Abstract
Background:
Epstein-Barr virus (EBV) is linked to multiple cancers, including classical Hodgkin lymphoma (cHL), endemic Burkitt lymphoma (eBL), nasopharyngeal carcinoma (NPC), and extranodal natural killer/T-cell lymphoma (NKTCL).
Methods:
Anti-EBV IgG and IgA antibody responses targeting 202 sequences from 86 EBV proteins were measured using the same EBV whole proteome array across four case-control studies investigating EBV-positive cHL, eBL, NPC and NKTCL (407 cases/620 controls). We grouped EBV-targeted antibodies into pathways by immunoglobulin type (IgA and IgG) and life cycle stage (latent, immediate early lytic, early lytic, late lytic, and glycoprotein) and evaluated their association with each cancer type. In an additional analysis, we focused on the subset of 46 individual antibodies representing the top candidates for each cancer and compared their associations across the four cancer types using multivariable linear regression models.
Results:
IgA antibody responses targeting all EBV life cycle stages were associated with NPC but limited to anti-early lytic stage for cHL. NPC and eBL were associated with IgG antibodies across the viral life cycle; cHL with antibodies in the early lytic, late lytic and glycoprotein stages; and NKTCL with antibodies in the latent, immediate early lytic and early lytic phases. EBNA3A, BBLF1, BDLF4, and BLRF2 IgG antibodies were associated with all cancer types.
Conclusion:
Our observed similarities and differences across four EBV-associated cancers may inform EBV-related oncogenesis.
Impact:
Understanding the comparative humoral immune response across EBV-related cancers may aid in identifying shared etiologic roles of EBV proteins and inform unique pathogenic processes for each cancer.
Keywords: Epstein-Barr virus, humoral immune response, nasopharyngeal carcinoma, lymphoma, antibodies
Introduction
Epstein-Barr virus (EBV) is a ubiquitous gamma-herpesvirus transmitted through oral secretions and found in over 90% of the global population (1). In 1964, EBV became the first virus to be etiologically linked to human cancer when it was discovered in tumor cells obtained from an African child with Burkitt lymphoma (BL), and it was subsequently found to be epidemiologically associated with pediatric Burkitt lymphoma in Uganda (2). Since then, EBV has been implicated in both Hodgkin and non-Hodgkin lymphomas, as well as in epithelial cancers of the gastrointestinal tract and nasopharynx (1,3). EBV-related cancers exhibit unique ethnic and geographical distributions, although the reason for this remains unclear (4). EBV is unique since it causes cancers that affect distinct cell types, including epithelial cells, lymphocytes, and mesenchymal cells (5).
EBV infection initiates in the oropharyngeal mucosal epithelium (6) and persists latently in B cells (7), with periodic phases of viral reactivation and lytic replication (7). Host factors such as age at primary infection with EBV (8), smoking (9), and host genetics (10) might influence the levels of humoral immune (antibody) responses against EBV. The molecular diversity (i.e., individual EBV antigen complexes) of the humoral immune response is thought to reflect host exposure to EBV proteins expressed during the different stages of viral latency, reactivation, and replication, which may differ among EBV disease entities linked to the pathogenic process (11). EBV infection in cells occurs as latent or lytic infection (12). Latent infection occurs almost exclusively in B cells and involves expression of a limited set of EBV genes (EBNA1 and EBV-encoded small RNAs [EBERs]) to maintain viral episomes in the cells. Lytic infection occurs in B cells and epithelial tissue, where the virus expresses many genes with a goal of producing viral progeny. Lytic replication involves temporal phases that are labeled as immediate early, early, and late cycles, during which specific EBV proteins are synthesized and released. Lytic replication ends when the infected cell bursts to release viral progeny that infect other cells in the same host or are transmitted to a new host, usually through saliva (13). Immunoglobulin A (IgA) antibodies to EBV are indicative of infection at mucosal surfaces and wane relatively rapidly upon control of lytic infection (14,15). In the context of tumorigenesis, these antibodies are believed to be enhanced by the reactivation of latent B cells that shed EBV along mucosal surfaces. In contrast to IgA, immunoglobulin G (IgG) antibodies reflect systemic exposure to EBV infection in circulating B cells. Since IgG antibody levels tend to wane very gradually over time, they reflect longer-term exposure.
The molecular profiles associated with IgG and IgA responses to EBV in the context of diverse EBV-associated diseases have received relatively little scientific attention (11). Due to technological constraints, earlier studies focused on testing antibodies against only a limited number of EBV proteins or EBV-encoded antigen complexes (11,15). However, although only a few EBV-encoded proteins are expressed during viral latency, approximately 100 open reading frames are translated by EBV during a full lytic replication. To comprehensively assess the complexity of antibody responses against EBV, our group has developed a custom EBV array targeting 202 sequences representing 86 proteins expressed in the complete EBV proteome and all known splice variants (at the time of array printing). High coverage was achieved across the five EBV strains, with 97% of the predicted sequences for each train represented on the microarray at ≥99% homology. The protein array includes three synthetic EBNA1, VCAp18, and EAD multi-epitope peptides; 85% (169) of the 199 predicted sequences represented complete transcripts from genes <1000bp in length, and 30 predicted sequences represented linear segments from eight EBV genes >1000bp. Comparison with the outputs from a well-established enzyme-linked immunosorbent assay (ELISA) showed high correlations between the microarray IgA output and previously generated ELISA data for IgA antibodies against VCAp18 and EBNA1 (Spearman coefficients = 0.76 and 0.79, respectively; p < 0.01). We utilized this assay to assess and compare serological responses for EBV-related nasopharyngeal carcinoma (NPC) (16), classical Hodgkin lymphoma (cHL) (17), endemic Burkitt lymphoma (eBL) (18), and extranodal NK/T-cell lymphoma (NKTCL) (19). cHL was defined as EBV-positive when EBV latent membrane antigen (LMP)-1 was detected using immunohistochemical staining and/or EBERs were detected using in situ hybridization of tumor biopsies. Herein, we integrated the datasets from studies of each of these malignancies to investigate how EBV proteins targeted by antibody responses compare across the distinct cancer types, to further elucidate EBV’s potential mechanisms of action in tumorigenesis.
Methods
Data and Specimen Collection
Data from various case-control studies were assessed to identify similarities and differences in anti-EBV serologic profiles across NPC, eBL, EBV-positive cHL, and NKTCL. Details regarding these studies are provided in the Supplementary Methods S1. Briefly, the NPC analysis was based on a study including 67 early-stage NPC cases and 175 community controls (frequency matched to cases on sex, age, and region) in northern Taiwan between 1991 and 1994 (16). NPC cases were recruited from two large referral hospitals in Taiwan and controls were selected randomly from the Taiwanese National Household Registration system. The eBL analysis was based on 150 eBL cases (age range, 0–17 years) and 150 apparently healthy controls (frequency matched on sex, age, and enrollment period) recruited in Ghana between 1965 and 1994 (18). All cases were recruited in one hospital in Ghana, and controls were selected from neighbors nearest to the home of the cases. The cHL analysis was based on 139 EBV-positive cHL cases and 141 population-based controls from the United Kingdom (20,21) and from Denmark and Sweden (22) enrolled in 1993–1997 and 1999–2002, respectively (17). Samples from the UK were derived from two population-based case–control studies (Scotland and Newcastle Epidemiological study of Hodgkin’s Disease [SNEHD] (23) and Young adult Hodgkin disease and Haematological malignancy Case Control Study [YHHCCS] (24)), as well as three cHL cases collected from a case series study (Investigation of The Cause of Hodgkin lymphoma [ITCH]. All cases and controls were resident in Scotland or the north of England. In the SNEHD study, eligible cases were residents aged 16–74 years diagnosed with cHL, and controls were randomly selected from computerized regional primary care lists. In the YHHCCS study, eligible cases were residents aged 16–24 years diagnosed with cHL, and controls were randomly selected from people registered with general practitioners in the study area. ITCH is an ongoing study focused on collection of newly diagnosed cHL cases aged ≥15 years in the west of Scotland and previously also in the northern region of England (21). Samples from Denmark and Sweden were collected as part of the Scandinavian Lymphoma Etiology (SCALE) study, which was based on the entire population aged 18–74 years living in Denmark and Sweden (22). Patients newly diagnosed with cHL were identified through a rapid case ascertainment system, and controls were randomly sampled from continuously updated computerized population registers. In the present study, we selected cHL cases with known tumor EBV status and controls who were frequency matched to EBV-positive cHL cases on sex, age, and study area. The cases and controls for cHL have been pooled into one case-control study to address the anti-EBV serologic profiles in patients with cHL stratified by EBV tumor status; this approach has been described in detail previously(25). Finally, the NKTCL analysis was based on 51 NKTCL cases and 154 control adults collected as part of the AsiaLymph, a multi-center hospital-based case-control study in Hong Kong and Taiwan conducted between 2012 and 2017 (19). Eligible cases were aged 18–79 years at diagnosis and living in the geographic region served by the partnering hospital, and controls were selected from patients seen at the same hospital for diseases/conditions that were not associated with risk factors under study.
We obtained written informed consent from the patients and the studies were conducted in accordance with Declaration of Helsinki. All contributing studies were approved by regional scientific ethics committees and data protection agencies.
Serum or plasma samples were probed against a custom EBV protein microarray designed to target IgG and IgA antibodies against a total of 202 EBV sequences and analyzed as described previously (16). All laboratory technicians were blinded to case-control status. A total of 199 EBV protein sequences representative of nonredundant open reading frames and predictive splice variants in 86 proteins from five viral strains (AG876, Akata, B96–8, Mutu, and Raji) were included, in addition to three synthetic EBV peptides (VCAp18, EBNA1, and EAD) commonly used for EBV serological studies (16,26–29) as well as positive and negative array controls. Details of the testing method have been published (16,18,19,25) and are summarized in the Supplementary Methods S1 section.
For each sample tested, four spots were blank or without DNA (no-DNA) to assess test background reactivity. Raw fluorescence intensities were corrected for spot-specific background; corrected data were transformed using variance stabilizing normalization in Gmine (30) and output was standardized to person-specific background (normalized value divided by mean + 1.5 standard deviation of the four “no DNA” spots). Subsequently, analyses were based on standardized fluorescence intensities. We included a random subset of blinded duplicates from participants in each study (n = 80 NPC, 25 cHL, 25 BL, and 25 NKTCL) to assess across-batch variability of the assay. Antibodies with coefficients of variation (CVs) >20% were excluded from analyses focusing on individual antibodies.
Statistical Analysis
To identify potential similarities and differences in the associations between immunoglobulin class (IgA and IgG) and/or stage of viral life cycle (latent, immediate early lytic, early lytic, late lytic, and glycoprotein) and risk of the four cancer types of interest, all 202 sequences (corresponding to 86 EBV proteins) across the EBV proteome were grouped by life cycle stage and immunoglobulin type, and compared across cancer types (Supplementary Table S1). Since our included studies were conducted in different periods and populations, and samples were tested in different batches, we did not conduct analyses by pooling the data from various cancer types directly. Instead, two approaches were used to assess patterns of anti-EBV serologic profiles across cancer types to account for heterogeneity of the populations and across-batch variability (i.e., the fact that samples were tested in different batches and periods).
In the first approach (Statistical Analysis 1), we analyzed the 202 antibodies on the array grouped by EBV life cycle stage separately for each cancer type. Details on how markers were grouped into life cycle stage can be found in Supplementary Table S2. Briefly, for each cancer and for each antibody in a particular stage of the EBV life cycle, we fitted linear regression models that included case status (cancer vs. control), age group, and sex as the independent variables, with levels of antibodies (i.e., standardized fluorescence intensities) as the dependent variable (Supplementary Methods S1). We then used an adaptive rank truncated product statistic (31) to compute the test statistic, a combined p-value for each stage of the EBV life cycle based on the antibody-specific p-values for the estimated coefficients associated with case-control status. The p-value for this test statistic was then determined using a permutation approach. This approach minimizes the multiple testing burden, as it treats EBV life cycle stage and immunoglobin type (as opposed to the individual antibody) as the unit of analysis. Stages of the EBV life cycle were classified by grouping antibodies into five biologically relevant phases of viral infection (i.e., latent, immediate early lytic, early lytic, late lytic, and glycoprotein) separately for IgG and IgA antibodies (10 groups). Additional assessments by overall immunoglobulin class (IgA and IgG) were also conducted, resulting in 12 overall groups for the analysis. P values less than 0.004 were considered statistically significant (equivalent to Bonferroni-corrected P<0.05 for 12 tests). A more detailed description of these methods can be found in the Supplementary Methods S1 section.
For the second approach (Statistical Analysis 2), we examined whether the associations between individual antigen-specific antibody response and cancer risk were similar or different across the four EBV-associated cancer types. To focus on the strongest cancer-associated markers, we compared the effect sizes (i.e., fold changes) across cancer types for antibodies identified in the literature (16,18,25). The individual antibodies with the lowest statistically significant p-values for each cancer type were selected from an initial literature-derived list (based on the Bonferroni corrected p-value cutoff in the original literature). The resulting list was then narrowed down to the top twenty antibodies for NPC and eBL, seventeen antibodies for cHL (representing all significant antibodies after Bonferroni correction), and six antibodies for NKTCL (again, representing all significant antibodies after Bonferroni correction). Overlapping hits across cancers were then assessed, resulting in a final list of 46 antibodies (representing 27 EBV proteins). We then fitted a single linear regression model to all cancers combined, adjusting for sex, age group, region and study (i.e. original data source). Additionally, we included an interaction term for case-control status with each cancer type to determine whether associations differed by outcome, and we based the test of heterogeneity by cancer type on the p-value of the interaction term. Correction for multiple testing was not applied because using a p-value of 0.05 was a conservative choice to determine heterogeneity. For each cancer type, antibodies associated with a ≥1.3-fold change in risk were considered to be strongly associated, and those with a fold change between 1.1 and 1.2 were considered to be modestly associated. P-values < 0.05 were considered statistically significant.
Data Availability Statement
The data generated in this study are available from the corresponding author upon request.
Results
Complete Array Analysis by Stage of EBV Viral Life Cycle and Immunoglobulin Type Groupings (Statistical Analysis 1)
Characteristics of study participants are summarized in Supplementary Table S2. Cases and controls within each study had a similar distribution of age, sex, and region.
In the analysis examining immunoglobin type and stage of viral life cycle as the unit of analysis, we found that IgA antibodies, including those targeting each stage of the viral life cycle, were significantly associated with NPC (all p-values <0.004) (Figure 1A). IgA was also found to be significantly associated with EBV-positive cHL overall (p-value=5.98×10−4), but in contrast with NPC, the IgA association for cHL was limited to antibodies targeting antigens from the early lytic phase of EBV infection (p-value=1.78×10−4).
Figure 1:
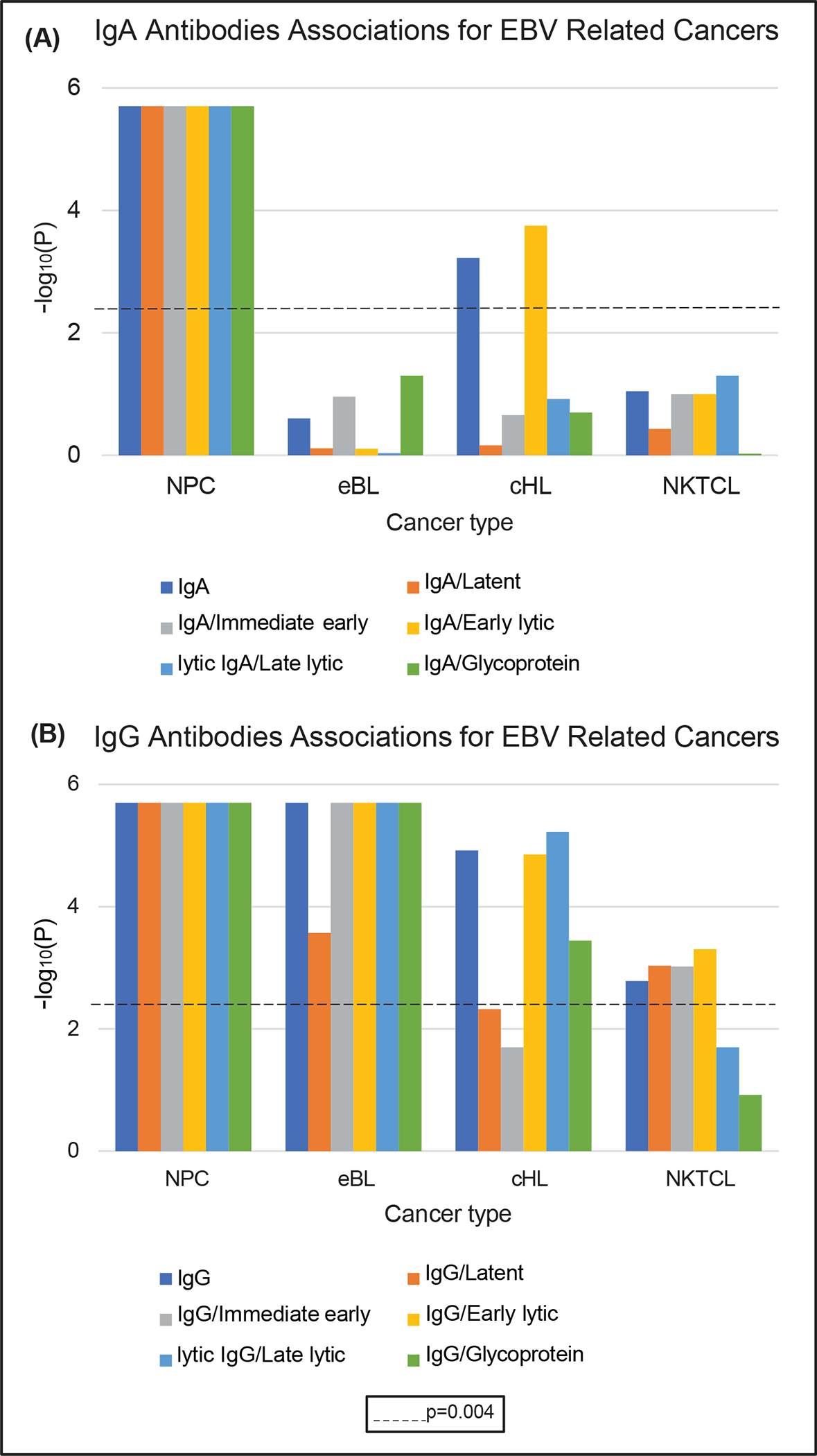
Summary of IgA and IgG antibodies associations for EBV-related cancer types (A) IgA antibodies associations depicting p-values on the −log10 scale by viral life cycle within each EBV-related cancer type, and (B) IgG antibodies associations depicting p-values on the −log10 scale by viral life cycle within each EBV-related cancer type. The dashed line represents the statistically significant p-value threshold (p=0.004; corresponding to correction for 12 tests) after Bonferroni correction.
Anti-EBV IgG antibodies were significantly associated with all four cancer types (all p-values <0.004; Figure 1B). NPC and eBL were associated with antibodies targeting EBV proteins from all stages of the viral cycle, whereas EBV-positive cHL and NKTCL were not. EBV-positive cHL was preferentially associated with IgG antibodies targeting early and late lytic proteins and glycoproteins (p-values=1.40×10−5, 6.00×10−6, and 3.60×10−4, respectively). In contrast, NKTCL was preferentially associated with IgG antibodies targeting immediate early lytic, early lytic, and latent proteins (p-values=9.52×10−4, 4.96×10−4, and 9.28×10−4, respectively).
Individual A-priori Marker Analysis (Statistical Analysis 2)
We next evaluated individual markers representing the strongest associations observed for each cancer type to better characterize the EBV proteins that might be driving the associations with viral life cycle stage described above. Antibody seropositivity rates varied considerably by isotype, antibody, and cancer type among the 46 individual antibodies examined for this study (Supplementary Table S3). A formal statistical assessment of similarities and differences observed across cancers for each antibody is presented in Tables 1 and 2. Individual antibodies with evidence for similar associations with all four cancer types evaluated (p-value for heterogeneity > 0.05) were IgG antibodies targeting EBNA-3A (latent protein; two variants), LMP-1 (latent protein; one variant), BBLF1 (early lytic protein; two variants), BDLF4 (early lytic protein), BDRF1(late lytic protein), BPLF1 (late lytic protein; one variant), BLRF2 (late lytic protein), BVRF2 (late lytic protein), BBRF3 (glycoprotein), BDLF3 (glycoprotein; two variants), and BILF2 (glycoprotein) (Table 1).
Table 1:
Antibodies with No Evidence of Heterogeneity for Associations with the Four EBV-related Cancers Evaluateda
| Marker and Array Sequence, by Stage of EBV Life Cycle | Ig Class | Heterogeneity across cancer types | Nasopharyngeal Carcinoma | Endemic Burkitt Lymphoma | EBV-positive classical Hodgkin Lymphoma | Extranodal NK/T-cell Lymphoma | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| p-value for heterogeneityb | Fold Changec | p-valued | Fold Changec | p-valued | Fold Changec | p-valued | Fold Changec | p-valued | ||
|
| ||||||||||
| Latent | ||||||||||
| EBNA3A YP_001129463.1-80447-82888 |
IgG | 0.55 | 1.2 | 0.01 | 1.1 | <.0001 | 1.1 | 0.002 | 1.2 | <.0001 |
| EBNA3A YP_401669.1-80382-82877 |
IgG | 0.47 | 1.1 | 0.01 | 1.0 | 0.2 | 1.0 | 0.83 | 1.3 | <.0001 |
| LMP1 YP_401722.1-168507-167702 |
IgG | 0.09 | 1.1 | 0.01 | 1.0 | 0.6 | 1.1 | 0.0001 | 1.1 | 0.05 |
| Early Lytic | ||||||||||
| BBLF1 AFY97956.1-108555-108328 |
IgG | 0.74 | 1.1 | 0.0001 | 1.2 | <.0001 | 1.1 | <.0001 | 1.1 | 0.004 |
| BBLF1 YP_001129480.1-109516-109289 |
IgG | 0.83 | 1.1 | <.0001 | 1.2 | <.0001 | 1.1 | <.0001 | 1.1 | 0.01 |
| BDLF4 YP_001129488.1-117560-116883 |
IgG | 0.91 | 1.1 | 0.0004 | 1.1 | <.0001 | 1.1 | 0.007 | 1.1 | 0.01 |
| Late Lytic | ||||||||||
| BDRF1 AFY97974.1-136284-137321 |
IgG | 0.15 | 1.2 | 0.0004 | 1.2 | 0.0001 | 1.2 | <.0001 | 1.2 | 0.003 |
| BPLF1 CAA24839.1-71527-62078-2 |
IgG | 0.26 | 1.2 | <.0001 | 1.1 | <.0001 | 1.1 | 0.0007 | 1.1 | 0.01 |
| BLRF2 YP_001129461.1-76771-77259 |
IgG | 0.70 | 1.1 | 0.0008 | 1.1 | <.0001 | 1.1 | <.0001 | 1.1 | 0.02 |
| BVRF2 YP_001129501.1-136465-138282 |
IgG | 0.22 | 1.0 | 0.33 | 1.2 | <.0001 | 1.1 | 0.0002 | 1.1 | 0.13 |
| Glycoprotein | ||||||||||
| BBRF3 YP_001129479.1-107679-108896 |
IgG | 0.27 | 1.1 | 0.0008 | 1.2 | <.0001 | 1.1 | <.0001 | 1.1 | 0.29 |
| BDLF3 YP_001129490.1-119605-118901 |
IgG | 0.06 | 1.2 | <.0001 | 1.2 | <.0001 | 1.1 | <.0001 | 1.1 | 0.02 |
| BDLF3 AFY97964.1-118644-117940 |
IgG | 0.14 | 1.2 | <.0001 | 1.2 | <.0001 | 1.1 | <.0001 | 1.1 | 0.02 |
| BILF2 YP_001129503.1-139063-138317 |
IgG | 0.49 | 1.2 | <.0001 | 1.2 | <.0001 | 1.1 | 0.0006 | 1.1 | 0.01 |
Table is ordered by stage of viral life cycle, followed by immunoglobulin type and viral protein in alphabetical order
P value denotes whether each marker’s effect on a given cancer significantly varies across the four cancer types after adjusting for sex, age group, and geographic region. P value <0.05 is considered statistically significant.
All fold-change values are based on log2 scale.
P value is corresponding to t-test. P value <0.05 is considered statistically significant.
Table 2:
Antibodies with Evidence of Heterogeneity for Associations with the Four EBV-related Cancers Evaluateda
| Marker and Array Sequence, by Stage of EBV Life Cycle | Ig Class | Heterogeneity across cancer types | Nasopharyngeal Carcinoma | Endemic Burkitt Lymphoma | EBV-positive classical Hodgkin Lymphoma | Extranodal NK/T-cell Lymphoma | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| p-value for heterogeneityb | Fold Changec | p-valued | Fold Changec | p-valued | Fold Changec | p-valued | Fold Changec | p-valued | ||
|
| ||||||||||
| Latent | ||||||||||
| EBNA1 (synthetic peptide) | IgA | <.0001 | 2.2 | <.0001 | 1.0 | 0.91 | 1.0 | 0.28 | 1.1 | 0.1 |
| Early Lytic | ||||||||||
| BBLF1 YP_001129480.1-109516-109289 |
IgA | <.0001 | 1.2 | <.0001 | 1.0 | 0.09 | 1.2 | <.0001 | 1.1 | 0.1 |
| BBLF1 AFY97956.1-108555-108328 |
IgA | <.0001 | 1.2 | <.0001 | 1.0 | 0.43 | 1.2 | <.0001 | 1.1 | 0.15 |
| BMRF1 AFY97929.1-67486-68700 |
IgA | <.0001 | 1.5 | <.0001 | 1.0 | 0.44 | 1.0 | 0.51 | 1.1 | 0.01 |
| BMRF1 YP_001129454.1-67745-68959 |
IgA | <.0001 | 1.6 | <.0001 | 1.0 | 0.76 | 1.0 | 0.24 | 1.1 | 0.01 |
| LF2 YP_001129504.1-151808-150519 |
IgA | <.0001 | 1.8 | <.0001 | 1.0 | 0.66 | 1.0 | 0.97 | 1.0 | 0.94 |
| Thymidine kinase YP_001129497.1-133399-131576 |
IgA | <.0001 | 1.5 | <.0001 | 1.0 | 0.79 | 1.0 | 0.07 | 1.0 | 0.48 |
| Early Lytic | ||||||||||
| BBLF1 YP_001129480.1-109516-109289 |
IgA | <.0001 | 1.2 | <.0001 | 1.0 | 0.09 | 1.2 | <.0001 | 1.1 | 0.1 |
| BBLF1 AFY97956.1-108555-108328 |
IgA | <.0001 | 1.2 | <.0001 | 1.0 | 0.43 | 1.2 | <.0001 | 1.1 | 0.15 |
| BMRF1 AFY97929.1-67486-68700 |
IgA | <.0001 | 1.5 | <.0001 | 1.0 | 0.44 | 1.0 | 0.51 | 1.1 | 0.01 |
| BMRF1 YP_001129454.1-67745-68959 |
IgA | <.0001 | 1.6 | <.0001 | 1.0 | 0.76 | 1.0 | 0.24 | 1.1 | 0.01 |
| LF2 YP_001129504.1-151808-150519 |
IgA | <.0001 | 1.8 | <.0001 | 1.0 | 0.66 | 1.0 | 0.97 | 1.0 | 0.94 |
| Thymidine kinase YP_001129497.1-133399-131576 |
IgA | <.0001 | 1.5 | <.0001 | 1.0 | 0.79 | 1.0 | 0.07 | 1.0 | 0.48 |
| Late Lytic | ||||||||||
| BPLF1 CAA24839.1-71527-62078-2 |
IgA | <.0001 | 1.3 | <.0001 | 1.0 | 0.46 | 1.1 | 0.006 | 1.1 | 0.1 |
| BVRF1 YP_001129499.1-133954-13566 |
IgA | <.0001 | 1.3 | <.0001 | 1.0 | 0.18 | 1.0 | 0.84 | 1.1 | 0.1 |
| Latent | ||||||||||
| LMP1 AFY97906.1-168167-168081 |
IgG | <.0001 | 1.4 | <.0001 | 1.1 | <.0001 | 1.1 | 0.02 | 1.1 | 0.003 |
| EBNA3A AFY97915.1-80252-82747 |
IgG | 0.04 | 1.1 | 0.01 | 1.1 | <.0001 | 1.1 | 0.004 | 1.3 | <.0001 |
| Immediate Early Lytic | ||||||||||
| BRLF1 YP_001129468.1-93725-91908 |
IgG | <.0001 | 1.6 | <.0001 | 1.1 | 0.002 | 1.1 | 0.01 | 1.3 | 0.001 |
| BZLF1 YP_001129467.1-90855-90724 |
IgG | <.0001 | 1.7 | <.0001 | 1.2 | <.0001 | 1.1 | 0.004 | 1.1 | 0.03 |
| BZLF1 YP_001129467.1-91697-91197 |
IgG | <.0001 | 1.4 | <.0001 | 1.1 | 0.1 | 1.1 | 0.01 | 1.2 | <.0001 |
| BZLF1 CAA24861.1-102338-102210 |
IgG | 0.002 | 1.4 | <.0001 | 1.3 | <.0001 | 1.1 | 0.003 | 1.2 | 0.01 |
| Early Lytic | ||||||||||
| BALF2 YP_001129510.1-165796-162410-1 |
IgG | <.0001 | 1.5 | <.0001 | 1.1 | <.0001 | 1.1 | 0.0001 | 1.3 | <.0001 |
| BGLF2 YP_001129486.1-115415-114405 |
IgG | <.0001 | 1.9 | <.0001 | 1.1 | 0.01 | 1.0 | 0.08 | 1.0 | 0.31 |
| BHRF1 YP_001129442.1-42204-42779 |
IgG | 0.004 | 1.2 | <.0001 | 1.3 | <.0001 | 1.1 | 0.0001 | 1.1 | 0.01 |
| BLLF3 YP_001129459.1-76320-75484 |
IgG | <.0001 | 1.6 | <.0001 | 1.1 | 0.01 | 1.1 | 0.0003 | 1.1 | 0.05 |
| BMRF1 YP_001129454.1-67745-68959 |
IgG | <.0001 | 1.4 | <.0001 | 1.4 | <.0001 | 1.1 | 0.005 | 1.2 | <.0001 |
| BMRF1 AFY97929.1-67486-68700 |
IgG | <.0001 | 1.3 | <.0001 | 1.4 | <.0001 | 1.1 | 0.003 | 1.2 | 0.0002 |
| EAD_p47 (synthetic peptide) | IgG | <.0001 | 3.5 | <.0001 | 2.0 | <.0001 | 1.1 | 0.05 | 1.5 | 0.008 |
| LF2 YP_001129504.1-151808-150519 |
IgG | <.0001 | 1.9 | <.0001 | 1.0 | 0.1 | 1.1 | 0.001 | 1.1 | 0.23 |
| Thymidine kinase YP_001129497.1-133399-131576 |
IgG | <.0001 | 1.4 | <.0001 | 1.1 | 0.0005 | 1.2 | <.0001 | 1.2 | 0.0003 |
| Late Lytic | ||||||||||
| BBRF1 YP_001129476.1-102746-104587 |
IgG | <.0001 | 1.3 | <.0001 | 1.0 | 0.55 | 1.2 | <.0001 | 1.1 | 0.01 |
| BcLF1 AFY97965.1-125044-120899-1 |
IgG | 0.001 | 1.3 | <.0001 | 1.2 | <.0001 | 1.2 | <.0001 | 1.2 | 0.01 |
| BcLF1 CAA24794.1-137466-133321-1 |
IgG | 0.002 | 1.3 | <.0001 | 1.1 | <.0001 | 1.2 | <.0001 | 1.1 | 0.03 |
| BcLF1 YP_001129493.1-126005-121860-1 |
IgG | 0.008 | 1.2 | <.0001 | 1.2 | <.0001 | 1.2 | <.0001 | 1.1 | 0.15 |
| BFLF2 YP_001129443.1-44763-43807 |
IgG | 0.005 | 1.3 | <.0001 | 1.1 | 0.01 | 1.1 | 0.0001 | 1.0 | 0.41 |
| BFRF1 YP_001129446.1-46719-47729 |
IgG | <.0001 | 1.4 | <.0001 | 1.1 | 0.0002 | 1.1 | 0.0004 | 1.1 | 0.02 |
| BPLF1 YP_001129449.1-59370-49906-2 |
IgG | <.0001 | 1.3 | <.0001 | 1.1 | <.0001 | 1.1 | 0.0002 | 1.1 | 0.03 |
| BVRF1 YP_001129499.1-133954-135666 |
IgG | <.0001 | 1.3 | <.0001 | 1.3 | <.0001 | 1.0 | 0.18 | 1.2 | 0.001 |
Table is ordered by viral life cycle, followed by immunoglobulin type and viral protein in alphabetical order
P value denotes whether each marker’s effect on a given cancer significantly varies across the four cancer types after adjusting for sex, age group, and g eographic region. P value <0.05 is considered statistically significant.
All fold-change values are based on log2 scale.
P value is corresponding to t-test. P value <0.05 is considered statistically significant.
For the majority of the 46 antibodies evaluated, we observed statistically significant differences in associations across cancer types (Table 2). Seven of the nine IgA antibodies included in the individual marker analysis (representing six EBV proteins: EBNA1, BMRF1, LF2, thymidine kinase, BPLF1, and BVRF1) were strongly associated only with NPC (1.3 ≤ fold change ≤ 2.2). Two of the nine IgA markers evaluated (both representing BBLF1) were modestly associated with NPC and EBV-positive cHL (1.1 ≤ fold change ≤ 1.2), but not with eBL or NKTCL.
Of the two IgG latent markers (representing two EBV proteins) evaluated (Table 2), we noted strong evidence for heterogeneity across cancer types in association with LMP1 (an EBV protein considered an oncoprotein (32)), for which the association was stronger for NPC (1.4-fold change) than the other three EBV-positive malignancies evaluated (1.1-fold change).
We also noted strong evidence for cancer-specific heterogeneity for three of the four IgG markers in the immediate early lytic phase of EBV infection, representing EBV proteins BRLF1 (Rta) and BZLF1 (Zta), two EBV proteins essential for EBV lytic reactivation (33). For BRLF1, the association was strongest for NPC (1.6-fold change), but was also notable for NKTCL (1.3-fold change), whereas it was more modest for eBL and EBV-positive cHL (1.1-fold change). For the two BZLF1 markers that exhibited heterogeneity across cancers, the strongest evidence for association was again noted for NPC (1.7- and 1.4-fold-change), whereas associations were more modest for the three EBV-positive lymphomas evaluated (≤1.2-fold-change).
Among the nine IgG early lytic markers (representing eight EBV proteins) evaluated, we noted strong evidence for cancer-specific heterogeneity for eight markers representing seven EBV proteins, BALF2, BGLF2, BLLF3, BMRF1, EAD_p47, LF2, and thymidine kinase. Strong evidence for an association with NPC was observed for all seven of these proteins (1.3 ≤ fold change ≤ 3.5). Strong associations between these early lytic markers and other cancers were also observed for BMRF1 and eBL (fold change = 1.4); EAD_p47 and eBL (fold change = 2.0); BALF2 and NKTCL (fold change = 1.3); and EAD_p47 and NKTCL (fold change = 1.5).
Among the eight IgG late lytic markers (representing six EBV proteins) evaluated, we noted strong evidence for cancer-specific heterogeneity for four markers representing four EBV proteins: BBRF1, BFRF1, BPLF1, and BVRF1. Strong evidence for an association with NPC was observed for all four of these proteins (1.3 ≤ fold-change ≤ 1.4), whereas they typically were not associated with the other three cancer types, except for an association between BVRF1 and eBL (fold-change =1.3).
No strong evidence for heterogeneity across cancer types was observed for any of the four IgG glycoprotein markers evaluated (p-heterogeneity > 0.05 for all markers).
Discussion
To our knowledge, this is the first study to compare antibody responses to the complete EBV proteome across multiple EBV-related cancers, including an epithelial cancer, NPC, and three lymphomas, eBL, EBV-positive cHL, and NKTCL. We report several notable differences in the associations with anti-EBV antibodies across these cancer types. These differences may highlight important differences in the viral pathogenesis of the cancers evaluated. More specifically, our results demonstrate that while elevated anti-EBV IgA antibodies across all stages of the EBV life cycle were strongly associated with NPC, IgA associations were restricted to antibodies in the early lytic phase of EBV infection for EBV-positive cHL, and absent for eBL and NKTCL. We also report associations between anti-EBV IgG antibodies and each of the four tumor types evaluated; these associations were evident for antibodies targeting proteins from all stages of the EBV life cycle for NPC and eBL; restricted to early and late lytic proteins and glycoproteins for EBV-positive cHL; and restricted to latent, immediate early, and early lytic proteins for NKTCL. In addition, we observed homogeneity of the association between several IgG antibodies and risk of all four cancer types evaluated. Such similarities may point to shared etiologic roles of specific EBV proteins.
Anti-EBV IgA antibodies, which are known to indicate a recent infection or reactivation of latent EBV in B cells along mucosal surfaces, have repeatedly been shown to be informative biomarkers for EBV-related epithelial tumors such as NPC (34–36). In this study, IgA antibodies were almost exclusively elevated in NPC cases, and these associations spanned all stages of the EBV life cycle. These results further support the role of EBV chronic reactivation at mucosal sites in NPC etiology.
We found that anti-EBV IgA antibodies targeting proteins involved in the early lytic phase of EBV infection were associated with EBV-positive cHL. This association was driven by BBLF1, a finding that warrants replication. Consistent with our findings, previous studies have reported elevated levels of IgA antibodies against other early and late lytic proteins such as EA and VCA complex antigens among EBV-positive cHL patients (37,38). Together with the knowledge that IgA antibodies are indicative of recent infection (39), our finding suggests that EBV infection at mucosal surfaces could play an important part in cHL pathology. Our retrospective study design does not allow us to determine whether the strong immune response to early lytic proteins reflects mucosal infection occurring before or as a consequence of the tumor; however, the etiologic hypothesis is further supported by long-standing epidemiological evidence indicating that delayed viral exposure (as indicated by a diagnosis of infectious mononucleosis, for example) increases the risk of EBV-positive cHL, particularly among younger adults (40,41), and that cHL in older adults may be attributable to senescence of EBV-specific immunity (41,42). Although the evidence linking prior EBV infection to subsequent EBV-positive cHL is strongest for young adults after mononucleosis, it is paradoxical that this is the age group in which the proportion of EBV positivity in cHL is the lowest. This paradox underscores recent observations suggesting that EBV-positive cHL and EBV-negative cHL are distinct biological groups based on molecular landscape, and the possibility that EBV-negative cHL may be the dominant tumor in young adults, whereas EBV-positive cHL predominates in younger and elderly people (43,44).
Our findings of homogeneity of the association between IgG antibodies and risk of all four EBV-related cancers highlights the shared roles of specific EBV proteins in the etiology of EBV-related cancers. For example, the consistent cancer-related IgG differences included a latent phase target essential for EBV persistence in the B-cell system and in modulating B-cell lymphomagenesis (i.e., EBNA3A) (45,46). It is plausible that increased EBV-driven proliferation and differentiation increases the number of infected B cells and provides more opportunities for a cancer-initiating event. We also observed differences in IgG patterns across the four EBV-related cancers for EBV proteins expressed in the lytic phase, such as BBLF1, BDLF4 and BLRF2. The biological functions of these proteins remain to be elucidated, although they may be involved in the efficient production of EBV particles (BBLF1) (47), expression of viral late lytic gene (BDLF4) (48), and production of viral capsid antigen (BLRF2) (49).
Of the IgG latent markers assessed, we noted strong heterogeneity across cancer types for LMP1, which was preferentially associated with NPC. Previous studies have shown LMP1 to be significantly correlated with the expression of proteins that mediate angiogenesis, invasion, and metastasis in the context of NPC (50–52); these biological effects have been seen even at very low levels, resulting in the promotion of tumor progression (53,54). LMP1 can be detected in the tissue of nearly all premalignant or preinvasive NPC samples (55,56), as well as malignant tumors (55), suggesting that its presence is not simply a reflection of long-term systemic exposure to EBV in B-lymphocytes.
Antibodies against immediate-early lytic proteins, which are transcription factors associated with initiation of viral replication, were strongly associated with NPC, with robust responses to BRLF1 (Rta) and BZLF1 (Zta); the latter was also strongly associated with eBL. In contrast, patients with EBV-positive cHL, compared with controls, did not appear to mount differential IgG antibodies against these immediate early lytic proteins. Although the mechanism of action is unknown, previous studies have shown that both Rta and Zta are crucial in EBV latent-to-lytic cycle switching (57). Thus, the observed lack of association with cHL may suggest lower levels of EBV reactivation in EBV-positive cHL than other EBV-related cancers.
In line with these findings, early and late lytic proteins targeted by IgG antibodies were most strongly associated with NPC and, to a lesser degree, eBL and EBV-positive cHL. While BRLF1 and BZLF1 work synergistically to enable a cascade of viral gene expression (33), the BMRF1 protein—a DNA polymerase processivity factor found in our study to be associated with NPC and eBL—has been identified as one of the primary early phosphoproteins that enables EBV lytic replication (58–62). BALF2, a single-stranded DNA binding protein significantly associated with NPC in our study, is similarly enacted, working at the replication forks during the lytic phase of the EBV DNA replication (62–64). Taken together, the expression of these proteins suggests productive viral replication in NPC, eBL, and EBV-positive cHL pathology.
In our study, patients with NK/T-cell lymphoma appeared have distinct EBV serologic patterns characterized by a lack of association with IgG antibodies to late lytic proteins and glycoproteins. NK and T cells are typically not permissive of EBV infection, although a recent study suggested that EBV can infect mature peripheral T cells via binding of EBV glycoprotein gp350 to the cellular membrane protein CD21 (65). Therefore, EBV’s role in NKTCL compared to B-cell lymphomas may differ following initial infection. Our findings suggest an abortive EBV lytic cycle in NKTCL, similar to that seen in EBV-positive gastric cancer (66), highlighting a possible similarity of EBV pathogenesis between these two EBV-associated malignancies.
The results presented in this study should be interpreted in light of several limitations. Although all the samples were run using the same standardized serological EBV microarray assay, participants in different studies varied by average age, sex, geographical region, sample collection date, and study size. The relatively young age of onset is particularly striking for eBL, which arises in children who have presumably experienced substantially fewer EBV re-infections and reactivation events than adults. We do not have reason to believe that length of biospecimen storage would impact the results, because antibodies are known to be stable proteins (67), and comparisons across cancer types were conducted after adjustment for sex, age group, and region. In addition, we used estimates obtained for each cancer that largely controlled for across-batch variability. However, our results using Statistical Approach 1 (ARTP pathway analysis) do not yield a measure of the strength of an association; instead, they assess only the statistical significance of an association with antibodies related to a given stage of the EBV life cycle.
All of the included studies were retrospective in design, with modest sample sizes; therefore, larger longitudinal studies are required to replicate these findings. Additionally, we did not adjust for host and environmental characteristics such as host genetics, smoking, and family history of cancer, that have been shown to be associated with levels of anti-EBV antibodies and risk of EBV-related cancers. For eBL, exposure to malaria could be an important co-factor for EBV in the etiology of eBL; however, we did not have data to comprehensively evaluate the impact of malaria infection. For cHL, various sources were used to form a single case-control study and controls were selected by frequency-matching on sex and age distribution of cHL cases by geographic region, which may have resulted in some uncontrolled confounding. It is unclear, however, whether residual confounding remained after frequency matching on age, sex and geography given that other than these three factors, there are no other risk factors known to be associated with both anti-EBV antibodies and cHL. The samples used in these studies were sources from limited populations/geographical areas; therefore, future studies should consider the potential impact of ancestral and environmental heterogeneity. The array used in this study was not designed to detect antibodies to conformational protein epitopes, preventing us from investigating potential associations between these cancers and transcripts that require glycosylation, including surface glycoproteins relevant to virus neutralization. Finally, a limitation of all antibody-based studies is that not only does a given antigen need to be expressed, but it also needs to be immunogenic or even immunodominant.
In conclusion, we compared and contrasted antibody responses against a comprehensive list of 86 in vitro expressed and array-spotted EBV proteins across four distinct EBV-related cancers: NPC, EBV-positive cHL, eBL and NKTCL. We noted some similarities among the associations observed for these four cancer types, which may aid in identifying shared etiologic roles of EBV proteins; as well as differences across these tumor types, which may inform unique pathogenic processes for each of these cancers.
Supplementary Material
Acknowledgements
This work was supported by the Intramural Research Program of National Cancer Institute (NCI), USA. We are grateful to the study subjects without whom this work would not be possible. We thank Dr. Ellen T. Chang for comments on the manuscript.
Funding:
A. Hildesheim was awarded funding from the Intramural Research Program of the National Cancer Institute
Footnotes
Conflict of Interest disclosure: The authors declare no potential conflicts of interest
References
- 1.Cohen JI. Epstein–Barr virus infection. New England Journal of Medicine 2000;343(7):481–92. [DOI] [PubMed] [Google Scholar]
- 2.Epstein MA, Achong BG, Barr YM. Virus Particles in Cultured Lymphoblasts from Burkitt’s Lymphoma. Lancet 1964;1(7335):702–3. [DOI] [PubMed] [Google Scholar]
- 3.Young LS, Rickinson AB. Epstein–Barr virus: 40 years on. Nature Reviews Cancer 2004;4(10):757–68. [DOI] [PubMed] [Google Scholar]
- 4.Khan G, Fitzmaurice C, Naghavi M, Ahmed LA. Global and regional incidence, mortality and disability-adjusted life-years for Epstein-Barr virus-attributable malignancies, 1990–2017. BMJ Open 2020;10(8):e037505 doi 10.1136/bmjopen-2020-037505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ko Y-H. EBV and human cancer. Experimental & molecular medicine 2015;47(1):e130–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tugizov SM, Herrera R, Palefsky JM. Epstein-Barr virus transcytosis through polarized oral epithelial cells. Journal of virology 2013;87(14):8179–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatton OL, Harris-Arnold A, Schaffert S, Krams SM, Martinez OM. The interplay between Epstein–Barr virus and B lymphocytes: implications for infection, immunity, and disease. Immunologic research 2014;58(2–3):268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winter JR, Taylor GS, Thomas OG, Jackson C, Lewis JEA, Stagg HR. Predictors of Epstein-Barr virus serostatus in young people in England. BMC Infect Dis 2019;19(1):1007 doi 10.1186/s12879-019-4578-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu FH, Xiong D, Xu YF, Cao SM, Xue WQ, Qin HD, et al. An epidemiological and molecular study of the relationship between smoking, risk of nasopharyngeal carcinoma, and Epstein-Barr virus activation. J Natl Cancer Inst 2012;104(18):1396–410 doi 10.1093/jnci/djs320. [DOI] [PubMed] [Google Scholar]
- 10.Sallah N, Miley W, Labo N, Carstensen T, Fatumo S, Gurdasani D, et al. Distinct genetic architectures and environmental factors associate with host response to the gamma2-herpesvirus infections. Nat Commun 2020;11(1):3849 doi 10.1038/s41467-020-17696-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Middeldorp JM. Epstein-Barr Virus-Specific Humoral Immune Responses in Health and Disease. Curr Top Microbiol Immunol 2015;391:289–323 doi 10.1007/978-3-319-22834-1_10. [DOI] [PubMed] [Google Scholar]
- 12.Cohen JI. Epstein-Barr virus infection. N Engl J Med 2000;343(7):481–92 doi 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- 13.Kieff E Epstein-Barr virus and its replication. Fields virology: Lippincott-Raven Publishers; 1996. p 1898–959. [Google Scholar]
- 14.Woof JM, Kerr MA. The function of immunoglobulin A in immunity. J Pathol 2006;208(2):270–82 doi 10.1002/path.1877. [DOI] [PubMed] [Google Scholar]
- 15.Coghill AE, Hildesheim A. Epstein-Barr virus antibodies and the risk of associated malignancies: review of the literature. Am J Epidemiol 2014;180(7):687–95 doi 10.1093/aje/kwu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coghill AE, Pfeiffer RM, Proietti C, Hsu WL, Chien YC, Lekieffre L, et al. Identification of a Novel, EBV-Based Antibody Risk Stratification Signature for Early Detection of Nasopharyngeal Carcinoma in Taiwan. Clin Cancer Res 2018;24(6):1305–14 doi 10.1158/1078-0432.CCR-17-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, Jarrett RF, Hjalgrim H, Proietti C, Chang ET, Smedby KE, et al. Evaluation of the antibody response to the EBV proteome in EBV-associated classical Hodgkin lymphoma. Int J Cancer 2020;147(3):608–18 doi 10.1002/ijc.32741. [DOI] [PubMed] [Google Scholar]
- 18.Coghill AE, Proietti C, Liu Z, Krause L, Bethony J, Prokunina-Olsson L, et al. The Association between the Comprehensive Epstein–Barr Virus Serologic Profile and Endemic Burkitt Lymphoma. Cancer Epidemiology and Prevention Biomarkers 2020;29(1):57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Sarathkumara YD, Chan JKC, Kwong YL, Lam TH, Ip DKM, et al. Characterization of the humoral immune response to the EBV proteome in extranodal NK/T-cell lymphoma. Sci Rep 2021;11(1):23664 doi 10.1038/s41598-021-02788-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarrett R, Krajewski A, Angus B, Freeland J, Taylor P, Taylor G, et al. The Scotland and Newcastle epidemiological study of Hodgkin’s disease: impact of histopathological review and EBV status on incidence estimates. Journal of clinical pathology 2003;56(11):811–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson PC, McAulay KA, Montgomery D, Lake A, Shield L, Gallagher A, et al. Modeling HLA associations with EBV-positive and -negative Hodgkin lymphoma suggests distinct mechanisms in disease pathogenesis. Int J Cancer 2015;137(5):1066–75 doi 10.1002/ijc.29467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smedby KE, Hjalgrim H, Melbye M, Torrang A, Rostgaard K, Munksgaard L, et al. Ultraviolet radiation exposure and risk of malignant lymphomas. J Natl Cancer Inst 2005;97(3):199–209 doi 10.1093/jnci/dji022. [DOI] [PubMed] [Google Scholar]
- 23.Alexander FE, Lawrence DJ, Freeland J, Krajewski AS, Angus B, Taylor GM, et al. An epidemiologic study of index and family infectious mononucleosis and adult Hodgkin’s disease (HD): evidence for a specific association with EBV+ve HD in young adults. International journal of cancer Journal international du cancer 2003;107(2):298–302 doi 10.1002/ijc.11156. [DOI] [PubMed] [Google Scholar]
- 24.Alexander FE, Jarrett RF, Lawrence D, Armstrong AA, Freeland J, Gokhale DA, et al. Risk factors for Hodgkin’s disease by Epstein-Barr virus (EBV) status: prior infection by EBV and other agents. Br J Cancer 2000;82(5):1117–21 doi 10.1054/bjoc.1999.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, Jarrett RF, Hjalgrim H, Proietti C, Chang ET, Smedby KE, et al. Evaluation of the antibody response to the EBV proteome in EBV-associated classical Hodgkin lymphoma. International Journal of Cancer 2019;147(3):608–618 doi 10.1002/ijc.32741 [DOI] [PubMed] [Google Scholar]
- 26.Vigil A, Davies DH, Felgner PL. Defining the humoral immune response to infectious agents using high-density protein microarrays. Future Microbiol 2010;5(2):241–51 doi 10.2217/fmb.09.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doolan DL. Plasmodium immunomics. Int J Parasitol 2011;41(1):3–20 doi 10.1016/j.ijpara.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies DH, Duffy P, Bodmer JL, Felgner PL, Doolan DL. Large screen approaches to identify novel malaria vaccine candidates. Vaccine 2015;33(52):7496–505 doi 10.1016/j.vaccine.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z, Coghill AE, Pfeiffer RM, Proietti C, Hsu WL, Chien YC, et al. Patterns of Interindividual Variability in the Antibody Repertoire Targeting Proteins Across the Epstein-Barr Virus Proteome. J Infect Dis 2018;217(12):1923–31 doi 10.1093/infdis/jiy122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Proietti C, Zakrzewski M, Watkins TS, Berger B, Hasan S, Ratnatunga CN, et al. Mining, visualizing and comparing multidimensional biomolecular data using the Genomics Data Miner (GMine) Web-Server. Sci Rep 2016;6:38178 doi 10.1038/srep38178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu K, Li Q, Bergen AW, Pfeiffer RM, Rosenberg PS, Caporaso N, et al. Pathway analysis by adaptive combination of P-values. Genet Epidemiol 2009;33(8):700–9 doi 10.1002/gepi.20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knecht H, Bachmann E, Brousset P, Sandvej K, Nadal D, Bachmann F, et al. Deletions within the LMP1 oncogene of Epstein-Barr virus are clustered in Hodgkin’s disease and identical to those observed in nasopharyngeal carcinoma. Blood 1993;82(10):2937–42. [PubMed] [Google Scholar]
- 33.Flemington EK, Goldfeld AE, Speck SH. Efficient transcription of the Epstein-Barr virus immediate-early BZLF1 and BRLF1 genes requires protein synthesis. J Virol 1991;65(12):7073–7 doi 10.1128/JVI.65.12.7073-7077.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coghill AE, Hsu WL, Pfeiffer RM, Juwana H, Yu KJ, Lou PJ, et al. Epstein-Barr virus serology as a potential screening marker for nasopharyngeal carcinoma among high-risk individuals from multiplex families in Taiwan. Cancer Epidemiol Biomarkers Prev 2014;23(7):1213–9 doi 10.1158/1055-9965.EPI-13-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henle G, Henle W. Epstein-Barr virus-specific IgA serum antibodies as an outstanding feature of nasopharyngeal carcinoma. International journal of cancer Journal international du cancer 1976;17(1):1–7 doi 10.1002/ijc.2910170102. [DOI] [PubMed] [Google Scholar]
- 36.Chien Y-C, Chen J-Y, Liu M-Y, Yang H-I, Hsu M-M, Chen C-J, et al. Serologic markers of Epstein–Barr virus infection and nasopharyngeal carcinoma in Taiwanese men. New England Journal of Medicine 2001;345(26):1877–82. [DOI] [PubMed] [Google Scholar]
- 37.Mueller N, Evans A, Harris NL, Comstock GW, Jellum E, Magnus K, et al. Hodgkin’s disease and Epstein-Barr virus. New England Journal of Medicine 1989;320(11):689–95. [DOI] [PubMed] [Google Scholar]
- 38.Chang ET, Zheng T, Lennette ET, Weir EG, Borowitz M, Mann R, et al. Heterogeneity of risk factors and antibody profiles in Epstein-Barr virus genome-positive and-negative Hodgkin lymphoma. The Journal of infectious diseases 2004;189(12):2271–81. [DOI] [PubMed] [Google Scholar]
- 39.Macpherson AJ, McCoy KD, Johansen FE, Brandtzaeg P. The immune geography of IgA induction and function. Mucosal Immunol 2008;1(1):11–22 doi 10.1038/mi.2007.6. [DOI] [PubMed] [Google Scholar]
- 40.Armstrong A, Alexander F, Cartwright R, Angus B, Krajewski A, Wright D, et al. Epstein–Barr virus and Hodgkin’s disease: further evidence for the three disease hypothesis. Leukemia 1998;12(8):1272–6. [DOI] [PubMed] [Google Scholar]
- 41.Vockerodt M, Cader FZ, Shannon-Lowe C, Murray P. Epstein-Barr virus and the origin of Hodgkin lymphoma. Chinese journal of cancer 2014;33(12):591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jarrett R, Gallagher A, Jones D, Alexander F, Krajewski A, Kelsey A, et al. Detection of Epstein-Barr virus genomes in Hodgkin’s disease: relation to age. Journal of clinical pathology 1991;44(10):844–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flavell KJ, Biddulph JP, Constandinou CM, Lowe D, Scott K, Crocker J, et al. Variation in the frequency of Epstein-Barr virus-associated Hodgkin’s disease with age. Leukemia 2000;14(4):748–53 doi 10.1038/sj.leu.2401724. [DOI] [PubMed] [Google Scholar]
- 44.Jarrett RF, Gallagher A, Jones DB, Alexander FE, Krajewski AS, Kelsey A, et al. Detection of Epstein-Barr virus genomes in Hodgkin’s disease: relation to age. Journal of clinical pathology 1991;44(10):844–8 doi 10.1136/jcp.44.10.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saha A, Robertson ES. Epstein-Barr virus-associated B-cell lymphomas: pathogenesis and clinical outcomes. Clin Cancer Res 2011;17(10):3056–63 doi 10.1158/1078-0432.CCR-10-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allday MJ, Bazot Q, White RE. The EBNA3 Family: Two Oncoproteins and a Tumour Suppressor that Are Central to the Biology of EBV in B Cells. Curr Top Microbiol Immunol 2015;391:61–117 doi 10.1007/978-3-319-22834-1_3. [DOI] [PubMed] [Google Scholar]
- 47.Hung CH, Chiu YF, Wang WH, Chen LW, Chang PJ, Huang TY, et al. Interaction Between BGLF2 and BBLF1 Is Required for the Efficient Production of Infectious Epstein-Barr Virus Particles. Front Microbiol 2019;10:3021 doi 10.3389/fmicb.2019.03021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe T, Narita Y, Yoshida M, Sato Y, Goshima F, Kimura H, et al. The Epstein-Barr Virus BDLF4 Gene Is Required for Efficient Expression of Viral Late Lytic Genes. J Virol 2015;89(19):10120–4 doi 10.1128/JVI.01604-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duarte M, Wang L, Calderwood MA, Adelmant G, Ohashi M, Roecklein-Canfield J, et al. An RS motif within the Epstein-Barr virus BLRF2 tegument protein is phosphorylated by SRPK2 and is important for viral replication. Plos One 2013;8(1):e53512 doi 10.1371/journal.pone.0053512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horikawa T, Sheen TS, Takeshita H, Sato H, Furukawa M, Yoshizaki T. Induction of c-Met proto-oncogene by Epstein-Barr virus latent membrane protein-1 and the correlation with cervical lymph node metastasis of nasopharyngeal carcinoma. Am J Pathol 2001;159(1):27–33 doi 10.1016/S0002-9440(10)61669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horikawa T, Yang J, Kondo S, Yoshizaki T, Joab I, Furukawa M, et al. Twist and epithelial-mesenchymal transition are induced by the EBV oncoprotein latent membrane protein 1 and are associated with metastatic nasopharyngeal carcinoma. Cancer Res 2007;67(5):1970–8 doi 10.1158/0008-5472.CAN-06-3933. [DOI] [PubMed] [Google Scholar]
- 52.Horikawa T, Yoshizaki T, Sheen TS, Lee SY, Furukawa M. Association of latent membrane protein 1 and matrix metalloproteinase 9 with metastasis in nasopharyngeal carcinoma. Cancer 2000;89(4):715–23 doi . [DOI] [PubMed] [Google Scholar]
- 53.Lo AK, Dawson CW, Lung HL, Wong KL, Young LS. The Role of EBV-Encoded LMP1 in the NPC Tumor Microenvironment: From Function to Therapy. Front Oncol 2021;11:640207 doi 10.3389/fonc.2021.640207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, et al. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A 2007;104(41):16164–9 doi 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benders AA, Tang W, Middeldorp JM, Greijer AE, Thorne LB, Funkhouser WK, et al. Epstein-Barr virus latent membrane protein 1 is not associated with vessel density nor with hypoxia inducible factor 1 alpha expression in nasopharyngeal carcinoma tissue. Head Neck Pathol 2009;3(4):276–82 doi 10.1007/s12105-009-0148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet 2019;394(10192):64–80 doi 10.1016/S0140-6736(19)30956-0. [DOI] [PubMed] [Google Scholar]
- 57.Heilmann AM, Calderwood MA, Johannsen E. Epstein-Barr virus LF2 protein regulates viral replication by altering Rta subcellular localization. J Virol 2010;84(19):9920–31 doi 10.1128/JVI.00573-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen LW, Lin LS, Chang YS, Liu ST. Functional analysis of EA-D of Epstein-Barr virus. Virology 1995;211(2):593–7 doi 10.1006/viro.1995.1443. [DOI] [PubMed] [Google Scholar]
- 59.Kiehl A, Dorsky DI. Cooperation of EBV DNA polymerase and EA-D(BMRF1) in vitro and colocalization in nuclei of infected cells. Virology 1991;184(1):330–40 doi 10.1016/0042-6822(91)90849-7. [DOI] [PubMed] [Google Scholar]
- 60.Kiehl A, Dorsky DI. Bipartite DNA-binding region of the Epstein-Barr virus BMRF1 product essential for DNA polymerase accessory function. J Virol 1995;69(3):1669–77 doi 10.1128/JVI.69.3.1669-1677.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li JS, Zhou BS, Dutschman GE, Grill SP, Tan RS, Cheng YC. Association of Epstein-Barr virus early antigen diffuse component and virus-specified DNA polymerase activity. J Virol 1987;61(9):2947–9 doi 10.1128/JVI.61.9.2947-2949.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakayama S, Murata T, Murayama K, Yasui Y, Sato Y, Kudoh A, et al. Epstein-Barr virus polymerase processivity factor enhances BALF2 promoter transcription as a coactivator for the BZLF1 immediate-early protein. J Biol Chem 2009;284(32):21557–68 doi 10.1074/jbc.M109.015685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fixman ED, Hayward GS, Hayward SD. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol 1995;69(5):2998–3006 doi 10.1128/JVI.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsurumi T EBV replication enzymes. Curr Top Microbiol Immunol 2001;258:65–87 doi 10.1007/978-3-642-56515-1_5. [DOI] [PubMed] [Google Scholar]
- 65.Smith NA, Coleman CB, Gewurz BE, Rochford R. CD21 (Complement Receptor 2) Is the Receptor for Epstein-Barr Virus Entry into T Cells. Journal of virology 2020;94(11) doi 10.1128/JVI.00428-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song L, Song M, Camargo MC, Van Duine J, Williams S, Chung Y, et al. Identification of anti-Epstein-Barr virus (EBV) antibody signature in EBV-associated gastric carcinoma. Gastric Cancer 2021;24(4):858–67 doi 10.1007/s10120-021-01170-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Argentieri MC, Pilla D, Vanzati A, Lonardi S, Facchetti F, Doglioni C, et al. Antibodies are forever: a study using 12–26-year-old expired antibodies. Histopathology 2013;63(6):869–76 doi 10.1111/his.12225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available from the corresponding author upon request.


