Abstract
Humans have had a major influence on the dissemination of crops beyond their native range, thereby offering new hybridization opportunities. Characterizing admixed genomes with mosaic origins generates valuable insight into the adaptive history of crops and the impact on current varietal diversity. We applied the ELAI tool—an efficient local ancestry inference method based on a two-layer hidden Markov model to track segments of wild origin in cultivated accessions in the case of multiway admixtures. Source populations—which may actually be limited and partially admixed—must be generally specified when using such inference models. We thus developed a framework to identify local ancestry with admixed source populations. Using sequencing data for wild and cultivated Coffea canephora (commonly called Robusta), our approach was found to be highly efficient and accurate on simulated hybrids. Application of the method to assess elite Robusta varieties from Vietnam led to the identification of an accession derived from a likely backcross between two genetic groups from the Congo Basin and the western coastal region of Central Africa. Admixtures resulting from crop hybridization and diffusion could thus lead to the generation of elite high-yielding varieties. Our methods should be widely applicable to gain insight into the role of hybridization during plant and animal evolutionary history.
Keywords: local ancestry inference, ELAI, crops, admixture, crop diffusion, Coffea canephora
Significance.
Local ancestry inference (LAI) has been widely investigated in humans to decipher genomic and evolutionary history, yet it is less commonly used in crops. Here we applied this approach to study mosaic genome origins in cultivated Coffea canephora (Robusta) accessions. We have proposed a new method to derive source populations for this analysis based on ancestral genotype frequencies estimated from native populations. Validation using simulated hybrids and ancestry deconvolution of the cultivated accessions revealed this approach to be promising for genomic studies of C. canephora as well as other crops.
Introduction
Human history and migrations have markedly impacted crop dispersal patterns worldwide (Khoury et al. 2016). Cultivated plants have gradually, over time, undergone diffusion beyond their centers of origin (Meyer et al. 2012), while exchanging genetic material from their original wild relatives via hybridization. For example, Australian wheat cultivars are the result of a multiway admixture of lineages of European, African, and American origins (Joukhadar et al. 2017). Intraspecific admixture is a key factor in population diversity, adaptation, and evolution (Goetz et al. 2014; Rius and Darling 2014). Cultivated individuals may, therefore, express novel phenotypic variability, for example, beneficial ear traits in domesticated emmers (Nave et al. 2019; Oliveira et al. 2020), or higher antioxidant activity in a new litchi cultivar (Zhao et al. 2020; Hu et al. 2022).
Population genetics tools have been widely developed to gain insight into admixture (Pritchard et al. 2000; Padhukasahasram 2014). STRUCTURE (Pritchard et al. 2000), ADMIXTURE (Alexander et al. 2009), or other tools like sNMF (Frichot et al. 2014) are commonly used to infer global admixture proportions and ancestry per individual in a population. Yet individuals presenting the same global ancestry might differ with respect to admixture patterns at the chromosome level. More recent advances in sequencing and the application of statistical and computing technologies have also enhanced ancestry inference at this chromosome level based on LAI approaches. At a fine genome scale, the local ancestry provides better information on the mosaic genetic origin of admixed individuals, that is enhancing knowledge of demographic histories, facilitating detection of adaptive introgression, deciphering complex traits, and mapping underlying genes in admixed populations (Padhukasahasram 2014; Thornton and Bermejo 2014; Mani 2017; Shriner 2017; Geza et al. 2019). For instance, admixture mapping based on LAI of a three-way admixed human population identified a genomic region in native American ancestry linked to Alzheimer's disease (Norris et al. 2020; Horimoto et al. 2021).
Numerous LAI tools have been developed. In most of them, genotypes from putative ancestral populations (so-called “source populations”) are used as a reference to infer the local ancestry of admixed individuals or tested individuals (Sankararaman et al. 2008). Most LAI models are based on hidden Markov models (HMM), including SABER (Tang et al. 2006), LAMP-LD (Baran et al. 2012), and ELAI (Guan 2014), which predict the classification of ancestries in hidden states by monitoring genotypes of the source populations (Baran et al. 2012). Other methods implement strategies based on principal component analysis, for example, PCADMIX (Brisbin et al. 2012), Markov chain Monte Carlo (Chromopainter, Lawson et al. 2012), random forest (RFMix, Maples et al. 2013), K-means (EILA, Yang et al. 2013), and other clustering methods (Wu et al. 2021). LAI models can also be categorized into linkage disequilibrium (LD)-based and non-LD-based models (Geza et al. 2019). Most LAI software requires phased genotypes (Wu et al. 2021), which are not always accurately estimated with short-read data obtained via next-generation sequencing (NGS) technologies (Garg et al. 2021). Some tools need biological information such as genetic and physical mapping data, recombination rates, and admixture generations, or statistical parameters such as hidden states and misfitting probabilities (Geza et al. 2019).
According to previous studies comparing the performance of several commonly used software programs (Cottin et al. 2019; Schubert et al. 2020; Molinaro et al. 2021), ELAI (Guan 2014) has proven to be one of the most efficient ancestry inference tools for dealing with unphased data. This two-layer HMM and LD-based method use source populations to predict the classification in two hidden state layers—haplotypes in the lower-layer and ancestries in the upper-layer. Therefore it does not require haplotype phasing, but only prior assumptions regarding the number of haplotype clusters and admixture generations, which are needed for hidden state modeling. ELAI has been applied in studies of different plants. For instance: in perennial plants such as aspen, local ancestry signals obtained using ELAI have generated insight into the local adaptation and demographic history of European varieties (Rendón-Anaya et al. 2021). In annual crops such as wheat, ELAI has also been applied to analyze gene flow from wild emmers to bread wheat (Zhou et al. 2020).
LAI tools perform more accurately when using source populations with a large sample size and high differentiation level (Cottin et al. 2019), whereas small, unbalanced structure or admixed source populations might cause erroneous ancestry assignment (Shringarpure and Xing 2014; Molinaro et al. 2021). In practice, it is often challenging to have a perfect sampling design across source populations (Hübner and Kantar 2021). Here we propose a solution to this problem of unbalanced and admixed individuals from source populations.
Coffea canephora Pierre ex A. Froehner, or so-called Robusta coffee, is an allogamous diploid species with high genetic and phenotypic differentiation (Berthaud 1986; Montagnon et al. 1992; Gomez et al. 2009; Cubry et al. 2013; Mérot-L’Anthoëne et al. 2019; Kiwuka et al. 2021; de Aquino et al. 2022). This species originates from central and western Africa (Davis et al. 2006), corresponding to two major genetic groups, that is Congolese and Guinean groups, respectively (Montagnon et al. 1992, 1998a; Cubry et al. 2013). The Congolese group consists of five well-described subgroups: group A in Benin and Gabon, group E in the Democratic Republic of the Congo (DRC), group C in Cameroon and the western Central Africa region, group B in eastern Central African Republic (CAR), and group O in Uganda; and two recently described groups, G in Angola and R in southern DRC (Mérot-L’Anthoëne et al. 2019). The Guinean group corresponds to group D (Mérot-L’Anthoëne et al. 2019).
While most of the crop species were domesticated during the Neolithic period, over the past 12,000 years (Harlan 1971; Larson et al. 2014), Robusta coffee cultivation and diffusion is much more recent. Robusta coffee has attracted interest as a potential cash crop since the late 19th century (Berthaud 1986), and the species has only become globally widespread since 1900 (Montagnon et al. 1998b). Coffee research stations and breeding centers have been set up since the early 20th century, in Java, Indonesia (1900–1930), and then DRC (1930–1960), and Central Africa (1960 onward) (Cramer 1957; Montagnon et al. 1998b). Coffea canephora breeding programs for varietal improvement have mainly been based on heterosis of crosses between the Congolese subgroup E and subgroup A or the Guinean group D (Leroy et al. 1993; Montagnon et al. 1998a; Oliveira et al. 2018).
Due to the gametophytic self-incompatibility of Robusta and its intense breeding history, cultivated populations might have experienced a high level of admixture, resulting in hybrids with complex mosaic genomes. The local ancestry deconvolution approach to cultivated Robusta varieties could help gain insight into their genetic makeup and trace back their origins. This novel approach would facilitate the development of admixture mapping—that is, associating the phenotype with ancestry segments in coffee—a technique that is sometimes more powerful than conventional genome-wide association studies (GWAS) (Horimoto et al. 2021). Several key traits for coffee breeding, such as drought tolerance, that is, a polygenic trait that is also considered to be a feature in the Congolese group A (Marraccini et al. 2012; Vieira et al. 2013).
Robusta coffee was first introduced to Vietnam in the early 20th century, probably from the Congo via France (Nogent-sur-Marne acclimation gardens) or via Java (coffee breeding center) (Vanden Abeele et al. 2021). In cultivated coffee, it takes 5–8 years to reach the maximum productive stage (Wrigley 1988), and it has been estimated to be even up to 20 years (Moat et al. 2019, Nab and Maslin 2020). Therefore, given 100 years of cultivation, the number of generations is not expected to be higher than 20. Even though the current Vietnamese Robusta varieties are mostly supposed to have originated from Java, their ancestral genetic groups are still largely unclear. Since materials of the Javanese breeding program mainly come from DRC, Uganda, and Gabon (Montagnon et al. 1998b), the accessions historically introduced in Vietnam may have Congolese origins and putatively some extent of intergroup admixture.
In this study, we implemented the ELAI approach on cultivated C. canephora with unbalanced and admixed native reference populations. We inferred ancestral genotypic frequencies for these native populations to build perfect source populations for ELAI. We assessed and validated our new approaches with simulated hybrids. We finally applied an optimal framework to a set of elite accessions cultivated in the Central Highlands of Vietnam to determine their mosaic genome origins.
Results
Characterization of Genetic Groups
A total of 11,919,576 high-quality biallelic SNPs were obtained in all of the 55 African and 10 Vietnamese individuals. We assessed the structure of African native groups by performing genetic structure analysis using sNMF on a set of 1,191,957 randomly picked SNPs.
The African individuals were classified into five groups (supplementary figs. S1 and S2, Supplementary Material online) that could be linked to geographical origins: a West African group with accessions from Guinea, Ghana, and Côte d’Ivoire (group D), a group with accessions from Cameroon (group C), a group with accessions from Gabon and Angola (group AG), an East African group with accessions from Uganda and CAR (group OB), and finally the last group consisted of DRC accessions (group ER). Most pairwise FST values between the five genetic groups were high and ranged from 0.39 to 0.55, except for the FST = 0.22 between ER and OB (supplementary table S1, Supplementary Material online). This strong structuring was also confirmed by principal component analysis (PCA) analysis (supplementary fig. S3, Supplementary Material online).
ELAI Accuracy Assessment
The size of the African reference set was small, with an unequal number of individuals (unbalanced structure), while 15 individuals presented some extent of admixture (>20% admixture) (supplementary fig. S2, Supplementary Material online). We built near-perfect source populations for the five groups based on ancestry genotype frequencies. All of them had perfect ancestry coefficients (>97%) relative to their respective groups, as expected (supplementary fig. S4, Supplementary Material online). The artificial source populations were then used for the assessment of ELAI performance in detecting simulated hybrids (supplementary fig. S5, Supplementary Material online).
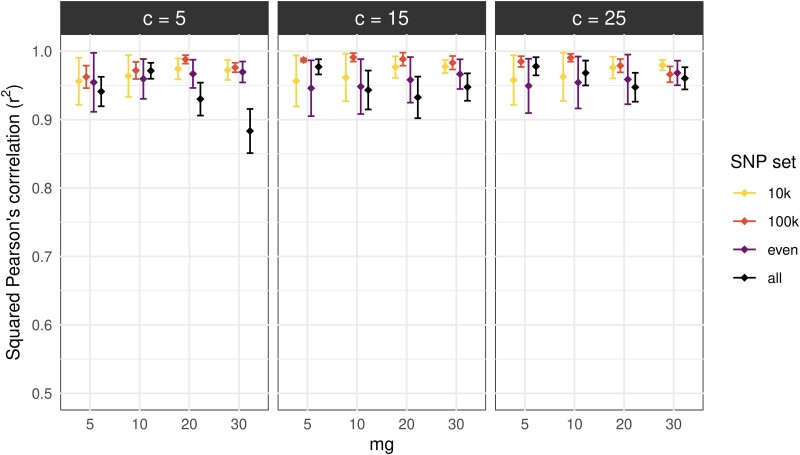
Using simulated hybrids, we found that our approach achieved accurate inferences with high correlations (r2) ranging from 0.859 to 0.997 (fig. 1), regardless of the set of parameters used. The lowest squared correlation (r2 = 0.859) corresponded to ELAI runs using all SNPs with c (number of lower clusters) = 5 and mg (number of admixture generations) = 30. All other ELAI runs had r2 values >0.9. The number of lower clusters and admixture generations did not have marked impacts on the ELAI accuracy, except when all SNPs were used. Conversely, the use of higher parameter values and SNP numbers increased the ELAI run time and memory usage (supplementary table S2, Supplementary Material online).
Fig. 1.
ELAI accuracy in inferring local ancestry in simulated hybrids. The plot shows correlations between the true local ancestry dosage and ELAI dosages with different parameter numbers: number of lower clusters (c = 5, 15, and 25), number of admixture generations (mg = 5, 10, 20, and 30), and different SNP sets (10 K SNPs, 100 K SNPs, evenly distributed SNPs—1 SNP/5 kb, and all SNPs—1 M SNPs). Each point represents an average of correlations computed for the three simulated hybrids, and error bars represent the standard deviation.
Among the four SNP sets tested, the overall accuracy was higher for ELAI runs with the 100 K SNP set, while slightly decreasing with the other SNP sets, 10 K SNPs, evenly distributed 1 SNP/5 kb SNPs, and all SNPs (when mg = 20 and 30), respectively. These results were in line with the fact that ELAI accounts for the background LD when detecting the haplotype structure, so a higher number of SNPs provides more haplotype information and thus greater ancestry assessment accuracy. However, using whole-chromosome SNPs did not improve but instead slightly reduced the ELAI accuracy as it might cause background noise or false inference with short segment lengths (<500 kb).
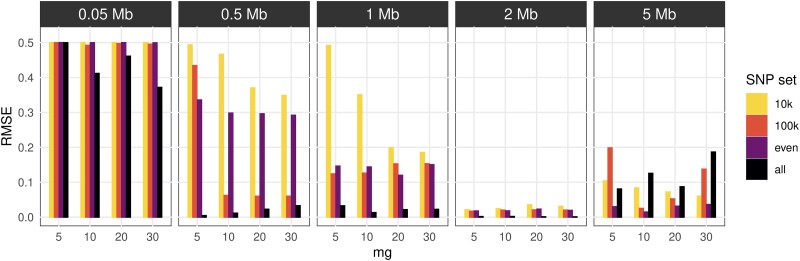
Despite the high r2 values, some false dosages were observed in the simulated introgression segments where the ancestry switched on both alleles compared to the flanking sequence. We computed the root mean square error (RMSE) between true and estimated dosages in the homozygous introgressed regions and compared it among a range of introgression sizes (0.05, 0.5, 1, 2, and 5 Mb).
Compared with the larger introgression tracts, RMSE values were higher for introgression tracts <1Mb, except for some runs using 100 K SNPs and the whole SNP set (fig. 2 and supplementary fig. S6, Supplementary Material online).
Fig. 2.
Error in detecting simulated introgression tracts of different lengths in simulated hybrids. RMSE was calculated between the true dosage and ELAI dosages of the simulated hybrids, for different homozygous introgressed segment lengths (0.05, 0.5, 1, 2, and 5 Mb). Each panel shows, for each tested length, the RMSE values (y-axis) for ELAI runs with the numbers of lower clusters (c = 5), different numbers of generations (mg = 5, 10, 20, and 30) on the x-axis, and four different SNP sets (10 K SNPs, 100 K SNPs, evenly distributed SNPs—1 SNP/5 kb, and all SNPs—1 M SNPs).
RMSE values of around 0.5 in some cases indicated that it was likely that only one haplotype had been correctly assigned along the introgression tracts. For introgression sizes 500 kb, 1 Mb, and especially 2 Mb, ELAI runs using whole-chromosome SNPs had the lowest RMSE values (<0.03), thereby indicating that the estimation was highly accurate. We observed an RMSE increase in larger introgressions (5 Mb) associated with false inferences of small fragments inside longer introgressed fragments. For 5 Mb admixture tracts, the least erroneous inferences (RMSE ranging from 0.01 to 0.04) were obtained in runs using the even SNPs set with c = 5 or 25, and in all, SNP sets with c = 15 (supplementary fig. S6, Supplementary Material online).
In summary, the ELAI method was highly accurate in assessing the ancestry deconvolution of the artificial hybrids using the simulated source populations, with good confidence for admixture tracts of >1 Mb length. The required parameters (number of lower clusters and admixture generations) only had a minor effect on the detection, while the SNPs chosen for analysis had more marked impacts on the admixture segment inference accuracy. The validation enabled us to define the parameters for the application of the method on cultivated Robusta individuals.
Optimized Framework and Application for Inference of the Local Ancestry of the Tested Vietnamese Robusta Cultivated Accessions
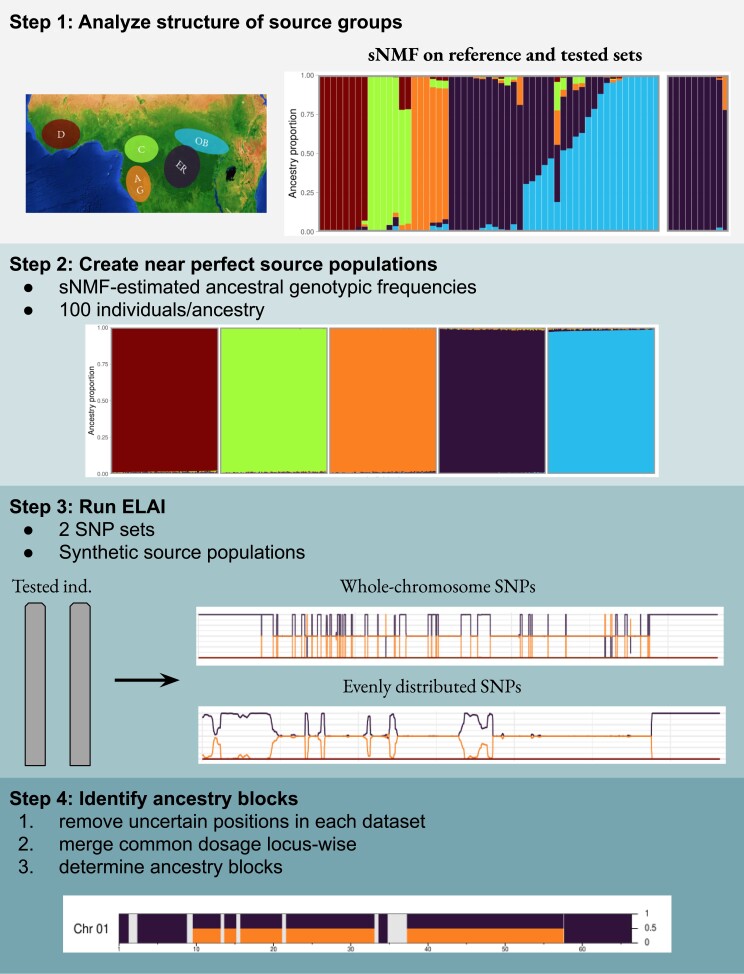
Based on our validation and optimization results using simulated hybrids, we developed an LAI framework that encompassed ELAI to efficiently study the admixture origin in Robusta coffee (fig. 3). For each chromosome, we performed ELAI using two SNP datasets (a set of evenly distributed SNPs, and another of whole-chromosome SNPs), with simulated ancestral groups serving as source populations. The lower cluster number was set at five as this factor did not influence the detection but did reduce the run time and memory usage (supplementary table S2, Supplementary Material online). We set the number of admixture generations at 20, which reflected the maximum possible number of generations of the cultivated accessions. Common results between the two datasets (evenly distributed SNPs and whole-chromosome SNPs) were then considered as the final LAI.
Fig. 3.
Framework for LAI of cultivated C. canephora. ELAI was performed for each individual chromosome and involved three main steps. Step 1: analyzing the genetic structure of the ancestral group, by performing sNMF on the reference set and tested set. Step 2: simulating source populations based on sNMF-estimated ancestral genotypic frequencies. Step 3: running ELAI on the tested individuals using two marker sets, that is whole-chromosome SNPs and evenly distributed SNPs. Step 4: merging the ancestry dosages inferred in the two SNP sets to determine the final consensus inference of the target chromosome.
The results of the two SNP sets were pooled in three steps. First, as the theoretical dosage of a given ancestry at an SNP locus is either 0, 0.5, or 1 but the dosage inferred by ELAI can be any value between 0 and 1, we approximated the ELAI-inferred dosage at each SNP with respect to the theoretical values, that is the dosage was set at 0 if the inferred dosage was in the [0, 0.1) range, at 0.5 if it was in the (0.4, 0.6) range, at 1 if it was in the (0.9, 1] range, or classified as “undetermined” otherwise. Second, the approximated dosages in the two SNP sets were compared locus-wise and also classified as “undetermined” if they were not equal. Finally, ancestry blocks were determined if contiguous positions had the same dosage and the distance between adjacent positions was not >1 Mb; and <1 Mb segments were also classified as “undetermined”.
This framework was then applied for LAI of the Vietnamese accessions.
A total of 94% to >100% of the genome could be assigned (supplementary fig. S7, Supplementary Material online) for all of the ten tested Vietnamese accessions. Undetermined regions were due to disagreement between the results of the two datasets, or the uncertainty in the ELAI inferred dosage (ancestry dosages within 0.1–0.4 or 0.6–0.9 ranges, or ancestry tracts <1 Mb were treated as uncertainties). Some accessions represented the same undetermined region on chromosome 10 (supplementary fig. S7, Supplementary Material online) because this region was assigned with a dosage of 1 to the ER group by the even SNPs set but with a dosage of 0.5 for both ER and AG groups by the whole-chromosome SNP set.
Based on these ancestry blocks, the global ancestry inference of the tested individuals could be estimated as follows: for each ancestry, the overall proportion was the sum of all block dosages/genome assembly size (≈585 Mb), with the block dosage being the length of an ancestry block × ancestry inference for the block. The global ancestry coefficients detected by our ELAI framework were generally similar to the results estimated by sNMF (supplementary fig. S2, Supplementary Material online). Nine accessions presented >99% ancestry in the DRC-native ER group (supplementary fig. S7, Supplementary Material online). Two of these accessions, that is TR5 and TR15, had minor admixture proportions in one haplotype, that is 1.6 Mb of group AG on chromosome 4 and 1.2 Mb of group C on chromosome 10, respectively.
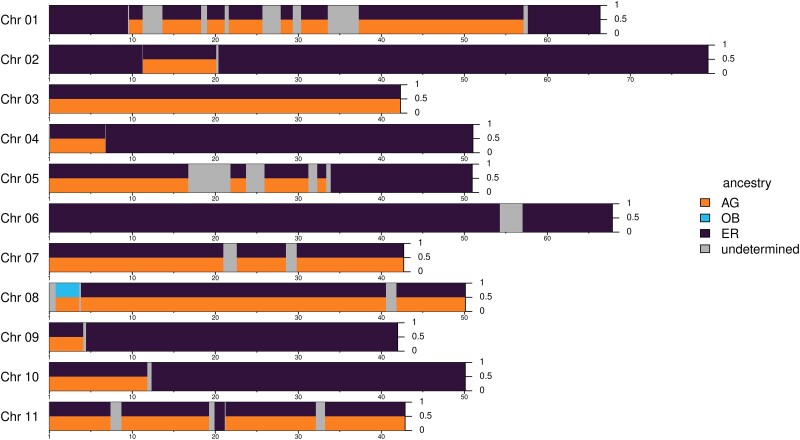
The TR6 accession genome consisted of segments from two ancestral groups, that is, ER and AG, with 72% and 22% of the genome, respectively (supplementary fig. S7, Supplementary Material online and fig. 4). The admixture patterns varied in different parts of genome: chromosomes 3, 7, 8, and 11 were completely heterozygous (excluding the undetermined segments); chromosomes 4, 9, and 10 had a single admixture tract at the terminal end of the chromosome; chromosome 1, 2, and 5 had admixture segments separately distributed along the chromosome; while, exceptionally, chromosome 6 did not present any admixture signal. A small haplotype fragment of 2.8 Mb length on chromosome 8 was assigned to group OB, which accounted for only 0.2% of the genome. The local ancestry pattern suggested that TR6 resulted from recombination events between individuals of the two main Congolese AG and ER subgroups, backcrossed with the ER group.
Fig. 4.
LAI of the Vietnamese TR6 accession. Each bar presents the consensus local ancestry dosage (y-axis) along the positions (x-axis) of each chromosome. The x-axis labels are in the Mb unit. The inferred ancestral groups—ER (from DRC), AG (from Benin, Gabon, and Angola), and OB (from Uganda and Central Africa), are denoted by colors. The gray portions are undetermined regions.
Discussion
An Optimized Framework to Infer Local Ancestry in Robusta Coffee
In this study, we developed an LAI framework implementing the ELAI method, which was performed with high accuracy for ancestry deconvolution of admixed individuals using derived source populations and with good confidence for admixture tracts of >1 Mb length.
In particular, we implemented a simple method to overcome the lack of bias in native reference individual sampling, which was efficient for ELAI on both simulated and real data inference. Our method was based on ancestral genotype frequencies, which can be directly obtained from unphased genotypic data using sNMF. To our knowledge, this is the first time that such an approach has been proposed for unphased data. Another method combining WINC-ChromoPainter with the non-negative least squares approach has been developed to analyze LAI when there are very few reference individuals and no source population simulations (Molinaro et al. 2021). A test of this method on real data showed that WINC was comparable to or could outperform ELAI in certain admixture scenarios when only two individuals per source population were used. However, WINC requires phased data and recombination maps, which are not always available for other datasets, especially our dataset. ELAI was later adapted to use a large number of admixed samples (a cohort set) to compensate for the lack of a pure source population (Zhou et al. 2016). The cohort set was down-weighted to not outweigh other training source sets. This method could be applicable if a large cohort sample size is available. We acknowledge that our simulation method did not preserve LD in the native populations, which might explain the misassignment or uncertainty in the inference of small ancestry tracts in admixed individuals. However, the uncertainty only accounted for a minor portion of the genome (<10%) and the global ancestry findings of our approach were close to the global ancestry obtained by sNMF.
We also assessed the number of haplotype clusters and admixture generations required by ELAI to predict the ancestry model. The Robusta coffee breeding program was launched about 100 years ago, which could be considered as relatively recent compared to most other crops, and therefore the maximum number of Robusta generations out of Africa and especially in Vietnam was estimated at most 20. We tested the methods with 5, 10, 20, and 30 generations. Guan (2014) found that a higher number of admixture generations improved the inference smoothness. In many studies using ELAI, the number of haplotype clusters is often set at 5-fold the number of source populations. These parameters were shown to have an impact on inference in human genomes (Guan 2014), but we did not clearly observe such an effect in our simulations. Even when the number of lower clusters was set at the number of source populations, so each source population was linked to only one haplotype in the higher clusters, we could still quite accurately infer the ancestry source. Our approach to generate ideal source populations will only keep LD linked to population structure. As we observed high genetic differentiation in C. canephora, we certainly had high LD linked to the population structure. Lower differentiation between ancestral populations might lead to lower performance of the approach we have proposed here.
ELAI has been shown to be a highly robust LAI tool (Cottin et al. 2019; Schubert et al. 2020; Molinaro et al. 2021). Indeed, our results obtained using simulated C. canephora hybrids also illustrated its high accuracy for detecting >1 Mb ancestry blocks, even with a small portion (1%) of whole-genome SNPs, that is 10 K SNPs out of the 1.1 M whole-genome SNPs on chromosome 1. Yet in our framework, we combined inferences obtained with SNP sets of two densities (whole-chromosome SNPs and evenly distributed SNPs, i.e., 203 and 1.8 SNPs per 10 kb, respectively) to enhance the inferred ancestry confidence.
Robusta Origin, Diffusion, and LAI
The native African Robusta individuals were classified into five groups that could be linked to geographical origins, and this genetic structure was perfectly congruent with previous study findings (Tournebize et al. 2022). The latest classification of C. canephora using 8.5 K SNP arrays (Mérot-L’Anthoëne et al. 2019) led to an eight-group classification, but the differentiation between groups O and B, E and R, and A and G was low, so they were clustered in this study and our previous study (Tournebize et al. 2022). The clustering of individuals of closely related groups was also due to bias toward one group when the other contained a small number of individuals.
Using source populations derived from these native groups, we ran our optimized framework method on a sample of ten elite cultivars from Vietnam. Inference of these test Robusta accessions revealed that all of them originated from the Congolese groups ER and AG. Nine accessions shared a common ancestry of group ER, and one likely came from a hybrid between the two Congolese ER and AG groups, backcrossed with ER. Previous studies on other Robusta accessions in Vietnam also identified their Congolese origin (Garavito et al. 2016; Akpertey et al. 2021). Garavito et al. (2016) used DArTseq SNPs and found six Vietnamese accessions from the Congolese E group (included in our ER group). Akpertey et al. (2021) used KASP (Kompetitive Allele Specific PCR) SNPs and found 33 Vietnamese accessions distinguished from Côte d’Ivoire and Togo accessions (putatively the Guinean group), but no reference for the Congolese groups was used in that study. These inferences are in line with historical coffee breeding data.
These accessions, which are recognized elite Robusta accessions in Vietnam, could serve as potential breeding materials for varietal improvement. The TR6 accession genome was found to be composed of two ancestral ER and AG groups, accounting for 72% and 22% of the genome, respectively. The Congolese genetic group E was previously found to present advantageous phenotypic characteristics such as good aroma and low acidity, high leaf rust resistance, but with susceptibility to drought and twig borers (Montagnon et al. 1998a). In contrast to group E, genetic group A has very high twig borer resistance and drought tolerance, but is only moderately resistant to leaf rust, and sometimes exhibits lower cup quality (Montagnon et al. 1998a). Hybridization of these two groups might produce accessions with heterosis characteristics combining these advantageous agronomic traits.
Inference of wild ancestry segments in the cultivated accessions could also enable downstream analyses such as admixture mapping of important traits, or genomic selection for breeding programs. Breeding strategies could now also be tailored for different purposes. Reciprocal recurrent selection between the Congolese group and Guinean group (group D) has been used to improve yield and vegetative vigor (Leroy et al. 1993; Montagnon et al. 1998b; Oliveira et al. 2018), while recurrent selection within hybrid populations was more effective for enhancing disease resistance (Alkimim et al. 2021). Therefore, studies on the genetic origin, especially the LAI of Robusta materials available in collections, could also boost the efficiency of coffee breeding programs.
This approach could also be adapted to other species when studying admixed populations with a low number of reference individuals.
Materials and Methods
Materials
We used two sets of accessions: (1) 55 previously sequenced wild C. canephora accessions from Africa (Tournebize 2017; Tournebize et al. 2022); and (2) ten newly sequenced cultivated C. canephora accessions from Vietnam. The wild African samples are representative of the native range of C. canephora. The cultivated samples from Vietnam are recognized as elite plants and conserved in the germplasm bank of the Western Highlands Agriculture & Forestry Science Institute (WASI).
Sequencing, Mapping, and SNP Calling
The ten Vietnamese individuals were sequenced using Illumina Hiseq X Ten PE 150 bp. The 55 samples from Africa were obtained from GenBank and analyzed with the new ten Vietnamese genomes. Variant calling was performed according to GATK Best Practices recommendations for germline short variant discovery using the TOGGLE framework (Monat et al. 2015). The reads were first mapped against the v1.8 reference genome (de Kochko and ACGC 2018) using BWA mem 0.7.2 (Li 2013), then sorted using Picard Tools 1.83 (https://broadinstitute.github.io/picard/) and SAMtools 0.1.3 (Danecek et al. 2021). Variants per sample were called in individual GVCFs (Genomic Variant Call Format) using GATK HaplotypeCaller 3.6, and consolidated using GATK CombineGVCFs, and then final variant calling was jointly performed in the whole GVCF set using GATK GenotypeGVCFs (Poplin et al. 2017). High-quality biallelic SNPs were obtained by applying the following filtering criteria: remove indels, consider only biallelic SNPs, remove clusters of at least 4 SNPs in 10 bp sliding windows, remove SNPs with QUAL <200, MQ0 > 4 & MQ0/DP >0.1, mean depth >100 or <10, and missing data >15%, by using GATK 4.0.0.0, BCFtools 1.9 (Danecek et al. 2021), and VCFtools 0.1.16 (Danecek et al. 2011).
Characterization of Population Structure
We characterized the genetic groups present in the native African Robusta coffee populations via genetic structure analysis with sNMF (Frichot et al. 2014), using the R package LEA (Frichot and François 2015). 10% randomly chosen SNPs were used to assess the number of genetic groups. The optimal number of ancestral groups (K) was determined using cross-entropy criteria over ten iterations with K ranging from 1 to 10. Individuals were assigned to a given cluster at an 80% ancestry threshold.
Pairwise FST between the genetic groups (restricted to individuals with >80% ancestry) was computed using the R package StAMPP (Pembleton et al. 2013). Hundred bootstraps across loci were performed to assess the significance. We also performed a PCA in the R package LEA (Frichot and François 2015).
Inference of Local Ancestry
We inferred the local ancestry of cultivated Robusta using ELAI (Guan 2014), which was suitable for our unphased data and did not require any additional biological information. The source populations must be specified when using ELAI. To overcome limitations due to the unbalanced structure of wild populations that include admixed individuals, we tested an alternative method using population structure analysis. This new method was validated by simulation of known hybrids, while the ELAI accuracy was assessed and optimal parameters determined. Based on these results, we built a framework to detect the local ancestry in Vietnamese-cultivated Robusta accessions.
Source Population
Genotypes from ancestral groups of wild accessions must be defined for the purpose of ELAI analysis (Guan 2014). Given the small and unbalanced sample size of the wild populations and some evidence of admixed ancestry between groups in the wild accessions, we generated new synthetic populations exhibiting genotypic frequencies similar to those estimated in the ancestral pools. To this end, we used ancestral genotypic frequencies inferred using the sNMF analysis (Frichot et al. 2014). For each chromosome, we ran sNMF on whole-chromosome SNPs of all the African and Vietnamese individuals, with ten iterations and the optimized K value. The best run over the ten iterations was chosen based on cross-entropy criteria. We applied the G function (R package LEA) on the result of the best run to retrieve a G matrix containing genotype frequencies inferred in each different ancestral group for all diploid SNPs. This G matrix was then used as a probability matrix for randomly choosing genotypes at each site by groups using the sample function in R (R Core Team 2022).
We checked if the generated genotypes were representative of the ancestral groups by performing a further sNMF analysis jointly on simulated and real accessions using random 100 K SNPs on chromosome 1 (with optimal K and ten iterations). The ancestral proportion of each simulated individual was obtained from the run with the lowest cross entropy.
Simulated Hybrids
We simulated hybrids with known admixture levels in order to determine optimal parameters and assess the accuracy of the ELAI approach with the simulated source populations in our C. canephora model. We chose two accessions from the wild African set (BGQ07 and 20738) representative of two divergent genetic groups to simulate three different hybrids with different admixed segment sizes. Each hybrid had admixture segments of different lengths (50 kb, 500 kb, 1 Mb, 2 Mb, and 5 Mb), and in homozygous (both alleles originating from one of the ancestral groups) and/or heterozygous form (one allele from each of the two ancestries). In the homozygous regions, the hybrid genotypes were copied from one respective progenitor, while at heterozygous loci each of two alleles was drawn randomly from the alleles of the two parents. Simulations were based on chromosome 1 SNPs.
Sets of ELAI Parameters
We determined the optimal ELAI parameters by running the software with varying parameter values, including the number of haplotype clusters (c), that is lower clusters, the number of admixture generations (mg), and the set of SNPs used for the analysis. We tested three values (5, 15, and 25) for the number of haplotype clusters, and four values (5, 10, 20, and 30) for the number of admixture generations. We also used four different sets of SNPs: (1) randomly selected 10 K SNPs, (2) randomly selected 100 K SNPs, (3) about 11 K SNPs resulting from randomly selecting one SNP in every nonoverlapping 5 kb window (referred to as the “even SNP set”), and (4) whole-chromosome SNPs, that is the “all SNP set” comprising 1,133,736 SNPs. The average SNP densities in the four datasets were 1.5, 15.1, 1.8, and 203.0 SNPs per 10 kb, respectively.
In summary, we performed 48 ELAI runs with different combinations of parameters, and each run used 20 expectation maximization (EM) steps (Guan 2014). To reduce the computational cost, for the run with whole-chromosome SNPs, the analysis was performed by splitting the chromosome into consecutive SNP chunks so that each subset contained a maximum of 100 K SNPs. The ELAI results obtained on the SNP subsets were then concatenated.
Assessment of the ELAI Inference Accuracy based on Simulated Hybrids
Each ancestry group was defined for the simulated hybrids as ancestry dosage = 1, where the two alleles were copied from the first parent (first genetic group); ancestry dosage = 0, where both alleles were from the second parent (second genetic group); and ancestry dosage = 0.5, where the alleles were derived from both parents (50% admixture). As we knew the true allelic dosages of the simulated hybrids, we could compare them to those inferred via ELAI.
The ELAI performance was assessed by correlation and root mean square error (RMSE) metrics. A correlation was the average Pearson's correlation between the estimated and true dosages. The RMSE between the estimated and true dosages of each admixture tract was calculated and averaged for different segment lengths (50 and 500 kb, and 1, 2, and 5 Mb).
Framework to Infer Local Ancestry in Vietnamese-Cultivated Robusta Coffee
Based on the ELAI validation data, a workflow (fig. 3) was developed to detect local ancestry in cultivated Robusta accessions and applied to the ten Vietnamese Robusta accessions. ELAI was performed for each chromosome, with the simulated ancestral populations as the ancestry source using the whole-chromosome SNP set and the evenly distributed SNP set. Three independent ELAI runs with 20 EM steps were conducted for each individual and SNP was set to obtain average results. The ELAI inferences of these sets were then combined to obtain the final ELAI.
Supplementary Material
Acknowledgments
T.V. was funded by Ph.D. grants from the French Embassy in Vietnam and the ARTS program (IRD). This research was funded by the Ministry of Science and Technology of Vietnam (MOST) under grant number NVQG-2020/ĐT.15. The authors acknowledge the ISO 9001 certified IRD i-Trop High Performance Computing (member of the South Green Platform) at IRD Montpellier for providing HPC resources that contributed to the research results reported in this paper. URL: https://bioinfo.ird.fr/-http://www.southgreen.fr.
Contributor Information
Tram Vi, UMR DIADE, Univ Montpellier, IRD, CIRAD, Montpellier, France; National Key Laboratory of Plant Cellular Biotechnology, Agricultural Genetics Institute, Hanoi, Vietnam.
Yves Vigouroux, UMR DIADE, Univ Montpellier, IRD, CIRAD, Montpellier, France.
Philippe Cubry, UMR DIADE, Univ Montpellier, IRD, CIRAD, Montpellier, France.
Pierre Marraccini, UMR DIADE, Univ Montpellier, IRD, CIRAD, Montpellier, France; UMR DIADE, CIRAD, Montpellier, France.
Ha Viet Phan, Western Highlands Agriculture & Forestry Science Institute, Buon Ma Thuot, Vietnam.
Giang Ngan Khong, National Key Laboratory of Plant Cellular Biotechnology, Agricultural Genetics Institute, Hanoi, Vietnam.
Valerie Poncet, UMR DIADE, Univ Montpellier, IRD, CIRAD, Montpellier, France.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online, Supplementary material online.
Author Contributions
V.P., Y.V., P.C., N.G.K., and T.V. designed the approach; V.P., P.M., and V.H.P. selected the Vietnamese coffee materials; P.C. and T.V. performed the mapping and SNP calling; T.V. performed the genetic structure and LAI analyses, all co-authors interpreted the results; T.V. wrote the first draft of the manuscript; V.P., Y.V., P.M., and P.C. commented and edited the manuscript, while all co-authors approved the manuscript.
Data Availability
The raw sequencing data of the African accessions were taken from NCBI SRA database under project accession number PRJNA803612 (Tournebize et al. 2022). The raw sequencing data of the Vietnamese accessions are available in NCBI SRA database under project accession number PRJNA950219 (this study). The software used for this study was downloaded from: https://github.com/haplotype/elai, https://github.com/bcm-uga/LEA, and other packages were available via IRD i-Trop HPC (South Green Platform).
Literature Cited
- Akpertey A, Padi FK, Meinhardt L, Zhang D. 2021. Effectiveness of single nucleotide polymorphism markers in genotyping germplasm collections of Coffea canephora using KASP assay. Front Plant Sci. 11:612593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkimim ER, et al. 2021. Designing the best breeding strategy for Coffea canephora: genetic evaluation of pure and hybrid individuals aiming to select for productivity and disease resistance traits. PLoS ONE. 16:e0260997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran Y, et al. 2012. Fast and accurate inference of local ancestry in Latino populations. Bioinform 28:1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthaud J. 1986. Les ressources genetiques pour l’amelioration des cafeires africains diploides. Paris: ORSTOM. [Google Scholar]
- Brisbin A, et al. 2012. PCAdmix: principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum Biol. 84:343–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottin A, Penaud B, Glaszmann J-C, Yahiaoui N, Gautier M. 2019. Simulation-based evaluation of three methods for local ancestry deconvolution of non-model crop species genomes. G3 (Bethesda). 10:569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer PJS. 1957, editors. A review of literature of coffee research in Indonesia. Turrialba: (Costa Rica: ): SIC Editorial, Inter American Institute of Agricultural Sciences. p. 128–140. [Google Scholar]
- Cubry P, De Bellis F, Pot D, Musoli P, Leroy T. 2013. Global analysis of Coffea canephora Pierre ex Froehner (Rubiaceae) from the Guineo-Congolese region reveals impacts from climatic refuges and migration effects. Genet Resour Crop Evol. 60:483–501. [Google Scholar]
- Danecek P, et al. 2011. The variant call format and VCFtools. Bioinform 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, et al. 2021. Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Govaerts R, Bridson DM, Stoffelen P. 2006. An annotated taxonomic conspectus of the genus Coffea (Rubiaceae). Bot J Linn Soc. 152:465–512. [Google Scholar]
- de Aquino SO, et al. 2022. Adaptive potential of Coffea canephora from Uganda in response to climate change. Mol Ecol. 31:1800–1819. [DOI] [PubMed] [Google Scholar]
- de Kochko A, ACGC . 2018. Deciphering the Allotetraploid Genome of Coffea arabica L. In: Plant and Animal Genome Conference XXVI; 2018 Januray 13–17; San Diego, CA, USA. [Google Scholar]
- Frichot E, François O. 2015. LEA: an R package for landscape and ecological association studies. Methods Ecol Evol. 6:925–929. [Google Scholar]
- Frichot E, Mathieu F, Trouillon T, Bouchard G, François O. 2014. Fast and efficient estimation of individual ancestry coefficients. Genet 196:973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavito A, Montagnon C, Guyot R, Bertrand B. 2016. Identification by the DArTseq method of the genetic origin of the Coffea canephora cultivated in Vietnam and Mexico. BMC Plant Biol. 16:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S, et al. 2021. Chromosome-scale, haplotype-resolved assembly of human genomes. Nat Biotechnol. 39:309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geza E, et al. 2019. A comprehensive survey of models for dissecting local ancestry deconvolution in human genome. Brief Bioinform. 20:1709–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz LH, Uribe-Bruce L, Quarless D, Libiger O, Schork NJ. 2014. Admixture and clinical phenotypic variation. Hum Hered. 77:73–86. [DOI] [PubMed] [Google Scholar]
- Gomez C, et al. 2009. Current genetic differentiation of Coffea canephora Pierre ex A. Froehn in the Guineo-Congolian African zone: cumulative impact of ancient climatic changes and recent human activities. BMC Evol Biol. 9:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y. 2014. Detecting structure of haplotypes and local ancestry. Genetics 196:625–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan JR. 1971. Agricultural origins: centers and noncenters. Science 174:468–474. [DOI] [PubMed] [Google Scholar]
- Horimoto ARVR, Xue D, Thornton TA, Blue EE. 2021. Admixture mapping reveals the association between native American ancestry at 3q13.11 and reduced risk of Alzheimer's Disease in Caribbean hispanics. Alzheimers Res Ther. 13:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, et al. 2022. Two divergent haplotypes from a highly heterozygous lychee genome suggest independent domestication events for early and late-maturing cultivars. Nat Genet. 54:73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner S, Kantar MB. 2021. Tapping diversity from the wild: from sampling to implementation. Front Plant Sci. 12:626565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukhadar R, Daetwyler HD, Bansal UK, Gendall AR, Hayden MJ. 2017. Genetic diversity, population structure and ancestral origin of Australian wheat. Front Plant Sci. 8:2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury CK, et al. 2016. Origins of food crops connect countries worldwide. Proc Royal Soc B. 283:1–9. [Google Scholar]
- Kiwuka C, et al. 2021. Genetic diversity of native and cultivated Ugandan Robusta coffee (Coffea canephora Pierre ex A. Froehner): climate influences, breeding potential and diversity conservation. PLoS One. 16:e0245965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, et al. 2014. Current perspectives and the future of domestication studies. Proc Natl Acad Sci U S A. 111:6139–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DJ, Hellenthal G, Myers S, Falush D. 2012. Inference of population structure using dense haplotype data. PLoS Genet. 8:e1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy T, Montagnon C, Charrier A, Eskes AB. 1993. Reciprocal recurrent selection applied to Coffea canephora Pierre. I. Characterization and evaluation of breeding populations and value of intergroup hybrids. Euphytica 67:113–125. [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. doi: 10.48550/arXiv.1303.3997. [DOI]
- Mani A. 2017. Local ancestry association, admixture mapping, and ongoing challenges. Circ Cardiovasc Genet. 10:e001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maples BK, Gravel S, Kenny EE, Bustamante CD. 2013. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet. 93:278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraccini P, et al. 2012. Differentially expressed genes and proteins upon drought acclimation in tolerant and sensitive genotypes of Coffea canephora. J Exp Bot. 63:4191–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérot-L’Anthoëne V, et al. 2019. Development and evaluation of a genome-wide coffee 8.5 K SNP array and its application for high-density genetic mapping and for investigating the origin of Coffea arabica L. Plant Biotechnol J. 17:1418–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RS, DuVal AE, Jensen HR. 2012. Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol. 196:29–48. [DOI] [PubMed] [Google Scholar]
- Moat J, Gole TW, Davis AP. 2019. Least concern to endangered: applying climate change projections profoundly influences the extinction risk assessment for wild Arabica coffee. Glob Chang Biol. 25:390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinaro L, et al. 2021. A chromosome-painting-based pipeline to infer local ancestry under limited source availability. Genome Biol Evol. 13:evab025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monat C, et al. 2015. TOGGLE: toolbox for generic NGS analyses. BMC Bioinform. 16:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnon C, Leroy T, Eskes A. 1998a. Amélioration variétale de Coffea canephora. 1 : critères et méthodes de sélection. Plantations: Recherche, Développement. p. 89–98. [Google Scholar]
- Montagnon C, Leroy T, Eskes A. 1998b. Amélioration variétale de Coffea canephora. 2 : les programmes de sélection et leurs résultats. Plantations, Recherche, Développement. http://agritrop.cirad.fr/390311/ (Accessed June 19, 2019).
- Montagnon C, Leroy T, Yapo A. 1992. Diversité génotypique et phénotypique de quelques groupes de caféiers (Coffea canephora pierre) en collection. Conséquences sur leur utilisation en sélection. Café, Cacao, Thé. 36:187–198. [Google Scholar]
- Nab C, Maslin M. 2020. Life cycle assessment synthesis of the carbon footprint of Arabica coffee: case study of Brazil and Vietnam conventional and sustainable coffee production and export to the United Kingdom. Geo: Geogr Environ. 7:e00096. [Google Scholar]
- Nave M, et al. 2019. Wheat domestication in light of haplotype analyses of the Brittle rachis 1 genes (BTR1-A and BTR1-B). Plant Sci. 285:193–199. [DOI] [PubMed] [Google Scholar]
- Norris ET, et al. 2020. Admixture-enabled selection for rapid adaptive evolution in the Americas. Genome Biol. 21:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira LNL, et al. 2018. Selection of Coffea canephora parents from the botanical varieties Conilon and Robusta for the production of intervarietal hybrids. Cienc Rural. 48:e20170444. [Google Scholar]
- Oliveira HR, Jacocks L, Czajkowska BI, Kennedy SL, Brown TA. 2020. Multiregional origins of the domesticated tetraploid wheats. PLoS One. 15:e0227148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhukasahasram B. 2014. Inferring ancestry from population genomic data and its applications. Front Genet. 5:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pembleton LW, Cogan NOI, Forster JW. 2013. StAMPP: an R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol Ecol Resour. 13:946–952. [DOI] [PubMed] [Google Scholar]
- Poplin R, et al. 2017. Scaling accurate genetic variant discovery to tens of thousands of samples. doi: 10.1101/201178. [DOI]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2022. R: A language and environment for statistical computing. Vienna: (Austria: ): R Foundation for Statistical Computing. https://www.R-project.org/ [Google Scholar]
- Rendón-Anaya M, et al. 2021. Adaptive introgression facilitates adaptation to high latitudes in European aspen (Populus tremula L.). Mol Biol Evol. 38:5034–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rius M, Darling JA. 2014. How important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol Evol. 29:233–242. [DOI] [PubMed] [Google Scholar]
- Sankararaman S, Sridhar S, Kimmel G, Halperin E. 2008. Estimating local ancestry in admixed populations. Am J Hum Genet. 82:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert R, Andaleon A, Wheeler HE. 2020. Comparing local ancestry inference models in populations of two- and three-way admixture. PeerJ 8:e10090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner D. 2017. Overview of admixture mapping. Curr Prot Human Genet. 94(1):1–23. http://onlinelibrary.wiley.com/doi/abs/10.1002/cphg.44 (Accessed February 18, 2022). [DOI] [PubMed] [Google Scholar]
- Shringarpure S, Xing EP. 2014. Effects of sample selection bias on the accuracy of population structure and ancestry inference. G3 (Bethesda). 4:901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Coram M, Wang P, Zhu X, Risch N. 2006. Reconstructing genetic ancestry blocks in admixed individuals. Am J Hum Genet. 79:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton TA, Bermejo JL. 2014. Local and global ancestry inference, and applications to genetic association analysis for admixed populations. Genet Epidemiol. 38:S5–S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R. 2017. Influence des variations spatio-temporelles de l’environnement sur la distribution actuelle de la diversité génétique des populations. Montpellier: (France: ): University of Montpellier. [Google Scholar]
- Tournebize R, et al. 2022. Ecological and genomic vulnerability to climate change across native populations of Robusta coffee (Coffea canephora). Glob Change Biol. 28:4124–4142. [DOI] [PubMed] [Google Scholar]
- Vanden Abeele S, et al. 2021. Genetic diversity of wild and cultivated Coffea canephora in northeastern DR Congo and the implications for conservation. Am J Bot. 108:2425–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira NG, et al. 2013. Different molecular mechanisms account for drought tolerance in Coffea canephora var. Conilon. Trop Plant Biol. 6:181–190. [Google Scholar]
- Wrigley G. 1988. Coffee–tropical agriculture series. Harlow: (UK: ): Longman Scientific & Technical. [Google Scholar]
- Wu J, Liu Y, Zhao Y. 2021. Systematic review on local ancestor inference from a mathematical and algorithmic perspective. Front Genet. 12:639877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JJ, Li J, Buu A, Williams LK. 2013. Efficient inference of local ancestry. Bioinform 29:2750–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Kun W, Kai W, Zhu J, Hu Z. 2020. Nutrient components, health benefits, and safety of litchi (Litchi chinensis Sonn.): a review. Compr Rev Food Sci Food Saf. 19:2139–2163. [DOI] [PubMed] [Google Scholar]
- Zhou Y, et al. 2020. Triticum population sequencing provides insights into wheat adaptation. Nat Genet. 52:1412–1422. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Zhao L, Guan Y. 2016. Strong selection at MHC in Mexicans since admixture. PLoS Genet. 12:e1005847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data of the African accessions were taken from NCBI SRA database under project accession number PRJNA803612 (Tournebize et al. 2022). The raw sequencing data of the Vietnamese accessions are available in NCBI SRA database under project accession number PRJNA950219 (this study). The software used for this study was downloaded from: https://github.com/haplotype/elai, https://github.com/bcm-uga/LEA, and other packages were available via IRD i-Trop HPC (South Green Platform).