Abstract
Transfer RNA fragments are proposed to regulate numerous processes in eukaryotes, including translation inhibition, epigenetic inheritance, and cancer. In the bacterium Salmonella enterica serovar Typhimurium, 5′ tRNA halves ending in 2′,3′ cyclic phosphate are proposed to bind the RtcR transcriptional activator, resulting in transcription of an RNA repair operon. However, since 5′ and 3′ tRNA halves can remain base paired after cleavage, the 5′ tRNA halves could potentially bind RtcR as nicked tRNAs. Here we report that nicked tRNAs are ligands for RtcR. By isolating RNA from bacteria under conditions that preserve base pairing, we show that many tRNA halves are in the form of nicked tRNAs. Using a circularly permuted tRNA that mimics a nicked tRNA, we show that nicked tRNA ending in 2′,3′ cyclic phosphate is a better ligand for RtcR than the corresponding 5′ tRNA half. In human cells, we show that some tRNA halves similarly remain base paired as nicked tRNAs following cleavage by anticodon nucleases. Our work supports a role for the RNA repair operon in repairing nicked tRNAs and has implications for the functions proposed for tRNA fragments in eukaryotes.
Keywords: nicked tRNAs, RNA repair, tRNA fragments
INTRODUCTION
Transfer RNA-derived fragments have been identified in all domains of life and are proposed to regulate myriad processes (Su et al. 2020). These fragments, which are often generated by cleavage in anticodon loops, accumulate during stresses such as starvation, hypoxia, oxidative stress, and virus infection (Lee and Collins 2005; Thompson et al. 2008; Fu et al. 2009; Yamasaki et al. 2009; Wang et al. 2013; Goodarzi et al. 2015) and are also detected in unstressed cells (Upton et al. 2021). Transfer RNA halves have been implicated in numerous processes, including translation inhibition (Ivanov et al. 2011; Blanco et al. 2016), epigenetic inheritance (Chen et al. 2016; Sharma et al. 2016) and cancer, where distinct tRNA halves function as tumor suppressors (Goodarzi et al. 2015) and promote metastasis (Liu et al. 2022).
Our laboratory previously described a role for tRNA 5′ halves as signaling molecules. In these studies, we examined the signals that activate transcription of the rtcBA RNA repair operon in Salmonella enterica serovar Typhimurium (S. Typhimurium). In many bacteria, such as E. coli, this operon encodes only RtcB, the RNA ligase that ligates tRNA halves after intron excision in metazoans and archaea, and RtcA, an RNA cyclase that converts 2′ or 3′ phosphate RNA ends to 2′,3′ cyclic phosphate (Genschik et al. 1998; Tanaka and Shuman 2011; Das and Shuman 2013; Burroughs and Aravind 2016). In other bacteria, such as S. Typhimurium, the operon includes an ortholog of the human Ro60 protein (called Rsr for Ro-sixty related) and two noncoding RNAs called YrlA and YrlB (Chen et al. 2013; Das and Shuman 2013; Burroughs and Aravind 2016). Operon transcription is regulated by RtcR, a σ54-dependent enhancer-binding protein consisting of an N-terminal ligand-binding domain adjacent to an AAA+ ATPase domain and a carboxy-terminal DNA-binding domain (Genschik et al. 1998). In other enhancer-binding proteins, ligand binding to the N-terminal domain results in oligomerization of the adjacent AAA+ ATPase domain, allowing the protein to interact with σ54 and activate transcription (Bush and Dixon 2012). In RtcR, the ligand-binding domain is a divergent form of the oligonucleotide-binding CRISPR-associated Rossman fold (CARF) domain, which has been best-characterized in type III CRISPR-Cas systems (Makarova et al. 2020). Binding of cyclic oligoadenylates to these CARF domains activates an adjacent ribonuclease (Kazlauskiene et al. 2017; Niewoehner et al. 2017).
Using a genetic screen to identify mutations that cause activation of the S. Typhimurium rsr-yrlBA-rtcBA operon, we found that mutations that cause both 5′ and 3′ tRNA halves to accumulate activate operon expression (Hughes et al. 2020). One class of mutants affected tRNA metabolism. Specifically, mutations in truA, which encodes the tRNA pseudouridine synthetase that modifies uridine to pseudouridine in the anticodon arm of many tRNAs (Cortese et al. 1974), pnp, which encodes polynucleotide phosphorylase, a 3′ to 5′ exoribonuclease that degrades aberrant tRNAs in E. coli (Li et al. 2002), and rtcB, which encodes the RNA ligase component of the operon, caused both accumulation of tRNA halves and operon transcription (Hughes et al. 2020). A second class of mutations were in genes whose inactivation leads to expression of the DNA damage-induced SOS response. This class included mutations in ruvA, a DNA-binding protein important for resolving Holliday junctions (Wyatt and West 2014), polA, which encodes DNA polymerase I, and recC, a component of the RecBCD complex that functions in repair of double-stranded DNA breaks (Dillingham and Kowalczykowski 2008). In these mutants, or when cells are treated with DNA damaging agents, one or more RecA-dependent anticodon nuclease(s) cleave many tRNAs in the anticodon loop (Hughes et al. 2020).
Our experiments and those of others support a model in which 5′ tRNA halves ending in 2′,3′ cyclic phosphate bind RtcR to activate operon expression. The 5′ tRNA halves that accumulate following DNA damage end in 2′,3′ cyclic phosphate, consistent with cleavage by a metal-independent nuclease (Hughes et al. 2020). In support of the hypothesis that an RNA ending in 2′,3′ cyclic phosphate is important for operon transcription, RtcA is required for operon activation in both E. coli and S. Typhimurium (Hughes et al. 2020; Kotta-Loizou et al. 2022), and overexpression of RtcA is sufficient to activate the operon in wild-type strains (Hughes et al. 2020). In addition, expression of mammalian cyclic nucleotide phosphodiesterase, which hydrolyzes 2′,3′ cyclic phosphates, down-regulates the operon (Duggal et al. 2022). We showed that the purified RtcR CARF domain binds 5′ tRNA halves ending in 2′,3′ cyclic phosphate with more than 25-fold higher affinity than the same RNAs ending in 3′ hydroxyl or 3′ phosphate. Binding of 5′ tRNA halves ending in 2′,3′ cyclic phosphate to full-length RtcR resulted in formation of oligomers that contained both RtcR and tRNA halves (Hughes et al. 2020). A role for 5′ tRNA halves is supported by the observation that ectopic expression of toxins that cleave specific tRNAs in the anticodon loop activated expression of the E. coli operon (Engl et al. 2016). However, since 5′ and 3′ tRNA halves can remain base paired after cleavage by anticodon nuclease(s) (Kaufmann and Amitsur 1985; Nandakumar et al. 2008), it remained possible that the 5′ tRNA fragments activated the operon in the form of nicked tRNAs.
Here we report that nicked tRNAs are ligands for RtcR. By isolating RNA from S. Typhimurium under conditions that preserve intermolecular base pairing, we show that many tRNA halves are present in the form of nicked tRNAs. Using a circularly permuted tRNA that mimics a nicked tRNA, we show that, although a 5′ tRNA fragment ending in 2′,3′ cyclic phosphate is a better ligand for the isolated CARF domain than the corresponding nicked tRNA, the nicked tRNA is a better ligand for full-length RtcR and increases RtcR oligomerization. We demonstrate that in human cells, where there are many reports of tRNA halves acting as regulatory molecules, some tRNA halves remain base paired in the form of nicked tRNAs following stress-induced cleavage by anticodon nuclease(s). Our work supports a role for the rsr-yrlBA-rtcBA operon in repair of nicked tRNAs and may have implications for the functions proposed for tRNA halves in eukaryotes.
RESULTS AND DISCUSSION
Many 5′ and 3′ pre-tRNA halves remain base paired in S. Typhimurium
To determine the extent to which tRNA halves remained base paired, we asked whether RNA isolation protocols that maintain base pairing between S. cerevisiae U4 and U6 snRNAs (Li and Brow 1996; Pannone et al. 1998) also maintain base pairing of tRNA halves in S. Typhimurium. RNA was isolated from wild-type, ΔtruA and ΔruvA S. Typhimurium cells using cold phenol, fractionated in denaturing or native gels and northern analyses performed to detect tRNA halves that accumulate in unstressed ΔtruA and ΔruvA cells (Hughes et al. 2020). Since similar tRNA halves accumulate in S. Typhimurium following treatment with DNA damaging agents (Hughes et al. 2020), we also isolated RNA from cells incubated with the interstrand crosslinker mitomycin C (MMC). Because previous experiments revealed that both specific tRNA cleavage and operon induction were unaffected in strains lacking the abundant periplasmic endoribonuclease RNase I (Hughes et al. 2020), all experiments were carried out in strains lacking RNase I (Δrna) to reduce nonspecific nicking of tRNAs during cell lysis.
Northern blotting to detect specific tRNAs revealed that, although tRNA halves were prominent species when the RNA was fractionated in denaturing gels, these same tRNA halves were far less prominent when the same RNA was fractionated in native gels (Fig. 1). Specifically, 5′ halves of tRNAPhe(GAA), tRNALeu(UAG), tRNATrp(CCA), and tRNAHis(GUG) were detected in ΔruvA strains and in all strains after MMC treatment when the RNA was heated to 95°C and fractionated in denaturing gels (Fig. 1A,B,E,F). As described (Hughes et al. 2020), 3′ halves were also detected (Fig. 1I,J). Similar levels of tRNA halves were detected when the heating step was omitted (Fig. 1A,B,E,F,I,J, lane 7). In contrast, when the same RNA was fractionated in native gels, primarily full-length tRNAs were detected (Fig. 1C,D,G,H,K,L).
FIGURE 1.
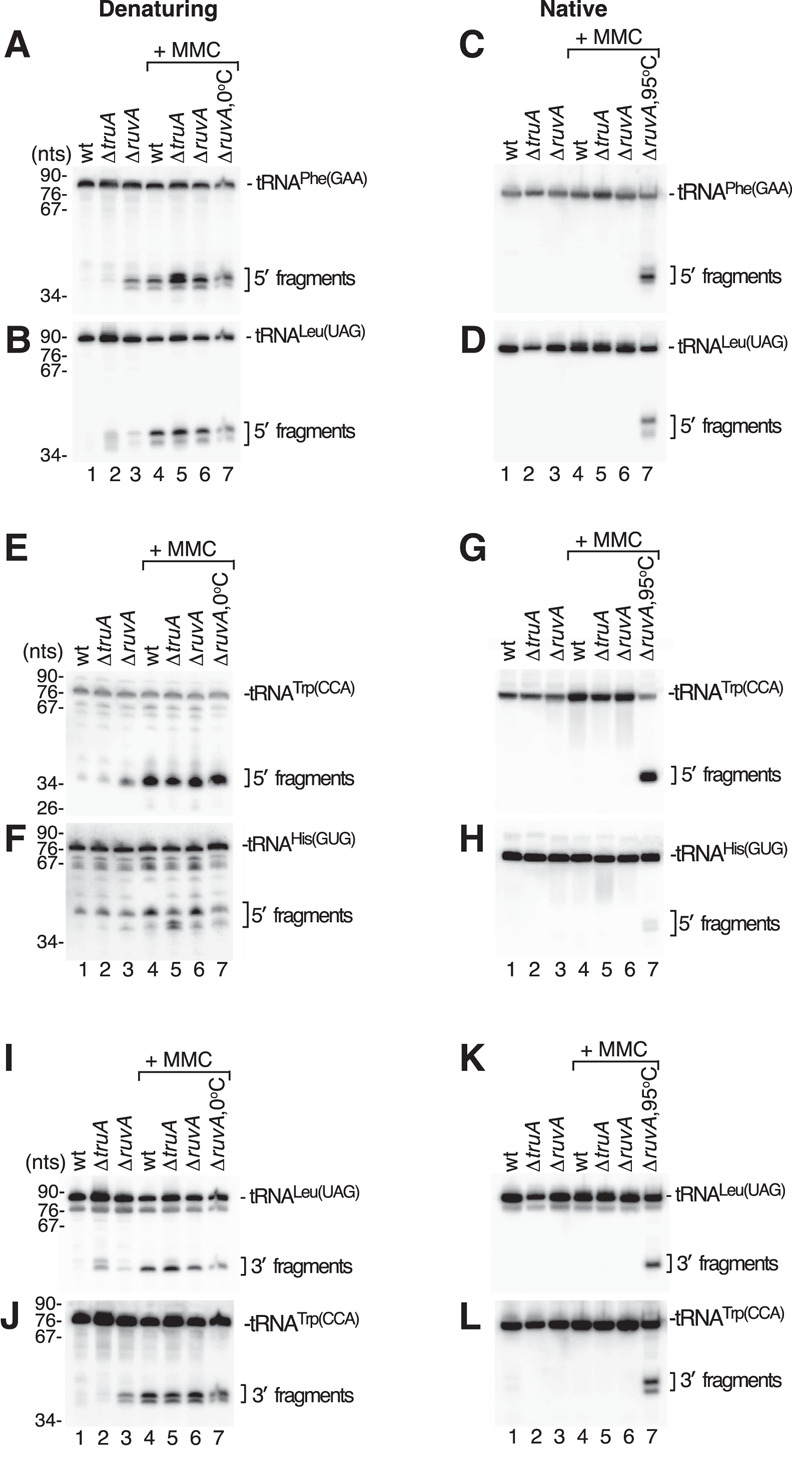
Many tRNA fragments in S. Typhimurium are base paired in cells. (A,B) RNA was isolated from the indicated rnaΔ strains using cold (4°C) phenol/chloroform/isoamyl alcohol, subjected to ethanol precipitation, and resuspended in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl. After mixing with equal volumes of 95% formamide, 5 mM EDTA, and 0.02% bromophenol blue, the RNA was heated to 95°C for 5 min, fractionated in denaturing gels, and subjected to northern blotting to detect 5′ halves of tRNAPhe(GAA) (A) and tRNALeu(UAG) (B). In lane 7, the heating step was omitted. (C,D) The resuspended RNA described in A–B was mixed with 1/3 vol of 25% glycerol and 0.02% bromophenol blue, fractionated in native gels, and analyzed by northern blotting to detect 5′ halves of tRNAPhe(GAA) (C) and tRNALeu(UAG) (D). In lane 7, the RNA was heated to 95°C for 5 min before loading. (E,F) After heating and fractionation in denaturing gels as in A–B, the indicated RNAs were subjected to northern blotting to detect 5′ halves of tRNATrp(CCA) (E) and tRNAHis(GUG) (F). (G,H) The same RNAs as in E and F were fractionated in native gels and subjected to northern blotting to detect 5′ halves of tRNATrp(CCA) (G) and tRNAHis(GUG) (H). (I–L) The northern blots in A–B and C–D were reprobed to detect 3′ halves of tRNALeu(UAG) and tRNATrp(CCA) after fractionation in denaturing (I,J) or native gels (K,L).
To determine if the tRNA fragments detected in denaturing gels remained base paired as nicked tRNAs in native gels, we heated the RNA before fractionating in native gels. A similar strategy was used to demonstrate base pairing between U4 and U6 snRNAs (Hashimoto and Steitz 1984; Brow and Guthrie 1988). Upon heating to 95°C, both 5′ and 3′ tRNA halves became prominent (Fig. 1C,D,G,H,K,L, lane 7). These data indicate that at least some 5′ and 3′ tRNA halves remain base paired after cleavage but are separated upon either heating or fractionation in denaturing gels.
A circularly permuted nicked tRNA ending in 2′,3′ cyclic phosphate is a better ligand for RtcR than the corresponding 5′ tRNA half
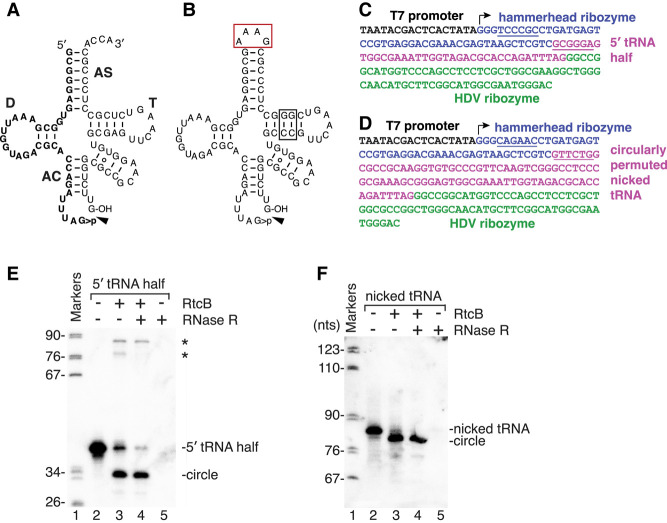
Our finding that some 5′ and 3′ tRNA halves are base paired in cells raised the question of whether nicked tRNAs might also be ligands for RtcR. Initial attempts to produce a nicked tRNA by annealing 5′ halves ending in 2′,3′ cyclic phosphate with 3′ halves containing 5′-OH revealed that duplex formation was extremely inefficient, possibly because intramolecular structures formed by the individual halves competed with duplex formation. We therefore generated a circularly permuted tRNA for binding studies. To this end, we replaced the ACCA at the 3′ end of tRNALeu(UAG) with a tetraloop, allowing us to make the nicked tRNA as a single transcript beginning and ending in the anticodon loop (Fig. 2A,B). We chose this tRNA because, of several 5′ tRNA halves tested, 5′ tRNALeu(UAG) halves bound most efficiently to both the RtcR CARF domain and full-length RtcR (Hughes et al. 2020). Because the RNA folding program Mfold (Zuker 2003) predicted that an alternative structure could form in wild-type tRNALeu(UAG), we mutated two base pairs in the T stem to stabilize the canonical cloverleaf secondary structure and prevent formation of the alternative structure (Fig. 2B). By using a hammerhead ribozyme to generate a 5′-OH and the hepatitis delta virus (HDV) ribozyme to generate a 2′,3′ cyclic phosphate (Fig. 2D), the resulting transcript resembled a tRNA that has undergone cleavage by a metal-independent anticodon nuclease. As a control, we synthesized a 5′ tRNA half using the same strategy (Fig. 2C). As expected, both the tRNA half and the circularly permuted tRNA, both of which contain 5′-OH and 2′,3′ cyclic phosphate, could be ligated by E. coli RtcB to form circles resistant to degradation with the exoribonuclease RNase R (Fig. 2E,F).
FIGURE 2.
Generation of a circularly permuted nicked tRNA. (A) Sequence of S. Typhimurium tRNALeu(UAG) following cleavage by a metal-independent anticodon nuclease. The D, T, and anticodon (AC) arms are labeled, as is the acceptor stem (AS). The 5′ tRNALeu(UAG) half ending in 2′,3′ cyclic phosphate is in bold type. (B) A circularly permuted nicked tRNALeu(UAG) was generated by converting the ACCA at the 3′ end to a tetraloop (red box). To stabilize the T arm, two base pairs were altered (black box). (C,D) To generate the 5′ tRNALeu(UAG) half (C) or the circularly permuted nicked tRNA (D), a DNA fragment was constructed consisting of a T7 promoter (black), hammerhead ribozyme (blue), the sequence of 5′ tRNALeu(UAG) half (C, pink) or the circularly permuted tRNA (D, pink), followed by an HDV ribozyme (green). To generate the hammerhead ribozyme cleavage site, the ribozyme sequence contains 6 nt (underlined in blue) complementary to the first 6 nt of the circularly permuted tRNA (underlined in pink). (E,F) Following incubation of the 5′ tRNALeu(UAG) half (E) or the circularly permuted nicked tRNA (F) without (lanes 2,5) or with RtcB (lanes 3,4), the reaction was incubated without (lanes 2,3) or with RNase R (lanes 4,5). Lane 1, molecular size markers (nt). Bands denoted with asterisks in E may represent products of intermolecular ligation.
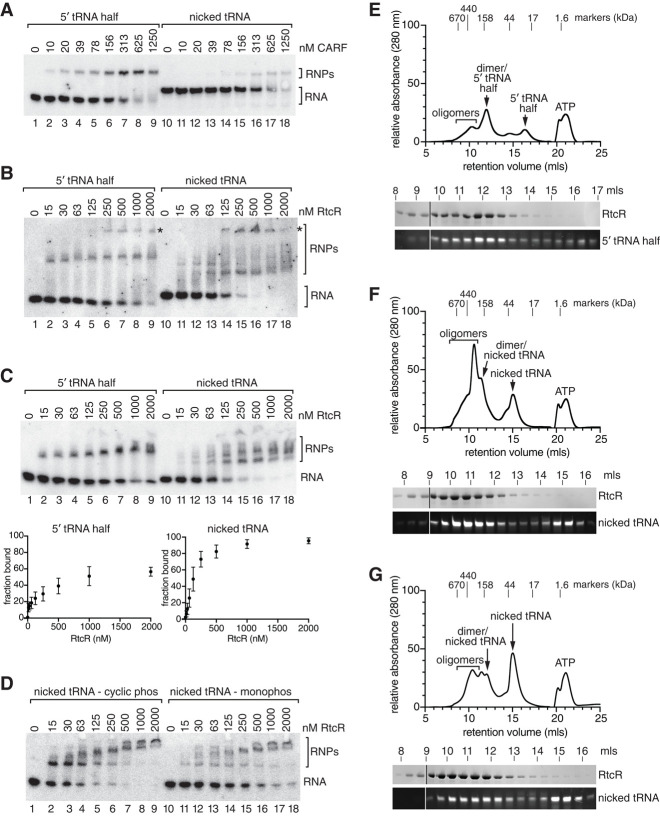
We used electrophoretic mobility shift assays (EMSAs) to compare binding of the isolated CARF domain and the full-length RtcR to the circularly permuted nicked tRNALeu(UAG) and the 5′ tRNA half ending in 2′,3′ cyclic phosphate. Because the need for the circularly permuted nicked tRNA to begin with 5′-OH and end with 2′,3′ cyclic phosphate precluded end-labeling of the RNA, we used northern blotting to detect binding. For the CARF domain, the 5′ tRNA half ending in 2′,3′ cyclic phosphate was the better ligand. Specifically, although the amount of CARF domain required to shift 50% of the tRNALeu(UAG) 5′ half was similar to the previously determined Kd of 195 nM (Hughes et al. 2020), more than 625 nM was required to shift 50% of the nicked tRNA into RNPs (Fig. 3A).
FIGURE 3.
The circularly permuted nicked tRNA is a better ligand for RtcR binding and oligomerization than the 5′ tRNA half. (A,B) tRNALeu(UAG) 5′ tRNA halves ending in 2′,3′ cyclic phosphate (lanes 1–9) or the circularly permuted nicked tRNA (lanes 10–18) were incubated with the indicated concentrations of CARF domain (A) or RtcR (B). RNA–protein complexes were separated from naked RNA in 6% (A) or 4% (B) polyacrylamide (80:1 acrylamide:bisacrylamide) native gels. In B, asterisks denote potential oligomers. (C) Gel shifts were performed as in B, except that RNA–protein complexes were separated from naked RNA by fractionation in discontinuous 3%–4% polyacrylamide (80:1 acrylamide:bisacrylamide)/2.5% glycerol gels (top panels). Bottom panels show quantification of three independent experiments using the 5′ tRNA half ending in 2′,3′ cyclic phosphate (left panel) or the circularly permuted nicked tRNA (right panel). (D) The circularly permuted nicked tRNA ending in cyclic phosphate (lanes 1–9) or the same RNA ending in monophosphate (lanes 10–18) was incubated with the indicated concentrations of full-length RtcR. RNA–protein complexes were separated from naked RNA as in B. (E–G) 24 µM RtcR dimer was incubated with 48 µM 5′ tRNA halves (E), nicked tRNAs ending in 2′,3′ cyclic phosphate (F), or nicked tRNAs after treatment with acid to open the cyclic phosphate (G) and separated by size exclusion chromatography. The presence of RtcR and RNA in each of the peaks was determined by fractionating proteins in SDS-PAGE (top panel) and RNA in denaturing polyacrylamide gels (bottom panel). Both RtcR and RNAs present in the fractions were analyzed in multiple gels that were joined at the lines.
In contrast, the nicked tRNA was the better ligand for full-length RtcR. Based both on our size exclusion chromatography (Hughes et al. 2020) and other AAA+ ATPase-containing bacterial enhancer binding proteins (Bush and Dixon 2012), the unliganded RtcR is expected to be a dimer. Upon ligand binding to the N-terminal CARF domain, the RtcR AAA+ ATPase domain is expected to oligomerize (Bush and Dixon 2012). As described (Hughes et al. 2020), binding of the tRNALeu(UAG) 5′ half resulted in formation of a band likely to be the RtcR dimer bound to RNA (Fig. 3B, left panel), and a larger complex at the top of the gel that could represent oligomerization (asterisk). In contrast, when the nicked tRNA was used in the binding experiments, we detected a heterogeneous mixture of RNP species that migrated both slower and faster than those detected when the ligand was the tRNALeu(UAG) 5′ half (Fig. 3B, brackets). The largest complex at the top of the gel also became more prominent (Fig. 3B, right panel, asterisk). Although the multiple species made quantitation difficult, the fact that the nicked tRNA was entirely shifted into RNPs at 500 nM RtcR, while some of the tRNALeu(UAG) 5′ half remained unbound at 2 µM RtcR, indicated that the nicked tRNA was the better ligand.
Because the native gels used in our analyses did not resolve all the RtcR-containing RNPs, we tried several other native gel protocols to better detect these complexes. We were most successful when we replaced the 4% polyacrylamide (80:1 acrylamide:bisacrylamide)/2.5% glycerol gels used in Figure 3A,B with discontinuous 3%–4% polyacrylamide (80:1 acrylamide:bisacrylamide)/2.5% glycerol gels. While these gels did not entirely eliminate the heterogeneity, they did resolve the RtcR/RNPs into several discrete bands, some of which could represent oligomers (Fig. 3C).
To test whether the 2′,3′ cyclic phosphate contributed to binding of the circularly permuted nicked tRNA to RtcR, we treated the tRNA with acid, which opens the cyclic phosphate to form RNAs ending in 3′ or 2′ monophosphate (Lund and Dahlberg 1992). As expected if the cyclic phosphate is an important determinant of binding, all RNP complexes were reduced in levels (Fig. 3D). Thus, as previously demonstrated for binding of tRNALeu(UAG) 5′ halves to RtcR (Hughes et al. 2020), RtcR is able to discriminate between nicked tRNAs containing 2′,3′ cyclic phosphate and those in which the 5′ half ends in monophosphate.
To examine oligomerization, we performed size exclusion chromatography. For these experiments, we mixed RtcR with a twofold excess of either the 5′ tRNA half ending in cyclic phosphate, the circularly permuted nicked tRNA ending in cyclic phosphate, or the nicked tRNA ending in monophosphate. For the 5′ tRNA half, we detected the two complexes described previously, migrating at ∼150 and ∼400 kDa (Fig. 3E), consistent with dimeric (119 kDa) and hexameric (358 kDa) forms of RtcR bound to one (12 kDa) or three (36 kDa) tRNA halves, respectively (Hughes et al. 2020). Fractionation of these peaks in denaturing gels revealed that, as expected, both RtcR and 5′ tRNA halves were present (Fig. 3E). As expected for an experiment in tRNA excess, we also detected a peak corresponding to unbound 5′ tRNA halves (Fig. 3E). In the presence of the circularly permuted nicked tRNA ending in cyclic phosphate, we detected a large peak migrating at ∼300 kDa with two shoulders that peaked at ∼160 and ∼400 kDa. All three peaks contained both RtcR and the nicked tRNA (Fig. 3F). As the slower migrating shoulder likely represents the RtcR dimer bound to the 27.5 kDa nicked tRNA, and the faster migrating shoulder is consistent with a hexamer, the prominent peak at ∼300 kDa may correspond to an intermediate in hexamer formation. In contrast, for the nicked tRNA ending in monophosphate, the largest peak was the unbound tRNA, and the RtcR-containing peaks were all reduced compared to those detected when the nicked tRNA ended in cyclic phosphate (Fig. 3G). Efforts to obtain precise masses using size exclusion chromatography-multiple angle light scattering (SEC-MALS) and mass photometry have thus far been unsuccessful, due to the tendency of the complex to dissociate under the conditions used. Despite these limitations, the large increase in RtcR RNPs detected by both EMSA and size exclusion chromatography with the nicked tRNA ending in 2′,3′ cyclic phosphate, compared to those formed with either the 5′ tRNA half ending in 2′,3′ cyclic phosphate or the nicked tRNA ending in monophosphate, reveals that the nicked tRNA ending in 2′,3′ cyclic phosphate is more effective at binding RtcR and triggering oligomerization.
Some tRNA halves remain base paired following cleavage in human cells
In human cells, tRNA halves accumulate during oxidative and other types of environmental stress. To determine whether some of these tRNA halves remain base paired, we examined the tRNA halves that accumulate following treatment of human HEK293A cells with sodium arsenite, a common inducer of oxidative stress and tRNA cleavage (Yamasaki et al. 2009; Emara et al. 2010; Saikia et al. 2012; Elkordy et al. 2018). Following incubation with 0.5 M sodium arsenite for either 30 min or 2 h, cells were lysed by sonication and RNA extracted with cold phenol and fractionated in either denaturing or native gels. Northern analysis was performed to detect 5′ halves of tRNAGly(GCC), tRNAHis(GUG), tRNASer(GCU), and tRNA(iMet)CAT, all of which have been described to accumulate under these conditions (Yamasaki et al. 2009; Akiyama et al. 2022).
Northern blotting revealed that while 5′ halves of tRNAGly(GCC), tRNAHis(GUG), tRNASer(GCU), and tRNA(iMet)CAT were all detected when the RNA was fractionated in denaturing gels (Fig. 4A,B,E,F), 5′ halves of tRNAGly(GCC), tRNAHis(GUG), and tRNASer(GCU) were strongly reduced when the same RNA was fractionated in native gels (Fig. 4C,D,G). Because 5′ halves of these three tRNAs became detectable when the RNA was first heated to 95°C (Fig. 4C,D,G, lane 4), these tRNA halves were likely base paired in the original RNA. For these three tRNAs, in the absence of heating, we also detected diffuse bands that migrated with reduced mobility in native gels (Fig. 4C,D,G, lane 3). Although these could represent less stably base paired 5′ halves that dissociate during gel fractionation, quantitation revealed that these species accounted for 32% (tRNASer(GCU)), 40% (tRNAGly(GCC)), and 71% (tRNAHis(GUG)) of the counts of the discretely migrating RNAs that were detected with heating. Thus, at minimum, 29 to 68% of these 5′ fragments were base paired before gel fractionation. As in bacteria, 3′ halves of these tRNAs behaved similarly (Fig. 4I–L). For tRNAiMet(CAT) RNA, 5′ halves were also detected in the native gel and did not increase significantly upon heating, suggesting these halves may be present as discrete fragments in cells (Fig. 4H, compare lanes 3 and 4). We conclude that, as in bacteria, at least some tRNA halves remain base paired following cleavage in the anticodon arm.
FIGURE 4.
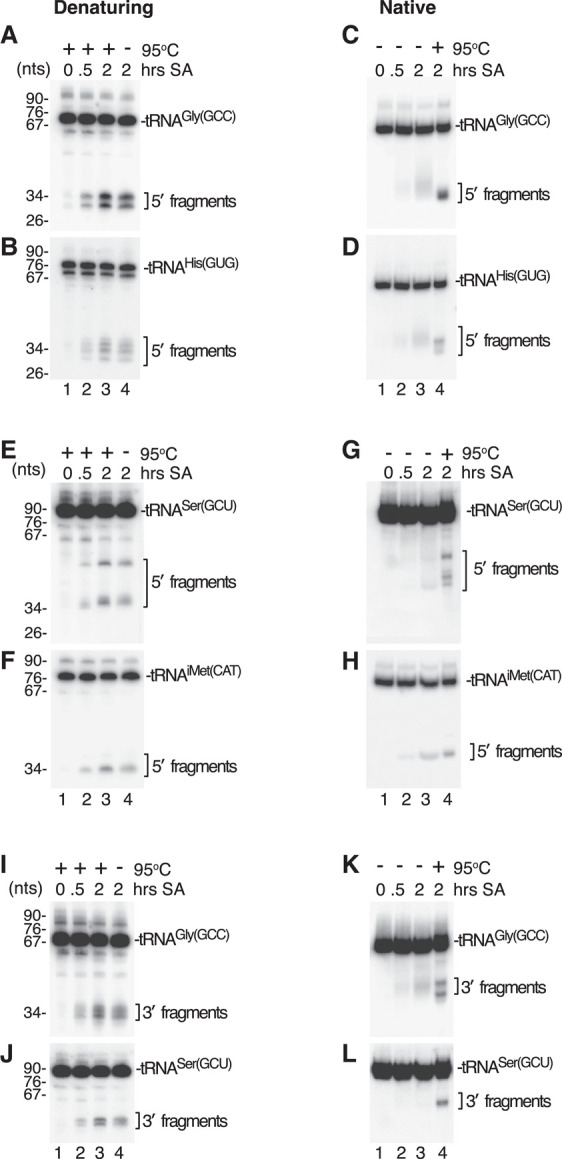
Some human tRNA fragments remain base paired in cells. (A,B) After incubating HEK293A cells with 0.5 M sodium arsenite (SA) for the indicated times, cells were lysed and RNA isolated as described in Materials and Methods and resuspended in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl. After mixing with an equal volume of 95% formamide, 5 mM EDTA and 0.02% bromophenol blue, the RNA was heated to 95°C for 5 min, fractionated in denaturing gels and subjected to northern blotting to detect 5′ halves of tRNAGly(GCC) (A) and tRNAHis(GUG) (B). In lane 4, the heating step was omitted. (C,D) After resuspending in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, the RNA was mixed with 1/3 vol of 25% glycerol and 0.02% bromophenol blue. After fractionation in native gels, northern blotting was performed to detect 5′ halves of tRNAGly(GCC) (C) and tRNAHis(GUG) (D). In lane 4, the RNA was heated to 95°C for 5 min before loading. (E,F) After heating and fractionation in denaturing gels as in A–B, the indicated RNAs were subjected to northern blotting to detect 5′ halves of tRNASer(GCU) (E) and tRNAiMet(CAT) (F). In lane 4, the heating step was omitted. (G,H) The same RNAs as in E and F were fractionated in native gels followed by northern blotting to detect 5′ halves of tRNASer(GCU) (G) and tRNAiMet(CAT) (H). In lane 4, the RNA was heated to 95°C for 5 min before loading. (I–L) The northern blots in A–B and C–D were reprobed to detect 3′ halves of tRNAGly(GCC) and tRNASer(GCU) after fractionation in denaturing I,J or native gels (K,L).
Conclusions
Our demonstration that tRNA halves can remain base paired to form nicked tRNAs has implications for the functions that have been proposed for these fragments. Since some nicked tRNAs may not withstand low temperature phenol extraction and subsequent native gel analysis, we consider the fraction of tRNA halves that remains base paired under these conditions to be a conservative estimate of the actual fraction that is base paired in cells.
In bacteria, our finding that nicked tRNAs bind RtcR and trigger its oligomerization expands the ligands that can activate the rsr-yrlBA-rtcBA operon. Because the nicked tRNA binds RtcR with higher affinity than the corresponding 5′ tRNA half, we consider it likely that nicked tRNA is the preferred ligand in cells. Since 5′ halves can bind RtcR and trigger oligomerization, these ligands may also contribute to operon activation. There also may be stresses, in S. Typhimurium or other bacteria, in which tRNA halves are predominantly unpaired. The fact that both 5′ halves and nicked tRNAs can bind RtcR and trigger oligomerization to different extents could also be used to fine-tune operon activation. Definitive proof of the roles of nicked tRNAs, tRNA fragments, and possibly other RNAs as RtcR ligands will require appropriate experiments addressing their roles in vivo. Moreover, the fact that oligomerization is relatively inefficient in our purified system suggests that additional components may contribute to operon activation.
Our findings that (i) nicked tRNAs are RtcR ligands, and that (ii) nicked tRNAs are the predominant form of tRNA fragments under conditions of operon activation, supports the hypothesis that a major role of the operon is to repair damaged tRNAs. In this scenario, the accumulation of nicked tRNAs that are RtcB substrates would trigger operon transcription, increasing the RtcB available to ligate these tRNAs. Such a role could be important for maintaining efficient protein synthesis and/or for resolving an as yet unexplored RNA damage response.
Although the operon also encodes RtcA, which converts 2′ or 3′ monophosphate RNA ends to 2′,3′ cyclic phosphate (Genschik et al. 1998; Das and Shuman 2013), the findings that 5′ tRNA halves and nicked tRNAs ending in monophosphate are poor RtcR ligands and that RtcA is required for operon activation in both wild-type and ΔtruA strains (Hughes et al. 2020; Kotta-Loizou et al. 2022) supports a model in which nicked tRNAs containing 2′,3′ cyclic phosphate are preferred ligands. These data imply that in strains such as S. Typhimurium 14028s, where leaky transcription results in low levels of RtcA in unstressed wild-type cells, those nicked tRNAs generated by metal-dependent anticodon nucleases (which generate 5′ fragments ending in 3′ phosphate) become RtcR ligands after 3′ end cyclization by RtcA. However, in E. coli and those S. Typhimurium strains where operon expression is more tightly regulated, operon activation may preferentially occur after tRNA cleavage by metal-independent endonucleases.
In human cells, our experiments reveal that at least some 5′ tRNA halves that are proposed to function as isolated halves are base paired with their 3′ counterparts to form nicked tRNAs. For example, 5′ tRNAGly(GCC) halves induce stress granules when transfected into cells (Emara et al. 2010), inhibit translation of reporter mRNAs in reticulocyte lysates (Akiyama et al. 2020) and are important for tRNA transcription in zebrafish (Chen et al. 2021). Additionally, 5′ tRNAGly(GCC) fragments in mouse sperm were reported to repress transcription of endogenous retroelements in embryos (Sharma et al. 2016). Our finding that most 5′ halves of this tRNA exist as nicked tRNA in HEK293A cells, coupled with an independent report in human U-2 OS cells (Costa et al. 2023), suggests that, for these and other tRNA halves proposed to have novel functions, it will be important to determine the fraction that is unpaired in the cells under study.
MATERIALS AND METHODS
Bacterial strains, cell culture, and treatments
All S. Typhimurium mutants were derived from strain 14028S (Jarvik et al. 2010) and described previously (Hughes et al. 2020). The specific strains used in this study were XC1410 (flag3X-rsr Δrna), XC1428 (flag3X-rsr Δrna ΔtruA), and XC1421 (flag3X-rsr Δrna ΔruvA) (Hughes et al. 2020). S. Typhimurium strains was grown in Luria-Bertani broth at 37°C. For mitomycin C treatment, cells were grown to OD600 = 0.8 and incubated with 1.5 µM mitomycin C for 2 h. The construction and growth of E. coli BL21(DE3) strains expressing the CARF domain and full-length RtcR was described by Hughes et al. (2020). HEK293A cells (Thermo Fisher) were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (Gibco), 2 mM L-Glutamine, 100 U/mL Penicillin-Streptomycin and treated with 0.5 M sodium arsenite (Sigma-Aldrich) for 30 min and 2 h.
Total RNA preparation
All procedures were performed at 4°C. Bacterial pellets were resuspended in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl and extracted with phenol:chloroform:isoamyl alcohol (25:24:1) by vortexing in the presence of glass beads (0.1 mm, Biospec Products). RNA was precipitated with 2.5 volumes ethanol and resuspended in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl. HEK293A cell pellets were resuspended in 200 µL 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, lysed using a Bioruptor Plus (Diagenode) at the low intensity setting, and RNA extracted with phenol:chloroform:isoamyl alcohol (25:24:1). The RNA was precipitated with 2.5 volumes ethanol and resuspended in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl.
Northern blotting
For northern analyses, RNA was resuspended in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl and either mixed with an equal volume of 95% formamide, 5 mM EDTA, and 0.02% bromophenol blue, heated to 95°C for 5 min and separated in 8% polyacrylamide/8.3 M urea gels (“denaturing” gels) or mixed with 1/3 vol of 25% glycerol and 0.02% bromophenol blue, fractionated in 8% native polyacrylamide gels (“native” gels), and transferred to Hybond N+ membranes (Cytiva) in 0.5× TBE at 400 mA for 1 h. Blots were hybridized with [32P]-labeled oligonucleotides as described (Tarn et al. 1995). Radioactive signals were detected using a Typhoon FLA 7000 Phosphorimager (GE Healthcare). The fraction of base paired tRNA in native gels was quantitated using ImageQuant TL analysis software (GE Healthcare). As we successfully detected both full-length tRNAs and nicked tRNAs in northern blots of native gels, at least a fraction of these tRNAs may be denatured during transfer. Oligonucleotide probes for S. Typhimurium tRNAs were:
5′ tRNAPhe(GAA): 5′-TTTCAATCCCCTGCTCTACCGACTGAGC TATCCGGGC-3′
5′ tRNALeu(UAG): 5′-ACCTAAATCTGGTGCGTCTACCAATTTC GCCACTCCCGC-3′
3′ tRNALeu(UAG): 5′-AGGCGAGACTTGAACTCGCACACCTTGC GGCGCCAGAACCTAAA-3′
5′ tRNATrp(CCA): 5′-TTTGGAGACCGGTGCTCTACCAATT-3′
3′ tRNATrp(CCA): 5′-ACTCGAACTCCCAACACCCGGTTTTGGAG-3′
5′ tRNAHis(GUG): 5′-ATCACAATCCAGGGCTCTACCAACTGAGC TA-3′
Probes for human tRNAs were:
5′ tRNAGly(GCC): 5′-GAGAATTCTACCACTGAACCACCCATGC-3′
5′ tRNAHis(GUG): 5′-CGCAGAGTACTAACCACTATACGATCACG GC-3′
5′ tRNASer(GCU): 5′-ATCGCCTTAACCACTCGGCCACCTCGTC-3′
5′ tRNAiMet(CAU): 5′-CAGCACGCTTCCGCTGCGCCACTCTGCT-3′
3′ tRNAGly(GCC): 5′-GCCGGGAATCGAACCCGGGCCTCCCGCG-3′
3′ tRNASer(GCU): 5′-GATTCGAACCCACGCGTGCAGAGCACAAT GGATT-3′
Generation of in vitro–transcribed RNAs for binding experiments
To make 5′ tRNALeu(UAG) halves and the circularly permuted nicked RNAs for binding experiments, gBlocks (Integrated DNA Technologies) were synthesized that contained an upstream hammerhead ribozyme, which generates 5′-OH, and a downstream HDV ribozyme which generates 2′,3′ cyclic phosphate (Gao and Zhao 2014). The sequences of the gBlocks are given in Figures 2C and 2D. After amplifying the tRNALeu(UAG) 5′ half-encoding gBlock DNA with 5′-TAATACGACTCACTATAGGGTCCCGCCTG-3′ and 5′-GTCCCATTCGCCATGCCGAAG-3′ and the nicked tRNA-encoding gBlock with 5′-TAATACGACTCACTATAGGGCAGAACCTG and 5′-GTCCCATTCGCCATGCCGAAG-3′, the resulting templates were transcribed with T7 RNA polymerase. Reactions (8 mL) contained 4 µg/mL DNA template, 2.5 mM rNTPs, and 0.5 mg/mL T7 RNA polymerase, in 40 mM Tris-HCl (pH 7.5), 10 mM DTT, 4 mM spermidine, 25 mM MgCl2, 0.01% Triton X100, 1 U/mL inorganic pyrophosphatase at 37°C for 3 h. Afterward, the MgCl2 concentration was increased to 0.12 M and the reaction incubated at 60°C for 30 additional min to ensure ribozyme cleavage. The RNA was fractionated in 8% (nicked tRNA) or 10% (5′ tRNA half) polyacrylamide/8.3 M urea gels and eluted in 20 mM Tris-HCl (pH 7.5), 0.3 M NaOAc, 1 mM EDTA, and 0.1% SDS. Afterward, the eluate was passed through a 0.22 µm filter (Millipore) and the RNA subjected to ethanol precipitation. To open the 2′,3′ cyclic phosphate, the nicked tRNA was diluted to 0.4 µg/µL in 10 mM HCl and incubated on ice for 3 h (Honda et al. 2016). Afterward, 1/10 volume of 3 M sodium acetate was added and the RNA precipitated with three volumes of ethanol.
RtcB ligation and RNase R treatment
The ligase reaction containing 50 mM Tris-HCl, pH 8.0, 2 mM MnCl2, 0.1 mM GTP, 5 nM RNA (5′ tRNALeu(UAG) half or circularly permuted nicked tRNALeu(UAG)), and 0.75 µM E. coli RtcB (New England Biolabs) was incubated at 37°C for 1 h. For RNase R treatment, the reaction was supplemented with 20 mM Tris-HCl (pH 8.0), 100 mM KCl, 0.1 mM MgCl2, 0.35 U/µL RNase R (LGC, Biosearch Technologies), and incubated at 37°C for an additional 1 h. Following phenol:chloroform extraction and ethanol precipitation, the RNA was fractionated in 6% polyacrylamide/8.3 M urea gels.
RNA binding assays
RNA was resuspended in 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM MgCl2, 0.05% Tween 20, refolded by heating to 95°C for 2 min, frozen on dry ice and thawed on ice. Refolded RNAs were mixed with RtcR or CARF domain in 10 µL binding buffer, incubated at 4°C for 30 min and at room temperature for 30 min. RtcR binding reactions included 1 mM ATP. For CARF domain EMSAs, reactions were fractionated in 6% polyacrylamide (80:1 acrylamide:bisacrylamide)/5% glycerol gels. For RtcR EMSAs, reactions were fractionated in 4% polyacrylamide (80:1 acrylamide:bisacrylamide)/2.5% glycerol gels or 3%–4% discontinuous polyacrylamide (80:1 acrylamide:bisacrylamide)/2.5% glycerol gels, in which the top (0.5 cm) contained 3% polyacrylamide, 0.5 mM ATP and 100 mM NaCl. Gels were run at 4°C, 5 V/cm for 20 min, then 10 V/cm in 0.5× TBE (50 mM Tris, 45 mM boric acid, 1.25 mM EDTA) until the bromophenol blue migrated 3 cm (Fig. 3A,B), 1 cm (Fig. 3C), or 2 cm (Fig. 3D). The RNA was transferred to Hybond N+ membranes and northern blotting performed as described above.
Protein purification
Purification of the CARF domain and full-length RtcR was as described previously (Hughes et al. 2020), except that a Superdex 16/600 column was used for the final gel filtration.
Size exclusion chromatography
RtcR–RNA complexes were formed by mixing 24 µM RtcR dimer and 48 µM refolded RNA in 100 µL gel filtration buffer (20 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM MgCl2, 0.05% Tween 20, 1 mM ATP). After incubating on ice for 30 min and at room temperature for 30 min, the reaction was injected onto a Superdex 200 Increase 10/300 column equilibrated with gel filtration buffer. The column flow rate was 0.25 mL/min and the elution was monitored by measuring absorbance at 280 nm. Peak fractions were analyzed by fractionating RNA in 8% polyacrylamide/8.3 M urea gel and staining with GelRed (Biotium), while proteins were fractionated in 4%–12% SDS-PAGE and stained with Coomassie blue.
ACKNOWLEDGMENTS
We thank Marco Boccitto for the sodium arsenite–treated HEK293A cells and Eugene Valkov for much helpful advice regarding size exclusion chromatography. We also thank Soyeong Sim, Colin Wu, and Yuanyuan Leng for comments on the manuscript. This work was supported by the Intramural Research Program, Center for Cancer Research, National Cancer Institute, National Institutes of Health (Project ZIA BC011757).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079575.122.
Freely available online through the RNA Open Access option.
REFERENCES
- Akiyama Y, Kharel P, Abe T, Anderson P, Ivanov P. 2020. Isolation and initial structure-functional characterization of endogenous tRNA-derived stress-induced RNAs. RNA Biol 17: 1116–1124. 10.1080/15476286.2020.1732702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama Y, Lyons SM, Fay MM, Tomioka Y, Abe T, Anderson PJ, Ivanov P. 2022. Selective cleavage at CCA ends and anticodon loops of tRNAs by stress-induced RNases. Front Mol Biosci 9: 791094. 10.3389/fmolb.2022.791094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortes-Garrido R, Gkatza N, et al. 2016. Stem cell function and stress response are controlled by protein synthesis. Nature 534: 335–340. 10.1038/nature18282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA, Guthrie C. 1988. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature 334: 213–218. 10.1038/334213a0 [DOI] [PubMed] [Google Scholar]
- Burroughs AM, Aravind L. 2016. RNA damage in biological conflicts and the diversity of responding RNA repair systems. Nucleic Acids Res 44: 8525–8555. 10.1093/nar/gkw722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush M, Dixon R. 2012. The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol Mol Biol Rev 76: 497–529. 10.1128/MMBR.00006-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Taylor DW, Fowler CC, Galan JE, Wang HW, Wolin SL. 2013. An RNA degradation machine sculpted by Ro autoantigen and noncoding RNA. Cell 153: 166–177. 10.1016/j.cell.2013.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, et al. 2016. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351: 397–400. 10.1126/science.aad7977 [DOI] [PubMed] [Google Scholar]
- Chen L, Xu W, Liu K, Jiang Z, Han Y, Jin H, Zhang L, Shen W, Jia S, Sun Q, et al. 2021. 5′ half of specific tRNAs feeds back to promote corresponding tRNA gene transcription in vertebrate embryos. Sci Adv 7: eabh0494. 10.1126/sciadv.abh0494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese R, Landsberg R, Haar RA, Umbarger HE, Ames BN. 1974. Pleiotropy of hisT mutants blocked in pseudouridine synthesis in tRNA: leucine and isoleucine-valine operons. Proc Natl Acad Sci 71: 1857–1861. 10.1073/pnas.71.5.1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa B, Li Calzi M, Castellano M, Blanco V, Cuevasanta E, Litvan I, Ivanov P, Witwer K, Cayota A, Tosar JP. 2023. Nicked tRNAs are stable reservoirs of tRNA halves in cells and biofluids. Proc Natl Acad Sci 120: e2216330120. 10.1073/pnas.2216330120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U, Shuman S. 2013. 2′-Phosphate cyclase activity of RtcA: a potential rationale for the operon organization of RtcA with an RNA repair ligase RtcB in Escherichia coli and other bacterial taxa. RNA 19: 1355–1362. 10.1261/rna.039917.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS, Kowalczykowski SC. 2008. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev 72: 642–671. 10.1128/MMBR.00020-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal Y, Kurasz JE, Fontaine BM, Marotta NJ, Chauhan SS, Karls AC, Weinert EE. 2022. Cellular effects of 2′,3′-cyclic nucleotide monophosphates in gram-negative bacteria. J Bacteriol 204: e0020821. 10.1128/JB.00208-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkordy A, Mishima E, Niizuma K, Akiyama Y, Fujimura M, Tominaga T, Abe T. 2018. Stress-induced tRNA cleavage and tiRNA generation in rat neuronal PC12 cells. J Neurochem 146: 560–569. 10.1111/jnc.14321 [DOI] [PubMed] [Google Scholar]
- Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. 2010. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 285: 10959–10968. 10.1074/jbc.M109.077560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engl C, Schaefer J, Kotta-Loizou I, Buck M. 2016. Cellular and molecular phenotypes depending upon the RNA repair system RtcAB of Escherichia coli. Nucleic Acids Res 44: 9933–9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. 2009. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 583: 437–442. 10.1016/j.febslet.2008.12.043 [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhao Y. 2014. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol 56: 343–349. 10.1111/jipb.12152 [DOI] [PubMed] [Google Scholar]
- Genschik P, Drabikowski K, Filipowicz W. 1998. Characterization of the Escherichia coli RNA 3'-terminal phosphate cyclase and its ς54-regulated operon. J Biol Chem 273: 25516–25526. 10.1074/jbc.273.39.25516 [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. 2015. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161: 790–802. 10.1016/j.cell.2015.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto C, Steitz JA. 1984. U4 and U6 RNAs coexist in a single small nuclear ribonucleoprotein particle. Nucleic Acids Res 12: 3283–3293. 10.1093/nar/12.7.3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Morichika K, Kirino Y. 2016. Selective amplification and sequencing of cyclic phosphate-containing RNAs by the cP-RNA-seq method. Nat Protoc 11: 476–489. 10.1038/nprot.2016.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KJ, Chen X, Burroughs AM, Aravind L, Wolin SL. 2020. An RNA repair operon regulated by damaged tRNAs. Cell Rep 33: 108527. 10.1016/j.celrep.2020.108527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. 2011. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 43: 613–623. 10.1016/j.molcel.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvik T, Smillie C, Groisman EA, Ochman H. 2010. Short-term signatures of evolutionary change in the Salmonella enterica serovar typhimurium 14028 genome. J Bacteriol 192: 560–567. 10.1128/JB.01233-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann G, Amitsur M. 1985. Host transfer RNA cleavage and reunion in T4-infected Escherichia coli CTr5x. Nucleic Acids Res 13: 4333–4341. 10.1093/nar/13.12.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskiene M, Kostiuk G, Venclovas C, Tamulaitis G, Siksnys V. 2017. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357: 605–609. 10.1126/science.aao0100 [DOI] [PubMed] [Google Scholar]
- Kotta-Loizou I, Giuliano MG, Jovanovic M, Schaefer J, Ye F, Zhang N, Irakleidi DA, Liu X, Zhang X, Buck M, et al. 2022. The RNA repair proteins RtcAB regulate transcription activator RtcR via its CRISPR-associated Rossmann fold domain. iScience 25: 105425. 10.1016/j.isci.2022.105425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K. 2005. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem 280: 42744–42749. 10.1074/jbc.M510356200 [DOI] [PubMed] [Google Scholar]
- Li Z, Brow DA. 1996. A spontaneous duplication in U6 spliceosomal RNA uncouples the early and late functions of the ACAGA element in vivo. RNA 2: 879–894. [PMC free article] [PubMed] [Google Scholar]
- Li Z, Reimers S, Pandit S, Deutscher MP. 2002. RNA quality control: degradation of defective transfer RNA. EMBO J 21: 1132–1138. 10.1093/emboj/21.5.1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Mei W, Padmanaban V, Alwaseem H, Molina H, Passarelli MC, Tavora B, Tavazoie SF. 2022. A pro-metastatic tRNA fragment drives Nucleolin oligomerization and stabilization of its bound metabolic mRNAs. Mol Cell 82: 2604–2617.e2608. 10.1016/j.molcel.2022.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. 1992. Cyclic 2′,3′ phosphates and nontemplated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNA's. Science 255: 327–330. 10.1126/science.1549778 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Timinskas A, Wolf YI, Gussow AB, Siksnys V, Venclovas C, Koonin EV. 2020. Evolutionary and functional classification of the CARF domain superfamily, key sensors in prokaryotic antivirus defense. Nucleic Acids Res 48: 8828–8847. 10.1093/nar/gkaa635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Schwer B, Schaffrath R, Shuman S. 2008. RNA repair: an antidote to cytotoxic eukaryal RNA damage. Mol Cell 31: 278–286. 10.1016/j.molcel.2008.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewoehner O, Garcia-Doval C, Rostol JT, Berk C, Schwede F, Bigler L, Hall J, Marraffini LA, Jinek M. 2017. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 548: 543–548. 10.1038/nature23467 [DOI] [PubMed] [Google Scholar]
- Pannone B, Xue D, Wolin S. 1998. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J 17: 7442–7453. 10.1093/emboj/17.24.7442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, Anderson P, Pan T, Hatzoglou M. 2012. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem 287: 42708–42725. 10.1074/jbc.M112.371799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, et al. 2016. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351: 391–396. 10.1126/science.aad6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Wilson B, Kumar P, Dutta A. 2020. Noncanonical roles of tRNAs: tRNA fragments and beyond. Annu Rev Genet 54: 47–69. 10.1146/annurev-genet-022620-101840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Shuman S. 2011. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J Biol Chem 286: 7727–7731. 10.1074/jbc.C111.219022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Yario TA, Steitz JA. 1995. U12 snRNA in vertebrates: evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of introns. RNA 1: 644–656. [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Lu C, Green PJ, Parker R. 2008. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14: 2095–2103. 10.1261/rna.1232808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton HE, Ferguson L, Temoche-Diaz MM, Liu XM, Pimentel SC, Ingolia NT, Schekman R, Collins K. 2021. Low-bias ncRNA libraries using ordered two-template relay: serial template jumping by a modified retroelement reverse transcriptase. Proc Natl Acad Sci 118: e2107900118. 10.1073/pnas.2107900118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X. 2013. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther 21: 368–379. 10.1038/mt.2012.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt HD, West SC. 2014. Holliday junction resolvases. Cold Spring Harb Perspect Biol 6: a023192. 10.1101/cshperspect.a023192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P. 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42. 10.1083/jcb.200811106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]