Abstract
VIM-1 is a new group 3 metallo-β-lactamase recently detected in carbapenem-resistant nosocomial isolates of Pseudomonas aeruginosa from the Mediterranean area. In this work, VIM-1 was purified from an Escherichia coli strain carrying the cloned blaVIM-1 gene by means of an anion-exchange chromatography step followed by a gel permeation chromatography step. The purified enzyme exhibited a molecular mass of 26 kDa in sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and an acidic pI of 5.1 in analytical isoelectric focusing. Amino-terminal sequencing showed that mature VIM-1 results from the removal of a 26-amino-acid signal peptide from the precursor. VIM-1 hydrolyzes a broad array of β-lactam compounds, including penicillins, narrow- to expanded-spectrum cephalosporins, carbapenems, and mechanism-based serine-β-lactamase inactivators. Only monobactams escape hydrolysis. The highest catalytic constant/Km ratios (>106 M−1 · s−1) were observed with carbenicillin, azlocillin, some cephalosporins (cephaloridine, cephalothin, cefuroxime, cefepime, and cefpirome), imipenem, and biapenem. Kinetic parameters showed remarkable variability with different β-lactams and also within the various penam, cephem, and carbapenem compounds, resulting in no clear preference of the enzyme for any of these β-lactam subfamilies. Significant differences were observed with some substrates between the kinetic parameters of VIM-1 and those of other metallo-β-lactamases. Inactivation assays carried out with various chelating agents (EDTA, 1,10-o-phenanthroline, and pyridine-2,6-dicarboxylic acid) indicated that formation of a ternary enzyme-metal-chelator complex precedes metal removal from the zinc center of the protein and revealed notable differences in the inactivation parameters of VIM-1 with different agents.
Production of β-lactamases is the most common mechanism of bacterial resistance to β-lactam antibiotics. Among the β-lactam-degrading enzymes, metallo-β-lactamases are notable for their substrate profile, which always includes carbapenems and often most classes of β-lactams, and for their resistance to mechanism-based β-lactamase inactivators (3, 4, 20). Owing to these features, the emergence of acquired metallo-β-lactamases in clinical isolates of major gram-negative pathogens, such as members of the family Enterobacteriaceae and Pseudomonas aeruginosa, is a most worrying development in the field of microbial drug resistance (12).
VIM-1 is a new molecular class B metallo-β-lactamase which has recently been identified in carbapenem-resistant isolates of Pseudomonas aeruginosa from various Italian hospitals (11, 13, 21) and from Greece (27). Like the blaIMP gene, which was the first acquired metallo-β-lactamase determinant found in nosocomial isolates of various gram-negative species in Japan (1, 7, 8, 16, 24, 25) and, more recently, even outside Japan (G. Cornaglia, M. L. Riccio, A. Mazzariol, P. Piccoli, L. Lauretti, R. Fontana, and G. M. Rossolini, Letter, Lancet 353:899–900; T. H. Koh, G. S. Babini, N. Woodford, L. H. Sng, L. M. Hall, and D. M. Livermore, Letter, Lancet 353:2162; K. Lee, Y. Chong, H. B. Shin, and D. Yong, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. E-85, p. 193, 1998), the blaVIM-1 gene is carried on an integron-borne gene cassette (11) and has the potential for spreading among major bacterial pathogens. For this reason, blaVIM-1 is potentially a very dangerous resistance determinant from the clinical standpoint.
In this work, a protocol for purification of the VIM-1 enzyme was developed, and the functional properties of the purified enzyme were investigated.
MATERIALS AND METHODS
Purification of VIM-1.
The VIM-1 enzyme was purified from Escherichia coli(pBCLL/39H) (11) grown aerobically in 2 liters of brain heart infusion broth (Difco Laboratories, Detroit, Mich.) containing chloramphenicol (60 μg/ml) for 18 h at 37°C. The cells were harvested by centrifugation, washed twice with 30 mM sodium cacodylate buffer (pH 6.3), resuspended in 100 ml of the same buffer supplemented with 100 μM ZnCl2 (CBZ), and disrupted by sonication (five times for 30 s each time at 60 W). Cell debris was removed by high-speed centrifugation (105,000 × g for 60 min at 4°C), and the clarified supernatant was loaded on a Q-Sepharose FF column (2.6 by 26 cm; Amersham-Pharmacia Biotech, Uppsala, Sweden) previously equilibrated with CBZ. After the column was washed with the same buffer, the bound proteins were eluted by a linear NaCl gradient (0 to 0.7 M) in CBZ (flow rate, 3 ml/min). The fractions containing carbapenemase activity were pooled, dialyzed against 30 mM HEPES buffer (pH 7.5), concentrated 10-fold by ultrafiltration, and loaded on a Superdex 75 column (1.6 by 75 cm; Amersham-Pharmacia Biotech) equilibrated and eluted with 30 mM HEPES (pH 7.5). The β-lactamase-containing elution peak was concentrated at 1 mg/ml by ultrafiltration and stored at −80°C until it was used. The protein concentration in solution was assayed with a commercial kit (Bio-Rad [Richmond, Calif.] protein assay), with bovine serum albumin used as a standard.
During the purification procedure the presence of carbapenemase activity was monitored using 100 μM meropenem as a substrate in 30 mM HEPES (pH 7.5) at 30°C.
Protein electrophoretic techniques.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to the method of Laemmli (9) using final acrylamide concentrations of 12 and 5% (wt/vol) for the separating and stacking gels, respectively. After electrophoresis, the protein bands were stained with Coomassie brilliant blue R-250. Analytical isoelectric focusing of the purified protein was performed in a precast polyacrylamide gel containing ampholites (pH range, 3.5 to 9.5) (Ampholine PAGplate; Amersham Pharmacia Biotech) using a Multiphor II Apparatus (Pharmacia-LKB, Uppsala, Sweden). The gels were focused at 0.1 W/cm2 for 3 h at 10°C. The β-lactamase activity was revealed by overlaying the gel with a filter paper soaked with 0.25 mM nitrocefin (Unipath, Milan, Italy) as described previously (11).
N-terminal sequencing.
The N-terminal sequence of the purified VIM-1 protein was determined using a gas phase sequencer (Procise-492; Applied Biosystems, Foster City, Calif.) after resuspension of the protein (50 pmol) in a 0.1% (vol/vol) trifluoroacetic acid solution and loading the sample onto a polyvinylidene difluoride membrane (Millipore Corp., Bedford, Mass.).
Determination of kinetic parameters.
Substrate hydrolysis by the purified enzyme was monitored by following the absorbance variation at 30°C, using a lambda 2 spectrophotometer (Perkin-Elmer, Rahway, N.J.) equipped with thermostatically controlled cells. Azlocillin was from Sigma Chemical Co. (St. Louis, Mo.), and mezlocillin was from Bayer AG (Leverkusen, Germany). The wavelengths and changes in extinction coefficients used in the spectrophotometric assays were 240 nm and −1,130 M−1 cm−1 with azlocillin and 235 nm and −1,100 M−1 cm−1 with mezlocillin. For other β-lactam compounds, the sources and the wavelengths and changes in extinction coefficients used in the spectrophotometric assays were as described previously (10, 19). The experimental conditions (reaction buffer and volume) used in the spectrophotometric assays were as described previously (10). The steady-state kinetic parameters (Km and catalytic constant [kcat]) were determined under initial-rate conditions using the Hanes-Woolf plot (23). Km values lower than 20 μM were measured as inhibition constants (Kis) in a competitive model, using 50 μM nitrocefin as the reporter substrate. The Ki value was determined by the plot of Vo/Vi versus I, yielding a line whose slope is Kms/(Kms + S) · Ki, where Vo and Vi are the initial rates of nitrocefin hydrolysis in the absence and presence of the inhibitor, respectively, I is the inhibitor concentration, S is the reporter substrate concentration, and Kms the Michaelis constant of the enzyme for the reporter substrate.
Inactivation by chelating agents.
Inactivation of VIM-1 by divalent ion chelators was studied in 50 mM HEPES (pH 7.5) at 30°C, using 50 μM nitrocefin as the reporter substrate, in the presence of different concentrations of EDTA, 1,10-o-phenanthroline, and pyridine-2,6-dicarboxylic (dipicolinic) acid (Sigma) in a total volume of 0.5 ml. Since the inactivation time courses followed pseudo-first-order kinetics, the rate constants characterizing the inactivation of the enzyme were calculated from the dependence of the pseudo-first-order constant ki upon the chelating agent concentration on the basis of the following model:
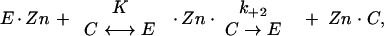 |
in which E · Zn, C, E · Zn · C, E, and Zn · C are the metalloenzyme, the chelator, a ternary metalloenzyme-chelator complex, the apoenzyme, and the metal-chelator complex, respectively; K represents the dissociation constant of the E · Zn · C ternary complex; and k+2 is the individual rate constant for the dissociation of E · Zn · C into E + Zn · C, a step which, under our experimental conditions, appeared to be essentially irreversible. The individual values of K and k+2 were determined by fitting the value of ki, the pseudo-first-order inactivation rate constant, to the equation
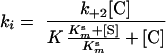 |
where [S] and Kms were the concentration and the Km value of the reporter substrate, respectively.
RESULTS
Purification and structural properties of the VIM-1 enzyme.
The VIM-1 enzyme was purified from a lysate of E. coli DH5α(pBCLL/39H), which carries the cloned blaVIM-1 gene on a multicopy plasmid vector (11), by means of an anion-exchange chromatography step on Q-Sepharose followed by a gel permeation chromatography step on Superdex 75. Approximately 1.15 mg of purified enzyme was obtained per liter of culture using the above-described protocol. The degree of purity, evaluated by SDS-PAGE, was higher than 95% (Fig. 1), and the overall yield of the purification protocol was 17% (Table 1).
FIG. 1.
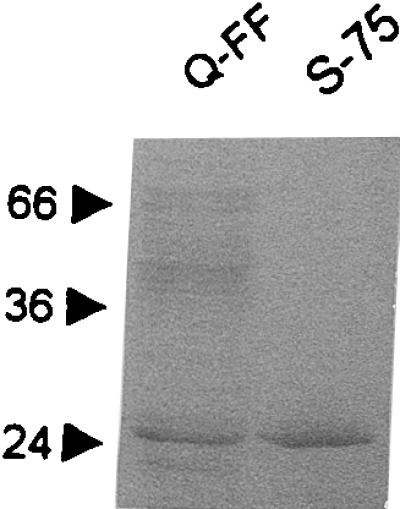
SDS-PAGE analysis of the fractions containing carbapenemase activity eluted from the Q-Sepharose FF columns (Q-FF) and from the Superdex 75 column (S-75). Protein mass standards are reported in kilodaltons on the left.
TABLE 1.
Summary of the purification steps of the VIM-1 enzyme produced by E. coli DH5α(pBCLL/39H)a
| Purification step | Total protein (mg) | Sp act (U/mg of protein) | Total activity (U) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 797 | 1,064 | 848,008 | 100 | 1 |
| Q-Sepharose FF eluate | 31 | 13,886 | 430,466 | 51 | 13 |
| Superdex 75 eluate | 2.3 | 61,538 | 141,537 | 17 | 58 |
One unit of activity is defined as the amount of enzyme hydrolyzing 1 nmol of meropenem per min under the conditions described in Materials and Methods.
In SDS-PAGE, the VIM-1 polypeptide migrated with an apparent molecular mass of approximately 26 kDa (Fig. 1). The isoelectric pH of purified VIM-1, determined by analytical isoelectric focusing, was 5.1 (data not shown). The N-terminal sequence of purified VIM-1 was found to be NH2-GEPSG, indicating that mature VIM-1 is generated following the cleavage of a 26-amino-acid signal peptide from the precursor (11). According to this information, and assuming no further posttranslational modification, the calculated molecular mass of VIM-1 is 25,322 Da.
Kinetic parameters of VIM-1 with various β-lactam compounds.
The kinetic parameters of VIM-1, including Km, kcat, and the kcat/Km ratio, were determined with several different β-lactam compounds.
Under the experimental conditions adopted, the enzyme hydrolyzed all the tested compounds except aztreonam (Table 2). Individual kinetic parameters were remarkably variable for different β-lactams, with up to 1,000-fold differences in the values of Km, kcat, and the kcat/Km ratio. The highest values of the kcat/Km ratio (>106 M−1 · s−1) were found with carbenicillin, azlocillin, imipenem, biapenem, and some cephalosporins (nitrocefin, cephaloridine, cephalothin, cefuroxime, cefepime, and cefpirome). The lowest values of the kcat/Km ratio (<105 M−1 · s−1) were observed with penicillin G, ampicillin, temocillin, ceftazidime, and the serine-β-lactamase inactivators sulbactam, tazobactam, and β-iodopenicillanate. Considerable variability was observed within the various β-lactam subfamilies (penam, cephem, and carbapenem compounds), resulting in no clear preference of VIM-1 for any of these groups (Table 2). Among carbapenems, VIM-1 exhibited relatively low kcat values but very high affinities for imipenem and biapenem, eventually resulting in high kcat/Km ratios (Table 2). This behavior was quite different from that of other metallo-β-lactamases (Table 3).
TABLE 2.
Kinetic parameters of the VIM-1 enzyme with various β-lactam compoundsa
| β-Lactam | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| Penicillin G | 841 ± 70 (520) | 29 ± 2 (320) | 3.4 × 104 (6.2 × 105) |
| Ampicillin | 917 ± 83 (200) | 37 ± 3 (950) | 4.0 × 104 (4.8 × 106) |
| Carbenicillin | 75 ± 5 NA | 167 ± 15 NA | 2.2 × 106 (2.0 × 104) |
| Piperacillin | 3,500 ± 278 NA | 1,860 ± 166 NA | 5.3 × 105 (7.2 × 105) |
| Azlocillin | 123 ± 11 NA | 1,525 ± 125 NA | 1.2 × 107 NA |
| Mezlocillin | 346 ± 27 NA | 255 ± 20 NA | 7.4 × 105 NA |
| Ticarcillin | 1,117 ± 100 (740) | 452 ± 36 (1.1) | 4.1 × 105 (1.5 × 103) |
| Temocillin | 22 ± 2 (>2,000) | 0.5 ± 0.04 NA | 2.3 × 104 (<1.0 × 102) |
| Nitrocefin | 17 ± 1 (27) | 95 ± 8 (63) | 5.6 × 106 (2.3 × 106) |
| Cephaloridine | 30 ± 3 (22) | 313 ± 26 (53) | 1.0 × 107 (2.4 × 106) |
| Cephalothin | 53 ± 4 (21) | 281 ± 24 (48) | 5.3 × 106 (2.3 × 106) |
| Cefoxitin | 131 ± 11 (8) | 26 ± 2 (16) | 2.0 × 105 (2.0 × 106) |
| Cefuroxime | 42 ± 4 (37) | 324 ± 29 (8) | 7.7 × 106 (2.2 × 105) |
| Cefotaxime | 247 ± 19 (4) | 169 ± 14 (1.3) | 6.8 × 105 (3.3 × 105) |
| Ceftazidime | 794 ± 69 (44) | 60 × 5 (8) | 7.6 × 104 (1.8 × 105) |
| Cefepime | 145 ± 13 (11) | 549 ± 48 (7) | 3.8 × 106 (6.4 × 105) |
| Cefpirome | 287 ± 25 (14) | 707 ± 56 (9) | 2.5 × 106 (6.4 × 105) |
| Imipenem | 1.5 ± 0.1 (39) | 2.0 ± 0.1 (46) | 1.3 × 106 (1.2 × 106) |
| Meropenem | 48 ± 3 (10) | 13 ± 1 (5) | 2.7 × 105 (5.0 × 105) |
| Biapenem | 7.5 ± 0.6 (28) | 8.5 ± 0.7 (160) | 1.1 × 106 (5.7 × 106) |
| Aztreonam | >1,000 (>1,000) | <0.01 (>0.01) | <1.0 × 102 (<1.0 × 102) |
| Sulbactam | 194 ± 17 (>1,000) | 10 ± 1 (>13.7) | 5.2 × 104 (1.3 × 104) |
| Tazobactam | 337 ± 30 (>1,000) | 5.3 ± 0.4 NA | 1.6 × 104 (4.0 × 102) |
| β-Iodopenicillanate | 165 ± 16 (>1,000) | 1.2 ± 0.1 (>3.98) | 7.3 × 103 (3.9 × 103) |
TABLE 3.
Comparison of the kinetic parameters of VIM-1 and other metallo-β-lactamases with imipenem
| Enzyme | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) | Reference |
|---|---|---|---|---|
| VIM-1 | 1.5 | 2.0 | 1.3 × 106 | This study |
| VIM-2 | 10 | 9.9 | 9.9 × 105 | 18 |
| IMP-1 | 39 | 46 | 1.2 × 106 | 10 |
| Bc-II | >1,000 | >100 | 1.3 × 105 | 5 |
| BlaB | 370 | 350 | 9.5 × 105 | 22 |
| IND-1 | 198 | NAa | NA | 2 |
| MET-1 | >500 | >48 | 6.0 × 104 | 15 |
| CcrA | 270 | 200 | 7.4 × 105 | 31 |
| CphA | 90 | 170 | 1.9 × 106 | 5 |
| AsbM1 | 230 | 71 | 3.1 × 105 | 30 |
| ImiS | 180 | 160 | 8.9 × 105 | 28 |
| L1 | 100 | 75 | 7.5 × 105 | 6 |
NA, not available.
Interaction of VIM-1 with chelating agents.
The activity of VIM-1 was inhibited by chelating agents, including EDTA, 1-10-o-phenanthroline, and dipicolinic acid. The inactivation time courses followed pseudo-first-order kinetics, and the inactivation rates increased with the chelating agent concentrations in a hyperbolic manner (data not shown). This indicated that the above-mentioned compounds did not act by simply scavenging the free metal ions and that the formation of a ternary enzyme-metal-chelator complex precedes the removal of the metal from the zinc center of the protein.
Although the models of the enzyme interaction with chelators were similar, a remarkable variability was observed in the inactivation parameters of different agents, with EDTA behaving as a poor inactivator and 1,10-o-phenanthroline apparently being the most efficient inactivator (Table 4).
TABLE 4.
Kinetic parameters for the inactivation of the purified VIM-1 enzyme by chelating agentsa
| Chelating agent | K (mM) | k+2 (s−1) | k+2/K (M−1 · s−1) |
|---|---|---|---|
| EDTA | >10 | NDa | ND |
| 1,10-o-Phenanthroline | 0.013 ± 0.001 | 0.0028 ± 0.0002 | 215 |
| Dipicolinic acid | 0.234 ± 0.018 | 0.0054 ± 0.0004 | 23 |
The K and k+2 values represent the means of three measurements ± standard deviations. ND, not determined.
DISCUSSION
Results of this study showed that VIM-1 exhibits an exceedingly broad substrate specificity, which includes virtually all β-lactams except monobactams. Based on these properties, therefore, VIM-1 can be assigned to subgroup 3a of group 3 of the functional classification of β-lactamases (3, 4, 20). This behavior emphasizes the clinical significance of this new mobile β-lactamase.
Determination of the kinetic parameters of the purified enzyme with several substrates allowed us to make comparisons with other metallo-β-lactamases and revealed some interesting features of VIM-1.
In penicillins, the structure of the C-6 side chain apparently exerts a remarkable influence on the substrate susceptibility to the enzyme. In fact, carbenicillin behaved as a good substrate, while hydrolysis of penicillin G and ampicillin was approximately 50-fold less efficient (Table 2). This behavior is quite different from that of IMP-1, which exhibits a marked preference for penicillin G and ampicillin compared to carboxy-penicillins (10), and also from that of the Bc-II, CcrA, and L1 enzymes, which hydrolyze the above-mentioned compounds with similar efficiencies (6, 31). Another relevant difference was observed with temocillin, which was recognized with a relatively high affinity and hydrolyzed by VIM-1 (Table 2) but is not a substrate for either IMP-1 (10) or Bc-II (5). The activity of VIM-1 on temocillin was confirmed by the fact that the MIC of temocillin for E. coli DH5α(pBCLL/39H), which carries a recombinant plasmid containing the blaVIM-1 cassette and produces the VIM-1 enzyme (11), is increased remarkably compared to that for DH5α harboring the empty cloning vector (>256 versus 8 μg/ml) (G. M. Rossolini, M. L. Riccio, and J.-D. Docquier, unpublished results). This indicates that the presence of an α-methoxy group at position C-6 does not prevent the interaction of penicillins with VIM-1.
Cephalosporins were usually good substrates for VIM-1 (values of kcat/Km were often higher than 106 M−1 · s−1), with somewhat lower efficiencies being observed with oxymino-cephalosporins (cefotaxime and ceftazidime) and cefoxitin (Table 2). The difference in the kinetic constants of cephalothin and cefoxitin suggests that the presence of an α-methoxy group at position C-7 reduces the susceptibility of cephalosporins to VIM-1, although it does not prevent interaction and hydrolysis. Again, this behavior is different from that of IMP-1, which is not affected by the presence of an α-methoxy group at position C-7 (10), while it resembles that of Bc-II, which, however, is more severely affected by this type of modification (6).
Also, carbapenems were good substrates for VIM-1 (Table 2), with values of kcat/Km overall similar to those reported for other metallo-β-lactamases (Table 3). Interestingly, the highly efficient degradation of imipenem by VIM-1 (kcat/Km, ≈106 M−1 · s−1) was dependent on a very high affinity for the substrate associated with relatively low turnover rates, which is a strategy different from that adopted by the other group 3 enzymes, which exhibit comparable kcat/Km ratios resulting from higher turnover rates and remarkably lower affinities (Table 3).
Most recently, an allelic variant of the blaVIM-1 gene, named blaVIM-2, whose product exhibits 93% amino acid identity to mature VIM-1, was identified in a P. aeruginosa clinical isolate from France (18) and in P. aeruginosa clinical isolates from Italy (Rossolini et al., unpublished). Comparison of the kinetic parameters of VIM-1 determined in this work with those reported for VIM-2 (18) revealed similar kcat/Km ratios with several substrates (carbapenems, piperacillin, ticarcillin, narrow-spectrum cephalosporins, and cefotaxime) but notable differences with penicillin G, cefuroxime, ceftazidime, cefepime, and cefpirome (compared to those reported for VIM-2, the kcat/Km ratios of VIM-1 with these substrates were 34-fold lower, 14-fold higher, 12-fold lower, 127-fold higher, and 36-fold higher, respectively). This suggests that at least some of the structural differences between VIM-1 and VIM-2 could be functionally relevant, although a comparative kinetic analysis of the two enzymes under identical experimental conditions would be necessary for a definitive confirmation.
The mode of interaction of VIM-1 with chelating agents appeared to follow a mechanism similar to that observed with other metallo-β-lactamases (10, 22), in which the formation of a ternary enzyme-metal-chelator complex precedes the removal of the metal from the zinc center of the protein. Similar to other group 3 enzymes (10, 22), VIM-1 appeared to be more susceptible to 1,10-o-phenanthroline and dipicolinic acid than to EDTA. However, unlike BlaB and IMP-1, which are overall more susceptible to dipicolinic acid than to 1,10-o-phenanthroline (10, 22), VIM-1 appeared to be more susceptible to the latter agent.
The potential clinical significance of VIM-1 and the uniqueness of some of its functional properties render this enzyme a most interesting candidate for further studies of the molecular structure and the interaction with known metallo-β-lactamase inhibitors (14, 17, 26, 29).
ACKNOWLEDGMENTS
This work was supported by the European research network on metallo-β-lactamases within the Training and Mobility of Researchers (TMR) Program (contract no. FMRX-CT98-0232) and by grant no. 9906404271 from MURST ex-40%.
Footnotes
Mailing address for Gian Maria Rossolini: Dipartimento di Biologia Molecolare, Sez. di Microbiologia, Università di Siena, Via Laterina, 8, 53100-Siena, Italy. Phone: 39 0577 233327. Fax: 39 0577 233325. E-mail: rossolini@unisi.it.
REFERENCES
- 1.Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankun R, Ohsuka S, Kato N, Ohta M. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellais S, Léotard S, Poirel L, Naas T, Nordmann P. Molecular characterization of a carbapenem-hydrolyzing β-lactamase from Chryseobacterium (Flavobacterium) indologenes. FEMS Microbiol Lett. 1999;171:127–132. doi: 10.1111/j.1574-6968.1999.tb13422.x. [DOI] [PubMed] [Google Scholar]
- 3.Bush K. Metallo-β-lactamases: a class apart. Clin Infect Dis. 1998;27(Suppl. 1):S48–S53. doi: 10.1086/514922. [DOI] [PubMed] [Google Scholar]
- 4.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felici A, Amicosante G. Kinetic analysis of extension of substrate specificity with Xanthomonas maltophilia, Aeromonas hydrophila, and Bacillus cereus metallo-β-lactamases. Antimicrob Agents Chemother. 1995;39:192–199. doi: 10.1128/aac.39.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felici A, Amicosante G, Oratore A, Strom R, Ledent P, Joris B, Fanuel L, Frère J-M. An overview of the kinetic parameters of class B β-lactamases. Biochem J. 1993;291:1a. doi: 10.1042/bj2910151. , S. Ohsuka, R. Wacharotayankun, N. Kato, and M. Ohta. 1995. Plasmid-mediated dissemination of the metallo-β-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob. Agents Chemother. 39:824–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 10.Laraki N, Franceschini N, Rossolini G M, Santucci P, Meunier C, de Pauw E, Amicosante G, Frère J-M, Galleni M. Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-β-lactamase IMP-1 produced by Escherichia coli. Antimicrob Agents Chemother. 1999;43:902–906. doi: 10.1128/aac.43.4.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lauretti L, Riccio M L, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini G M. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999;43:1584–1590. doi: 10.1128/aac.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livermore D M. Acquired carbapenemases. J Antimicrob Chemother. 1997;39:673–676. doi: 10.1093/jac/39.6.673. [DOI] [PubMed] [Google Scholar]
- 13.Mazzariol A, Cornaglia G, Piccoli P, Lauretti L, Riccio M L, Rossolini G M, Fontana R. Carbapenem-hydrolyzing β-lactamases in Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis. 1999;18:455–456. doi: 10.1007/s100960050320. [DOI] [PubMed] [Google Scholar]
- 14.Nagano R, Adachi Y, Imamura H, Yamada K, Hashizume T, Morishima H. Carbapenem derivatives as potential inhibitors of various β-lactamases, including class B metallo-β-lactamases. Antimicrob Agents Chemother. 1999;43:2497–2503. doi: 10.1128/aac.43.10.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Hara K, Haruta S, Sawai T, Tsunoda M, Iyobe S. Novel metallo-β-lactamase mediated by a Shigella flexneri plasmid. FEMS Microbiol Lett. 1998;162:201–206. doi: 10.1111/j.1574-6968.1998.tb12999.x. [DOI] [PubMed] [Google Scholar]
- 16.Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, Yoshimura F, Kato N. Molecular characterization of an enterobacterial metallo-β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother. 1994;38:71–78. doi: 10.1128/aac.38.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payne D J, Bateson J H, Gasson B C, Khushi T, Proctor D, Pearson S C, Reid R. Inhibition of metallo-β-lactamases by a series of thiol ester derivatives of mercaptophenylacetic acid. FEMS Microbiol Lett. 1997;157:171–175. doi: 10.1111/j.1574-6968.1997.tb12769.x. [DOI] [PubMed] [Google Scholar]
- 18.Poirel L, Naas T, Nicholas D, Collet L, Bellais S, Cavallo J-D, Nordmann P. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase, and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother. 2000;44:891–897. doi: 10.1128/aac.44.4.891-897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prosperi-Meys C, Llabres G, de Seny D, Paul Soto R, Hernandez Valladares M, Laraki N, Frère J-M, Galleni M. Interaction between class B β-lactamases and suicide substrates of active-site serine β-lactamases. FEBS Lett. 1999;443:109–111. doi: 10.1016/s0014-5793(98)01689-5. [DOI] [PubMed] [Google Scholar]
- 20.Rasmussen B A, Bush K. Carbapenem-hydrolyzing β-lactamases. Antimicrob Agents Chemother. 1997;41:223–232. doi: 10.1128/aac.41.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossolini G M, Riccio M L, Cornaglia G, Pagani L, Lagatolla C, Selan L, Fontana R. Carbapenem-resistant Pseudomonas aeruginosa with acquired blaVIM metallo-β-lactamase determinants, Italy. Emerg Infect Dis. 2000;6:312–313. doi: 10.3201/eid0603.000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossolini G M, Franceschini N, Riccio M L, Mercuri P S, Perilli M, Galleni M, Frère J-M, Amicosante G. Characterization and sequence of the Chryseobacterium (Flavobacterium) meningosepticum carbapenemase: a new molecular class B β-lactamase showing a broad substrate profile. Biochem J. 1998;332:145–152. doi: 10.1042/bj3320145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segel I H. Biochemical calculations. 2nd ed. New York, N.Y: John Wiley & Sons; 1976. pp. 236–241. [Google Scholar]
- 24.Senda K, Arakawa Y, Nakashima K, Ito H, Ichiyama S, Shimokata K, Kato N, Ohta M. Multifocal outbreaks of metallo-β-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum β-lactams, including carbapenems. Antimicrob Agents Chemother. 1996;40:349–353. doi: 10.1128/aac.40.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, Ohsuka S, Shimokata K, Kato N, Ohta M. PCR detection of metallo-β-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum β-lactams. J Clin Microbiol. 1996;34:2909–2913. doi: 10.1128/jcm.34.12.2909-2913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toney J H, Fitzgerald P M, Grover-Sharma N, Olson S H, May W J, Sundelof J G, Vanderwall D E, Cleary K A, Grant S K, Wu J K, Kozarich J W, Pompliano D L, Hammond G G. Antibiotic sensitization using biphenyl tetrazoles as potent inhibitors of Bacteroides fragilis metallo-β-lactamase. Chem Biol. 1998;5:185–196. doi: 10.1016/s1074-5521(98)90632-9. [DOI] [PubMed] [Google Scholar]
- 27.Tsakris A, Pournaras S, Woodford N, Palepou M-F I, Babini G S, Douboyas J, Livermore D M. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J Clin Microbiol. 2000;38:1290–1292. doi: 10.1128/jcm.38.3.1290-1292.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh T R, Gamblin S, Emery D C, MacGowan A P, Bennett P M. Enzyme kinetics and biochemical analysis of ImiS, the metallo-β-lactamase from Aeromonas sobria 163a. J Antimicrob Chemother. 1996;37:423–431. doi: 10.1093/jac/37.3.423. [DOI] [PubMed] [Google Scholar]
- 29.Walter M W, Felici A, Galleni M, Paul Soto R, Adlington R M, Baldwin J E, Frère J-M, Gololobov M, Schofield C J. Trifluoromethyl alcohol and ketone inhibitors of metallo-β-lactamases. Bioorg Med Chem. 1996;6:2455–2458. [Google Scholar]
- 30.Yang Y, Bush K. Biochemical characterization of the carbapenem-hydrolyzing β-lactamase AsbM1 from Aeromonas sobria AER 14M: a member of a novel subgroup of metallo-β-lactamases. FEMS Microbiol Lett. 1996;137:193–200. doi: 10.1111/j.1574-6968.1996.tb08105.x. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y, Rasmussen B A, Bush K. Biochemical characterization of the metallo-β-lactamase CcrA from Bacteroides fragilis TAL3636. Antimicrob Agents Chemother. 1992;36:1155–1157. doi: 10.1128/aac.36.5.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]


