Abstract
High-level azole resistance in the Darlington strain of Candida albicans was investigated by gene replacement in C. albicans and expression in Saccharomyces cerevisiae. We sequenced the ERG11 gene, which encodes the sterol C14α-demethylase, from our copy of the Darlington strain. Both alleles contained the histidine for tyrosine substitution at position 132 (Y132H) reported in Darlington by others, but we also found a threonine-for-isoleucine substitution (I471T) not previously reported in the C. albicans ERG11. The encoded I471T change in amino acids conferred azole resistance when overexpressed alone and increased azole resistance when added to the Y132H amino acid sequence in an S. cerevisiae expression system. Replacement of one copy of ERG11 in an azole-susceptible strain of C. albicans with a single copy of the Darlington ERG11 resulted in expression of the integrated copy and a modest increase in azole resistance. The profound azole resistance of the Darlington strain is the result of multiple mutations.
One of the major mechanisms of azole resistance in Candida albicans has been mutations in the gene, ERG11 (ERG16), encoding the azole target enzyme P450-dependent sterol C14α- demethylase (CYP51A1). The principal approach to understanding the mechanism of azole resistance has been to sequence the ERG11 gene from azole-resistant isolates (20, 25). To date, most of the differences in sequences from azole-resistant C. albicans strains appear to have been in deduced amino acids 105 to 165, 266 to 287, and 405 to 488 (14). CYP51A1 extracted from an azole-resistant strain has been shown to have enzymatic activity less susceptible to inhibition by azole (12). The mechanism by which the peptide sequence alters enzyme affinity for azoles remains a matter of conjecture (19). Exploration of the role of gene dosage, allelic polymorphisms in the ERG11 gene, and contribution of individual amino acid substitutions has received less attention. However, the roles of five different amino acid substitutions, alone and in combination, have been elucidated by site-directed mutagenesis of the C. albicans ERG11 gene, using overexpression in Saccharomyces cerevisiae (19).
Several different azole-resistant isolates of C. albicans were obtained from a patient named Darlington, who received long-term azole therapy for chronic mucocutaneous candidiasis (22). The Darlington isolates in different culture collections have dissimilar sterol contents, restriction fragment length polymorphisms, and azole susceptibilities either due to repeated passage over many years or, perhaps, due to collection from the patient at different times (8, 17). No pretreatment, azole-susceptible isolate from this patient is available for comparison. It has been reported that azole efflux pump activity in one isolate of the Darlington strain is not impaired (1). The mechanism of azole resistance of the Darlington strain has been investigated but never fully explained.
Although the Darlington strain has been known to have an abnormal sterol content (8), we recently reported that we could restore normal sterol content by ERG3 replacement (15). However, restoration of normal ergosterol content did not restore azole susceptibility. We therefore focused our efforts on the Darlington ERG11 gene, which we cloned and sequenced. Two amino acid substitutions (Y132H, I471T) were found in the ERG11 open reading frame (ORF), one of which (Y132H) was reported previously in Darlington (3). To assess the significance of these substitutions in the homologous species, we replaced one of the two copies of the ERG11 gene in an azole-susceptible isolate of C. albicans with a copy of the Darlington ERG11 gene. To estimate the effects of Y132H and I471T individually, the ERG11 gene with either or both mutations was expressed in a fluconazole-susceptible pdr5 mutant of S. cerevisiae.
MATERIALS AND METHODS
Fungal strains and culture conditions.
The azole-resistant C. albicans strain Darlington (NCPF3310) was a gift of Christopher Hitchcock (7, 8). B311 was from the authors' collection (15). C. albicans strains SC5314 (URA3/URA3) and CAF2-1 (Δura3:: imm434/URA3) and the ura3 mutant CAI4 (Δura3::imm434/Δura3::imm434) were kindly provided by W. A. Fonzi (4). All strains were maintained on yeast extract (1%)–peptone (2%)–dextrose (2%) (YEPD) agar.
S. cerevisiae DKYI (Δura3 Δhis Δlys Δtrp Δleu Δpdr5) was a gift from Scott Moye-Rowley (10). YEp351G, a gift from Reed Wickner, is a 2μm-based vector that contains a GAL1,10 promoter (26). S. cerevisiae was cultured in yeast nitrogen base (YNB) medium (Difco, Detroit, Mich.) supplemented with lysine (30 mg per ml) and uracil, histidine, leucine, and tryptophan (each at 20 mg per ml) and containing 2% glucose. For azole susceptibility testing by the Etest, S. cerevisiae cells were grown in the same medium but in the presence of 2% galactose and 1% raffinose.
Probes.
DNA was isolated from mechanically disrupted yeast cells as described previously (5). PCR was performed by using standard conditions (18) with a thermal cycler (Minicycler; MJ Research, San Francisco, Calif.) for 25 cycles and with annealing temperatures of 50 to 55°C, depending on the primers. A 1.58-kb PCR product containing the ERG11 ORF was obtained by using C. albicans B311 genomic DNA as the template, Taq DNA polymerase (Roche Molecular Biochemicals, Indianapolis, Ind.), and the following primers: P-1878 (5′-GCGGATCCTATGGCTATTGTTGAAACTGTC-3′) and P-1879 (5′-ACGCGTCGACAATTAAAACATACAAGTTTCTCTTTT-3′). These primers contain BamHI (5′-) and SalI (3′-) restriction sites. A 0.5-kb PCR product from the middle of the ERG11 ORF was used for Southern blotting and was obtained by using pDarERG11URA3 (see below) as the template, Taq polymerase, and the following primers: 5′-CCTCATTATTGGAGACGTGATGC-3′ and 5′-GAATTAGCTTTGGCAGCAGCAG-3′. PCR products were labeled with [α-32P]dCTP by random priming (Prime It II; Stratagene, Cedar Creek, Tex.).
Southern analysis.
Southern analysis was performed by the methodology described previously (18). Restriction endonucleases were obtained from New England Biolabs, Beverly, Mass., unless otherwise indicated. The Southern blots were probed with the 0.5-kb C. albicans ERG11 fragment unless otherwise stated, using a hybridization temperature of 65°C and final washing of the membrane in 0.2× SSC (0.3 M NaCl plus 0.03 M sodium citrate) with 0.1% sodium dodecyl sulfate. To quantitate signal intensities, the blots were exposed to Storage Phosphor Screens (Molecular Dynamics, Sunnyvale, Calif.) for 3 h, and the screens were scanned with the PhosphorImager 445 SI (Molecular Dynamics) scanner. The scanned images were quantitated with ImageQuant software (Molecular Dynamics). Quantitative volume data for the same-sized rectangular square on the blot image obtained with each probe were used for analysis.
Clones and plasmid constructs.
A 3.5-kb EcoRV, HindIII fragment from Darlington was cloned from a size-selected genomic library through colony hybridization with the 1.58-kb B311 ERG11 probe and was cloned into pBSK [pBluescript SK II+; Stratagene] as pDarERG11. A 2.0-kb fragment containing the C. albicans URA3 gene was cloned into a blunt-ended XbaI site and the XhoI site of pDarERG11 to obtain pDarERG11URA3.
The ERG11 ORF was obtained from CAI4 and Darlington by using genomic DNA as the template, Taq+ DNA polymerase (Stratagene), and primers P-1878 and P-1879, described above. PCR products were ligated into YeP351G by using the BamHI and SalI restriction sites, giving plasmids pCA14 and pDAR, respectively. Plasmid DNA was propagated in Escherichia coli DH10B (Gibco BRL, Gaithersburg, Md.) grown at 37°C on Luria-Bertani broth or agar containing 50 μg of ampicillin per ml. The ERG11 ORFs in pDAR, pCAI4, and pDarERG11URA3 were sequenced in their entirety by using a rhodamine terminator sequencing reaction, run on an ABI Prizm 377 sequencer (Perkin-Elmer, Foster City, Calif.). The base sequence was analyzed with GCG software (Genetics Computer Group, Madison, Wis.).
To address the effect of different ERG11 base sequences on the S. cerevisae susceptibility test system, fragments of the Darlington and CAI4 ERG11 ORFs were combined. The NdeI fragment from pDarERG11URA3, which encodes the Y132H mutation, was ligated into NdeI-digested pCAI4 to create pCAI4-132H. Similarly, the NdeI fragment of pCAI4 was ligated into NdeI-digested pDAR, creating pDAR-471T. In the latter plasmid, the Y132H mutation has been removed but the sequence encoding the threonine at position 471 remains. Introduction of the mutations and maintenance of the authentic sequence were corroborated by DNA sequencing.
Plasmids pCAI4, pDAR, pCAI4-132H, and pDAR-471T were transformed into DKY1 by the lithium acetate method (6). PCR amplification of the transformants with primer pairs P-1878 and P-1879 confirmed the presence of the anticipated 1.6-kb product.
Replacement of the ERG11 gene in CAI4.
CAI4 cells were electroporated with the 5.5-kb XbaI-XhoI fragment from pDarERG11URA3 under conditions described previously (21). Electroporated cells were spread on minimal medium and incubated at 37°C for 7 days. Approximately two URA-positive colonies were obtained per microgram of DNA. Transformants were verified by Southern blotting with the 0.5-kb ERG11 probe.
Drug susceptibility assay.
The MICs of fluconazole for C. albicans isolates were measured by a modification of the National Committee for Clinical Laboratory Standards M27-A microdilution protocol (16). Instead of RPMI 1640 medium, which is usually used in this method, YNB (Difco) containing 2% glucose was used in order to parallel the Etest conditions described below. For the CAI4 strain, uridine (200 μg/ml) was added. Flat-bottom 96-well microtiter plates containing the cell suspension and serial dilutions of fluconazole were incubated for 2 to 7 days at 30°C, the contents of the wells were mixed by pipetting, and the plates were scanned with a microtiter reader (Dynatech, Herndon, Va.) at 450 nm. The results at 4 days were selected for presentation because day 4 was the first day that all drug-free wells had sufficient growth. Incubation was continued for a total of 7 days to permit better growth of the mutant strain, CA14. Fluconazole was a generous gift of Pfizer Inc. (New York, N.Y.).
The fluconazole susceptibilities of the S. cerevisiae transformants were assessed on agar plates by using drug-impregnated paper strips laid on inoculated agar (Etest; AB BIODISK, Solna, Sweden). S. cerevisiae cells were grown in YNB broth containing 2% galactose and 1% raffinose at 30°C with constant agitation for 2 days and were diluted to a density of 2 × 105 cells per ml. Plates with the same medium in 2% agar were inoculated with cotton swabs saturated with the cell suspension. Fluconazole strips were placed on the centers of the agar plates. The plates were incubated for 5 days at 30°C. Leucine (50 μg/ml) was added for the untransformed strain DKY1.
RT-PCR.
To confirm the transcription of the integrated Darlington ERG11 gene in transformant 12 (described below), double-stranded cDNA was obtained by reverse transcription (RT)-PCR (Life Technologies, Gaithersburg, Md.) by using as the template poly(A)+ RNA harvested from an overnight growth of C. albicans. RNA was extracted from C. albicans cells with the FastRNA kit (Bio 101, Vista, Calif.). K18 (5′-GAAAAAACTCATGGGGTTGC-3′) and G04 (5′-ATAATCAGGGTCAGGCAC-3′) served as PCR primers. The amplified double-stranded cDNA was digested with BsrI, and Southern analysis was performed with a 0.98-kb PCR product (K18-G04) as a probe.
RESULTS
Sequence of ERG11 from Darlington strain.
The Darlington genomic clone pDarERG11 was found to contain the anticipated 1,584-bp ERG11 ORF, a 1,235-bp 5′- flanking sequence, and a 1,430-bp 3′-flanking sequence. Analysis of the ERG11 ORF sequence revealed two differences in the encoded amino acids from the published ERG11 sequence (11). A T-to-C change at position 394 encoded a substitution of tyrosine for histidine, a sequence also reported by Favre et al. (3) in their Darlington strain. We did not find the T214C or G1349A substitutions which they reported. However, we did find a T1412C base substitution, coding for threonine, not isoleucine, at amino acid 471. The Y132H and the I471T mutations could be confirmed on Southern analysis of genomic DNA because they created new restriction sites for RcaI (Roche Molecular Biochemicals) and BsrI, respectively (Fig. 1). Southern analysis of genomic DNA from the Darlington strain was done by digesting the DNA with NspV (Life Technologies) and probing with the 1.58-kb ERG11 gene. NspV cuts within the 214TTC (Phe72) site of the Darlington ERG11 gene, giving on Southern analysis only the anticipated 2.8-kb fragment (data not shown). NspV would not be predicted to cut the 214CTC (Leu72) sequence reported by Favre et al. (3). Our Darlington strain is clearly different from theirs. Their strain (ATCC 64124) was deposited in a different national culture collection by a different author of the publication (22) compared with that in which our strain (strain NCPF3310) was deposited and may have been obtained at a different time in the patient's course. The existence of differences between Darlington strains in various culture collections has been noted previously (8, 17).
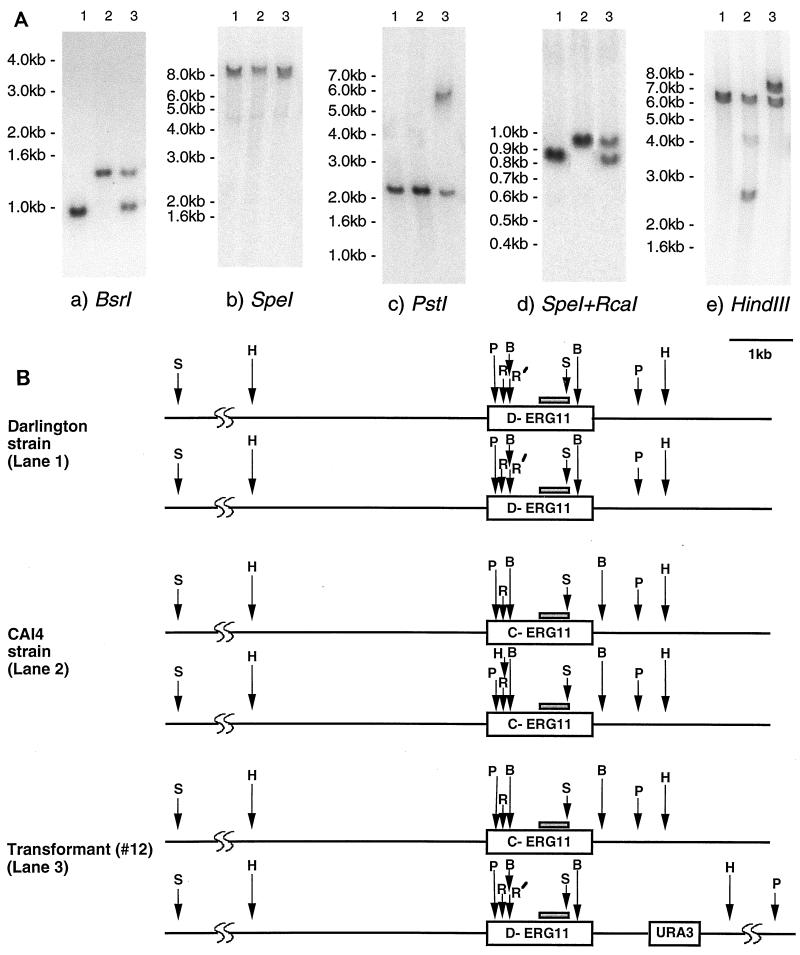
FIG. 1.
(A) Southern analysis for the Darlington strain (lanes 1), the CAI4 strain (lanes 2), and transformant 12 (lanes 3). DNA was digested with the indicated enzymes and was probed with the 0.5-kb ERG11 probe for BsrI, SpeI, PstI, and SpeI plus RcaI. The Southern blot of the HindIII digest was probed with the 1.58-kb ERG11 fragment. (B) Restriction maps of the Darlington and CAI4 strains as well as transformant 12. The following abbreviations indicate restriction sites: B, BsrI; S, SpeI; P, PstI, and R and R′, RcaI. Small boxes indicate the locations of the sequences that hybridized with the probe. The large boxes labeled D-ERG11 and C-ERG11 indicate the locations of the ERG11 ORFs from the Darlington strain and the CAI4 strain, respectively.
Two different sequences were obtained from pCAI4 clones on multiple PCRs. One was identical to the published sequence from SC5314, the parent of CAI4 (11). The other clones contained A348T and A381C substitutions, creating HindIII and RcaI sites but also encoding D116E and K128T changes in the encoded amino acids. The ERG11 gene from SC5314 is known to be heterozygous at a HindIII site within the ORF (11).
Chromosomal replacement of the ERG11 gene in C. albicans CAI4.
Southern analysis of CAI4 cells transformed with pDarERG11URA3 revealed a transformant in which one allele was replaced by a single copy of the Darlington ERG11, as shown in Fig. 1A and diagrammed in Fig. 1B. The presence of T1412C in both copies of the Darlington ERG11 ORF created a 1.0-kb fragment when the BsrI digest of genomic DNA was probed with the 0.5-kb ERG11 fragment (Fig. 1A, part a, lane 1). In CAI4, the BsrI site lies in the 3′-flanking region, creating a 1.4-kb fragment in both alleles on Southern analysis (Fig. 1A, part a, lane 2). Transformant 12 showed both 1.4- and 1.0-kb fragments (Fig. 1A, part a, lane 3), indicating that CAI4 had acquired a copy of the Darlington ERG11 gene. By phosphorimaging quantitation, the ratios of the 1.4- and 1.0-kb fragments in ERG11 transformant 12 was 1.0. This ratio suggested that transformant 12 had one ERG11 allele replaced by a single integrated copy of the Darlington ERG11. These data also indicated that Darlington and CAI4 are homozygous at the Bsrl sites flanking the 0.5-kb probe.
Additional Southern analysis was performed with SpeI, PstI, SpeI plus RcaI, and HindIII digests. The XbaI-XhoI fragment of pDarERG11URA3 used to transform CAI4 contained a SpeI site in the 5′ multiple cloning site. An additional SpeI site is contained in the ERG11 ORF. On SpeI digestion and Southern analysis (Fig. 1), transformant and the parent strains contain a large SpeI fragment, at least 7 kb. In transformant 12 (Fig. 1A, part b, lane 3), the lack of a new SpeI site from the pDarERG11URA3 fragment indicated that the 5′ end had been lost but that the location of the integrated copy was approximately the same as that in the host strain.
Host strain CAI4 contained a PstI site in the 3′-flanking region, creating a 2.2-kb fragment on Southern blotting (Fig. 1A, part c, lane 2). Transformant 12 appears to have lost this PstI site, as it did the SpeI site, but the SpeI site was retained in the CAI4 sequence (Fig. 1A, part c, lane 3). This site was now shifted downstream the expected distance by the presence of the URA3 gene, creating a 5.5-kb fragment. By phosphorimaging quantitation, the ratio of the 2.2- and 5.5-kb fragments in transformant 12 was 1.0, consistent with replacement with a single copy of the Darlington ERG11 gene.
To analyze homozygosity at the sequence encoding 132H, double digestion of genomic DNA with SpeI and RcaI was done. The A394C base substitution created a second RcaI restriction site in the Darlington ERG11 gene, designated R′ in Fig. 1B. In the Darlington and CAI4 ERG11 ORFs, one SpeI site was located in the ORF. Darlington and CAI4 were homozygous at the SpeI restriction site (Fig. 1A, part b, lane 1 and 2). Darlington (Fig. 1A, part d, lane 1) and CAI4 (Fig. 1A, part d, lane 2) showed single bands of 0.84 and 0.93 kb, respectively, on the double digest. Transformant 12 (Fig. 1A, part d, lane 3) showed both the 0.84- and the 0.93-kb bands, suggesting that this strain is heterozygous at this locus. By phosphorimaging quantitation, the ratio of the 0.93- and 0.84-kb bands in transformant 12 was 1.0, consistent with replacement by a single copy of the Darlington ERG11 gene.
Southern analysis was done with genomic DNAs from Darlington, CAI4, and transformant 12 digested with HindIII by using the 1.58-kb ERG11 probe. Darlington showed only one fragment approximately 6.5 kb (Fig. 1A, part e, lane 1). CAI4 showed 6.5-, 4.0-, and 2.5-kb fragments (Fig. 1A, part e, lane 2), indicating that CAI4 but not Darlington is heterozygous at this restriction site. Transformant 12 showed 6.5- and 7.5-kb fragments but lost the 4.0- and 2.5-kb fragments (Fig. 1A, part e, lane 3). By phosphorimaging quantitation, the ratio of the 7.5- and 6.5-kb fragments in transformant 12 was approximately 1.0. These results suggested that the allele of the CAI4 ERG11 gene which contained the HindlII restriction site was replaced by the Darlington ERG11 gene in transformant 12.
On Southern analysis of transformant 12 with the C. albicans URA3 probe, both the 5.5-kb fragment obtained with PstI and the 7.5-kb fragment obtained with HindIII hybridized with the probe, verifying the presence of the integrated URA3, as predicted from Fig. 1B (data not shown).
RT-PCR.
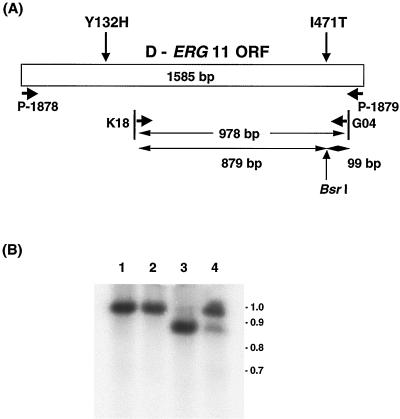
To examine the transcription of the integrated Darlington ERG11 gene, RT-PCR was performed. After extracting RNA from C. albicans, DNase was used to digest the remaining DNA and PCR was performed with primers P-1878 and P-1879. No products were found, indicating no detectable contamination with genomic DNA (data not shown). Genomic DNA from Darlington was used as a positive control. After the RT reaction, PCR was performed with primers K18 and G04. In this PCR product, one BsrI site lies in the Darlington ERG11 gene, as shown in Fig. 2A. Southern analysis (Fig. 2B) of a BsrI digest showed the following: strains SC5314 (lane 1) and CAI4 (lane 2) gave a 1.0-kb fragment and strain Darlington (lane 3) gave a 0.9-kb fragment. Transformant 12 (lane 4) expectedly showed both 1.0- and 0.9-kb fragments. This result showed that transcription of the integrated Darlington ERG11 had occurred in transformant 12. Although the 0.9-kb band was less intense, relative transcription of the two alleles could not be determined by the method used.
FIG. 2.
(A) Diagram of the Darlington strain ERG11 ORF. The primers used for PCR were P1878, P1879, K18, and G04. The product obtained by PCR with primers K18 and G04 was used as a probe for Southern analysis with BsrI. (B) Southern blot of RT-PCR product. Double-stranded cDNA was obtained by RT with poly(A)+ RNA of C. albicans as the template, followed by PCR amplification with K18 and G04 as primers. The amplified double-stranded cDNA was digested with BsrI, and Southern analysis was performed with a 0.98-kb PCR product (obtained with primers K18 and G04) as a probe. BsrI-digested amplified cDNA of SC5314 and CAI4 showed a 1.0-kb single band (lanes 1 and 2). The cDNA of Darlington showed a 0.9-kb single band (lane 3). Transformant 12 showed both 1.0- and 0.9-kb bands (lane 4). Numbers on the right are in kilobases.
Antifungal susceptibility of the C. albicans strains.
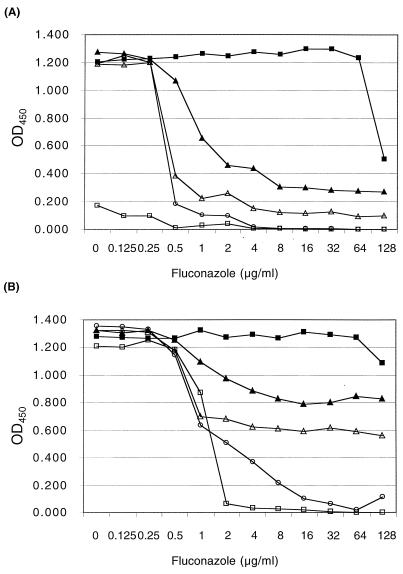
The drug susceptibility of each C. albicans strain was compared over 7 days in microtiter wells. Optical densities from day 4 and day 7 of incubation are shown in Fig. 3. Darlington is clearly the most resistant strain, with transformant 12 being slightly less susceptible than the three host strains (Fig. 3A). CAI4 did not grow well. At day 7, transformant 12 and SC5314 increased their turbidities. Transformant 12 was less susceptible than the host strains at all time points (Fig. 3B).
FIG. 3.
Fluconazole growth inhibition in C. albicans at day 4 (A) and day 7 (B). Data are for C. albicans isolates CAI4 (open squares), CAF2-1 (open circles), SC5314 (open triangles), Darlington (filled squares), and transformant 12 (filled triangles). OD450, optical density at 450 nm.
Site-directed mutagenesis.
Etest results at 5 days for DKY1 and the five transformants indicated that the MIC was 0.25 μg/ml for DKY1, 0.064 μg/ml for DKY1 transformed with the empty plasmid, 1.5 μg/ml for DKY1 transformed with pCAI4, >256 μg/ml for DKY1 transformed with pDAR, 16 μg/ml for DKY1 transformed with pCAI4-132H, and 6 μg/ml for DKY1 transformed with pDAR-471T. In this expression system, both the Y132H and I471T mutations increased the level of resistance, but fluconazole resistance was greatest in the presence of both mutations.
DISCUSSION
Marichal and coworkers (14) have recently catalogued the ERG11 sequences in azole-resistant isolates of C. albicans, including those listed in abstracts. They have pointed out that the 29 amino acid substitutions are clustered in three areas: deduced amino acids 105 to 165, 266 to 287, and 405 to 488. Of the first two amino-terminal areas, there is substantial evidence for only one substitution, a histidine for a tyrosine at amino acid 132, in conferring azole resistance. However, three such substitutions appear to be important in the carboxy-terminal area. Because multiple amino acid substitutions are usually present in any one isolate, the contribution of a single substitution to azole resistance is not elucidated by sequence data alone. Nor is it clear from many of the sequence data whether the strains are homozygous at the locus of a base change. There have been two different approaches to determining the effect of changing a single deduced amino acid. One has been to express the C. albicans ERG11 in S. cerevisiae (19) and the other has been to use a patient's sequential and probably congenic isolates that show increased azole resistance (23). Overexpression with a GAL1 promoter, similar to what was done here, was used to show that Y132H, S405F, G464S, and R467K increased the level of azole susceptibility of an azole-susceptible pdr5 S. cerevisiae mutant (19). Concerned about the possible toxicity of overexpressed genes, Favre and colleagues (3) used a different S. cerevisiae expression system. They used an upstream area of the S. cerevisiae ERG11 gene as a promoter for the C. albicans ERG11 (3). The promoter and a C. albicans ERG11 were inserted into pRS416, a plasmid with a stable low copy number. The plasmid was transformed into an S. cerevisiae host which was moderately azole resistant. These workers replaced the C. albicans ERG11 CTG at position 787 with TCT to allow correct coding for serine in S. cerevisiae, a precaution that we did not take (9). However, this precaution made no difference in azole resistance (3). One of the four ERG11 genes which they sequenced and which they found conferred an increased level of azole resistance in S. cerevisiae was from the Darlington strain. Their nucleic acid sequence from the Darlington ERG11 encoded the same Y132H substitution which we also found. They also found F72L and G450E but not I471T (3). Favre and coworkers (3) did not use site-directed mutagenesis, as Sanglard and coworkers (19) did, to determine the significance of the two to four amino acid substitutions in their isolates or determine whether their strains were heterozygous at the ERG11 locus.
Our results with the Y132H and I471T substitutions in the S. cerevisiae expression system agree with those of Sanglard et al. (19), in that the effect of substitutions can be additive. The MICs for transformants overexpressing ERG11 with Y132H and I471T substitutions alone were 16 and 6 μg/ml, respectively, while the the MIC for the transformant with both substitutions was at least 128 μg/ml. While the I471T substitution has not been reported previously, it is close to the G464S (13) and R467K (24) substitutions, both of which have been noted in azole-resistant isolates.
Our study addressed two other aspects of azole resistance not previously reported. One is the effect of gene dosage in the diploid species C. albicans. Replacement of one copy of ERG11 in an azole-susceptible C. albicans strain with that from an azole-resistant isolate caused only a moderate increase in azole resistance. This supports the conjecture that Y132H must be encoded in both copies of the ERG11 gene to cause resistance (2). We did not, however, do the opposite experiment of replacing one copy of ERG11 in an azole-resistant isolate with ERG11 from a susceptible isolate, nor did we attempt to quantify the relative transcription of the two genes in our transformant 12. The other aspect studied here was the homozygosity at the loci encoding 132H and 471T in Darlington, as determined by Southern analysis.
Continuing study of the changes in ERG11 which lead to azole resistance may produce a better understanding of the topology of this enzyme and, it is hoped, may lead to drug designs which will maintain the usefulness of drug classes that use the ERG11 gene product as the antifungal target.
ACKNOWLEDGMENT
This work was partially supported by an unrestricted grant from Pfizer, Inc.
REFERENCES
- 1.Albertson G D, Niimi M, Cannon R D, Jenkinson H F. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob Agents Chemother. 1996;40:2835–2841. doi: 10.1128/aac.40.12.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai K, Tsuchimori N, Okonogi K, Perfect J R, Gotoh O, Yoshida Y. Formation of azole-resistant Candida albicans by mutation of sterol 14-demethylase P450. Antimicrob Agents Chemother. 1999;43:1163–1169. doi: 10.1128/aac.43.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Favre B, Didmon M, Ryder N S. Multiple amino acid substitutions in lanosterol 14α-demethylase contribute to azole resistance in Candida albicans. Microbiology. 1999;145:2715–2725. doi: 10.1099/00221287-145-10-2715. [DOI] [PubMed] [Google Scholar]
- 4.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujimura H, Sakura Y. Simplified isolation of chromosomal and plasmid DNA from yeast. BioTechniques. 1993;14:538–539. [PubMed] [Google Scholar]
- 6.Geitz D, St. Jean A, Woods R A, Schiestl R H. Improved methods for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hitchcock C A, Barrett-Bee K J, Russell N J. Inhibition of 14 alpha-sterol demethylase activity in Candida albicans Darlington does not correlate with azole resistance. J Med Vet Mycol. 1987;25:329–333. [PubMed] [Google Scholar]
- 8.Howell S A, Mallet A I, Noble W C. A comparison of the sterol content of multiple isolates of the Candida albicans Darlington strain with other clinically azole-sensitive and resistant strains. J Appl Bacteriol. 1990;69:629–696. doi: 10.1111/j.1365-2672.1990.tb01564.x. [DOI] [PubMed] [Google Scholar]
- 9.Jute T H, Osawa S. CUG codons in Candida spp. J Mol Evol. 1996;42:321–322. doi: 10.1007/BF02198859. [DOI] [PubMed] [Google Scholar]
- 10.Katzmann D J, Burnett P E, Golin J, Mahe Y, Moye-Rowley W S. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol Cell Biol. 1994;14:4653–4661. doi: 10.1128/mcb.14.7.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai M H, Kirsh D R. Nucleotide sequence of cytochrome P450 L1A1 (lanosterol 14 alpha-demethylase) from Candida albicans. Nucleic Acids Res. 1989;17:804. doi: 10.1093/nar/17.2.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamb D C, Kelly D E, Schunck W H, Shyadehi A Z, Akhtar M, Lowe D J, Baldwin B C, Kelly S L. The mutation T315A in Candida albicans sterol 14alpha-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J Biol Chem. 1997;272:5682–5688. doi: 10.1074/jbc.272.9.5682. [DOI] [PubMed] [Google Scholar]
- 13.Löffler J, Kelly S L, Hebart H, Schumacher U, Lass-Flörl C, Einsele H. Molecular analysis of Cyp51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol Lett. 1997;151:263–268. doi: 10.1111/j.1574-6968.1997.tb12580.x. [DOI] [PubMed] [Google Scholar]
- 14.Marichal P, Koyamans L, Willemsens S, Bellens D, Verhasselt P, Luyten W, Borgers M, Ramaekers F C S, Odds F C, vanden Bossche H. Contribution of mutations in the cytochrome P450 14α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology. 1999;145:2701–2713. doi: 10.1099/00221287-145-10-2701. [DOI] [PubMed] [Google Scholar]
- 15.Miyazaki Y, Gerber A, Miyazaki H, Falconer D, Parkinson T, Hitchcock C, Grimberg B, Nyswaner K, Bennett J E. Cloning, sequencing, expression and allelic sequence diversity of ERG3 (C-5 sterol desaturase gene) in Candida albicans. Gene. 1999;236:43–51. doi: 10.1016/s0378-1119(99)00263-2. [DOI] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeast. Approved standard M27-A. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1995. [Google Scholar]
- 17.Pearce M A, Howell S A. Restriction fragment length polymorphism analysis of azole-resistant and azole-susceptible Candida albicans strains. J Clin Microbiol. 1991;29:1364–1367. doi: 10.1128/jcm.29.7.1364-1367.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Sanglard D, Isher F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14 alpha-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanden Bossche H, Marichal P, Gorrens J, Bellens D, Moereels H, Janssen P A. Mutation in cytochrome P450-dependent 14-demethylase results in decreased affinity for azole antifungals. Biochem Soc Trans. 1990;18:56–59. doi: 10.1042/bst0180056. [DOI] [PubMed] [Google Scholar]
- 21.Varma A, Edman J C, Kwon-Chung K J. Molecular and genetic analysis of URA5 transformants of Cryptococcus neoformans. Infect Immun. 1992;60:1101–1108. doi: 10.1128/iai.60.3.1101-1108.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warnock D W, Johnson E M, Richardson M D, Vickers C F. Modified response to ketoconazole of Candida albicans from a treatment failure. Lancet. 1983;i:642–643. doi: 10.1016/s0140-6736(83)91809-3. [DOI] [PubMed] [Google Scholar]
- 23.White T C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White T C. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14-α-demethylase in Candida albicans. Antimicrob Agents Chemother. 1997;41:1488–1494. doi: 10.1128/aac.41.7.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White T C, Marr K A, Bowden R A. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wickner R B. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]