FIGURE 1.
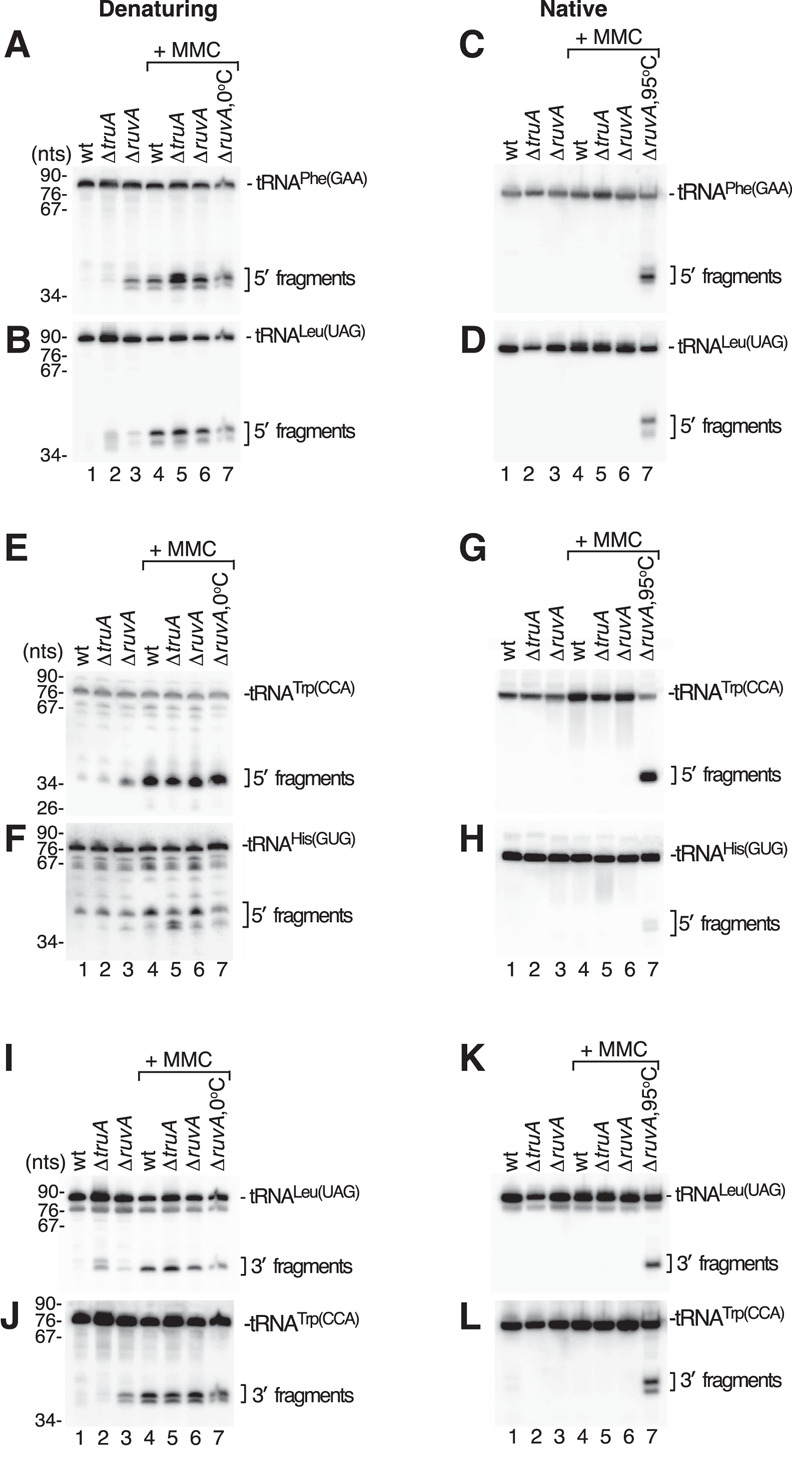
Many tRNA fragments in S. Typhimurium are base paired in cells. (A,B) RNA was isolated from the indicated rnaΔ strains using cold (4°C) phenol/chloroform/isoamyl alcohol, subjected to ethanol precipitation, and resuspended in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl. After mixing with equal volumes of 95% formamide, 5 mM EDTA, and 0.02% bromophenol blue, the RNA was heated to 95°C for 5 min, fractionated in denaturing gels, and subjected to northern blotting to detect 5′ halves of tRNAPhe(GAA) (A) and tRNALeu(UAG) (B). In lane 7, the heating step was omitted. (C,D) The resuspended RNA described in A–B was mixed with 1/3 vol of 25% glycerol and 0.02% bromophenol blue, fractionated in native gels, and analyzed by northern blotting to detect 5′ halves of tRNAPhe(GAA) (C) and tRNALeu(UAG) (D). In lane 7, the RNA was heated to 95°C for 5 min before loading. (E,F) After heating and fractionation in denaturing gels as in A–B, the indicated RNAs were subjected to northern blotting to detect 5′ halves of tRNATrp(CCA) (E) and tRNAHis(GUG) (F). (G,H) The same RNAs as in E and F were fractionated in native gels and subjected to northern blotting to detect 5′ halves of tRNATrp(CCA) (G) and tRNAHis(GUG) (H). (I–L) The northern blots in A–B and C–D were reprobed to detect 3′ halves of tRNALeu(UAG) and tRNATrp(CCA) after fractionation in denaturing (I,J) or native gels (K,L).
