Abstract
Objective:
To evaluate the performance of an internally developed and previously validated artificial-intelligence (AI) algorithm for magnetic resonance (MR)-derived total kidney volume (TKV) in autosomal dominant polycystic kidney disease (ADPKD) when implemented in clinical practice.
Patients and Methods:
The study included adult patients with ADPKD seen by a nephrologist at our institution between November 2019 and January 2021 and undergoing an MR imaging examination as part of standard clinical care. Thirty-three nephrologists ordered MR imaging, requesting AI-based TKV calculation for 170 cases in these 161 unique patients. We tracked implementation and performance of the algorithm over one year. All cases (N=170) were reviewed by a radiologist and a radiology technologist (RT) for quality and accuracy. Manual editing of algorithm output occurred at radiologist or RT discretion. Performance was assessed by comparing AI-based and manually edited segmentations via measures of similarity and dissimilarity to ensure expected performance. We analyzed ADPKD severity class assignment of algorithm-derived versus manually edited TKV to assess impact.
Results:
Clinical implementation was successful. AI-algorithm based segmentation showed high levels of agreement and was non-inferior to interobserver variability and other methods for determining TKV. Of manually edited cases (N=84), the AI-algorithm TKV output showed a small mean volume difference of −3.3%. Agreement for disease class between AI-based and manually edited segmentation was high (5 cases differed).
Conclusion:
Performance of an AI algorithm in real-life clinical practice can be preserved if there is careful development and validation and if the implementation environment closely matches the development conditions.
Introduction
With the rapid advancement and increasing availability of artificial intelligence (AI) algorithms in medicine and in radiology specifically, there has been growing interest and investigation into their potential clinical implementation. Much of the literature to date is focused on pre-implementation topics, including algorithm development and validation, usually in a controlled setting far removed from the clinical workflow. Full clinical implementation has not yet been widely achieved among radiology practices as it requires not only algorithm development and validation, but also integration into an already complex clinical imaging environment. Process evaluations regarding translating AI innovations from discovery and validation to an integrated component of the clinical workflow are currently lacking. This process involves new challenges, including how the algorithm is ordered, how it is triggered, how it is routed, how it is monitored, and how to educate all those who will be involved at various stages of the workflow. It is important that real-life performance, which exposes the process to a myriad of unpredictable variables, matches that of a more controlled pre-implementation environment.
At our institution we have investigated a previously-validated AI algorithm for magnetic resonance (MR)-derived measurement of total kidney volume (TKV) in autosomal dominant polycystic kidney disease (ADPKD) in clinical practice. ADPKD is the most common genetic cause of chronic kidney disease (CKD) and TKV is an important prognostic biomarker 1–10. Along with age, TKV reliably predicts eGFR decline and is used to identify patients who would benefit from specific novel therapies 5, 11. The process of clinical implementation of an AI algorithm, such as MR-derived measurement of TKV in ADPKD, involves multiple intersecting systems and people, including but not limited to patients, imaging equipment, technologists, digital data, radiologists, and referring clinicians. Successful exam ordering, image acquisition, algorithm processing, output reporting, and continuous quality assurance are all necessary for successful execution of the AI assisted workflow.
The potential for clinical implementation of AI algorithms is what drives scientific inquiry in this field but remains an under-studied step. The purpose of this study is to evaluate the performance of an internally developed and previously validated AI algorithm for TKV in ADPKD when implemented in clinical practice.
Patients and Methods
The study was performed with IRB approval. Details regarding this AI algorithm have been published previously 12–19. Referring providers had the option to order AI-based TKV measurements when placing an abdominal imaging exam order (Table 1). One sequence in the exam, a routine clinical single-shot fast spin echo coronal sequence, was used by the AI algorithm (Seimens HASTE or GE SSFSE with fat saturation) for TKV calculation. , AI-based segmented images were first reviewed by a Medical Image Analyst (a certified CT or MR technologist with extra training and expertise in 3D image analysis and anatomic segmentation) and either accepted without any manual editing if AI segmentation was deemed to be optimal by visually comparing the output segmentation overlay to the organ borders slice-by-slice (“pass”) or manually edited (“rework”). This step was performed despite prior algorithm validation due to our commitment to extract and evaluate real-life performance metrics. A second quality check of the output was performed by the reading radiologist. This radiologist could trigger the manual rework pathway if the RT had not, or the radiologist could accept the algorithm output or the RT-triggered manually edited segmentation if it had already been reworked. Segmentations were then approved and used to provide a report of right, left, and total kidney volumes.
Table 1.
Scanner, location, and demographic information.*
| Pass (N=86) | Rework (N=84) | ||
|---|---|---|---|
|
Scanner Information
| |||
| Manufacturer – no. (%) | |||
| Model – no. (%)† | |||
| GE MEDICAL SYSTEMS | 48 (55.8) | 56 (66.7) | |
| Optima MR450w | 27 (31.4) | 23 (27.4) | |
| Signa HDxt | 12 (14) | 20 (23.8) | |
| DISCOVERY MR750w | 6 (7) | 12 (14.3) | |
| DISCOVERY MR450 | 1 (1.2) | 1 (1.2) | |
| DISCOVERY MR750 | 2 (2.3) | 0 (0) | |
| SIEMENS | 38 (44.2) | 28 (33.3) | |
| Skyra | 19 (22.1) | 14 (16.7) | |
| MAGNETOM Vida | 11 (12.8) | 9 (10.7) | |
| Aera | 7 (8.1) | 4 (4.8) | |
| MAGNETOM Sola | 1 (1.2) | 1 (1.2) | |
| Field Strength – no. (%) | |||
| 1.5 T | 48 (55.8) | 49 (58.3) | |
| 3 T | 38 (44.2) | 35 (41.7) | |
| Slice Thickness – no. (%) | |||
| 4 mm | 72 (83.7) | 71 (84.5) | |
| 5 mm | 14 (16.3) | 13 (15.5) | |
|
Location Information
| |||
| Mayo Clinic Location – no. (%) | |||
| Rochester | 67 (77.9) | 67 (79.8) | |
| Arizona | 8 (9.3) | 11 (13.1) | |
| Florida | 11 (12.8) | 6 (7.1) | |
| Demographic Information | |||
| Sex – no. (%) | |||
| F | 49 (57) | 62 (73.8) | |
| M | 37 (43) | 22 (26.2) | |
| Age – yr | 43.4±13.5 | 47±15.4 | |
| Race – no. (%) | |||
| White | 78 (90.7) | 79 (94) | |
| Asian | 3 (3.5) | 2 (2.4) | |
| Black or African American | 0 (0) | 1 (1.2) | |
| Other | 3 (3.5) | 1 (1.2) | |
| Unknown | 2 (2.3) | 1 (1.2) | |
| Height – cm | 173±9.9 | 169.7±9.5 | |
| Weight – kg | 82.3±19.6 | 76.7±17.8 | |
| BMI | 23.7±2 | 23±2 | |
| Kidney Disease Subtype – no. (%) | |||
| Typical | 80 (93) | 70 (83.3) | |
| Atypical | 2 (2.3) | 6 (7.1) | |
| Unknown | 4 (4.7) | 8 (9.5) | |
Plus-minus values are means±SD
Percentages for the manufacturer model are of the whole not the specific manufacturer.
For inclusion, patients were required to be over 18 years of age, have a previous diagnosis of ADPKD, and have an MR imaging examination ordered as part of standard clinical care. Patient International Classification of Disease (ICD)-10 and ICD-9 diagnosis codes were extracted from a Mayo Clinic internal database to confirm ADPKD diagnosis. A small subset of patients where ADPKD diagnosis could not be confirmed were grouped as ‘other’, including cystic and non-cystic kidney disease, non-PKD patients as well as kidney transplant patients, and autosomal recessive PKD diagnoses. ADPKD can be classified into two main subclassifications based on presentation: typical or atypical 7, 20. Typical diffuse cystic ADPKD is classified by utilizing height-adjusted TKV and age to identify patients with the highest risk of disease progression 21. The 5-group classification scale ranges from least severe (class 1A) to most severe (class 1E). ADPKD subtype and classification were assigned by a trained observer according to previous criteria 21. Demographic information, including age, sex, race, and ethnicity was collected from DICOM metadata and/or an internal patient database. Patient-related kidney function data, including estimated glomerular filtration rate (eGFR), serum creatinine, blood urea nitrogen (BUN), and albumin/creatinine ratio were also extracted. All patient research authorizations were confirmed prior to inclusion in the study.
Statistical analysis
Statistical analyses were performed to determine both the performance of the AI-based segmentation tool compared to manually edited AI segmentation and any potential variables which may have been associated with a manually edited segmentation. The Shapiro-Wilk test (SciPy v1.5.4) was used to determine if data were normally distributed. All statistical analyses were performed using Python (v3.8.3) and the following modules: SciPy (v1.5.4), statmodels (v0.12.2), pydicom (v2.1.1), SimpleITK (v2.0.2), seaborn (v.0.11.0) and matplotlib (v.3.2.2).
Algorithm performance
Algorithm performance was determined via comparison of AI-based and manually edited AI segmentations for the manually edited data only. Common image metrics of similarity (Dice coefficient [two times the area of overlap divided by the total number of pixels in both segmentations; minimum value(0), maximum(1)] and Jaccard index [size of intersection divided by size union; minimum value(0), maximum(1)]) and dissimilarity (volume difference, percent volume difference, surface distance [mean of all distances between every surface voxel across segmentations; values close to zero represent perfect overlap], and Hausdorff distance [greatest of all distances between all points between segmentations; values close to zero represent perfect overlap]) were computed (SimpleITK; v2.0.2). Bland-Altman plots (pingouin v0.4.12) were constructed to look at agreement, fixed bias, and any outliers, while linear regression (SciPy v1.5.4) assessed correlation between AI and manually edited AI TKV measurements. A scatter plot of the Dice coefficient versus corrected AI TKV was constructed to determine if kidney volume was related to AI-based segmentation performance. Finally, a one-sided Welch’s t-test was computed to determine if the AI-based segmentation was non-inferior to manually edited AI segmentation (SciPy v1.5.4). Power calculations were performed to determine the sample size needed to observe a delta value for the non-inferiority test. Tests were run across a range of clinically relevant delta values to arrive at a minimum significant delta of non-inferiority 22.
Pass versus rework comparisons
Scanner characteristics, patient demographics, and disease severity markers were investigated for association with either an AI-based segmentation accept or manually edited rework pathway. A chi-square test of independence compared distributions across accept and rework workflows for discrete variables (SciPyv1.5.4). Additionally, a two-sided Kolmogorov-Smirnov test was used to test for distributional differences between images which were accepted or sent for rework for continuous variables (SciPyv1.5.4). No adjustment for multiple comparisons was performed.
Results
Participants and Imaging
From November 2019 to January 2021, a total of 33 nephrologists across three sites within our institution ordered MR imaging, requesting AI-based TKV calculation for 170 cases in 161 unique patients. There were 7 patients that were imaged at different times throughout the study. Two patients were imaged three times, while the remaining 5 were imaged twice. For these cases, the time span between exams was 184 ± 82 days (minimum was 105 days). Of the total 170 cases, output of AI-based segmentation in 86 cases was accepted without manual editing (‘pass’), while 84 cases were manually edited (‘rework’). The workflow diagram can be seen in Figure 1. In total, 12 Medical Image Analysts and 49 radiologists were involved in this study. The mean patient age was 45.2 ± 14.5 years and 65.3% were female. Nephrologist-confirmed ADPKD subtype was typical in 88.2% of patients and atypical in 4.7%. The remaining patients (7.1%) were excluded from classification for non-PKD, kidney transplant or autosomal recessive-PKD. Images were acquired across two scanner manufacturers, GE Medical (61%) and Siemens (39%), and 9 different models in total. Coronal Half-Fourier Acquisition Single-shot Turbo spin Echo (HASTE, Siemens) or Single-Shot Fast Spin Echo (SSFSE, GE) scan protocols were used. Images were collected across two field strengths, 1.5T (57.1%) and 3T (42.9%), and two different slice thicknesses, 4mm (84.1%) and 5mm (15.9%). Breakdown of scanner, site location, and demographics across pathways can be seen in Table 1.
Figure 1: Workflow diagram illustrating the steps and decisions points the clinicians (beige), radiologists (light blue), MR technologists (brown), and Image Analysts (pink) played in the study.
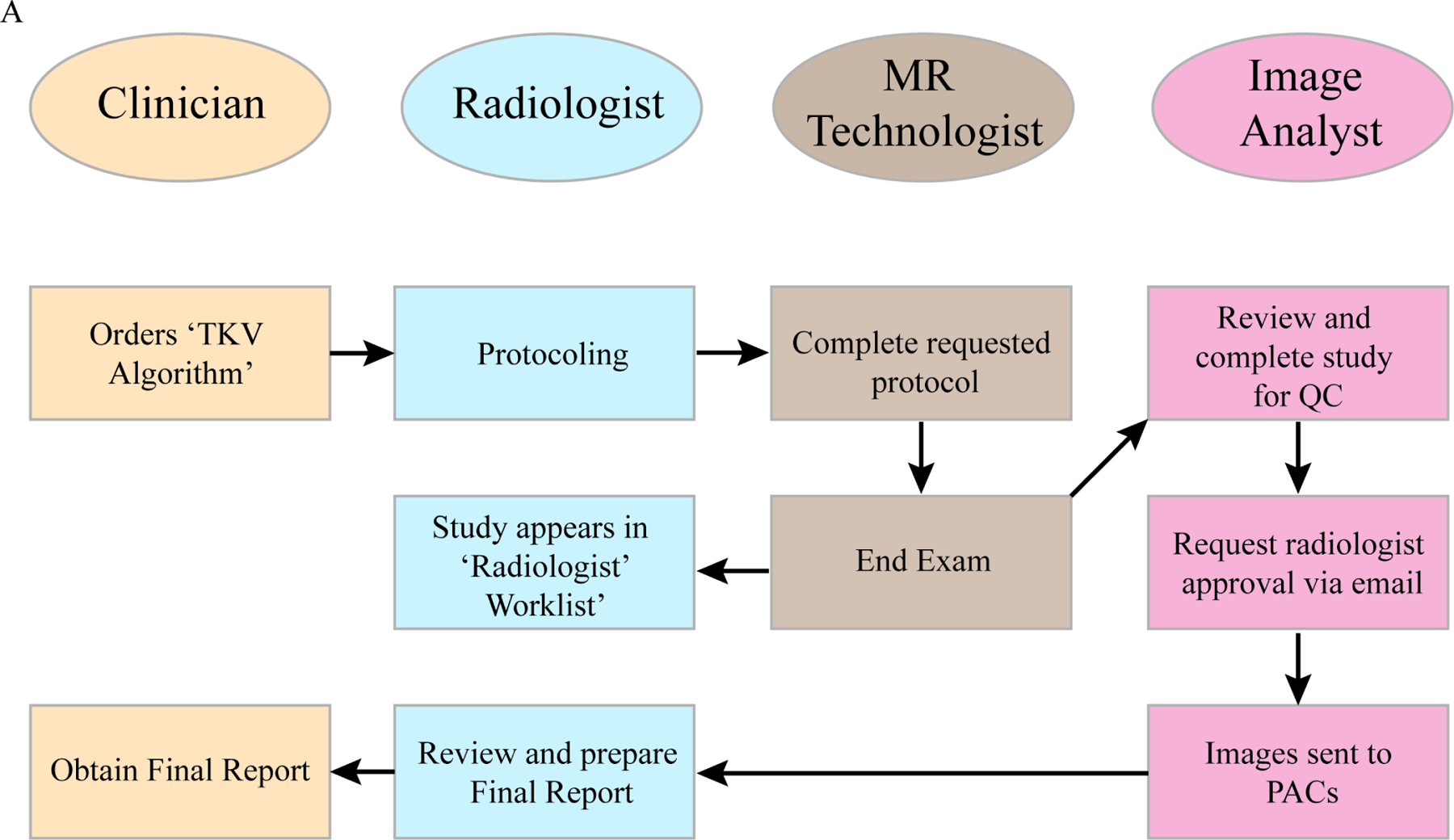
An arrow indicates the sequence of steps and the direction of the workflow. The clinician is positioned at the start and end of the workflow.
Algorithm Performance
To determine how well the AI algorithm for TKV performed, AI- and manually edited AI segmentations were compared. Most commonly, these corrections were minor segmentation alterations. Exemplar MR images with TKV segmentation overlays (AI or manually edited) of maximum (Dice = 0.99, Fig. 2A), minimum (Dice = 0.77, Fig. 2B) and median (Dice = 0.98, Fig. 2C) are shown in Figure 2. Dice coefficients are shown in Table 2. The mean TKV difference was –34.0 cc (range –413.8 cc to 415.4 cc) and mean percent difference was –3.3% (range – 41.0% to 22.2%) as shown in Table 2, Figures 3A and 3B. AI and manually edited TKVs (cc) were highly correlated with a small volumetric offset, suggesting that most rework cases involved very minor corrections (slope = 1.0, intercept = −41.08, r2 = 0.99, P < .0001, Figure 3C). Furthermore, the intraclass correlation coefficient between AI and manually edited TKV (cc) indicated excellent agreement (ICC = 0.997). Dice scores were more variable with smaller corrected AI TKVs (Figure 3D). The mean Jaccard index was 0.926 (range 0.63 – 0.99, Table 2), the mean Hausdorff distance was 30.51 mm (range 5.27 – 174.29), and the mean surface difference was 1.68 mm (range 0.06–18.43) (Figure 3E). Finally, to confirm the AI approach was non-inferior to previous non-AI assisted segmentation approaches 12, a non-inferiority test was conducted. The non-inferiority test was powered to a percent delta of 4.97% (2.5% one-sided type 1 error: 80% power). The percent TKV difference between AI and manually edited TKV was non-inferior at a minimum percent delta of 4.80% and non-inferior to previously determined inter-rater percent delta (6.21%), stereology percent delta (9.12%), and ellipsoid percent delta (22.27%) values (Figure 3F, inter-rater P < .001, stereology P < .0001, ellipsoid P < 0.0001) 12. Only 7.05% (12/170) of the total cases recorded differences outside the inter-rater delta (6.21%) range, yielding an approximation of the performance of the algorithm without a rework pathway.
Figure 2: Example images with AI-generated and Medical Image Analyst corrected segmentations.
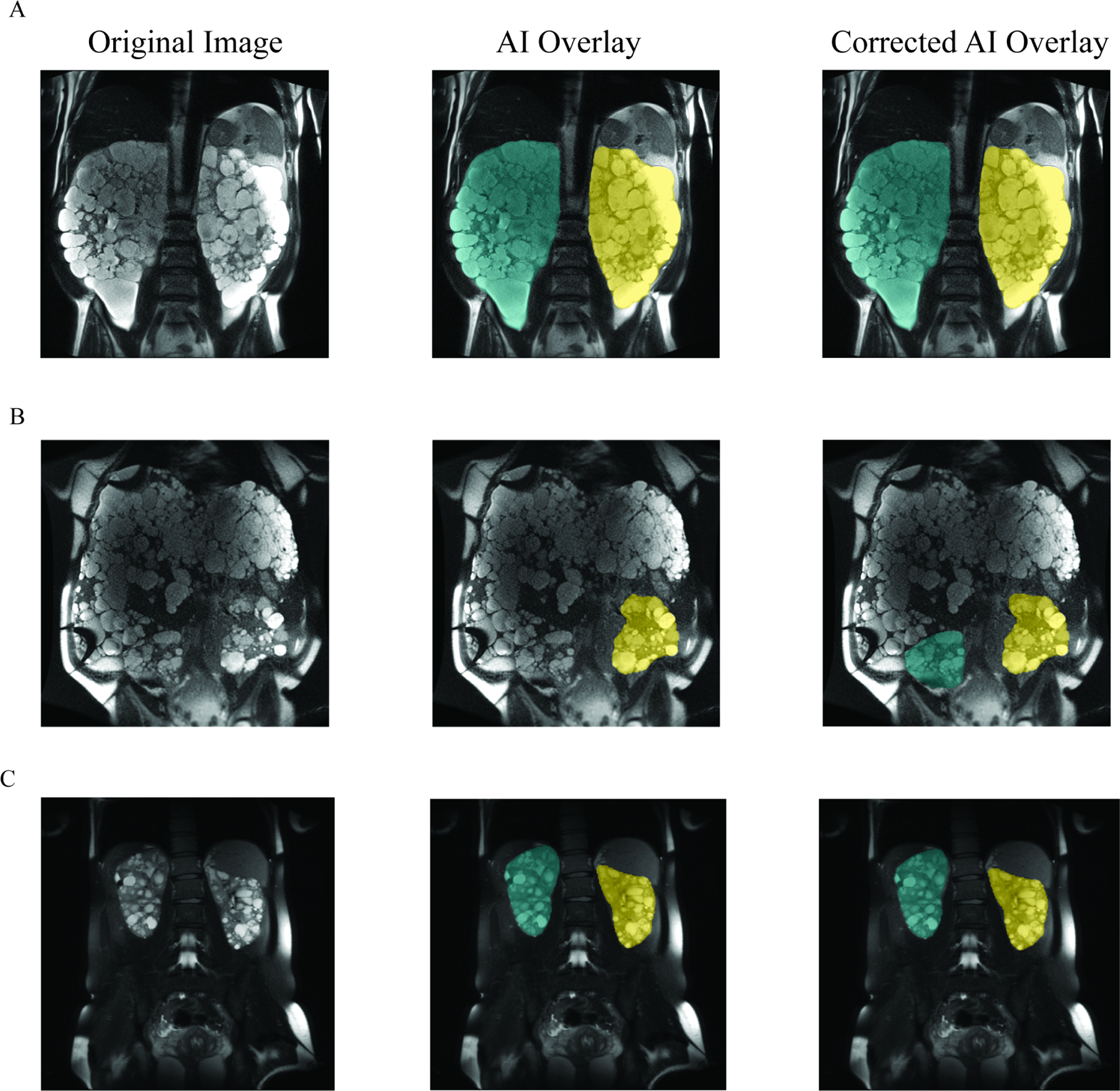
Panel A shows the original CT image (left), original image with AI-generated TKV segmentation overlay (middle), original image plus medical image analyst corrected AI overlay (right) from a case with max Dice score (0.99). Left kidney segmentation is shown in yellow and right kidney segmentation is shown in green. Panel B shows minimum Dice score (0.77) example. Panel C shows median Dice score (0.98) example.
Table 2:
Similarity and dissimilarity metrics between initial AI segmentation image and reworked image
| Mean | Minimum | Median | Maximum | |
|---|---|---|---|---|
| Dice | 0.959 | 0.774 | 0.977 | 0.999 |
| Jaccard | 0.926 | 0.631 | 0.956 | 0.997 |
| Difference | −34.004 | −413.843 | −13.201 | 415.419 |
| Percent Difference | −3.318 | −41.003 | −1.445 | 22.151 |
| Hausdorff Distance – mm | 30.512 | 5.265 | 20.258 | 174.289 |
| Mean Surface Distance – mm | 1.679 | 0.061 | 0.750 | 18.433 |
Figure 3: Overall performance of AI-generated TKV segmentation compared to Medical Image Analyst corrected AI-generated TKV segmentation.
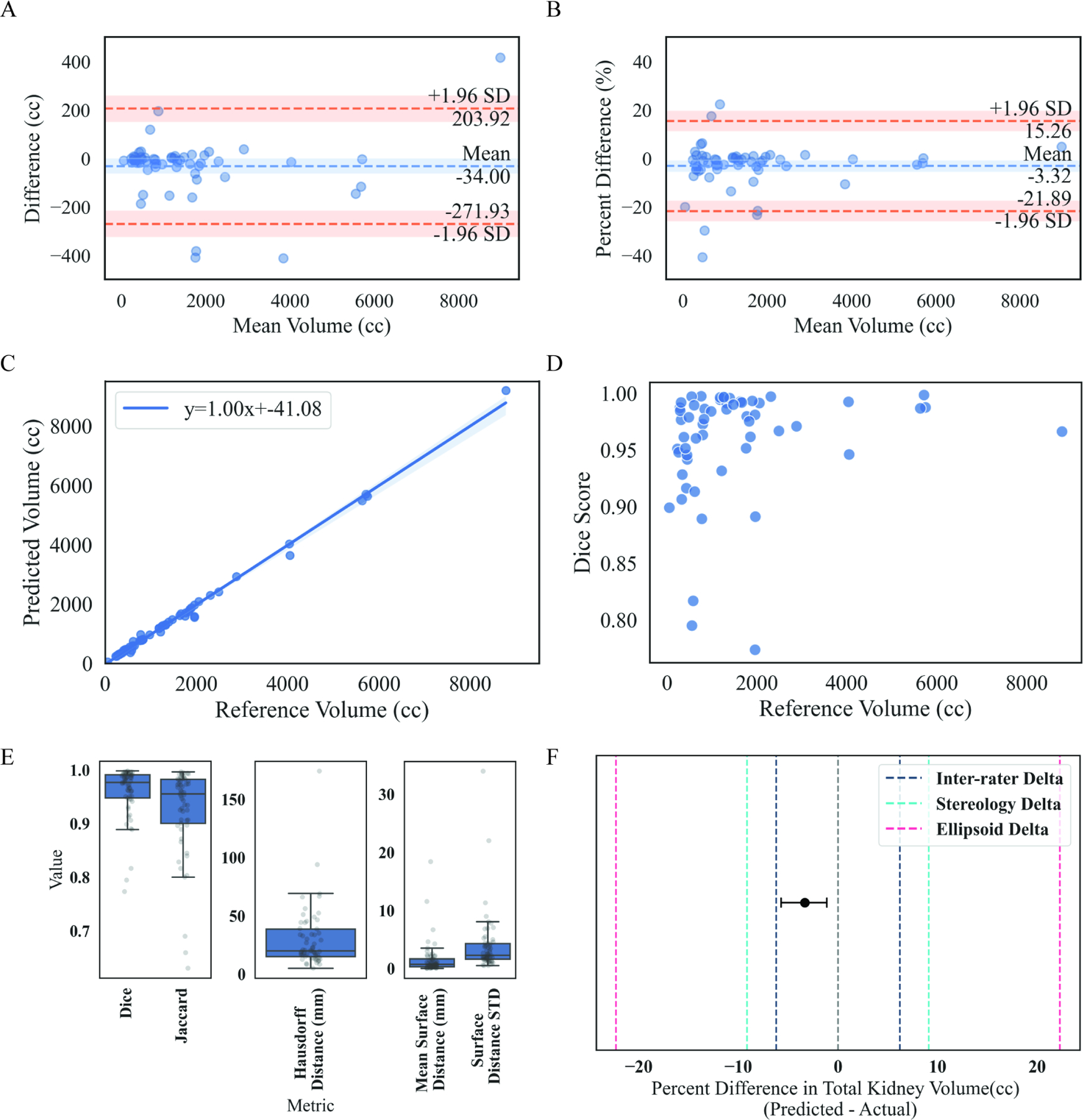
Panel A shows Bland-Altman plots to evaluate absolute agreement between AI-generated segmentation and medical image analyst corrected AI-generated segmentation. Mean difference between measures (blue dashed line); 95% CI for mean difference (shaded blue band); 95% limits of agreement (orange dashed line; average ± 1.96 standard deviation of difference); 95% CI for limits of agreement (shaded orange band). Panel B is the same as Panel A, but for percent difference between AI-generated TKV and medical image analyst corrected AI-generated TKV. Panel C shows a linear regression of highly correlated AI-generated TKV, and medical image analyst corrected AI-generated TKV (slope=1.00, intercept=−41.08, r2 = 0.99, P <.0001). Panel D shows a scatter plot of medical image analyst corrected AI-generated TKV (cc) by Dice score. Panel E shows box plots with individual case scatter of similarity and dissimilarity metrics including dice, jaccard, hausdorff distance (mm), mean surface distance (mm) and surface distance standard deviation. Panel F shows a non-inferiority plot of the mean percent difference (± 95% CI) between AI TKV and corrected AI TKV (gray dashed line = zero difference between methods; dark blue dashed line represents delta acquired from prior inter-rater agreement study; teal dashed line represents delta acquired from stereology measurements; pink dashed line represents delta acquired from ellipsoid measurements. Mean AI TKV and corrected AI TKV difference is non-inferior to inter-rater, stereology and ellipsoid deltas (One sided t-test; inter-rater P<.0001, stereology P<.0001, ellipsoid P<.0001) 12.
These results indicate that our algorithm performs well and is non-inferior to manual Medical Image Analyst-corrected segmentations at an experimentally derived and clinically relevant delta value.
Determining factors associated with rework pathway
To identify factors associated with a case being sent for rework, we compared scanner information across pass and rework pathways. No significant differences in scanner manufacturer (p – 𝑣𝑎𝑙𝑢e = 0.20), manufacturer model (p – value= 0.43), field strength (p – value = 0.86), slice thickness (p – value > 0.99), and pixel spacing (p – value = 0.25) were observed (Supplemental Table 1). Comparison of additional imaging parameters, including repetition time, echo time/train length, flip angle, percent sampling, image size, number of images in acquisition, field of view, and patient position were all not significantly different (Supplemental Table 2).
Furthermore, patient demographic factors across pass and rework pathways were compared. Age (p – value = 0.26), body mass index (BMI) (p – value = 0.06), race/ethnicity (p – value = 0.64), and study imaging date (p – value= 0.08 Supplemental Figure 1A) were all not significantly different across AI (pass) and corrected AI (rework) pathways (Supplemental Table 1). Sex was the only measure we found that was significantly different across AI (pass) and corrected AI (rework) pathways (p – value = 0.03, Supplemental Table 1). Females were overrepresented in the corrected AI pathway (73.8%) versus the AI pathway (57.0%) with significantly lower BMI (F [mean ± SD] = 22.42 ± 1.68; M [mean ± SD] = 25.11 ± 1.42; KS Test, stat = 0.779, P < .0001; Supplemental Figure 1A) and smaller total kidney volumes (F [mean ± SD] = 1299.69 ± 1072.60; M [mean ± SD] = 2153.25 ± 1835.19; KS Test, stat = 0.26, P = .008; Supplemental Figure 1B) compared to males.
Typical ADPKD is classified at the Mayo Clinic utilizing height-adjusted TKV and age to identify patients with the highest risk of disease progression 21. The 5-group classification scale ranges from least severe (class 1A) to most severe (class 1E). The pre-rework and post-rework classifications were compared to determine changes in classification and degree of change. Only a small percentage of rework cases changed classification assignment after rework (10.4%). Agreement between pre-rework and post-rework classification across all cases was high (weighted Cohen’s kappa = 0.86; Supplemental Figure 1C). Pre-rework and post-rework classification agreement was higher in females (weighted Cohen’s kappa = 0.90; Supplemental Figure 1C) than males (weighted Cohen’s kappa = 0.74; Supplemental Figure 1D). Overall, no re-classification changes of greater than one class were observed (Supplemental Figure 1B).
Kidney function was assessed by estimated glomerular filtration rate (GFR), serum creatinine, blood urea nitrogen (BUN) and albumin/creatinine ratio 6, 11. We evaluated whether kidney disease severity was associated with images routing to the rework pathway (Supplemental Table 3). TKV distributions were not significantly different between pass and rework groups (p – value= 0.23). Measurements of eGFR (p – value = 0.87), creatinine (p – value = 0.56, Supplemental Table 3), BUN (p – value= 0.81), and albumin/creatinine ratio (p – value = 0.45) were not significantly different between pass and rework pathways.
Discussion
Advances in artificial intelligence (AI) in medicine remain weighted toward algorithm development and validation with large-scale clinical implementation still unrealized. Barriers to broad clinical adoption of AI algorithms include poor understanding of the steps involved in their implementation within a practice and a lack of data on their real-world performance. Coordinated interdisciplinary efforts to integrate algorithms into clinical workflows are necessary to drive the work of AI scientists to their full potential and to utilize algorithms for their intended purpose.
We have demonstrated the potential for successful clinical implementation of an AI algorithm into a complex radiology practice which required coordination of technical deployment, education of interdisciplinary stakeholders, extraction of real-life performance metrics, and analysis of impact on the intended clinical question. Our internally developed algorithm for MR-derived measurement of TKV in ADPKD was effectively integrated and performed as expected in the real-life clinical setting, proving to be non-inferior to non-AI assisted segmentation. In addition, without the AI tool, manual processing takes 60–90 minutes. Even in cases needing editing, the final metrics were now obtained in only a few minutes.
Technical deployment
Technical deployment of the algorithm into the clinical workflow relied upon an integrated IT team that could set up image filtering and routing rules based on specific inclusion criteria. In this study, routing rules were set up based on the MR series description, thereby only sending a single series for AI processing. Images moved downstream through our institutional orchestration engine 23, and eventually to the Medical Image Analysts for review before output routing to the radiologist and the PACS.
Education of stakeholders
Communication and education for those involved in the AI algorithm clinical implementation are critical to success, both prior to any change and throughout implementation. For our algorithm, those primarily involved in the clinical workflow are the MRI-ordering clinician (nephrologist), the radiologist protocolling and interpreting the exam, including report of algorithm output, the MR technologist acquiring the images, and the Medical Image Analysts responsible for review and possible segmentation editing.
Educational materials were developed for each role. Learning modules were available electronically and included both text and graphic presentation of the background, rationale, and steps involved for algorithm implementation. Leaders from each stakeholder group (physicians and technologists) were identified to disseminate the information and act as resources for questions. For example, the radiologist proponent sent informational emails with links to modules, presented information at divisional meetings (including history of pre-implementation algorithm validation), communicated with residents and fellows, and fielded inquiries from radiologists and trainees in real time as cases arose in the clinical practice. Throughout the educational efforts two messages were critical to adoption of this initiative: first, an emphasis on real-world patient benefits of this algorithm’s implementation and second, a reassurance that despite the inherent discomfort that accompanies workflow change, it would not be onerous for the radiologist.
Performance metrics
Our extraction of real-life performance metrics relied on review of each AI-based segmentation by a Medical Image Analyst and a radiologist. While half of the cases (84/170, 49.4%) during the study period were manually edited, the mean percent volume difference was just −3.3%, indicating that corrections were minor. This also indicated that the technologists had a very low threshold for editing. Therefore, the 50% which were not reworked were accepted at a very high standard. The percent TKV difference between AI-based segmentation and manually edited segmentation was non-inferior to previously determined inter-rater difference and to other clinically accepted methods for determining TKV (e.g., stereology-based and/or ellipsoid-based measurements).
Bias Analysis
We investigated the rework cases where the class changed pre/post rework to determine if there was an underlying characteristic which led to the class being changed. The variables investigated included manufacturer, scanner model, field strength, location, sex, age, race, height, weight, continuous BMI, discrete BMI interpretation, algorithm TKV value (cc), eGFR (mL/min/BSA), creatinine (mg/dL), BUN (mg/dL), and presence of PLD. As rework caused a shift in class for seven out of the 67 reworked cases, little concrete information was felt to likely result from this investigation. In all cases, histograms were generated. For the continuous variables, the values observed for the rework individuals where a class change occurred tended to be distributed throughout without obvious clustering in a given region. and we did investigate in more detail the influence of PLD. In particular, for the 7 cases which switched image class, 4 of the cases had PLD (2 with severe PLD), and 3 did not have PLD. Also note that PLD prevalence in patients affected by PKD is ~70%. We do feel that severe PLD can often cause issues with, for example, assigning adjacent cysts to the right kidney or liver.
Impact on intended clinical question
Another critical step in assessing the success of algorithm implementation is the analysis of its impact on intended clinical questions. TKV as an imaging biomarker in ADPKD is a valuable major variable for assignment of a disease severity class (5 groups, 1A to 1E), a reliable and widely utilized predictor of future eGFR decline, and an important determinant of eligibility for certain therapies. In our study, the agreement for disease class assignment between AI-based segmentation and manually edited segmentation was high (with only 5 cases being assigned a different class). In the few cases of re-classification from manual editing, no changes greater than one class occurred. Given that the AI-based segmentations were shown to be non-inferior to inter-rater difference and other methods of TKV calculation, we would expect a similar rate of reclassification if those methods were similarly investigated.
Next steps
While AI algorithm discovery, development, and initial validation can occur in isolation of a practice’s clinical workflow and real-time patient care, the application of these algorithms for true clinical impact cannot. Future work will include implementation of a workflow where the radiologist first reviews the cases and then triggers a ‘pass’ or ‘rework’ pathway, as well as the incorporation of additional analytics (e.g., liver segmentation for total liver volume assessment).
Conclusion
Performance of an AI algorithm in a large radiology clinical practice can be preserved if careful attention is paid to validation of the algorithm during development and if the implementation environment closely matches the development conditions.
Supplementary Material
Figure 4: Comparing study date distributions between pass and rework and comparing classification of typical ADPKD pre- or post-rework pathway.
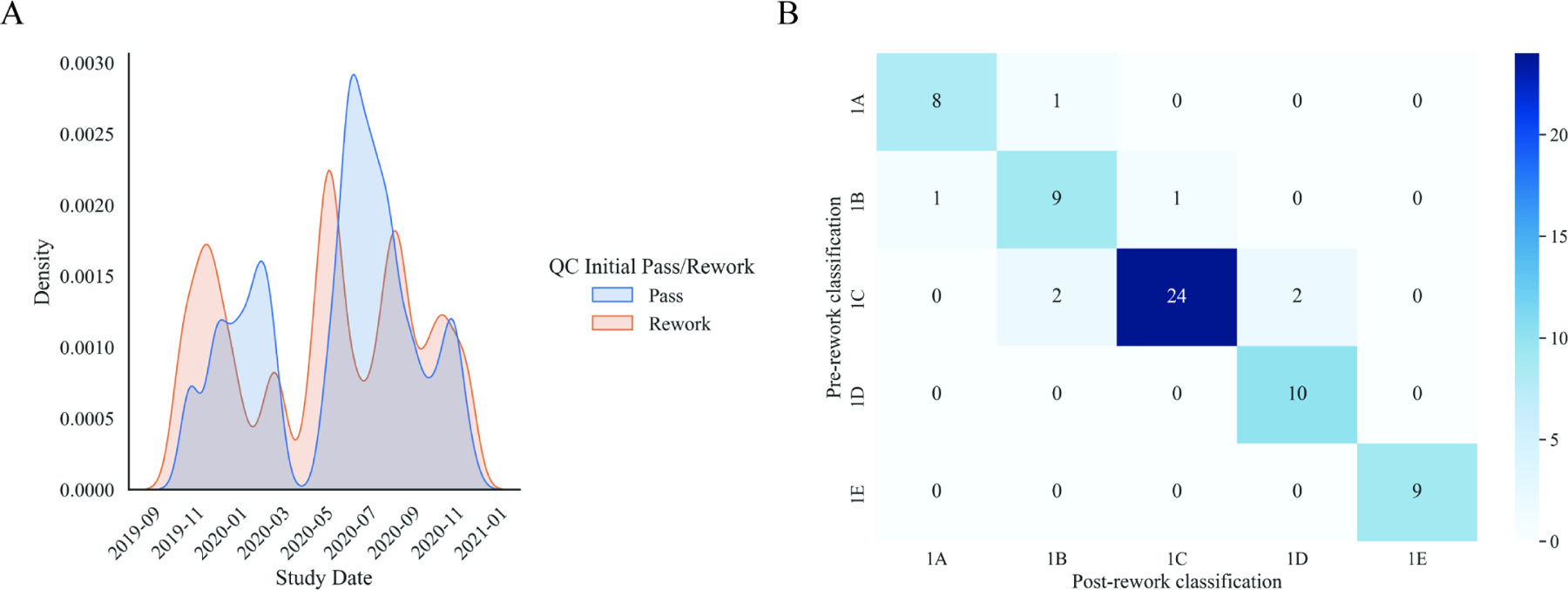
Panel A shows kernel density estimated distributions of study dates between pass (blue) and rework (orange) pathways that are not significantly different (two-sample Kolmogorov-Smirnov test, P=.08). Panel B represents an agreement heatmap between AI (pre-rework) and corrected AI (post-rework) typical ADPKD classification for all patients (weighted Cohen’s kappa= 0.86). Diagonal represents perfect agreement. The darker the shade of blue represents greater counts.
Acknowledgments
The authors acknowledge the assistance of Lucy Bahn, Ph.D. in the preparation of this manuscript.
Funding:
This project and publication were supported in part by funding from the Department of Radiology’s Framework for AI Software Technology (FAST) at Mayo Clinic. Research reported in this publication was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under Award Numbers K01DK110136, and R03DK125632. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations:
- AI
artificial intelligence
- TKV
total kidney volume
- ADPKD
autosomal dominant polycystic kidney disease
- RT
radiology technologist
- CKD
chronic kidney disease
- eGFR
estimated glomerular filtration rate
- DICOM
digital imaging and communications in medicine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: P.C.H. and V.E.T. have received research support from Otsuka. All other authors declare no competing interests.
CRediT Author Statement
TA Potretzke: Conceptualization, Investigation, Formal Analysis, Writing – Original Draft, Writing – Review & Editing
P Korfiatis: Conceptualization, Investigation, Formal Analysis, Writing – Review & Editing
DJ Blezek, FT Chebib, VE Torres: Conceptualization, Investigation, Formal Analysis, Writing – Review & Editing
PC Harris, MC Hogan, CW Bolan, K Sandrasegaran, A Kawashima, JD Collins, N Takahashi, RP Hartman, EE Williamson: Investigation, Formal Analysis, Writing – Review & Editing
ME Edwards, JR Klug, CJ Cook, AV Gregory: Data Curation, Formal Analysis, Writing – Review & Editing
BF King: Conceptualization, Investigation, Formal Analysis, Writing – Review & Editing
MR Callstrom: Supervision
BJ Erickson: Conceptualization, Investigation, Formal Analysis, Supervision, Writing – Review & Editing
TL Kline: Conceptualization, Investigation, Formal Analysis, Supervision, Writing – Original Draft
References
- 1.Gabow PA. Autosomal dominant polycystic kidney disease. N Engl J Med 1993;329:332–342. [DOI] [PubMed] [Google Scholar]
- 2.Harris PC, Torres VE. Polycystic kidney disease. Annu Rev Med 2009;60:321–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torres VE, Harris PC, Pirson Y. Autosomal dominant polycystic kidney disease. Lancet 2007;369:1287–1301. [DOI] [PubMed] [Google Scholar]
- 4.Grantham JJ, Chapman AB, Torres VE. Volume progression in autosomal dominant polycystic kidney disease: the major factor determining clinical outcomes. Clin J Am Soc Nephrol 2006;1:148–157. [DOI] [PubMed] [Google Scholar]
- 5.Fick-Brosnahan GM, Belz MM, McFann KK, Johnson AM, Schrier RW. Relationship between renal volume growth and renal function in autosomal dominant polycystic kidney disease: a longitudinal study. Am J Kidney Dis 2002;39:1127–1134. [DOI] [PubMed] [Google Scholar]
- 6.Grantham JJ, Torres VE, Chapman AB, et al. Volume progression in polycystic kidney disease. N Engl J Med 2006;354:2122–2130. [DOI] [PubMed] [Google Scholar]
- 7.Bae KT, Shi T, Tao C, et al. Expanded Imaging Classification of Autosomal Dominant Polycystic Kidney Disease. J Am Soc Nephrol 2020;31:1640–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bae KT, Tao C, Wang J, et al. Novel approach to estimate kidney and cyst volumes using mid-slice magnetic resonance images in polycystic kidney disease. Am J Nephrol 2013;38:333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kistler AD, Poster D, Krauer F, et al. Increases in kidney volume in autosomal dominant polycystic kidney disease can be detected within 6 months. Kidney Int 2009;75:235–241. [DOI] [PubMed] [Google Scholar]
- 10.King BF, Reed JE, Bergstralh EJ, Sheedy PF 2nd, Torres VE. Quantification and longitudinal trends of kidney, renal cyst, and renal parenchyma volumes in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 2000;11:1505–1511. [DOI] [PubMed] [Google Scholar]
- 11.Tangri N, Hougen I, Alam A, Perrone R, McFarlane P, Pei Y. Total Kidney Volume as a Biomarker of Disease Progression in Autosomal Dominant Polycystic Kidney Disease. Can J Kidney Health Dis 2017;4:2054358117693355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kline TL, Edwards ME, Korfiatis P, Akkus Z, Torres VE, Erickson BJ. Semiautomated Segmentation of Polycystic Kidneys in T2-Weighted MR Images. AJR Am J Roentgenol 2016;207:605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kline TL, Korfiatis P, Edwards ME, et al. Image texture features predict renal function decline in patients with autosomal dominant polycystic kidney disease. Kidney Int 2017;92:1206–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kline TL, Korfiatis P, Edwards ME, et al. Automatic total kidney volume measurement on follow-up magnetic resonance images to facilitate monitoring of autosomal dominant polycystic kidney disease progression. Nephrol Dial Transplant 2016;31:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregory AV, Anaam DA, Vercnocke AJ, et al. Semantic Instance Segmentation of Kidney Cysts in MR Images: A Fully Automated 3D Approach Developed Through Active Learning. J Digit Imaging 2021;34:773–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards ME, Blais JD, Czerwiec FS, Erickson BJ, Torres VE, Kline TL. Standardizing total kidney volume measurements for clinical trials of autosomal dominant polycystic kidney disease. Clin Kidney J 2019;12:71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards ME, Periyanan S, Anaam D, Gregory AV, Kline TL. Automated total kidney volume measurements in pre-clinical magnetic resonance imaging for resourcing imaging data, annotations, and source code. Kidney Int 2021;99:763–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kline TL, Edwards ME, Fetzer J, et al. Automatic semantic segmentation of kidney cysts in MR images of patients affected by autosomal-dominant polycystic kidney disease. Abdom Radiol (NY) 2021;46:1053–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards ME, Chebib FT, Irazabal MV, et al. Long-Term Administration of Tolvaptan in Autosomal Dominant Polycystic Kidney Disease. Clin J Am Soc Nephrol 2018;13:1153–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schönauer R, Baatz S, Nemitz-Kliemchen M, et al. Matching clinical and genetic diagnoses in autosomal dominant polycystic kidney disease reveals novel phenocopies and potential candidate genes. Genetics in Medicine 2020;22:1374–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irazabal MV, Rangel LJ, Bergstralh EJ, et al. Imaging classification of autosomal dominant polycystic kidney disease: a simple model for selecting patients for clinical trials. J Am Soc Nephrol 2015;26:160–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn S, Park SH, Lee KH. How to Demonstrate Similarity by Using Noninferiority and Equivalence Statistical Testing in Radiology Research. Radiology 2013;267:328–338. [DOI] [PubMed] [Google Scholar]
- 23.Erickson BJ, Langer SG, Blezek DJ, Ryan WJ, French TL. DEWEY: the DICOM-enabled workflow engine system. J Digit Imaging 2014;27:309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


