2020. Splicing kinetics and coordination revealed by direct nascent RNA sequencing through nanopores. Mol Cell 77: 985–998.e988. 10.1016/j.molcel.2019.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufu K, Livingstone MJ, Seebacher J, Gygi SP, Wilson SA, Reed R. 2010. ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev 24: 2043–2053. 10.1101/gad.1898610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen TJ, Eichhorn SW, Subtelny AO, Lin KS, McGeary SE, Gupta S, Bartel DP. 2020. The dynamics of cytoplasmic mRNA metabolism. Mol Cell 77: 786–799.e710. 10.1016/j.molcel.2019.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erber L, Luo A, Chen Y. 2019. Targeted and interactome proteomics revealed the role of PHD2 in regulating BRD4 proline hydroxylation. Mol Cell Proteomics 18: 1772–1781. 10.1074/mcp.RA119.001535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst RK, Bray M, Rekosh D, Hammarskjold ML. 1997a. Secondary structure and mutational analysis of the Mason-Pfizer monkey virus RNA constitutive transport element. RNA 3: 210–222. [PMC free article] [PubMed] [Google Scholar]
- Ernst RK, Bray M, Rekosh D, Hammarskjold ML. 1997b. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol Cell Biol 17: 135–144. 10.1128/MCB.17.1.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasken MB, Stewart M, Corbett AH. 2008. Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated protein, Mlp1, in mRNA export. J Biol Chem 283: 27130–27143. 10.1074/jbc.M803649200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82: 475–483. 10.1016/0092-8674(95)90436-0 [DOI] [PubMed] [Google Scholar]
- Folco EG, Lee CS, Dufu K, Yamazaki T, Reed R. 2012. The proteins PDIP3 and ZC11A associate with the human TREX complex in an ATP-dependent manner and function in mRNA export. PLoS ONE 7: e43804. 10.1371/journal.pone.0043804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90: 1051–1060. 10.1016/S0092-8674(00)80371-2 [DOI] [PubMed] [Google Scholar]
- Fribourg S, Braun IC, Izaurralde E, Conti E. 2001. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol Cell 8: 645–656. 10.1016/S1097-2765(01)00348-3 [DOI] [PubMed] [Google Scholar]
- Gales JP, Kubina J, Geldreich A, Dimitrova M. 2020. Strength in diversity: nuclear export of viral RNAs. Viruses 12: 1014. 10.3390/v12091014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73. 10.1016/S0092-8674(03)01026-2 [DOI] [PubMed] [Google Scholar]
- Gingras AC, Abe KT, Raught B. 2019. Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles. Curr Opin Chem Biol 48: 44–54. 10.1016/j.cbpa.2018.10.017 [DOI] [PubMed] [Google Scholar]
- Gordon JM, Phizicky DV, Neugebauer KM. 2021. Nuclear mechanisms of gene expression control: pre-mRNA splicing as a life or death decision. Curr Opin Genet Dev 67: 67–76. 10.1016/j.gde.2020.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant RP, Hurt E, Neuhaus D, Stewart M. 2002. Structure of the C-terminal FG-nucleoporin binding domain of Tap/NXF1. Nat Struct Biol 9: 247–251. 10.1038/nsb773 [DOI] [PubMed] [Google Scholar]
- Green DM, Johnson CP, Hagan H, Corbett AH. 2003. The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc Natl Acad Sci 100: 1010–1015. 10.1073/pnas.0336594100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarskjold ML, Heimer J, Hammarskjold B, Sangwan I, Albert L, Rekosh D. 1989. Regulation of human immunodeficiency virus env expression by the rev gene product. J Virol 63: 1959–1966. 10.1128/jvi.63.5.1959-1966.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe KL, Achuthan P, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J, et al. 2021. Ensembl 2021. Nucleic Acids Res 49: D884–D891. 10.1093/nar/gkaa942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U, Gundel M, Knoth A, Kerwitz Y, Rudel S, Wahle E. 2009. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J Biol Chem 284: 22803–22814. 10.1074/jbc.M109.018226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry-Voyer AM, Bergeron D, Yague-Sanz C, Baker B, Bachand F. 2020. PDCD2 functions as an evolutionarily conserved chaperone dedicated for the 40S ribosomal protein uS5 (RPS2). Nucleic Acids Res 48: 12900–12916. 10.1093/nar/gkaa1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumont CM, Daouda T, Laverdure JP, Bonneil E, Caron-Lizotte O, Hardy MP, Granados DP, Durette C, Lemieux S, Thibault P, et al. 2016. Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat Commun 7: 10238. 10.1038/ncomms10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Glaunsinger BA. 2009. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol 7: e1000107. 10.1371/journal.pbio.1000107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ES, Wolf EJ, Ihn SSJ, Smith HW, Emili A, Palazzo AF. 2020. TPR is required for the efficient nuclear export of mRNAs and lncRNAs from short and intron-poor genes. Nucleic Acids Res 48: 11645–11663. 10.1093/nar/gkaa919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ES, Smith HW, Wolf EJ, Guvenek A, Wang YE, Emili A, Tian B, Palazzo AF. 2022. ZFC3H1 and U1-70K promote the nuclear retention of mRNAs with 5′ splice site motifs within nuclear speckles. RNA 28: 878–894. 10.1261/rna.079104.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay JF, D'Amours A, Lemieux C, Lackner DH, St-Sauver VG, Bahler J, Bachand F. 2010. The nuclear poly(A)-binding protein interacts with the exosome to promote synthesis of noncoding small nucleolar RNAs. Mol Cell 37: 34–45. 10.1016/j.molcel.2009.12.019 [DOI] [PubMed] [Google Scholar]
- Lemieux C, Bachand F. 2009. Cotranscriptional recruitment of the nuclear poly(A)-binding protein Pab2 to nascent transcripts and association with translating mRNPs. Nucleic Acids Res 37: 3418–3430. 10.1093/nar/gkp207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux C, Marguerat S, Lafontaine J, Barbezier N, Bahler J, Bachand F. 2011. A pre-mRNA degradation pathway that selectively targets intron-containing genes requires the nuclear poly(A)-binding protein. Mol Cell 44: 108–119. 10.1016/j.molcel.2011.06.035 [DOI] [PubMed] [Google Scholar]
- Li HD, Funk CC, Price ND. 2020. iREAD: a tool for intron retention detection from RNA-seq data. BMC Genomics 21: 128. 10.1186/s12864-020-6541-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lleres D, Denegri M, Biggiogera M, Ajuh P, Lamond AI. 2010. Direct interaction between hnRNP-M and CDC5L/PLRG1 proteins affects alternative splice site choice. EMBO Rep 11: 445–451. 10.1038/embor.2010.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell 43: 624–637. 10.1016/j.molcel.2011.06.028 [DOI] [PubMed] [Google Scholar]
- Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. 1989. The HIV-1 Rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338: 254–257. 10.1038/338254a0 [DOI] [PubMed] [Google Scholar]
- Meola N, Domanski M, Karadoulama E, Chen Y, Gentil C, Pultz D, Vitting-Seerup K, Lykke-Andersen S, Andersen JS, Sandelin A, et al. 2016. Identification of a nuclear exosome decay pathway for processed transcripts. Mol Cell 64: 520–533. 10.1016/j.molcel.2016.09.025 [DOI] [PubMed] [Google Scholar]
- Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. 2006. An interaction between U2AF 65 and CF Im links the splicing and 3′ end processing machineries. EMBO J 25: 4854–4864. 10.1038/sj.emboj.7601331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldon A, Malapeira J, Gabrielli N, Gogol M, Gomez-Escoda B, Ivanova T, Seidel C, Ayte J. 2008. Promoter-driven splicing regulation in fission yeast. Nature 455: 997–1000. 10.1038/nature07325 [DOI] [PubMed] [Google Scholar]
- Morris KJ, Corbett AH. 2018. The polyadenosine RNA-binding protein ZC3H14 interacts with the THO complex and coordinately regulates the processing of neuronal transcripts. Nucleic Acids Res 46: 6561–6575. 10.1093/nar/gky446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz L, Davidson L, West S. 2015. Poly(A) polymerase and the nuclear poly(A) binding protein, PABPN1, coordinate the splicing and degradation of a subset of human pre-mRNAs. Mol Cell Biol 35: 2218–2230. 10.1128/MCB.00123-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro Gonzalez J, Zweig AS, Speir ML, Schmelter D, Rosenbloom KR, Raney BJ, Powell CC, Nassar LR, Maulding ND, Lee CM, et al. 2021. The UCSC Genome Browser database: 2021 update. Nucleic Acids Res 49: D1046–D1057. 10.1093/nar/gkaa1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekhai S, Jeang KT. 2006. Transcriptional and post-transcriptional regulation of HIV-1 gene expression: role of cellular factors for Tat and Rev. Future Microbiol 1: 417–426. 10.2217/17460913.1.4.417 [DOI] [PubMed] [Google Scholar]
- Nguyen D, Grenier St-Sauveur V, Bergeron D, Dupuis-Sandoval F, Scott MS, Bachand F. 2015. A polyadenylation-dependent 3′ end maturation pathway is required for the synthesis of the human telomerase RNA. Cell Rep 13: 2244–2257. 10.1016/j.celrep.2015.11.003 [DOI] [PubMed] [Google Scholar]
- Nicholson-Shaw AL, Kofman ER, Yeo GW, Pasquinelli AE. 2022. Nuclear and cytoplasmic poly(A) binding proteins (PABPs) favor distinct transcripts and isoforms. Nucleic Acids Res 50: 4685–4702. 10.1093/nar/gkac263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Berget SM. 1991. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev 5: 2086–2095. 10.1101/gad.5.11.2086 [DOI] [PubMed] [Google Scholar]
- Ogami K, Richard P, Chen Y, Hoque M, Li W, Moresco JJ, Yates JR III, Tian B, Manley JL. 2017. An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev 31: 1257–1271. 10.1101/gad.302604.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1: 376–386. 10.1074/mcp.M200025-MCP200 [DOI] [PubMed] [Google Scholar]
- Palancade B, Zuccolo M, Loeillet S, Nicolas A, Doye V. 2005. Pml39, a novel protein of the nuclear periphery required for nuclear retention of improper messenger ribonucleoparticles. Mol Biol Cell 16: 5258–5268. 10.1091/mbc.e05-06-0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik CM, Storti RV, Jacobson A. 1979. Fractionation and functional analysis of newly synthesized and decaying messenger RNAs from vegetative cells of Dictyostelium discoideum. J Mol Biol 128: 371–395. 10.1016/0022-2836(79)90093-7 [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Lee ES. 2018. Sequence determinants for nuclear retention and cytoplasmic export of mRNAs and lncRNAs. Front Genet 9: 440. 10.3389/fgene.2018.00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajanala K, Nandicoori VK. 2012. Localization of nucleoporin Tpr to the nuclear pore complex is essential for Tpr mediated regulation of the export of unspliced RNA. PLoS One 7: e29921. 10.1371/journal.pone.0029921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekosh D, Hammarskjold ML. 2018. Intron retention in viruses and cellular genes: detention, border controls and passports. Wiley Interdiscip Rev RNA 9: e1470. 10.1002/wrna.1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo F, Martinson HG. 2008. Functional coupling of last-intron splicing and 3'-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol Cell Biol 28: 849–862. 10.1128/MCB.01410-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JC, Hunter OV, Conrad NK. 2019. Kaposi's sarcoma-associated herpesvirus ORF57 protein protects viral transcripts from specific nuclear RNA decay pathways by preventing hMTR4 recruitment. PLoS Pathog 15: e1007596. 10.1371/journal.ppat.1007596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Poulsen MB, Olszewski P, Pelechano V, Saguez C, Gupta I, Steinmetz LM, Moore C, Jensen TH. 2012. Rrp6p controls mRNA poly(A) tail length and its decoration with poly(A) binding proteins. Mol Cell 47: 267–280. 10.1016/j.molcel.2012.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheiness D, Darnell JE. 1973. Polyadenylic acid segment in mRNA becomes shorter with age. Nat New Biol 241: 265–268. 10.1038/newbio241265a0 [DOI] [PubMed] [Google Scholar]
- Shu S, Wu HJ, Ge JY, Zeid R, Harris IS, Jovanovic B, Murphy K, Wang B, Qiu X, Endress JE, et al. 2020. Synthetic lethal and resistance interactions with BET bromodomain inhibitors in triple-negative breast cancer. Mol Cell 78: 1096–1113.e1098. 10.1016/j.molcel.2020.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silla T, Karadoulama E, Makosa D, Lubas M, Jensen TH. 2018. The RNA exosome adaptor ZFC3H1 functionally competes with nuclear export activity to retain target transcripts. Cell Rep 23: 2199–2210. 10.1016/j.celrep.2018.04.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silla T, Schmid M, Dou Y, Garland W, Milek M, Imami K, Johnsen D, Polak P, Andersen JS, Selbach M, et al. 2020. The human ZC3H3 and RBM26/27 proteins are critical for PAXT-mediated nuclear RNA decay. Nucleic Acids Res 48: 2518–2530. 10.1093/nar/gkz1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek S, Zeng Y, Bellur DL, Bergkessel M, Morris KJ, Deng Q, Duong D, Seyfried NT, Guthrie C, Staley JP, et al. 2016. The evolutionarily-conserved polyadenosine RNA binding protein, Nab2, cooperates with splicing machinery to regulate the fate of pre-mRNA. Mol Cell Biol 36: 2697–2714. 10.1128/MCB.00402-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. 2019. Polyadenylation and nuclear export of mRNAs. J Biol Chem 294: 2977–2987. 10.1074/jbc.REV118.005594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Wohlbold L, Khin NW, Izaurralde E, Patel DJ. 2011. Structure-function studies of nucleocytoplasmic transport of retroviral genomic RNA by mRNA export factor TAP. Nat Struct Mol Biol 18: 990–998. 10.1038/nsmb.2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsanov N, Samacoits A, Chouaib R, Traboulsi AM, Gostan T, Weber C, Zimmer C, Zibara K, Walter T, Peter M, et al. 2016. smiFISH and FISH-quant: a flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res 44: e165. 10.1093/nar/gkw784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudek A, Lloret-Llinares M, Jensen TH. 2018. The multitasking polyA tail: nuclear RNA maturation, degradation and export. Philos Trans R Soc Lond B Biol Sci 373: 20180169. 10.1098/rstb.2018.0169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Vagner C, Mattaj IW. 2000. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev 14: 403–413. 10.1101/gad.14.4.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra P, Iglesias N, Camblong J, Zenklusen D, Stutz F. 2005. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J 24: 813–823. 10.1038/sj.emboj.7600527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigington CP, Williams KR, Meers MP, Bassell GJ, Corbett AH. 2014. Poly(A) RNA-binding proteins and polyadenosine RNA: new members and novel functions. Wiley Interdiscipl Rev RNA 5: 601–622. 10.1002/wrna.1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JJ, Ritchie W, Ebner OA, Selbach M, Wong JW, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, et al. 2013. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154: 583–595. 10.1016/j.cell.2013.06.052 [DOI] [PubMed] [Google Scholar]
- Xie Y, Ren Y. 2019. Mechanisms of nuclear mRNA export: a structural perspective. Traffic 20: 829–840. 10.1111/tra.12691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yague-Sanz C, Duval M, Larochelle M, Bachand F. 2021. Co-transcriptional RNA cleavage by Drosha homolog Pac1 triggers transcription termination in fission yeast. Nucleic Acids Res 49: 8610–8624. 10.1093/nar/gkab654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV. 2012. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev 26: 1209–1223. 10.1101/gad.188037.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. 2004. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119: 381–392. 10.1016/j.cell.2004.09.029 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Orre LM, Johansson HJ, Huss M, Boekel J, Vesterlund M, Fernandez-Woodbridge A, Branca RMM, Lehtio J. 2018. Discovery of coding regions in the human genome by integrated proteogenomics analysis workflow. Nat Commun 9: 903. 10.1038/s41467-018-03311-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerman B, Ron M, Mikl M, Segal E, Ulitsky I. 2020. Gene architecture and sequence composition underpin selective dependency of nuclear export of long RNAs on NXF1 and the TREX complex. Mol Cell 79: 251–267.e256. 10.1016/j.molcel.2020.05.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.