Abstract
Three sets of carbapenem-resistant Serratia marcescens isolates have been identified in the United States: 1 isolate in Minnesota in 1985 (before approval of carbapenems for clinical use), 5 isolates in Los Angeles (University of California at Los Angeles [UCLA]) in 1992, and 19 isolates in Boston from 1994 to 1999. All isolates tested produced two β-lactamases, an AmpC-type enzyme with pI values of 8.6 to 9.0 and one with a pI value of approximately 9.5. The enzyme with the higher pI in each strain hydrolyzed carbapenems and was not inhibited by EDTA, similar to the chromosomal class A SME-1 β-lactamase isolated from the 1982 London strain S. marcescens S6. The genes encoding the carbapenem-hydrolyzing enzymes were cloned in Escherichia coli and sequenced. The enzyme from the Minnesota isolate had an amino acid sequence identical to that of SME-1. The isolates from Boston and UCLA produced SME-2, an enzyme with a single amino acid change relative to SME-1, a substitution from valine to glutamine at position 207. Purified SME enzymes from the U.S. isolates had β-lactam hydrolysis profiles similar to that of the London SME-1 enzyme. Pulsed-field gel electrophoresis analysis revealed that the isolates showed some similarity but differed by at least three genetic events. In conclusion, a family of rare class A carbapenem-hydrolyzing β-lactamases first described in London has now been identified in S. marcescens isolates across the United States.
Carbapenems are β-lactam antibiotics with broad antibacterial activity, increased stability to hydrolysis, and high rates of penetration through the bacterial outer membrane. Several mechanisms of resistance to carbapenems in gram-negative bacteria have been described and include the loss of outer membrane permeability and the production of β-lactam-hydrolyzing enzymes (2, 11, 16). Most carbapenem-hydrolyzing activity has been due to molecular class B metalloenzymes, such as CcrA in Bacteriodes fragilis (23) and IMP-1 in organisms such as Pseudomonas aeruginosa and Serratia marcescens (9, 20). The metalloenzymes, which contain two zinc atoms at the active site and which are distinguished by EDTA inhibition, were initially identified as chromosomal β-lactamases in Japan, Europe, and the United States (11, 16). In Japan, where carbapenems are used more frequently, IMP-1 has been found on plasmids (9). In Italy, the VIM-1 metallo-β-lactamase is located on an integron (10).
In addition to the zinc-based metalloenzymes, two groups of serine-based β-lactamases capable of carbapenem hydrolysis have emerged. The class D ARI enzyme from Acinetobacter baumannii has been found on plasmids in the United Kingdom, Argentina, Turkey, and Spain (7, 19). Three chromosomal class A β-lactamases that cause imipenem resistance have also been described: from Enterobacter cloacae, NMC-A in France (14) and IMI-1 in California (18), and from S. marcescens, SME-1 in England (12).
SME-1 was isolated from the imipenem-resistant S. marcescens strain S6, as well as strain S8, in London in 1982 (12). This chromosomal enzyme is capable of hydrolyzing penicillin, aztreonam, and cephalosporins in addition to imipenem. Since the discovery of SME-1, three sets of imipenem-resistant S. marcescens strains have been isolated from different regions of the United States. In 1985, a single imipenem-resistant isolate was discovered in Minnesota (A. A. Medeiros and R. S. Hare, Abstr. 26th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 116, 1986). Five isolates were collected from the University of California at Los Angeles (UCLA) in 1992 (J. P. Quinn, D. Miyashiro, J. Hindler, C. Holt, and K. Bush, Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-99, 1997), and 19 isolates were collected from the Deaconess Hospital (Boston) between 1994 and 1999 (Y. Carmeli, M. Samore, N. Troillet, M. Dube, L. Venkataraman, P. Degirolami, G. Eliopoulos, and K. Bush, Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-100, 1997). Isoelectric focusing of extracts from the United States strains revealed that they carried two β-lactamases—an AmpC-type enzyme with a pI of approximately 8.5 and one with a pI of approximately 9.5—similar to the pattern of enzymes seen in the London strain S. marcescens S6. Like SME-1 in the London strain S6, the carbapenem-hydrolyzing enzymes in the U.S. strains were assumed to be chromosomal, as no evidence of plasmids was found (12; J. P. Quinn, D. Miyashiro, J. Hindler, C. Holt, and K. Bush, Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-99, 1997). The purpose of this study was to compare the SME-type β-lactamases found in the United States to the London SME-1 enzyme.
(This study was presented in part at the 39th Interscience Conference on Antimicrobial Agents and Chemotherapy [A. M. Queenan, C. Torres-Viera, H. S. Gold, Y. Carmeli, G. M. Eliopoulos, R. C. Moellering, Jr., J. P. Quinn, J. Hindler, A. A. Medeiros, and K. Bush, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1466, 1999].)
MATERIALS AND METHODS
Bacterial strains.
The original S. marcescens clinical isolates and the Escherichia coli transformants harboring the SME-type β-lactamases are shown in Table 1. For locations at which multiple isolates were identified (UCLA and Boston), a representative isolate was chosen for analysis.
TABLE 1.
Bacterial strains
| Strain | Relevant properties | Reference or source |
|---|---|---|
| S. marcescens clincal isolates | ||
| S6 | 1982 Imipenem-resistant strain from London, producing SME-1 | 12 |
| 4176 | 1985 Imipenem-resistant strain from Minnesota (MN-2701) | Medeiros and Hare, 26th ICAAC |
| 4126 | 1992 Imipenem-resistant strain from UCLA (UCLA-1) | Quinn et al., 37th ICAAC |
| 4124 | 1995 Imipenem-resistant strain from Boston (no. 4) | Carmeli et al., 37th ICAAC |
| E. coli transformants | ||
| 4911 | Carries plasmid pIRS1, SME from 4176 cloned into pCR2.1 | This work |
| 4912 | Carries plasmid pIRS2, SME from 4126 cloned into pCR2.1 | This work |
| 4913 | Carries plasmid pIRS3, SME from S6 cloned into pCR2.1 | This work |
| 4914 | Carries plasmid pIRS4, SME from 4124 cloned into pCR2.1 | This work |
| E. coli One Shot | Recipient for sme genes cloned into pCR2.1 | Invitrogen |
| E. coli One Shot/pCR2.1 | Vector-only control for MIC testing | Invitrogen |
Antimicrobial agents.
Substrates for the hydrolysis assays were obtained from the following sources: cephaloridine, penicillin G, ampicillin, cefotaxime, and cefoxitin, Sigma Chemical Co. (St. Louis, Mo.); ceftazidime and clavulanic acid, U.S. Pharmacopoeia (Rockville, Md.); imipenem, Merck (Rahway, N.J.); meropenem, AstraZeneca (Wilmington, Del.); and tazobactam, Lederle Laboratories (Pearl River, N.Y.). All substrates were prepared fresh daily as 1-mg/ml stocks in 50 mM phosphate buffer (pH 7.0).
Susceptibility testing.
MICs were determined by the National Committee for Clinical Laboratory Standards broth microdilution method (13).
PCR.
Two sets of primers were designed to amplify SME-related DNA sequences. Primers of the first set, IRS1 and IRS2 (5′ AACGGCTTCATTTTTGTTTAG 3′ and 5′ GCTTCCGCAATAGTTTTATCA 3′), were complementary to bp 151 to 171 and 961 to 981, respectively, of the sequence of SME-1 and flanking DNA (12) and amplified an 830-bp intragenic fragment of SME-1. Primers of the second set, IRS5 and IRS6 (5′ AGATAGTAAATTTTATAG 3′ and 5′ CTCTAACGCTAATAG 3′), complemented bp 5 to 22 and 1128 to 1142 of the same sequence. These primers produced a PCR product that included the putative promoter, the ribosome binding site, and the entire open reading frame. The PCR program for IRS1 and IRS2 consisted of a lysis-denaturation step for 10 min at 95°C; 30 cycles of a 30-s denaturation step at 94°C, a 30-s annealing step at 58°C, and a 30-s extension step with Taq polymerase at 72°C; and a final 10-minute extension step at 72°C. The program for IRS5 and IRS6 included an annealing temperature of 50°C and an extension time of 60 s. Genomic DNA was the template for PCR (15).
DNA cloning and sequencing.
PCR of purified genomic DNA from imipenem-resistant S. marcescens yielded amplification products that were cloned into plasmid pCR2.1 and transformed into One Shot (competent Escherichia coli) using the protocol provided by the manufacturer of the TA cloning kit (Invitrogen, Carlsbad, Calif.). Screening for transformants was done using Luria-Bertani agar plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (80 μg/ml) and ampicillin (50 μg/ml) for the intragenic PCR product obtained by using primers IRS1 and IRS2 or imipenem (10 μg/ml) for selecting plasmids containing the complete open reading frame (cloned PCR products of IRS5 and IRS6). Sequencing of both strands was performed using primers T7 and M13 Reverse at the Molecular Biology Core Facility of the Dana-Farber Cancer Institute (Boston, Mass.). At least two cloned PCR products were sequenced for each template. The Clustal method (8) was used to perform alignments of deduced amino acid sequences with the MEGALIGN software program (DNASTAR, Inc., Madison, Wis.).
β-Lactamase purification.
The SME-type β-lactamases for the kinetic analysis were purified from the transformant E. coli strains 4911, 4912, 4913, and 4914, which were shown by isoelectric focusing to contain a TEM-1 β-lactamase from the cloning vector in addition to SME-type enzymes (Queenan et al., 39th ICAAC). Bacteria from a 1-liter overnight culture in Trypticase soy broth supplemented with 100 μg of ampicillin per ml were harvested by centrifugation and lysed with a freeze-thaw procedure (21). The supernatant was loaded onto a Sephadex G-100 column with 50 mM phosphate buffer (pH 7.0). Protein in peak fractions containing nitrocefin-hydrolyzing activity was precipitated with 90% ammonium sulfate; pellets were resuspended in 50 mM phosphate buffer (pH 7.0) and dialyzed in 1 liter of 25 mM Tris-HCl (pH 7.5) at 4°C with two buffer changes. The plasmid vector-derived TEM-1 (pI, 5.4) enzyme was separated from the SME-type β-lactamase (pI, ∼9.5) on a QAE Sephadex A-25 ion-exchange column with 25 mM Tris-HCl (pH 7.5). The protein concentrations of the purified β-lactamases were determined with the Pierce (Rockford, Ill.) bicinchoninic acid protein assay. The purity of the enzymes ranged from 90 to 92%, as determined by spot densitometry of a silver-stained 10% NuPAGE gel.
Kinetic studies.
Initial hydrolysis rates were measured on a Shimadzu UV-1601 spectrophotometer at 25°C with 50 mM phosphate buffer (pH 7.0) (21). Km and Vmax values were obtained by averaging results from Lineweaver-Burk, Eadie-Hofstee, Hanes-Woolf, and direct linear plot analyses. Substrates were assayed on at least two separate days, with cephaloridine included as a reference each day. Inhibition of hydrolysis was measured after 5 min of preincubation of enzyme with inhibitor in 10 μl of phosphate buffer (pH 7.0). Cephaloridine (360 μM) was the substrate for the inhibition studies. Inhibitor concentrations causing 50% inhibition (IC50s) were determined from inhibition graphs of percent control activity versus concentration of inhibitor. Ki values were calculated by the method of Cheng and Prusoff (6).
PFGE.
For pulsed-field gel electrophoresis (PFGE), a CHEF-DRII system (BioRad Laboratories, Hercules, Calif.) was used to analyze SpeI-digested DNA from the S. marcescens isolates. DNA was prepared with a Bio-Rad CHEF genomic DNA plug kit according to the manufacturer's protocol. Agarose (1.2%) gels were run in 0.5× Tris-borate-EDTA for 22 h at 190 V, with pulse times of 5 to 42 s. The imipenem-sensitive S. marcescens clinical strain SC 9782, from the Seattle Veterans Affairs Medical Center (3), was used as a control.
Nucleotide sequence accession number.
The sequence of sme-2 was assigned GenBank accession number AF275256.
RESULTS
Antimicrobial susceptibility.
The imipenem-resistant S. marcescens clinical isolates used in this study were collected from geographically diverse regions of the United States. As shown in Table 2, all of the clinical isolates had decreased susceptibility to imipenem, meropenem, aztreonam, and cefoxitin. The E. coli transformant strains in Table 2 contained the sme genes from the four clinical isolates inserted into cloning vector pCR2.1. The presence of the sme gene on a plasmid elevated the MICs of imipenem, meropenem, ceftazidime, and aztreonam compared to those for the recipient strain alone. A modest increase in MICs was detected for cefoxitin when the SME enzyme was present. The effect on penicillin MICs could not be evaluated due to the TEM-1 enzyme being present on the cloning vector.
TABLE 2.
Antimicrobial susceptibility of S. marcescens isolates and E. coli transformants with SME enzymes
| Location | Strain | β-Lactamase(s) | MIC (μg/ml) ofa:
|
||||
|---|---|---|---|---|---|---|---|
| IPM | MEM | ATM | FOX | CAZ | |||
| London | S. marcescens S6 | SME-1, AmpC | 512 | 64 | 128 | 32 | <0.25 |
| E. coli 4913 | SME-1, TEM-1 | 256 | 32 | >256 | 16 | 8 | |
| Minnesota | S. marcescens 4176 | SME-1, AmpC | 512 | 128 | 64 | 128 | 2 |
| E. coli 4911 | SME-1, TEM-1 | 256 | 32 | >256 | 32 | 8 | |
| UCLA | S. marcescens 4126 | SME-2, AmpC | 512 | 128 | 128 | 128 | 0.5 |
| E. coli 4912 | SME-2, TEM-1 | 256 | 64 | >256 | 32 | 8 | |
| Boston | S. marcescens 4124 | SME-2, AmpC | 512 | 128 | >256 | 64 | <0.25 |
| E. coli 4914 | SME-2, TEM-1 | 256 | 32 | >256 | 16 | 16 | |
| Controls | E. coli recipient | <0.5 | <0.5 | <0.25 | 4 | <0.25 | |
| E. coli recipient/pCR2.1 | TEM-1 | <0.5 | <0.5 | <0.25 | 8 | <0.25 | |
IPM, imipenem; MEM, meropenem; ATM, aztreonam; FOX, cefoxitin; CAZ, ceftazidime.
Sequence analysis.
Two protein sequences, initially designated SME-1 and SME-2, containing an isoleucine and a tyrosine, respectively, at position 245, have been determined for the same carbapenem-hydrolyzing enzyme from the London strain S. marcescens S6 (12; B. A. Rasmussen, D. Keeney, and C. Cohen, GenBank accession number U60295). Recently, this discrepancy has been resolved, and it has been determined that tyrosine is the correct amino acid for this position (P. Nordmann, personal communication). We now refer to the enzyme from the London strain S6 as SME-1.
The cloned sme genes from the U.S. isolates were sequenced from both strands of the plasmids from the E. coli transformants. The Minnesota strain MN-2701 produced an enzyme identical to the SME-1 enzyme from strain S6 (B. A. Rasmussen, D. Keeney, and C. Cohen, GenBank Accession number U60295). A modified enzyme was found in the Boston and UCLA strains. This new member of the SME family contained three nucleotide substitutions in the gene relative to SME-1—A for G at 258, A for T at 620, and A for G at 714—resulting in a single amino acid change, Glu for Val at position 207, according to the Ambler numbering system (1). As a result of the sequence data, we have designated the Minnesota enzyme SME-1 and the Boston and UCLA enzymes SME-2. The promoter region for the sme-2 gene was identical to the published sequence for the sme-1 gene (data not shown).
Biochemical characterization.
The SME β-lactamases were purified chromatographically from the E. coli transformants. As expected from the sequence analysis, all four of the cloned sme genes produced proteins of approximately 30 kDa when examined on a polyacrylamide gel, in accord with the 29.3-kDa protein predicted from the sequence (data not shown).
Kinetic data for the SME-1 and SME-2 β-lactamases are summarized in Table 3. Km and kcat values for each substrate were similar for both enzymes, indicating that the single amino acid change in SME-2 did not affect the substrate binding or hydrolysis rates. SME-1 and SME-2 hydrolyzed a variety of β-lactams from the penicillin, cephalosporin, monobactam, and carbapenem groups. The highest kcat values were obtained for cephaloridine, followed by ampicillin, aztreonam, and imipenem. Meropenem had a kcat value 10 times lower than that of imipenem.
TABLE 3.
Kinetic parameters of SME β-lactamases
| Enzymea |
kcat (s−1) for:
|
Relative kcat for:
|
Km (μM) for:
|
kcat/Km for:
|
||||
|---|---|---|---|---|---|---|---|---|
| SME-1 | SME-2 | SME-1 | SME-2 | SME-1 | SME-2 | SME-1 | SME-2 | |
| Substrate | ||||||||
| LOR | 980 ± 18 | 1081 ± 5 | 100 | 100 | 770 ± 30 | 859 ± 45 | 1.27 | 1.26 |
| PEN | 19.3 ± 1.1 | 21.3 ± 0.4 | 2.0 | 1.9 | 16.7 ± 6 | 17.7 ± 0.4 | 1.16 | 1.20 |
| IPM | 104 ± 4.2 | 136 ± 21 | 11 | 13 | 202 ± 38 | 313 ± 57 | 0.52 | 0.44 |
| MEM | 8.9 ± 0.6 | 7.3 ± 4.9 | 0.9 | 0.7 | 13.4 ± 3.2 | 9.6 ± 8.9 | 0.66 | 0.76 |
| AMP | 181 ± 13 | 204 ± 18 | 18 | 19 | 488 ± 47 | 609 ± 124 | 0.37 | 0.34 |
| ATM | 108 ± 21 | 140 ± 5 | 11 | 13 | 259 ± 23 | 277 ± 8.5 | 0.42 | 0.51 |
| FOX | <0.15 | <0.17 | <0.02 | <0.02 | NDb | ND | ND | ND |
| CTX | <0.98 | <1.0 | <0.10 | <0.09 | ND | ND | ND | ND |
| CAZ | <0.07 | <0.09 | <0.01 | <0.01 | ND | ND | ND | ND |
| Inhibitor | ||||||||
| CLA | NAc | NA | NA | NA | 0.19d | 0.18d | NA | NA |
| TZB | NA | NA | NA | NA | 0.11d | 0.12d | NA | NA |
LOR, cephaloridine; PEN, penicillin; IPM, imipenem; MEM, meropenem; AMP, ampicillin; ATM, aztreonam; FOX, cefoxitin; CTX, cefotaxime; CAZ, ceftazidime; CLA, clavulanic acid; TZB, tazobactam.
ND, not determined; hydrolysis was too slow.
NA, not available.
Ki. IC50s for SME-1 and SME-2, respectively: CLA, 0.28 and 0.25 μM; TZB, 0.16 and 0.17 μM.
The SME enzymes had the highest affinity, or the lowest Km, for penicillin and meropenem. Cefoxitin, cefotaxime, and ceftazidime were poor substrates, with hydrolysis rates too slow to obtain accurate Km values under these assay conditions. Taking the binding affinity into account, kcat/Km values showed that the catalytic efficiencies of both enzymes varied approximately threefold for all the hydrolyzable substrates.
Clavulanic acid inhibited the SME-1 and SME-2 β-lactamases, with Ki values of 0.19 and 0.18 μM, respectively (Table 3). Tazobactam was a slightly better inhibitor for these enzymes, with Ki values of 0.11 and 0.12 μM for the SME-1 and SME-2 enzymes respectively (Table 3). No inhibition was observed when the enzymes were preincubated with 2.5 μM EDTA at pH 7.0, as expected for serine-based β-lactamases.
PFGE analysis.
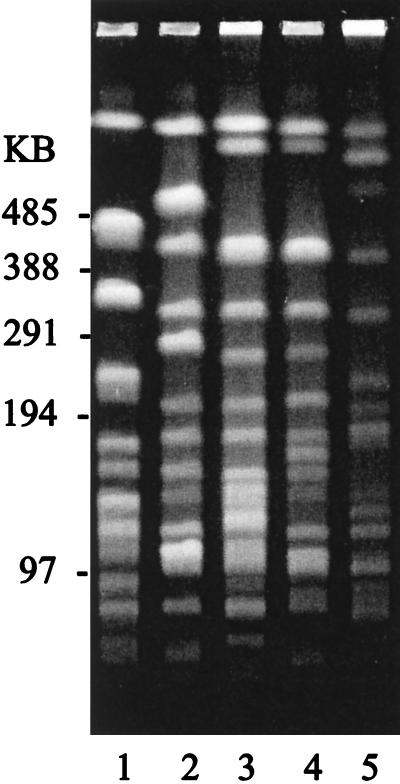
DNAs from the four imipenem-resistant S. marcescens strains and an imipenem-sensitive S. marcescens clinical strain were compared by PFGE (Fig. 1). There were at least seven band differences between any two of the imipenem-resistant strains. The imipenem-sensitive strain had a minimum of 17 band differences compared to any of the imipenem-resistant strains.
FIG. 1.
PFGE of SpeI-digested DNA from S. marcescens isolates. Lane 1, imipenem-sensitive strain SC 9782. Lanes 2 to 5, imipenem-resistant strains S6 (London), 4176 (Minnesota), 4124 (Boston), and 4126 (California), respectively. Markers are from a Bio-Rad lambda ladder.
DISCUSSION
Imipenem resistance due to class A serine-based β-lactamases is rare. Only three sets of enzymes have been reported in functional group 2f: NMC-A and IMI-1, both from E. cloacae, and SME-1, from S. marcescens (12, 14, 17). Each of these enzymes was found either as a single isolate or in a pair of clinical isolates from geographically diverse regions. In the United States, imipenem-resistant S. marcescens strains were isolated in Minnesota, California, and Boston over a period of 15 years. Preliminary testing suggested that the imipenem resistance was due to an enzyme similar to SME-1 (Medeiros and Hare, 26th ICAAC; Quinn et al., 37th ICAAC; Carmeli et al., 37th ICAAC).
The molecular relatedness of the β-lactamase genes cloned from the imipenem-resistant S. marcescens strains was investigated. The SME-1 enzyme and a single-amino-acid variant, SME-2, were identified in this study of U.S. isolates. The isolate from Minnesota produced an enzyme identical to the London SME-1 enzyme. SME-2, from the Boston and California strains, was distinguished by a single glutamic acid-for-valine substitution at position 207. This sequence change introduced another identical amino acid between SME-2 and the other molecular class A, functional group 2f enzymes that also contain a glutamic acid at position 207 (4). The SME enzymes shared 70% amino acid identity with NMC-A and IMI-1 (17).
SME-1 and SME-2 hydrolyzed a range of substrates, including penicillins, aztreonam, cephaloridine, and carbapenems. There was no detectable hydrolytic activity against ceftazidime, consistent with low ceftazidime MICs for the clinical isolates. A similar hydrolysis profile is seen for NMC-A and IMI-1. The relative hydrolysis rates for the SME enzymes purified in this study also compared well with those published for the other group 2f enzymes, where relative rates for cephaloridine and ampicillin were higher than those for imipenem (16). In contrast to previously published data for clavulanic acid inhibition of SME-1 (IC50, 14 μM [5]), the purified SME-1 and SME-2 enzymes in this study had IC50s similar to those published for NMC-A and IMI-1, in the range of 200 to 300 nM. The IC50 for tazobactam found in these experiments was 10-fold lower than that previously published for SME-1 (5). These differences may be due to the fact that nitrocefin was used as the reporter substrate in the earlier study (5) and that cephaloridine, at a sub-Km concentration, was used in this study.
The SME β-lactamases had a broad substrate spectrum, with hydrolysis of penicillins and early cephalosporins, in addition to carbapenems. This property could have been a factor in their emergence in strains isolated before imipenem was approved for clinical use in 1985. The original source of these enzymes remains unknown. It is unlikely that the SME β-lactamases evolved from an enzyme previously existing in the Serratia genome, as 16 imipenem-sensitive S. marcescens clinical isolates did not hybridize with an sme probe (H. S. Gold, unpublished data).
Initially, the rarity and geographically diverse locations of the imipenem-resistant S. marcescens strains suggested convergent evolution of the SME β-lactamases. This idea was tested by PFGE analysis of DNA from the four isolates. The results indicated that the four isolates differed by at least three genetic events (22). There was a degree of relatedness among these isolates of imipenem-resistant S. marcescens, as they shared more bands with each other than with an S. marcescens strain that lacked an SME enzyme. The PFGE results are consistent with global dissemination of a distinct S. marcescens subtype. SME-1 and SME-2, since they are not encoded on plasmids, are not readily transmissible, like the TEM and SHV extended-spectrum β-lactamases. The fact that these enzymes are chromosomally encoded is one reason for their rarity, although, as with all resistance genes, the possibility exists that their genes may be acquired by a mobile element. It appears that carbapenem-hydrolyzing β-lactamases have been present in the genetic environment for some time. It is likely that the increased frequency of carbapenem use will increase the frequency with which these β-lactamases are found in a clinical setting.
ACKNOWLEDGMENT
This work was supported in part by a grant from The R. W. Johnson Pharmaceutical Research Institute to the Beth Israel Deaconess Medical Center.
REFERENCES
- 1.Ambler R P, Coulson A F W, Frere J M, Ghuysen J M, Joris B, Forsman M, Levesque R C, Tiraby G, Waley S G. A standard numbering scheme for the class A β-lactamases. Biochem J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford P A, Urban C, Mariano N, Projan S J, Rahal J J, Bush K. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob Agents Chemother. 1997;41:563–569. doi: 10.1128/aac.41.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush K, Flamm R K, Ohringer S, Singer S B, Summerill R, Bonner D P. Effect of clavulanic acid on activity of β-lactam antibiotics in Serratia marcescens isolates producing both a TEM β-lactamase and a chromosomal cephalosporinase. Antimicrob Agents Chemother. 1991;35:2203–2208. doi: 10.1128/aac.35.11.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush K, Macalintal C, Rasmussen B A, Lee V J, Yang Y. Kinetic interactions of tazobactam with β-lactamases from all major structural classes. Antimicrob Agents Chemother. 1993;37:851–858. doi: 10.1128/aac.37.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng Y-C, Prusoff W H. Relation between the inhibition constant (Ki) and the concentration of inhibitor which causes fifty per cent inhibition (I50) of an enzymic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 7.Donald H M, Scaife W, Amyes S G B, Young H-K. Sequence analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob Agents Chemother. 2000;44:196–199. doi: 10.1128/aac.44.1.196-199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 9.Ito H, Arakawa Y, Ohsuka S, Wacharotayankun R, Kato N, Ohta M. Plasmid-mediated dissemination of the metallo-β-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob Agents Chemother. 1995;39:824–829. doi: 10.1128/aac.39.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lauretti L, Riccio M L, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini G M. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999;43:1584–1590. doi: 10.1128/aac.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livermore D M. Acquired carbapenemases. J Antimicrob Chemother. 1997;39:673–676. doi: 10.1093/jac/39.6.673. [DOI] [PubMed] [Google Scholar]
- 12.Naas T, Vandel L, Sougakoff W, Livermore D M, Nordmann P. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A β-lactamase, Sme-1, from Serratia marcescens S6. Antimicrob Agents Chemother. 1994;38:1262–1270. doi: 10.1128/aac.38.6.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 5th ed. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 14.Nordmann P, Mariotte S, Naas T, Labia R, Nicolas M H. Biochemical properties of a carbapenem-hydrolyzing β-lactamase from Enterobacter cloacae and cloning of the gene into Escherichia coli. Antimicrob Agents Chemother. 1993;37:939–946. doi: 10.1128/aac.37.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 16.Rasmussen B A, Bush K. Carbapenem-hydrolyzing β-lactamases. Antimicrob Agents Chemother. 1997;41:223–232. doi: 10.1128/aac.41.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasmussen B A, Bush K, Keeney D, Yang Y, Hare R, O'Gara C, Medeiros A A. Characterization of IMI-1 β-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob Agents Chemother. 1996;40:2080–2086. doi: 10.1128/aac.40.9.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen B A, Gluzman Y, Tally F P. Cloning and sequencing of the class B β-lactamase gene (ccrA) from Bacteroides fragilis TAL3636. Antimicrob Agents Chemother. 1990;34:1590–1592. doi: 10.1128/aac.34.8.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scaife W, Young H-K, Paton R H, Amyes S G B. Transferable imipenem-resistance in Acinetobacter species from a clinical source. J Antimicrob Chemother. 1995;36:585–586. doi: 10.1093/jac/36.3.585. [DOI] [PubMed] [Google Scholar]
- 20.Senda K, Arakawa Y, Nakashima K, Ito H, Ichiyama S, Shimokata K, Kato N, Ohta M. Multifocal outbreaks of metallo-β-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum β-lactams, including carbapenems. Antimicrob Agents Chemother. 1996;40:349–353. doi: 10.1128/aac.40.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sykes R B, Bonner D P, Bush K, Georgopapadakou N H. Azthreonam (SQ 26,776), a synthetic monobactam specifically active against aerobic gram-negative bacteria. Antimicrob Agents Chemother. 1982;21:85–92. doi: 10.1128/aac.21.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y, Rasmussen B A, Bush K. Biochemical characterization of the metallo-β-lactamase CcrA from Bacteroides fragilis TAL3636. Antimicrob Agents Chemother. 1992;36:1155–1157. doi: 10.1128/aac.36.5.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]